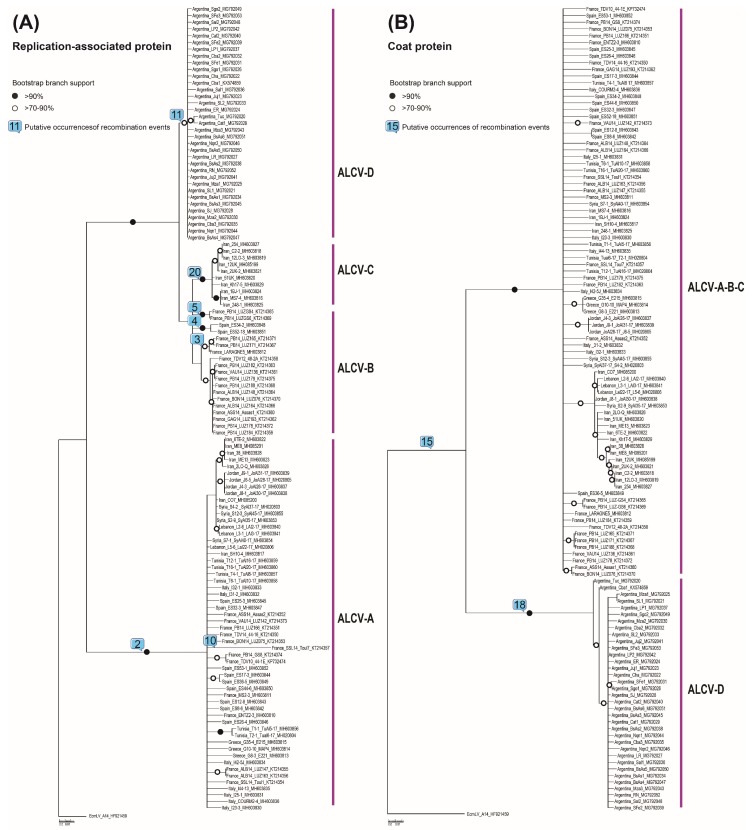
Figure 2.
Maximum-likelihood phylogenetic trees of predicted replication-associated protein (Rep) (A) and coat protein (CP) (B) amino acid sequences of 120 ALCV isolates, both rooted with Euphorbia caput-medusae latent virus CP and Rep amino acid sequences. Branches with less than 50% bootstrap support were collapsed. Branches associated with a black dot have bootstrap supports above 90% whereas those with white dots have bootstrap supports above 70%. Putative occurrences of recombination events (numbers within squares correspond to the number of the event as listed in Table 2) are depicted on branches of both phylogenetic trees.