Comprehensive understanding of an organism requires that we understand the contributions of most, if not all, of its genes. Classical genetic approaches to this issue have involved systematic deletion of each gene in the genome, with comprehensive sets of mutants available only for very-well-studied model organisms. We took a different approach, harnessing the power of in vivo transposition coupled with deep sequencing to identify >500,000 different mutations, one per cell, in the prevalent human fungal pathogen Candida albicans and to map their positions across the genome. The transposition approach is efficient and less labor-intensive than classic approaches. Here, we describe the production and analysis (aided by machine learning) of a large collection of mutants and the comprehensive identification of 1,610 C. albicans genes that are essential for growth under standard laboratory conditions. Among these C. albicans essential genes, we identify those that are also essential in two distantly related model yeasts as well as those that are conserved in all four major human fungal pathogens and that are not conserved in the human genome. This list of genes with functions important for the survival of the pathogen provides a good starting point for the development of new antifungal drugs, which are greatly needed because of the emergence of fungal pathogens with elevated resistance and/or tolerance of the currently limited set of available antifungal drugs.
KEYWORDS: Candida albicans, genome analysis, genomics, machine learning, phenotypic identification, transposons
ABSTRACT
Knowing the full set of essential genes for a given organism provides important information about ways to promote, and to limit, its growth and survival. For many non-model organisms, the lack of a stable haploid state and low transformation efficiencies impede the use of conventional approaches to generate a genome-wide comprehensive set of mutant strains and the identification of the genes essential for growth. Here we report on the isolation and utilization of a highly stable haploid derivative of the human pathogenic fungus Candida albicans, together with a modified heterologous transposon and machine learning (ML) analysis method, to predict the degree to which all of the open reading frames are required for growth under standard laboratory conditions. We identified 1,610 C. albicans essential genes, including 1,195 with high “essentiality confidence” scores, thereby increasing the number of essential genes (currently 66 in the Candida Genome Database) by >20-fold and providing an unbiased approach to determine the degree of confidence in the determination of essentiality. Among the genes essential in C. albicans were 602 genes also essential in the model budding and fission yeasts analyzed by both deletion and transposon mutagenesis. We also identified essential genes conserved among the four major human pathogens C. albicans, Aspergillus fumigatus, Cryptococcus neoformans, and Histoplasma capsulatum and highlight those that lack homologs in humans and that thus could serve as potential targets for the design of antifungal therapies.
INTRODUCTION
The complete set of genes that are essential for survival and growth of eukaryotes are currently known for only a few model eukaryotes, such as the budding yeast Saccharomyces cerevisiae and the fission yeast Schizosaccharomyces pombe (1–4). Gene essentiality is of critical interest in the case of pathogenic fungi; the set of essential genes that are conserved across pathogens, and not in their hosts, are candidate targets for broad-spectrum antifungal drugs. Genes specific for a smaller group of pathogens are candidate targets for more-specific applications. The identification of essential genes as antifungal targets for new classes of antifungals is critical because of the rapid emergence and spread of resistant or tolerant isolates and species in organisms treated with the currently available antifungal drugs (5–7).
Many human fungal pathogens lack a complete sexual cycle, making it difficult to perform classical genetic crosses that validate gene segregation. A classic example is Candida albicans, a member of the normal human microbiome and the most common cause of human fungal nosocomial infection (8). C. albicans generally grows as a heterozygous diploid organism. We recently identified C. albicans haploids, which arise via mitotic chromosome loss events rather than meiosis, providing a critical tool for the genetic analysis of this important pathogen (9).
In haploid model organisms, classic studies test gene essentiality by the analysis of meiotic segregants (2); linkage of a marker to the inability to grow as a haploid provides definitive proof of gene essentiality (10). Such approaches are not applicable to many pathogenic fungi, especially those that do not undergo conventional meiosis.
Much effort has been invested in constructing libraries of C. albicans mutant isolates via the use of directed deletions (11–14), induced deletions (15), or in vitro transposon (Tn) insertions (16–18) and by repression of expression from a single, regulatable copy of the gene of interest (19–21). In addition, the UAU1 system, which couples in vitro transposition with a double-selection scheme to select for homozygosis of the insertion allele (22), identified several hundred genes listed as “likely essential” or “possibly essential,” on the basis of failure to detect homozygosis (203 genes). Clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 drive systems make the gene deletion/disruption process more efficient (23–26). Yet all of those approaches rely upon transformation to generate each individual mutant, which often becomes the bottleneck for generating complete sets of mutant libraries. Despite all of these efforts, only 66 C. albicans genes are currently listed as “essential,” “essential for viability,” “essential for growth,” “essential protein,” or “plays an essential role during mitotic growth” under standard growth conditions in the Candida Genome Database (CGD) (27). However, such tests of essentiality are sensitive to growth conditions and the methods used to assess growth, leading to ambiguity in the literature regarding which C. albicans genes are essential for viability under laboratory conditions.
An alternative approach is to determine gene essentiality using in vivo transposition. In prokaryotes, transposon sequencing (TnSeq) involves the transformation of a transposon-transposase complex, which can generate millions of mutants in a single transformation, coupled with high-throughput sequencing that analyzes all of the transposon insertion sites (28–30). In a recent example, Tnseq phenotypic analysis of 32 bacterial species assigned >2,000 poorly annotated genes to specific functional groups (31). Importantly, while this approach is extremely efficient and valuable for genotype/phenotype analyses in prokaryotes, it cannot be used in eukaryotes.
In the model yeast S. cerevisiae, a heterologous maize Activator/Dissociation (Ac/Ds) element (32) was induced to transpose (33) and was harnessed to rapidly generate a large scale SATAY (SAturated Transposon Analysis in Yeast) library of insertion mutants (34). In the fission yeast S. pombe, genome saturating insertion mutagenesis performed with the Hermes transposon yielded >350,000 independent insertions (35). In the filamentous pathogenic fungus Aspergillus fumigatus, a smaller-scale analysis performed with the Impala transposon (from Fusarium oxysporum) identified 96 essential genes (36). These approaches identify recessive mutations in haploid organisms.
Here we generated a stable haploid C. albicans isolate carrying an Ac transposase/Ds-NAT1 two-element system (33) to implement an in vivo transposition approach for studying this important pathogen. We used the system to identify genes important for growth under standard laboratory conditions and developed a machine learning (ML) approach to infer essentiality/nonessentiality, i.e., the ability to grow under standard laboratory conditions, in an unbiased fashion. We also applied the ML approach to data from S. cerevisiae and S. pombe transposon studies and then utilized the results to identify a core set of orthologs essential in all three yeasts in deletion and transposon studies. We provide a comprehensive, genome-wide assessment of gene essentiality, a confidence measure of gene essentiality/nonessentiality on the basis of the range of studies that have been performed with each gene, and we highlight essential genes that are not conserved in humans and that can serve as potential targets for the development of new antifungal drugs.
RESULTS
Selection of a stable haploid strain carrying the Ac/Ds system.
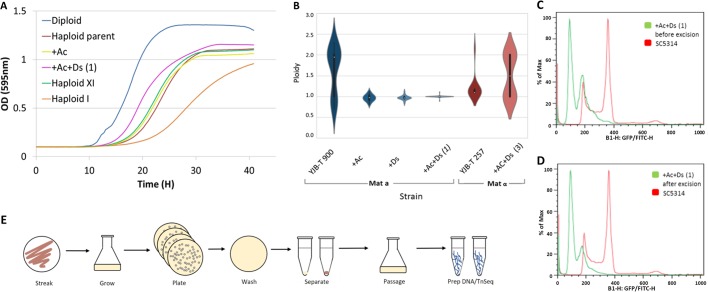
We constructed transposon insertion libraries in a haploid C. albicans isolate (YJB-T900) by sequential insertion of a codon-optimized Ac transposase (AcTPase4xCa) at the neutral NEUT5L locus and a Ds-NAT1 transposon such that it interrupted the ADE2 promoter region (37). After insertion of the Ac transposase, single colonies were checked for DNA content by flow cytometry. Among the colonies, 50% (6/12) were haploid and the others consisted of mixed populations of haploid and diploid cells. One all-haploid colony (YJB-T1792) was then transformed with Ds-NAT1. The DNA content was retested, and 58% (14/24) of the colonies were found to be haploid; one of them (YJB-T1081) was archived, restreaked, and rearchived (YJB-T1082). Like the original haploids, these haploids showed reduced virulence relative to strain SC5314, the heterozygous laboratory strain from which they were derived (see Fig. S1A in the supplemental material).
Virulence of haploid reads, transposon insertion read maps, and machine learning performance. Download FIG S1, TIF file, 0.2 MB (336.4KB, tif) .
Copyright © 2018 Shtifman Segal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Importantly, retesting of hundreds of single colonies from YJB-T1081 and YJB-T1082 from the archived stock always produced stable haploids, in contrast to its semistable parent (YJB-T900), in which 50% (12/24) of the colonies produced diploid subpopulations (Fig. 1B and C). Thus, YJB-T900 has improved stability relative to the originally identified haploids (9), and the Ac/Ds-NAT1 strains were consistently stable haploids and, like YJB-T900, exhibited improved growth relative to the originally isolated haploids (9) (Fig. 1A), making them ideal for transposon mutagenesis and the detection of recessive mutations.
Fig 1.
Strain and Tn library construction. (A) Growth curves for strains are indicated as follows: blue, diploid (SC5314); red, haploid MATa parent (YJB-T900); yellow, +Ac (YJB-T 1792); pink, +Ac+Ds(1) (YJB-T1081) (growth of YJB-T1082 [also +Ac+Ds] was indistinguishable from that of YJB-T1081); orange, haploid I (9); green, haploid XI (9). All growth curve analyses were performed in triplicate, with standard deviations ranging from 0 to 0.05. OD, optical density. (B) Summary of DNA content for multiple isolates for each strain (n = 24) [+Ds (YJB-T1794); MATα haploid parent YJB-T257; +Ac+Ds(3); YJB-T2743]. (C and D) Flow cytometric DNA content for strain YJB-T1081 before (C) and after (D) excision of Ds-NAT1. (E) Schematic of transposon mutant library preparation process.
Preparation and characterization of large-scale Ds-NAT1 insertion libraries.
Induction of the transposase (AcTPase4xCa) on maltose catalyzed excision of the Ds-NAT1 transposon, thereby restoring ADE2 expression and a shift from red (Ade negative [Ade−]) to white (Ade-positive [Ade+]) colonies (Fig. 1E). We generated libraries of C. albicans transposon (CaTn) mutants and passaged them twice to select for cells that had undergone Ds-NAT1 excision and reintegration. Flow cytometry of DNA content and PCR of the ADE2 locus of 96 Ade+ colonies (Fig. 1D) detected only those haploids that had undergone excision and reintegration.
Targeted sequencing identified CaTn insertions within the 11 libraries with the highest transposition frequencies (among 25 libraries initially prepared). Alignment of the resulting data (∼12 M to ∼32 M reads per library) (Table 1) with the reference strain sequence, SC5314 Assembly 22 haplotype A, identified the insertion sites relative to all annotated open reading frames (ORFs), functional RNAs, or transposons (see Table S1 in the supplemental material). Consistently, ∼one-third of the insertion sites (hits) were within annotated features and two-thirds fell in intergenic regions, with the proportion of annotated features that contained hits ranging from 55.5% in library 10 to 89.7% in library 3, yielding ∼2 to 14 hits per annotated feature (Table 1). Subsequent analysis focused on data combined from the three libraries (3, 7, and 11), each of which had an average of more than 1 hit/100 bp and approximately ≥10 hits per genomic feature (ORFs, noncoding RNAs, etc.). Together, these three libraries had ∼600,000 unique Ds-NAT1 insertions, with 33.2% of them hitting within annotated features and 66.8% of them in intergenic regions, on the CGD (38). A total of ∼95% of the annotated features included at least 1 Ds-NAT1 insertion site.
TABLE 1.
Transposition library sequence quantification
| Librarya | Total no. of reads (×106) |
Total no. of hits (×103) |
Mean no. of hits/100 bp |
% of hits in features |
% of intergenic hits |
% of features per hit |
Mean no. of hits per feature |
Mean no. of reads per feature (×103) |
Mean no. of reads per hit (×103) |
Mean no. of reads per hit in feature (×103) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 26.7 | 23.8 | 0.17 | 38.18 | 61.82 | 62.46 | 2.2 | 2.2 | 1.1 | 1 |
| 2 | 12.4 | 25.2 | 0.18 | 35.3 | 64.7 | 60.63 | 2.2 | 1.1 | 0.5 | 0.5 |
| 3* | 31.9 | 252.6 | 1.77 | 33.39 | 66.61 | 89.72 | 14.2 | 1.7 | 0.1 | 0.1 |
| 4 | 20.2 | 40.6 | 0.28 | 31.99 | 68.01 | 69.87 | 2.8 | 1.4 | 0.5 | 0.5 |
| 5 | 30.9 | 64.6 | 0.45 | 31.71 | 68.29 | 74.12 | 4.2 | 2.1 | 0.5 | 0.5 |
| 6 | 19.2 | 40.1 | 0.28 | 31.61 | 68.39 | 69.84 | 2.7 | 1.4 | 0.5 | 0.5 |
| 7* | 28.8 | 169 | 1.18 | 33.26 | 66.74 | 85.85 | 9.9 | 1.6 | 0.2 | 0.2 |
| 8 | 26.6 | 37 | 0.26 | 31.07 | 68.93 | 63.9 | 2.7 | 1.8 | 0.7 | 0.7 |
| 9 | 31.3 | 28.1 | 0.20 | 37.25 | 62.75 | 65.47 | 2.4 | 2.4 | 1.1 | 1 |
| 10 | 22.8 | 27.9 | 0.20 | 28.96 | 71.04 | 55.54 | 2.2 | 1.8 | 0.8 | 0.8 |
| 11* | 23.5 | 178.2 | 1.25 | 32.9 | 67.1 | 85.59 | 10.4 | 1.3 | 0.1 | 0.1 |
Asterisk indicates three libraries pooled for subsequent analysis.
Annotated features in C. albicans. Download Table S1, PDF file, 0.2 MB (245.6KB, pdf) .
Copyright © 2018 Shtifman Segal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Insertion sites exhibited a periodicity reminiscent of nucleosome occupancy. Comparison of log-read and hit counts on each chromosome for regions with lower and higher likelihoods of being occupied by a nucleosome revealed a consistent bias toward a higher frequency of insertions in regions with a lower likelihood of nucleosome occupancy (Table S2).
Number of insertions and reads in regions with low and high likelihood of nucleosome occupancy. Download Table S2, PDF file, 0.02 MB (16.7KB, pdf) .
Copyright © 2018 Shtifman Segal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
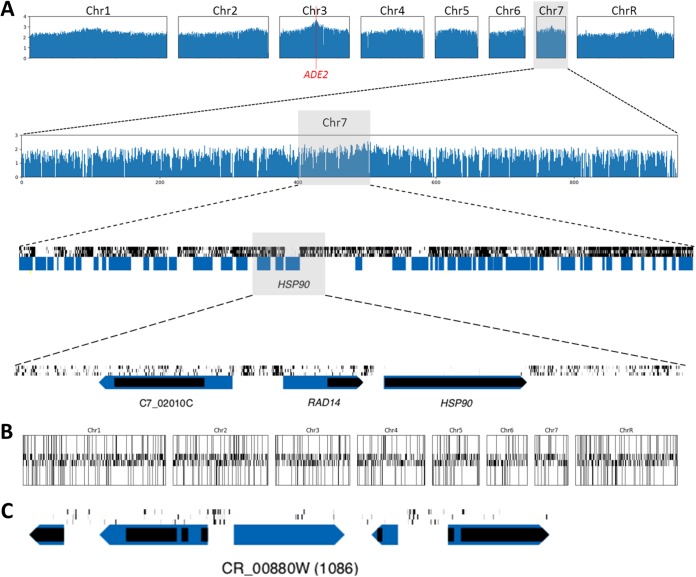
Ac/Ds transposons tend to reinsert near the donor site at high frequency (previously reviewed [34, 39, 40]). A bias for insertion in regions close to the original Ds-NAT1 insertion site at ADE2 was evident, with the highest density of hits (number of insertion sites within an ORF) and reads (total number of sequences per hit or ORF) within ∼100 kb of ADE2 on Chr3 (Fig. 2A; see also Fig. S1B, red bar). Despite this bias, genes very likely to be essential (those that sustained very few hits within the ORF and had many more hits detected in flanking intergenic regions [e.g., HSP90]) (Fig. 2A) were often evident. Yet intuitive visual analysis was not always sufficient to determine gene essentiality, especially in genome regions with lower hit density (see, e.g., Fig. 2C) as well as potential “domain-essential” genes that sustained hits only within a defined region of the coding sequence (see, e.g., Fig. 3A).
Fig 2.
Maps of transposon insertions at different scales. (A) Distribution of insertion sites (hits) for the pooled library used in this study (libraries 3, 7, and 11). In the top two rows, the 8 C. albicans chromosomes and Chr7 are in 10,000-bp bins, the y axis scale is log10, and the ADE2 gene is indicated by a red bar. In the lower two rows, 100-kb and 10-kb sections of Chr7 show three tracks of hit data in black, representing libraries 3, 7, and 11 (from top to bottom). Blue rectangles, predicted ORFs; black rectangles, recognized domains from protein information in CGD (38). The arrow points to the 3′ end of the gene. (B) Distribution of predicted essential and nonessential genes across the chromosomes. Essential genes are indicated with tall bars; nonessential genes are indicated with short bars; bars above and below the central axis represent genes transcribed on the Watson and Crick strands, respectively. (C) Example of low-hit region of CR_00880W gene.
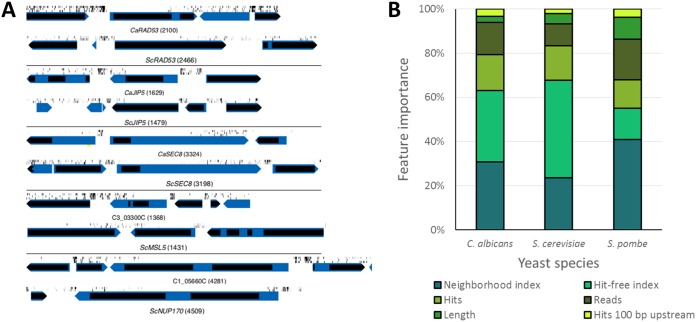
Fig 3.
(A) Examples of genes with hits in the C-terminal portion of the coding sequence for C. albicans and S. cerevisiae orthologs. Note that patterns are similar although S. cerevisiae genes sometimes have insertions in the extreme N-terminal coding sequence that are not evident in C. albicans orthologs. Yellow bars below the genes indicate introns. Similar maps for the other libraries are found in Fig. S2A and maps of all ORFs are available in Dataset 9 at https://doi.org/10.6084/m9.figshare.c.4251182. (B) Relative feature importance contributions to the random forest classifiers for all three model yeasts.
Hit maps of C. albicans and S. cerevisiae genes with conserved organization of essential domains, examples of misannotated C. albicans ORFs, and alignment of proteins that they predict. Download FIG S2, TIF file, 0.4 MB (760.8KB, tif) .
Copyright © 2018 Shtifman Segal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Prediction of gene essentiality on the basis of a machine learning approach.
To distinguish essential genes from nonessential ones in the transposon insertion data, we used a machine learning (ML) approach. Specifically, we constructed a random forest (RF) classifier with a set of features from the transposon data as follows: (i) the total number of hits per ORF, (ii) the total number of sequence reads per ORF, and (iii) the total number of hits within 100 bp 5′ to the ORF, as well as (iv) the ORF length, (v) the “neighborhood index” (the total number of hits per ORF normalized for the hits in surrounding intergenic sequences), and (vi) the longest hit-free region (normalized length of the largest ORF interval without hits) (Table 2). Training sets of presumed essential and nonessential genes were used to train a gene essentiality predictor that uses feature-based decision rules. Similar predictors were constructed for S. cerevisiae and S. pombe. Details are provided in Materials and Methods.
TABLE 2.
Input features for the machine learning classifier
| Feature | Definition |
|---|---|
| Hits | No. of insertion sites within the ORF |
| Reads | No. of reads within the ORF |
| Hits in promoter | No. of hits within 100 bp upstream of ORF start codon |
| ORF length | Total length of ORF coding sequence (intron-free) |
| Insertion indexa | No. of hits in the ORF divided by ORF length |
| Noncoding windowa | Noncoding sequence (including introns) within 10 kb up- and downstream of ORF |
| Neighborhood index (NI) | Insertion index normalized to the noncoding window (hits divided by length) |
| Hit-free interval (HFI) | Length of longest insertion-free interval divided by ORF length |
These features were input indirectly to calculate NI and HFI.
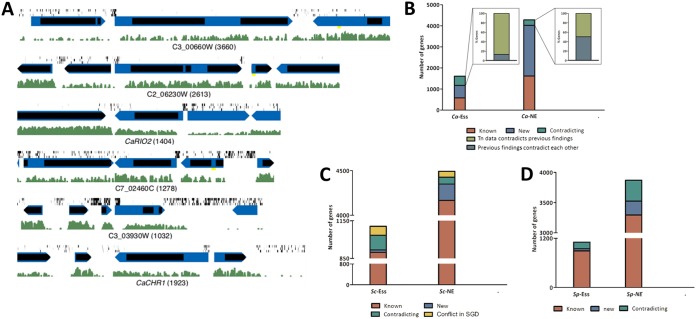
We assembled training sets of C. albicans genes (see Dataset 1A at https://doi.org/10.6084/m9.figshare.c.4251182) on the basis of two major assumptions: (i) that most of the 697 ORFs with essential orthologs in both S. cerevisiae and S. pombe (which diverged from each other over 300 million years ago [MYA] [41]) were likely to be essential in C. albicans and (ii) that the 759 ORFs that had been deleted, which included genes with and genes without orthologs in the model yeasts, were unlikely to represent essential genes. These sets were further filtered by manual inspection (see Materials and Methods). The resulting sets of genes were used to train a random forest classifier with 5-fold cross validation which showed that the predictions exhibited high accuracy (area under the receiver operating characteristic curve [AUC], 0.997) (Fig. S2C). A threshold of 0.8 yielded a false-positive rate (FPR) of 1% and a true-positive rate (TPR) of 92% (Table 3) and identified 1,610 C. albicans essential (CaTn-Ess) and 4,383 nonessential (CaTn-NE) genes among the 5,893 ORFs (see Dataset 2A at https://doi.org/10.6084/m9.figshare.c.4251182). This provided first-time data for a total of 3,697 genes (1,033 CaTn-Ess and 2,664 CaTn-NE) (Fig. 4B). The CaTn-Ess genes and the CaTn-NE genes were distributed relatively randomly across the 8 C. albicans chromosomes (Fig. 2B).
TABLE 3.
Cross-validation AUCs and thresholds chosen for prediction in each organism
| Organism and training data from: |
Cross validation AUC |
Threshold | FPRa | TPRb |
|---|---|---|---|---|
| C. albicans | 0.997 | 0.8 | 0.01 | 0.92 |
| S. cerevisiae | 0.989 | 0.67 | 0.02 | 0.89 |
| S. pombe | 0.966 | 0.62 | 0.04 | 0.80 |
False-positive rate.
True-positive rate.
Fig 4.
(A) Examples of genes with misannotated start codon positions. Symbols are as described in the Fig. 3 legend. Green histograms illustrate RNAseq expression levels (48). (B) Proportions of essential and nonessential genes compared to prior information are indicated. CaTn-Ess and CaTn-NE genes, as indicated. Red, genes with prior data indicating essentiality; blue, genes that were not previously tested for essentiality; green, genes with contradictory data. Dark green, genes with contradictory data prior to this study; light green, genes for which CaTn predictions contradicted at least one other report. (C) ScTn-Ess and ScTn-NE genes, as indicated. In yellow are genes that had conflicting annotations in SGD. (D) SpTn-Ess and SpTn-NE genes, as indicated.
To assess the reliability of the ML approach, similar analyses were performed with in vivo transposon insertion data for S. cerevisiae (34) and S. pombe (35), with training sets chosen from genes found to be essential or nonessential in deletion analyses in that organism (42, 43). The resulting accuracies (as reported by AUCs) were very high (Table 3).
The feature importance with respect to predicting gene essentiality with the random forest classifier differed somewhat between the three yeasts (Fig. 3B). The neighborhood index was the strongest predictor in C. albicans and S. cerevisiae. The “insertion-free region” was the most powerful predictor in S. pombe, where Hermes insertions were more evenly distributed, and was more important in C. albicans than in S. cerevisiae, possibly because S. cerevisiae tolerated insertions into interdomain regions and C. albicans did not. Not surprisingly, the nominal number of hits and number of reads per ORF also contributed substantially to the predictions, while ORF length and the number of hits within the 100 nucleotides (nt) 5′ to the start codon made only small contributions to predictions of essentiality in all three yeasts. Nonetheless, while ∼46% of the 1,610 CaTN-Ess genes had no hits in the 100-nt 5′ untranslated region (5′UTR), only ∼9% of the CaTn-NE group had none, suggesting that this feature has some predictive power in C. albicans. Importantly, the feature set used was sufficiently robust to allow high-accuracy predictions across all three yeasts (Table 3). Furthermore, using just the 66 “CGD essential” genes (see Dataset 3 at https://doi.org/10.6084/m9.figshare.c.4251182) together with the original training set of deleted C. albicans genes, the results were also quite strong (AUC 0.92). This suggests that the training set of known essential and nonessential genes does not need to be large and that criteria for hit patterns established for these three yeasts may be sufficient to predict essentiality in other organisms.
Comparison of deletion results (Δ) and transposon (Tn) predictions.
To test the accuracy of the ML classifier, we used data from either the S. cerevisiae SATAY (miniDs) data (34) or the S. pombe Hermes transposon data (35) (Table 3) together with training sets from comprehensive deletion studies in the same organism (42, 43) and compared the ML predictions (see Dataset 2B and C at https://doi.org/10.6084/m9.figshare.c.4251182) to the genome-wide deletion study conclusions (ScΔ and SpΔ). The ScTn study (34) analyzed by ML predicted 1,106 essential genes (there are 975 ScΔ-Ess genes in SGD). There was agreement for 92% (898) Sc-Ess genes and 98% (4166) Sc-NE genes (Fig. 4C). The SpTn study (35) analyzed by ML predicted 1,106 SpTn-Ess genes (there are 1,241 SpΔ-Ess genes in PomBase), with agreement for 72% (895) Sp-Ess genes and 95% (3,293) Sp-NE genes (similar to the predicted TP rate of 0.80) (Fig. 4D).
Disagreement between the deletion and transposon data can be due to the presence of secondary suppressors (44) and other issues in deletion strains (35), to conditional essentiality (45), and to strain-specific effects (11, 45, 46), as well as to the difficulty encountered in identifying all of the domain-essential genes (44) (see, e.g., Fig. 3A; more detail is provided below and in Fig. S4A).
Specific examples of C. albicans genes and their paralogs in S. cerevisiae and/or S. pombe. Download FIG S4, TIF file, 0.4 MB (697.5KB, tif) .
Copyright © 2018 Shtifman Segal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Domain-essential genes.
Domain-essential genes have many transposon insertions within a portion of the ORF and were defined in SATAY as having no hits in a >400-bp domain (Fig. 3A) (34). For the ML predictions, we defined a “hit-free interval length” feature as the longest region without hits, divided by the ORF length (Table 2). In C. albicans, unlike in S. cerevisiae, hit-free regions were evident in C-terminal coding regions and not in the N termini. For example, CaRAD53, a gene involved in DNA damage responses and filamentous growth in C. albicans (47), had no insertions in the first 1,402 bp of the coding sequence, which is predicted to include a protein kinase-like domain. This suggests that the kinase domain is likely important, or essential, for RAD53 function and that the C-terminal region may be dispensable (Fig. 3A). Similar domain patterns were seen for JIP5, SEC8, and MSL5 (34) (Fig. 3A). Domain-essential ORFs were found in all three yeast species on the basis of the Tn insertion patterns, and in a few ORFs, domain insertion patterns were conserved (e.g., Fig. S2A).
Refinement of transcription and translation start sites for several genes.
Unexpectedly, several genes appeared to be essential (very few hits within the ORF) and yet had high levels of insertions immediately upstream to and downstream of the start codon of the presumed ORF (Fig. 4A). In some cases, the S. cerevisiae ortholog (e.g, C7_02460C/ScNPA3) was essential and predicted to encode a shorter protein (Fig. S2C). Furthermore, in this example, data from transcriptome sequencing (RNAseq) (48) suggested that the transcription start site was 112 bp 3′ to the initiation codon (Fig. S2B) and that the predicted proteins aligned well from the downstream ATG codon (Fig. S2C). This demonstrates that transposon analysis can contribute to the identification of coding sequence boundaries.
“Core essential” and “core nonessential” genes.
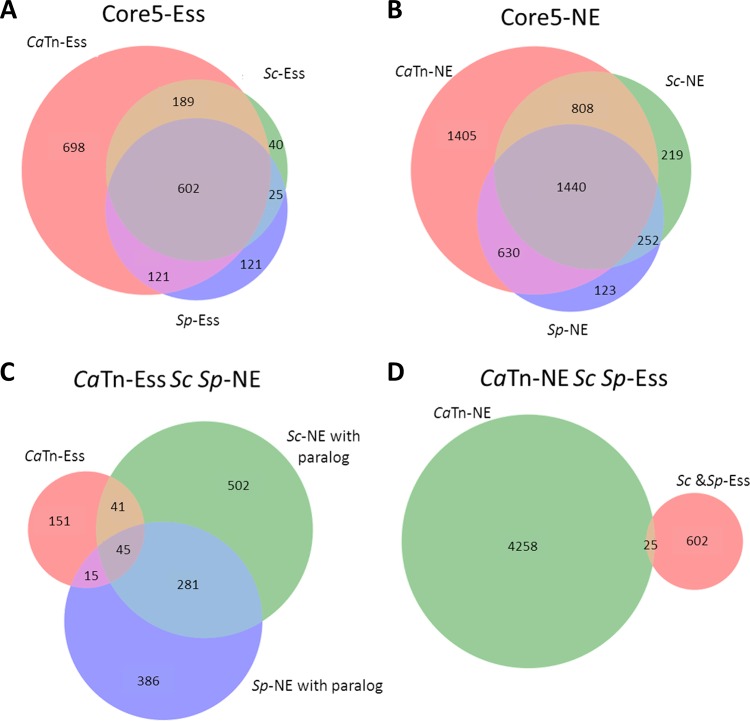
Overall, 694 genes were essential in all three Tn studies (ScTn/SpTn/CaTn) and 602 were essential in all 5 deletion and Tn studies (ScΔ/SpΔ/ScTn/SpTn/CaTn) (Core5-Ess) (Fig. 5A). Interestingly, 17 Core5 genes were not essential in a wild-type S. cerevisiae strain (Sigma 1278 b) (11), which highlights several notable points. First, as noted in several previous studies (11, 49, 50), the essentiality of some genes is strain background specific even within a given species. Second, any specific C. albicans haploid strain is likely is be carrying deleterious alleles that are recessive in the heterozygous diploid parent (9), consistent with the lower growth rates of haploids (even those that are stable) compared to heterozygous diploids (Fig. 1A). We assume that deleterious recessive alleles with no phenotype in diploids may have epistatic effects on a subset of genes; accordingly, some genes may appear essential in a haploid derivative of a wild-type heterozygous diploid strain. Third, ML provides a statistical inference; thus, a small proportion of false-positive and false-negative predictions are expected. The confidence score (see below) is designed to reduce this uncertainty further.
Fig 5.
Venn diagrams of the intersections of (A) Core5 essential genes; (B) Core5 nonessential genes; (C) S. cerevisiae and S. pombe paralogs with CaTnEss, ScSpNE group; and (D) CaTnEss versus ScEss and SpEss.
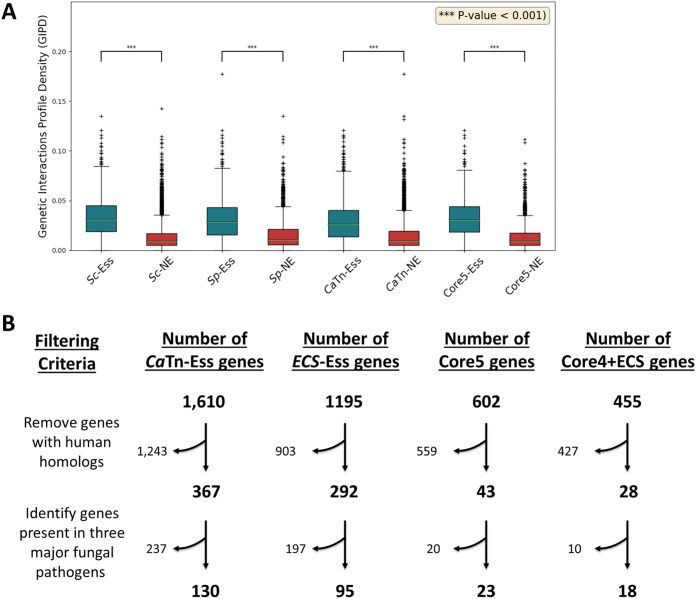
The majority of the core essential genes were enriched for fundamental eukaryotic processes such as RNA metabolism, regulation, organelle organization, ribosome biogenesis, and cell cycle (Fig. S3). In addition, the ScTn-Ess genes had a higher degree of genetic interaction profile density (GIPD)—representing the number of synthetic genetic interactions for a given gene divided by the number of interactions tested (51, 52) (Fig. 6A). Notably, this relationship held true for essential versus nonessential orthologs of those genes in S. pombe and C. albicans as well (Fig. 6A).
Fig 6.
Genetic interaction degree and number of essential genes conserved in pathogenic fungi and not in humans. (A) Genetic interaction degree, a measure of interactivity of C. albicans genes in experiments performed using SGA software (see Materials and Methods) plotted for essential (Ess) and nonessential (NE) genes/orthologs in C. albicans (CaTn) and S. cerevisiae (ScΔ and ScTn) and from S. pombe analyses (SpΔ and SpTn) as well as from the Core5 analyses (CaTn, ScΔ, ScTn, SpΔ, and SpTn). The y axis data represent the number of gene interactions normalized by the number of observations (GIPD) (46). The statistical significance of results of comparisons between Ess and NE genes was obtained using the Wilcoxon rank sum test (***, P < 0.001). (B) Four groups of essential genes of CaTn-Ess and Core5-Ess as described in the Fig. 5 legend as well as of genes of Ca-ECS and Core5-ECS-Ess (after filtering was performed using an essentiality confidence score) were then filtered for those without human homologs and then for those with homologs in the other three major pathogenic yeasts (A. fumigatus, C. neoformans, and H. capsulatum).
Gene ontology (GO) term analysis of Core5 essential genes and Core5 nonessential genes. Download FIG S3, TIF file, 0.2 MB (155.6KB, tif) .
Copyright © 2018 Shtifman Segal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Orthologs essential in C. albicans and not essential in both S. cerevisiae and S. pombe.
Among the genes with orthologs in all three yeasts, 252 were essential in C. albicans (CaTn-Ess) and not essential in the two model yeasts via both deletion and Tn studies (Core4-NE) (Fig. 5; see also Dataset 2D at https://doi.org/10.6084/m9.figshare.c.4251182). In S. cerevisiae, remnants of the whole-genome duplication provide redundancy/backup functions under some conditions and thus may not be essential individually, whereas their single-copy orthologs in C. albicans are essential (53, 54). For example, in S. cerevisiae, ScBDF1 and ScBDF2 are each dispensable and yet dependent upon the presence of the other (55). The single gene CaBDF1 was found to be essential both in CaTn analysis (see Dataset 2A at https://doi.org/10.6084/m9.figshare.c.4251182) and in classical deletion studies (56). Indeed, 101 of these 252 genes are found in single-copy form in C. albicans and have paralogs in at least one of the two model yeasts (57–59) (Fig. 5C). The remaining 151 genes that are essential in C. albicans and have no paralog in the model yeasts (Fig. 5C) were enriched for genes important for ATP synthesis via respiration and mitochondrial components (see Dataset 4 at https://doi.org/10.6084/m9.figshare.c.4251182), which highlights the rewiring of respiration and mitochondrial translation in C. albicans relative to the model yeasts (60). Several C. albicans genes involved in cell cycle progression were essential in C. albicans and nonessential in S. cerevisiae or vice versa (Fig. S4B).
Also of interest are obvious differences between insertion site frequencies in the three yeasts for MET6 and CLN3, which are CaTn-Ess and ScTn-NE (Fig. S4C and D). MET6 and CLN3 were also shown to be essential in classical C. albicans deletion studies (61–63), suggesting that both gene products participate in processes that have diverged significantly in humans, making it a potential target for antifungal drugs (discussed below).
Among the genes essential in both C. albicans and S. cerevisiae, but not in S. pombe, were three septins essential for bud neck function (CDC11, CDC12 and CDC3), a process very different in fission yeast than in the two budding yeasts, and several components of the DASH kinetochore complex that is present in S. cerevisiae and C. albicans but is not conserved in S. pombe (see Fig. S4E and Text S1 in the supplemental material). Genes essential in both C. albicans and S. pombe but not S. cerevisiae included those corresponding to a range of functions; an example is CR_01480W/AIM10/SPBC24C6.03, which encodes a mitochondrial tRNA synthesis and affects the stability of the mitochondrial genome and is dispensable in S. cerevisiae but essential in C. albicans and S. pombe.
Expanded supplemental figure legends with references. Download Text S1, DOCX file, 0.1 MB (56.7KB, docx) .
Copyright © 2018 Shtifman Segal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Candida-specific essential genes.
Comparisons among the three yeasts above necessarily involved analysis of genes with orthologs in at least two of the three yeasts. In addition to the essential genes that had orthologs in S. cerevisiae and/or S. pombe, there were 113 CaTn-Ess genes with no strict ortholog in either S. cerevisiae or S. pombe (38). Of these, 18 had sufficient similarity to S. cerevisiae genes to be annotated accordingly; 3 of those 18 were most similar to ScHSK3, ScDUO1, and ScSPC34, which encode components of the DASH complex (the outer kinetochore complex) that are essential in S. cerevisiae and wild-type C. albicans (64); these genes diverged rapidly and are not conserved in S. pombe, as noted above (65).
Of the 113 C. albicans essential genes with no homology or similarity to S. cerevisiae or S. pombe genes, 17 have no obvious orthologs among the two model yeasts or among the CUG clade species (see Dataset 5 at https://doi.org/10.6084/m9.figshare.c.4251182). None of them have been characterized directly, although transcripts have been detected for at least three of them under some growth conditions (48). By contrast, 55 of the CaTn-Ess genes had clear orthologs in all six pathogenic species related to C. albicans (see Dataset 5 at https://doi.org/10.6084/m9.figshare.c.4251182). The conservation of these genes supports the idea that they are important for the survival of the CUG group of fungi. It also suggests that future work should focus on the functions of these genes, as they have the potential to be clade-specific targets of antifungal therapies.
Orthologs not essential in C. albicans but essential in both model yeasts.
Of the 627 genes found to be essential in all deletion and transposon studies in the two model yeasts (ScΔ-Ess/ScTn-Ess/SpΔ-Ess/SpTn-Ess genes), 4% (25 genes) were predicted to be nonessential in C. albicans (Fig. 5D). Two of these (C7_02460C and C2_06230W) had misannotation of the start codon (Fig. 4A), and six had large insertion-free domains (Fig. S4F), suggesting that they could be domain essential. Gene ontology (GO) term analysis of these CaTn-NE/Sc-Ess and Sp-Ess genes is enriched in mRNA processing (false-discovery-rate [FDR]-corrected P value, 5.82e−06) and U2-type spliceosomes component (FDR-corrected P value, 1.5e−6).
Consistency among deletion, repression, and transposon insertion studies.
Prior to this study, 2 or more studies disagreed about essentiality for at least 190 (16%) of 1,183 genes, with new CaTn information for 2,996 genes with no prior experimental information; ∼13% (58/440 CaTn-Ess genes) correspond to at least one contradictory study (Fig. 4B). This highlights the difficulty in reaching definitive conclusions for every last gene.
To address this ambiguity, we calculated an essentiality confidence score (ECS) that captures available C. albicans essentiality information from deletion and repression data and CaTn studies (11–14, 19). The data from each study were considered equally (+1 for essential, −1 for nonessential), and the sum of their scores (net essentiality) for each gene was used with a logistic function to predict the likelihood of essentiality (using a 0-to-1 scale) (see Dataset 6 at https://doi.org/10.6084/m9.figshare.c.4251182; details in Materials and Methods). A caveat with respect to the ECS is that it assigns equal weight to each type of study. While the scheme can be generalized to learn individual weights from the different studies, we chose a simple scheme in order to avoid the potential biases that can stem from imperfect training data.
In total, the ECS identified 346/5,893 genes that were not clearly essential or nonessential (ECS = 0.5). Among these were primarily genes with two different outcomes (see Dataset 6 at https://doi.org/10.6084/m9.figshare.c.4251182). An interesting example is the riboflavin synthesis pathway in S. cerevisiae that requires RIB1, RIB7, RIB3, RIB4, RIB5, FMN1, and FAD1 (66), which have been shown to be essential in deletion and/or Tn studies, as are the S. pombe orthologs (Table S3; see also Dataset 2D at https://doi.org/10.6084/m9.figshare.c.4251182). Six of these (RIB1, RIB3, RIB4, RIB7, FMN1, and FAD1) were CaTn-Ess, and yet four (FMN1, RIB7, RIB3, and RIB4) were nonessential by repression analysis (19); thus, their ECS was 0.5 and their essentiality remains “unknown” (Table S3). We suggest that mutations in the riboflavin biosynthesis pathway may cause extremely slow growth, leading to equivocal results that depend on the criteria used to determine essentiality and the medium used for growth studies.
Riboflavin biosynthesis pathway analysis. Download Table S3, PDF file, 0.1 MB (67.9KB, pdf) .
Copyright © 2018 Shtifman Segal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Essential genes conserved in fungi and not humans.
Essential genes are thought to be good targets for antifungal therapy because their inactivation should kill the pathogen (21). However, the similarities between fungi and their animal hosts present a challenge to the development of antifungal drugs. In theory, preferred antifungal targets should be essential genes without human homologs that are conserved among pathogenic fungi. Accordingly, we examined sets of essential genes, including the CaTn-Ess, ECS-Ess, and Core5-Ess genes and the Core4+ECS-Ess genes (essential in all 3 yeasts using the ECS filter for Ca-Ess genes) and then examined those without human homologs. Among these genes, we then identified the subset with homologs in the other three major human pathogens: Aspergillus fumigatus (Eurotiomycetes, Ascomycota), Cryptococcus neoformans (Tremellomycetes, Basidiomycota), and Histoplasma capsulatum (Eurotiomycetes, Ascomycota). We found 130 CaTn-Ess genes and 95 ECS-Ess and 23 Core5-Ess and 18 Core4+ECS genes that have homologs in the other three major fungal pathogens and that do not have human homologs (Fig. 6B; see also Dataset 7 at https://doi.org/10.6084/m9.figshare.c.4251182).
Among the CaTn-Ess and ECS-Ess genes were 52 genes whose products are mitochondrial and that are involved in ATP synthesis and/or mitochondrial membrane function, 4 of the 5 genes encoding ERMES complex components (MDM10, MDM12, MDM34, and MMM1), and 7 genes encoding kinetochore components [SPC19, MTW1, ASK1, CaMAD1, CaSPC105, CaBIR1(CR_05100W), and CaKRR1]. In addition, components of the riboflavin pathway did not have human homologs and had homologs in the other pathogenic fungi (see Dataset 5 at https://doi.org/10.6084/m9.figshare.c.4251182). We suggest that genes in these groups have the potential to be high-priority candidates for broad-spectrum antifungal drug design.
DISCUSSION
This study leveraged a stable haploid isolate, a codon-optimized, inducible Ac transposase, strong selection of excision and reintegration events, and machine learning to double the number of predicted genes that are essential or nonessential for C. albicans survival under laboratory growth conditions. The ML classifier trained on the three yeasts studied here is highly accurate and has the potential to work across species. We also developed an essentiality confidence score that considers different types of mutation studies, thereby providing a useful perspective on the degree to which essentiality is conserved and, together with comparisons to predicted ORFs in other pathogenic fungi and in Homo sapiens, that identifies those essential genes that have the potential to be targets for antifungal drugs.
Genome-wide Ac/Ds mutagenesis in C. albicans haploids.
We leveraged two critical resources—a highly stable C. albicans haploid strain that does not autodiploidize and in vivo transposition using a modified Ac transposase/Ds-NAT1 two-element system. A strength of the Ac/Ds transposon is that, unlike Hermes or PiggyBac (67, 68), it does not have a strong insertion site preference either in maize or in other organisms, including S. cerevisiae (34, 37, 40, 69).
The in vivo transposition is extremely efficient: once a starting strain is engineered, no further transformation or homologous recombination steps are required. This is particularly useful for clinically relevant organisms where transformation and homologous recombination limit the transfer of deletion constructs to a new strain background (45, 70–73). In vivo transposition also obviates the inherent bias present when researchers select, and thereby limit, the sequences to be analyzed, e.g., regions between ORFs (e.g., misannotation of transcription start sites) (Fig. 4A) and potentially essential noncoding RNAs (Ca22chrRA: n = 1,421,741 to 1,424,356).
Ds insertion preferences were detected for the following three features of DNA: nucleosome occupancy, proximity to the excision site, and intergenic versus coding features. The nucleosome bias was weaker in C. albicans than in the SATAY system (34). This was likely due, at least in part, to differences in nucleosome occupancy between the diploid C. albicans strain used for nucleosome coverage data (74) and the haploid CaTn strain. Proximity to the initial site of excision was evident in our libraries and has been well documented in lower-throughput studies (39) and in SATAY (34), while it was far less evident when the site of Ds excision was on a plasmid. Unfortunately, autonomously replicating plasmids are not maintained well in C. albicans. We are currently constructing plasmids that may be useful for this purpose in the future (J. Berman, unpublished results).
Applying machine learning to in vivo transposon insertion data.
Previous transposon studies relied on statistical models specific to the organism and transposon, which may be advantageous when specialized information can be uniquely captured (28, 75) but are not generalizable and may be prone to error by the nature of the model specialization required. Here we applied an ML approach, which is more general and easier to implement and has the distinct advantage of being able to integrate an arbitrary number of data features (hits, reads, neighborhood, etc.). Interestingly, the classifier and feature set chosen were powerful enough to achieve accurate predictions across organisms—for example, the classifier for S. pombe achieved an AUC of 0.985 in the C. albicans benchmark (Table 4), despite the different transposon used (Hermes).
TABLE 4.
AUCs of across-species benchmarks
| Species | Across-species benchmark AUC |
||
|---|---|---|---|
| C. albicans | S. cerevisiae | S. pombe | |
| C. albicans | 0.981 | 0.938 | |
| S. cerevisiae | 0.993 | 0.942 | |
| S. pombe | 0.985 | 0.97 | |
A disadvantage of a supervised ML approach is the requirement for a reliable training set – prior knowledge of the essentiality status of a considerable number of genes. For determinations of gene essentiality, especially in a nonmeiotic organism, no training set can be perfect, due to the presence of conditionally essential genes and strain-specific genes (3). Even model yeast data are subject to artefacts (35, 44). Nonetheless, annotations of homologs from relatively distant evolutionary relatives, as well as training on a smaller subset of ~70 Ess and NE genes, were sufficient for strong predictions. Coupled with the possibility of training the classifier on a training set from a different organism, this approach should be applicable for the analysis of in vivo transposon data from other non-model organisms.
Similarities and differences in gene essentiality.
The Core5-Ess genes are primarily involved in central processes such as gene expression and cell cycle progression (see Fig. S3 in the supplemental material) and are more likely to be “hubs” (have a high number of genetic interactions) in synthetic genetic array analysis experiments (51) (Fig. 6A). This is consistent with the idea that essential genes are more frequently engaged in central processes that involve larger numbers of genetic partners than nonessential genes.
Genes nonessential in C. albicans and essential in both S. cerevisiae and S. pombe were U2-type spliceosome components (corrected P value, 1.5−6; false-discovery rate, 0.00%), despite the small number of predicted introns in C. albicans (361) and S. cerevisiae (273) relative to S. pombe (2,394). This is consistent with the loss of highly conserved snRNA binding proteins and changes in snRNA sequences within the spliceosome catalytic site in C. albicans relative to S. cerevisiae (76) and supports the idea that spliceosome components evolved rapidly in the hemiascomycete yeasts.
With SATAY, some ORFs were enriched for insertions within the N-terminal region of the coding sequence and caused gain-of-function mutations. This was not evident in either CaTn or SpTn data. We suggest that the larger size of both Hermes and the Ds-NAT1 may be less permissive with respect to spurious transcription initiation events that occur with the smaller Ds used in SATAY (34).
The different types of data (e.g., deletions and repression and transposon insertions in vitro and in vivo) provide a more complete view of gene functions than any single study. The ECS logistic function brings the list of genes with unclear essentiality to less than 6% (see Dataset 6 at https://doi.org/10.6084/m9.figshare.c.4251182). Potential inaccuracies in the in vivo transposon approach have several sources. First, DNA in cells that grow slowly or have died can still be amplified and inflate estimates of Tn-NE genes. Second, essential domains likely differ in a gene-specific manner that defies definitive categorization. Third, ML provides a statistical measure of the essentiality likelihood dependent on the training set quality, which is clearly imperfect. Finally, some C. albicans genes could be conditionally essential because of specific alleles that are present or absent in the haploid strain relative to the parental heterozygous diploid. There are likely to be one or more deleterious alleles in the haploid haplotype (77) as evidenced by their reduced growth rate and virulence relative to SC5314, the heterozygous diploid strain. Such allele-specific interactions could be akin to epistatic synthetic genetic interactions between a deleted allele and another partially functional allele elsewhere in the genome. Other genes may be conditional if they rendered C. albicans more sensitive to nourseothricin, which was used to select for the Ds-NAT1. Future studies of a range of haploid strains with different transposon markers have the potential to address this issue.
Identifying potential drug targets.
C. albicans represents a serious economic and health threat as a human pathogen (5), and the limited armamentarium of antifungal drugs is a major challenge. This work has identified genes essential in C. albicans and Core5-Ess genes that lack human homologs. One major group of these genes consists of components of the DASH kinetochore complex, emphasizing differences with human kinetochores (64). Importantly, 130 essential C. albicans genes have no human homologs but do have homologs in the other three major human fungal pathogens. We suggest that these 130 genes are of high priority as potential targets for antifungal drug design because they could target a larger set of fungal pathogens rather than be specific only to C. albicans or to the CUG clade (see Dataset 7 at https://doi.org/10.6084/m9.figshare.c.4251182). Notably, two of these genes, C1_01490W and C3_07550C, were important for infectivity in a mouse model of candidiasis (13, 78). C1_01490W encodes a plasma membrane protein (79) that is repressed by nitric oxide (80). Analysis of the repression collection using the systemic mouse model found 19 of the essential genes with no human homolog and with homologs in the 4 pathogens. These included several genes involved in the ergosterol biosynthesis pathway, a component of the DASH kinetochore complex, and MET6. They also include groups of genes enriched in mitochondrial membrane organization, drug metabolism, aromatic amino acid synthesis (ARO1, ARO2, and ARO7), and riboflavin biosynthesis (19). The riboflavin biosynthesis pathway is absent in mammals (81), and at least some of its components are essential in the three yeasts analyzed, suggesting that this pathway may be an interesting target for antifungals. Indeed, a recent study ranked the synthesis of riboflavin as being a rich source of antifungal targets (81).
The potential of in vivo transposition approaches for other purposes.
With essential genes identified, the next steps include analyzing the enrichment and depletion of genes in the existing pooled library analyzed here for those nonessential genes that are depleted or enriched under different growth and stress conditions. Meta-analysis of the results will establish the regulatory and metabolic networks that represent relevant host niches.
Another application is pooled synthetic genetic array analysis (51), which is performed by inducing transposition in strains carrying one or more mutations of interest and which will facilitate the detection of genetic interactions at the genome scale in an unbiased and relatively rapid and cost-effective manner. In addition, modifications to the transposon (39) can be engineered to add fluorescent protein or epitope tags to identify genes encoding proteins with specific cellular localization or protein-protein interactions. Inserting strong promoters (“activation tagging”), repressors, DNA binding proteins (e.g., lacI, TetR, or C. albicans DNA binding domains) or sequences that tether the target gene product to a specific cell structure can provide new functional insights as well. In vivo transposition also can guide domain structure-function studies, as elegantly demonstrated in SATAY (34).
In summary, analyzing large numbers of in vivo generated transposon mutants produced in a stable C. albicans haploid strain allows the rapid and efficient analysis of gene essentiality and the identification of potential antifungal drug targets. It also has the potential to greatly improve the amount and quality of phenotypic information available for studying non-model as well as model organisms.
MATERIALS AND METHODS
Plasmids and strains.
All C. albicans strains derived from strain YJB-T900 (GZY896; kindly provided by Guisheng Zeng and Yue Wang) and are listed in Table S4A in the supplemental material. YJB-T900 is a derivative of haploid XI (9), which was ultimately derived from laboratory strain SC5314. C. albicans was grown at 30°C in rich YPAD medium (1% yeast extract, 2% peptone, 2% glucose) under normal conditions and in YPAM medium (1% yeast extract, 2% peptone, 3% maltose) when AcTPase4xCa was induced. Transformants of C. albicans were selected in synthetic complete medium (SDC) (0.17% yeast nitrogen base with ammonium sulfate [Formedium], 2% glucose) supplemented with a dropout mix containing amino and nucleic acids except adenine or uridine, depending on the auxotrophic requirement for the selection (82).
Yeast strains and plasmids and primers used. Download Table S4, PDF file, 0.4 MB (360.1KB, pdf) .
Copyright © 2018 Shtifman Segal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
All media were supplemented with uridine (80 µg ml−1) or adenine (40 µg ml−1) except when used for selection of URA3 or ADE2 transformants. For solid media, 2% Bacto agar was added. Nat+ transformants were selected by plating the transformation mix on YPAD medium and replica plating the following day onto YPAD plates supplemented with 400 µg nourseothricin ml−1 (Jena Bioscience, Jena, Germany). All C. albicans transformations were performed following the haploid electroporation protocol (83). Escherichia coli strain DH-5alpha (Bio-labs Ltd.), and standard media and methods (84) were used for plasmid manipulations.
Yeast genomic DNA was isolated according to a previously described method (85).
Strain YJB-T1792 was constructed by directly transforming yeast with a fragment containing the AcTPase4xCa expression cassette together with a URA3 marker and flanking sequences from the C. albicans NEUT5L locus (86) from NaeI-digested BJB-T135/pKM300 (37) into strain YJB-T900. To integrate Ds-NAT1 into the ADE2 promoter, strain YJB-T1792 was transformed with NotI-digested BJB-T133/pRK402 (Table S4B). The correct integration of both the Ac-URA3 and Ds-NAT1 insertions was verified by PCR amplification of genomic DNA using primers BP104 and BP161 for the Ac-URA3 and primers BP117 and BP118, primers BP119 and BP120, and primers BP117 and BP120 for Ds-NAT1. Exact insertions were confirmed by Sanger sequencing of the amplified fragments generated with these primer sets. The final strain, YJB-T1081, which includes both Ac-URA3 and Ds-NAT1 in the ADE2 promoter, produces red colonies.
Ploidy verification by flow cytometry.
Flow cytometry was performed as described previously (9) using a MACSQuant flow cytometer (Miltenyi Biotec GmbH, Germany) and SYBR green (Lumiprobe) to stain DNA. Ploidy levels were determined relative to known diploid and haploid isolate data.
Growth analysis.
Strains were grown in SDC in a 96-well microtiter plate, and absorbance at 600 nm was measured every 15 min with a Tecan Infinite F200 Pro (Tecan, Switzerland) plate reader for 24 h. Haploid YJB-T900 parent and YJB-T1792 (Ac-only strain) and YJB-T1081/YJB-T1082 (Ac/Ds-NAT1 strain from same initial transformant) strains were grown in SDC medium and showed identical growth rates, which were lower than those of the diploid strain SC5314 and yet higher than those of haploid I (YJB12801) and haploid XI (YJB12881, which was the parent of YJB-T 900), haploid isolates previously studied (9) (Fig. 1A).
Virulence tests in mice.
A total of 15 C57BL/6J female mice, 6 to 8 weeks old, were purchased from Charles River UK Limited and maintained for 1 week at the Medical Research Facility (MRF) at the University of Aberdeen before the experiments were performed.
Candida albicans strains SC5314, YJBT1792, and YJBT1082 were grown overnight in SD-Ura minimal medium. The cells were washed three times with sterile phosphate-buffered saline (PBS), and suspensions of 2 × 106 cells/ml were made in sterile PBS. Five mice per group were randomly chosen for each Candida strain and injected with 100 µl of cell suspension (2 × 105 cells/mice) via the tail vein. The mice in each group were housed together in single individually ventilated cages (IVCs) in category II room facility at the MRF. Food and water were provided to mice ad libitum.
The mice were monitored daily for 2 weeks postinfection. Body weights were recorded each day, and mice were checked twice daily for any signs of clinical illness per the clinical scoring sheet (see Text S1 in the supplemental material). Mice showing a 20% weight loss or a clinical illness score of 2.5 were culled immediately by cervical dislocation. All surviving mice were culled at the end of study period.
The experimental design and protocol were approved by the study plan team at the MRF, and the experiments were performed under U.K. Home Office project license 70/8073 (to Gordon Brown).
Generation of insertion libraries.
A total of 109 YJB-T 1081 cells were grown in 25 ml of freshly prepared YPAM medium for ∼20 to 24 h to induce transposition events. Cells were collected by centrifugation (5 min at 1,000 × g, 20°C), washed twice with double-distilled water (ddH2O), and plated on ∼500 9-cm-diameter plates containing 25 ml of SDC-Ade+nourseothricin. Colonies in which transposon excision repaired the ADE2 gene appeared after 48 h. All colonies are then scraped off the plates using sterile ddH2O, pooled, washed, and frozen in 15% glycerol. To dilute any remaining Ade− or dead cells, ∼109 cells were inoculated in 25 ml SDC-Ade+nourseothricin medium for 48 h. The saturated culture was harvested by centrifugation (5 min, 1,000 × g), washed with sterile ddH2O, and the cell pellets were frozen (Fig. 1E).
Fractionation with Percoll.
To further separate Ade− cells from the pooled culture, we adopted a Percoll separation protocol (87). Percoll (GE17-0891-01) was diluted 9:1 (vol/vol) with 1.5 M NaCl. A Percoll gradient was formed using 10 ml of the Percoll solution in 15-ml tubes that were centrifuged at 13,000 rpm for 15 min at 20°C. Approximately 2 × 109 cells were pelleted, resuspended in 1 ml Tris buffer (pH 7.5), overlaid onto the preformed gradient, and centrifuged at 400 × g for 60 min in a tabletop centrifuge equipped with a swinging bucket rotor (Thermo Instruments) at 20°C. White cells were collected (Fig. 1E), washed once in 40 ml Tris buffer (pH 7.5), pelleted, and resuspended in ddH2O, and then yeast genomic DNA was isolated (85) to produce the insertion libraries.
Transposon insertion sequencing.
A 1-µg volume of genomic DNA from each insertion library was randomly sheared using a Covaris S2 device (Covaris Inc., Woburn, MA, USA). Single-end Illumina libraries were constructed by ligation of Illumina adapters (Table S4C) to sheared DNA. Enrichment of transposon/chromosomal junction regions was performed by PCR amplification with a 5′ biotinylated transposon enrichment primer (BP664) and adapter-specific PCR enrichment primer R_Tnseq_index (Table S4C). All the following transposon sequencing steps were performed as described previously (29, 30). The resulting transposon libraries were quantified on an Agilent 2200 TapeStation system using HS D1000 tape and sequenced using an Illumina NextSeq 500 system (Illumina, USA) high-output v2 kit. The resulting library insertion sequences are available at NCBI under project PRJNA490565 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA490565).
Analysis of transposon libraries. (i) Insertion site mapping.
Of the 11 C. albicans AcDs transposition libraries, the three with two or more insertions per 100 bp (Table 2), libraries 3, 7, and 11, were used for further analysis. Each library’s sequence FASTQ files were processed with cutadapt (88) (version 1.9.1) to remove the leading transposon sequence from the reads and possible trailing Illumina adapter. Reads that did not have the leading transposon sequence were discarded. The remaining reads were mapped onto the C. albicans reference genome using bowtie2 (89) (version 2.2.9) (using the “–very-sensitive” global alignment setting), and the output SAM files were compressed into the BAM format using SAMtools (90) (version 1.3.1). Reads that received a mapping quality score below 20 (i.e., probability of more than 1% of alignment to another region in the genome) were discarded. If an insertion location was attested by reads from both strands, it was counted as two separate insertion events; otherwise, it was counted as a single insertion event. We also observed that high-read-number insertions often would have single-read insertions adjacent to them, differing by only one nucleotide. We deemed this a sequencing error and counted every pair of immediately adjacent insertions as a single insertion. Further in the analysis, insertion sites from the 3 libraries were pooled and treated as a single data set (though they are shown separately in the figures), maintaining the constraint that every base pair was allowed at most two insertions. Numbers of insertions in all of the libraries were determined (see Dataset 8 at https://doi.org/10.6084/m9.figshare.c.4251182).
For S. cerevisiae, we used already-mapped insertion libraries WildType1 and WildType2 (34) and pooled them into one data set for further analysis. For S. pombe, we used sequencing data from references 35 and 94 (accession no. SRA043841.1 and SRR327340) and performed the mapping against version ASM294v2.30 of the S. pombe genome as described above for C. albicans, with the additional step of removing trailing bases that had a sequence quality score below 20 from the sequence file before mapping.
(ii) Selection of appropriate genes for analysis.
The assignment of hits to repetitive genomic regions is ambiguous. To find such regions, we simulated a FASTQ file representing all possible 131-bp-long reads from the C. albicans genome (the mode of read length distribution in the transposon libraries) and aligned it as described above. Consecutive regions with read alignments of mapping quality below 20 were ignored in further analysis, and genes that had more than 5% of such regions (142 genes overall) were discarded. For example, TEF1 and TEF2 (see Fig. S4G in the supplemental material) were excluded because their coding sequences are >85% identical. Additionally, in the case of C. albicans, some regions were not mapped at all, due either to deletions (e.g., URA3 and GAL1 were deleted during the construction of the strain) or to issues in sequencing. As in the case of the repetitive regions, we discarded 12 genes with >5% unmapped regions. Finally, for both training and prediction, only protein-coding genes were considered (marked as ORFs in CGD). Overall, 5,893 genes among 6,198 C. albicans ORFs and 6,620 annotated features in the CGD were used in the analysis (see Dataset 2A at https://doi.org/10.6084/m9.figshare.c.4251182).
A similar repetitive region analysis was performed for S. cerevisiae and S. pombe, with simulated reads with lengths of 75 and 40, respectively, matching the modes of the read lengths in their sequenced data sets (34, 35). Additionally, in S. cerevisiae, ORFs marked as “dubious” and ORFs that had no insertions in them and in a 20-kb window surrounding them were removed, resulting in 5,599 genes being used among 7,542 features (see Dataset 2B at https://doi.org/10.6084/m9.figshare.c.4251182). In S. pombe, 4,985 ORFs among 5,129 were used, 144 ORFs being excluded due to ≥5% genomic duplication (see Dataset 1C at https://doi.org/10.6084/m9.figshare.c.4251182).
(iii) Construction of the training sets.
We constructed “gold standard” training sets of genes that were likely to be essential or nonessential. Genes that had orthologs in both S. cerevisiae and S. pombe and that were consistently marked as essential (i.e., without contradicting evidence) in both organisms were considered likely to be essential (697 genes overall). In contrast, genes that were successfully deleted in a number of high-throughput deletion studies: (12–14) were considered to be likely nonessentials (759 genes overall). Three genes (CDC19, SGT1, and PWP1) were present in both data sets and were discarded. Because the assumptions upon which the training set were imperfect, as not all functions may be conserved, some of the reported deletion mutants might have acquired suppressors (3) and some screens for essentiality might have used different growth conditions. Thus, all 759 genes were visually inspected by three independent observers and we discarded 66 genes that were designated to be clear outliers by all of the observers. The complete training set and those genes that were manually excluded from it were determined (see Dataset 1A at https://doi.org/10.6084/m9.figshare.c.4251182).
For S. cerevisiae and S. pombe training sets, we used a similar construction method (697 essentials in S. cerevisiae and 689 in S. pombe), with the exception that the nonessential training set was constructed from orthologs in both S. cerevisiae and S. pombe that were marked in both by deletion studies (1,777 nonessentials in S. cerevisiae and 1,620 in S. pombe). Outliers were manually excluded as described above (in S. cerevisiae, 6 were discarded as false positives and 32 as false negatives; in S. pombe, 46 were discarded as false positives and 62 were discarded as false negatives).
(iv) Construction of a gene essentiality predictor.
We used the implementation of the random forest classifier (91) in the scikit-learn library (92) (version 0.18.1) with the default parameters. The classifier features are listed in Table 2. The classification quality was measured using the area under the receiver operating characteristic curve (AUC; values ranged from 0 to 1), which describes the sensitivity versus the specificity of the predictions (Fig. S1C). For all organisms, the AUCs were high, with the AUC value for C. albicans being an almost perfect 0.997, greatly outperforming the random expectation AUC of 0.5. For setting the classification threshold, we used a 5-fold cross validation setting. We chose a false-positive rate of 0.9%, yielding a true-positive rate of 92% and a threshold of 0.8 (Table 3). To assess the possibility of cross-organism predictions, we tested each classifier on training sets and features from the other two organisms (see Dataset 1B and C at https://doi.org/10.6084/m9.figshare.c.4251182). Feature importance was evaluated as the mean decrease in impurity (93) as reported by scikit-learn.
All of the code and required dependencies are available at https://github.com/berman-lab/transposon-pipeline.
Determining the essentiality confidence score (ECS).
To capture the contribution from all studies, we first assigned values to each gene in each data set, for each study independently, with scores of +1 for essential, 0 for no data, and −1 for nonessential, on the basis of the reported study results. This was done for deletion studies (11–14), for the newer repression study (19), and for the Tn study, where we assigned discrete “RF verdict” scores of +1 and −1 on the basis of the prediction verdict (RF score of ≥0.80, Ess; RF score of <0.80, NE) as described above.
The “net essentiality score” was then determined as the sum of all the values from all the other studies. The net essentiality scores ranged from +2 to −6 in discrete integer steps. (There are more deletion experiments, which can give scores of only −1 each, relative to the repression and CaTn experiments, which can give +1 and −1 results).
We then determined the essentiality confidence score (ECS) by applying a logistic function to the net essentiality score as follows:
where a = 1.55 was determined to achieve a value of >0.95 when x = 2 and a value of >0.99 when x = 3 and where Xi is the net essentiality score (sum of all studies) for a given gene. The resulting ECS range was 0 ≤ Xi ≤ 1 for each gene.
Nucleosome bias analysis.
The likelihood of mononucleosome occupancy in C. albicans was determined by mapping read depth from micrococcal nuclease experiments (74, 94) (accession no. SRR059732) as a measure of nucleosome occupancy likelihood (where higher numbers of reads correspond to a higher likelihood of nucleosome occupancy). For each chromosome, the median read depth was used to separate it into regions of high and low nucleosome occupancy likelihood. We then compared the numbers of TnSeq log reads and numbers of hits between the two region types for each chromosome in each high-insertion density library. Results are shown in Table S2 (Mann-Whitney U test yielded P value for every comparison, <10−6).
Sequences and annotations.
For C. albicans, the reference genome was haplotype A of Assembly 22, version s07-m01-r08 (38). For S. cerevisiae, the reference genome was R64-2-1 (95). For S. pombe, the reference genome was ASM294v2.30 (96). C. albicans ortholog and protein domain annotations were taken from the CGD (38). The S. cerevisiae feature annotations were downloaded from the SGD website (42). Essential and nonessential genes were called by collecting all phenotype annotations on the website and using only those that were annotated exclusively as either essential or nonessential. The S. pombe feature and essentiality annotations were downloaded from PomBase (43, 97).
Note that the specific transposons used in the three yeasts differed in a number of ways. The Ds used for C. albicans is 1,812 bp and includes the Nat1 ORF; the mini-Ds for SATAY is only ∼600 bp (34); and the Hermes transposon for S. pombe was ∼1,000 bp in length and included the ∼1,500-bp kanMX6 ORF (35). Notably, insertion of the mini-Ds in the SATAY study resulted in both loss-of-function and gain-of-function mutations (34), while only loss-of-function mutations were evident for the C. albicans Ds-NAT1 and the S. pombe Hermes insertion mutants.
Analysis of genetic interaction density.
Genetic interactions of S. cerevisiae genes were obtained using synthetic genetic array (51) and kindly provided by B. VanderSluis. For each gene, the number of interactions was normalized by the number of observations/experiments in which that gene was measured, which provided the GIPD score. GIPD scores for multiple alleles of the same gene were averaged. Negative genetic interactions were found to be more functionally informative (51). “Stringent” negative GIPD (nsGIPD) scores (98) were selected for further analysis in C. albicans and S. pombe orthologs, filtered by the use of the genetic threshold described by Costanzo et al (51). Essential and nonessential gene populations in each organism were compared using the Wilcoxon rank sum test.
Examining the conservation between human genes and fungal pathogens.
To determine the number of CaTn-Ess genes with homologs in humans or the major human pathogens, we conducted individual searches with each essential C. albicans gene for a homologous gene in the relevant proteome using BLASTP from NCBI’s BLAST+, version 2.3.0 (99), and an expectation value threshold of 1e−3 as recommended for searches for homologous sequences (100). We then compared proteins encoded by the CaTn-Ess genes to the proteomes of Aspergillus fumigatus af293 (Eurotiomycetes, Ascomycota), Cryptococcus neoformans H99 (Tremellomycetes, Basidiomycota), and Histoplasma capsulatum H88 (Eurotiomycetes, Ascomycota) using proteome data for the fungi obtained from FungiDB (http://fungidb.org/fungidb/) release 38 and human proteome data from the NCBI (Human Genome Assembly GRCh38.p12). To determine homologs, we used the same approach as described for human homologs using BLASTP from NCBI’s BLAST+, version 2.3.0 (100), using an expectation value threshold of 1e−3. Of course, we cannot rule out the possibility that distant orthologs were not detected with the stringent sequence similarities used here.
Data availability.
All of the code and required dependencies for analysis of the TnSeq data are available at https://github.com/berman-lab/transposon-pipeline.
Library insertion sequences are available at NCBI under project PRJNA490565 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA490565). Datasets S1 through S9 are available at https://doi.org/10.6084/m9.figshare.c.4251182.
ACKNOWLEDGMENTS
We thank Michael Bromley, Martin Kupiec, Kyle Cunningham, Aimee Dudley, Gareth Cromie, and Chad Myers for helpful discussions and Delyth Reid and Ivy Dambuza for advice and help with mouse experiments. We thank Aaron Mitchell, Agnes Michel, Benoit Kornmann, Brenda Andrews, and Benjamin VanderSluis for providing data prior to publications and Monica Mascarenas for help with Tn sequencing protocols and primer design. We thank Jon Binkley and Marek Skryzpek from CGD and Midori Harris and Valerie Wood from PomBase for providing useful data sets and Aaron Mitchell, Anna Selmecki, and CGD and PomBase staff members for helpful feedback on the manuscript.
This work was supported by European Research Council Advanced Award 340087 (RAPLODAPT) to J.B., the Dahlem Centre of Plant Sciences (DCPS) of the Freie Universität Berlin (R.K.), Israel Science Foundation grant no. 715/18 (R.S.), the Wellcome Trust (grants 086827, 075470, 101873, and 200208) and the MRC Centre for Medical Mycology (N006364/1) (N.A.R.G.).
Footnotes
Citation Segal ES, Gritsenko V, Levitan A, Yadav B, Dror N, Steenwyk JL, Silberberg Y, Mielich K, Rokas A, Gow NAR, Kunze R, Sharan R, Berman J. 2018. Gene essentiality analyzed by in vivo transposon mutagenesis and machine learning in a stable haploid isolate of Candida albicans. mBio 9:e02048-18. https://doi.org/10.1128/mBio.02048-18.
Contributor Information
Antonio Di Pietro, Universidad de Córdoba.
Nathan Springer, University of Minnesota.
Bernhard Hube, Friedrich Schiller University Jena.
REFERENCES
- 1.Forsburg SL. 2001. The art and design of genetic screens: yeast. Nat Rev Genet 2:659–668. doi: 10.1038/35088500. [DOI] [PubMed] [Google Scholar]
- 2.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, Dow S, Lucau-Danila A, Anderson K, André B, Arkin AP, Astromoff A, El Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian K-D, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Güldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kötter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang C-y, Ward TR, Wilhelmy J, Winzeler EA, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 3.Kim D-U, Hayles J, Kim D, Wood V, Park H-O, Won M, Yoo H-S, Duhig T, Nam M, Palmer G, Han S, Jeffery L, Baek S-T, Lee H, Shim YS, Lee M, Kim L, Heo K-S, Noh EJ, Lee A-R, Jang Y-J, Chung K-S, Choi S-J, Park J-Y, Park Y, Kim HM, Park S-K, Park H-J, Kang E-J, Kim HB, Kang H-S, Park H-M, Kim K, Song K, Song KB, Nurse P, Hoe K-L. 2010. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol 28:617–623. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winzeler EA, 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 5.Brown GD, Denning DW, Gow NAR, Levitz SM, Netea MG, White TC. 2012. Hidden killers: human fungal infections. Sci Transl Med 4:165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 6.Denning DW, Bromley MJ. 2015. How to bolster the antifungal pipeline. Science 347:1414–1416. doi: 10.1126/science.aaa6097. [DOI] [PubMed] [Google Scholar]
- 7.Sears D, Schwartz BS. 2017. Candida auris: an emerging multidrug-resistant pathogen. Int J Infect Dis 63:95–98. doi: 10.1016/j.ijid.2017.08.017. [DOI] [PubMed] [Google Scholar]
- 8.Kullberg BJ, Arendrup MC. 2015. Invasive candidiasis. N Engl J Med 373:1445–1456. doi: 10.1056/NEJMra1315399. [DOI] [PubMed] [Google Scholar]
- 9.Hickman MA, Zeng G, Forche A, Hirakawa MP, Abbey D, Harrison BD, Wang Y-M, Su C-h, Bennett RJ, Wang Y, Berman J. 2013. The 'obligate diploid' Candida albicans forms mating-competent haploids. Nature 494:55–59. doi: 10.1038/nature11865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McKee AH, Kleckner N. 1997. A general method for identifying recessive diploid-specific mutations in Saccharomyces cerevisiae, its application to the isolation of mutants blocked at intermediate stages of meiotic prophase and characterization of a new gene SAE2. Genetics 146:797–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y, Mallick J, Maqnas A, Sun Y, Choudhury BI, Côte P, Yan L, Ni T-j-h, Li Y, Zhang D, Rodríguez-Ortiz R, Lv Q-z, Jiang Y-y, Whiteway M. 2018. Chemogenomic profiling of the fungal pathogen Candida albicans. Antimicrob Agents Chemother 62:e02365-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Homann OR, Dea J, Noble SM, Johnson AD. 2009. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet 5:e1000783. doi: 10.1371/journal.pgen.1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noble SM, French S, Kohn LA, Chen V, Johnson AD. 2010. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet 42:590–598. doi: 10.1038/ng.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vandeputte P, Ischer F, Sanglard D, Coste AT. 2011. In vivo systematic analysis of Candida albicans Zn2-Cys6 transcription factors mutants for mice organ colonization. PLoS One 6:e26962. doi: 10.1371/journal.pone.0026962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michel S, Ushinsky S, Klebl B, Leberer E, Thomas D, Whiteway M, Morschhäuser J. 2002. Generation of conditional lethal Candida albicans mutants by inducible deletion of essential genes. Mol Microbiol 46:269–280. doi: 10.1046/j.1365-2958.2002.03167.x. [DOI] [PubMed] [Google Scholar]
- 16.Nobile CJ, Mitchell AP. 2009. Large-scale gene disruption using the UAU1 cassette. Methods Mol Biol 499:175–194. doi: 10.1007/978-1-60327-151-6_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oh J, Fung E, Schlecht U, Davis RW, Giaever G, St Onge RP, Deutschbauer A, Nislow C. 2010. Gene annotation and drug target discovery in Candida albicans with a tagged transposon mutant collection. PLoS Pathog 6:e1001140. doi: 10.1371/journal.ppat.1001140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uhl MA. 2003. Haploinsufficiency-based large-scale forward genetic analysis of filamentous growth in the diploid human fungal pathogen C. albicans. EMBO J 22:2668–2678. doi: 10.1093/emboj/cdg256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Meara TR, Veri AO, Ketela T, Jiang B, Roemer T, Cowen LE. 2015. Global analysis of fungal morphology exposes mechanisms of host cell escape. Nat Commun 6:6741. doi: 10.1038/ncomms7741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roemer T, Jiang B, Davison J, Ketela T, Veillette K, Breton A, Tandia F, Linteau A, Sillaots S, Marta C, Martel N, Veronneau S, Lemieux S, Kauffman S, Becker J, Storms R, Boone C, Bussey H. 2003. Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol Microbiol 50:167–181. doi: 10.1046/j.1365-2958.2003.03697.x. [DOI] [PubMed] [Google Scholar]
- 21.Xu D, Jiang B, Ketela T, Lemieux S, Veillette K, Martel N, Davison J, Sillaots S, Trosok S, Bachewich C, Bussey H, Youngman P, Roemer T. 2007. Genome-wide fitness test and mechanism-of-action studies of inhibitory compounds in Candida albicans. PLoS Pathog 3:e92. doi: 10.1371/journal.ppat.0030092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson RB. 2000. A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast 16:65–70. doi:. [DOI] [PubMed] [Google Scholar]
- 23.Min K, Ichikawa Y, Woolford CA, Mitchell AP. 2016. Candida albicans gene deletion with a transient CRISPR-Cas9 system. mSphere 1:e00130-16. doi: 10.1128/mSphere.00130-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen N, Mmf Q, Hernday AD. 2017. An efficient, rapid, and recyclable system for CRISPR-mediated genome editing in Candida albicans. mSphere 2:e00149-17. doi: 10.1128/mSphereDirect.00149-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shapiro RS, Chavez A, Porter CBM, Hamblin M, Kaas CS, DiCarlo JE, Zeng G, Xu X, Revtovich AV, Kirienko NV, Wang Y, Church GM, Collins JJ. 2018. A CRISPR-Cas9-based gene drive platform for genetic interaction analysis in Candida albicans. Nat Microbiol 3:73–82. doi: 10.1038/s41564-017-0043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vyas VK, Barrasa MI, Fink GR. 2015. A Candida albicans CRISPR system permits genetic engineering of essential genes and gene families. Sci Adv 1:e1500248. doi: 10.1126/sciadv.1500248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skrzypek MS, Binkley GBJ, Miyasato SR, Simison M, Sherlock G. 2018. Candida Genome Database. http://www.candidagenome.org/.
- 28.Langridge GC, Phan M-D, Turner DJ, Perkins TT, Parts L, Haase J, Charles I, Maskell DJ, Peters SE, Dougan G, Wain J, Parkhill J, Turner AK. 2009. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res 19:2308–2316. doi: 10.1101/gr.097097.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Opijnen T, Bodi KL, Camilli A. 2009. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods 6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Opijnen T, Camilli A. 2013. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat Rev Microbiol 11:435–442. doi: 10.1038/nrmicro3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price MN, Wetmore KM, Waters RJ, Callaghan M, Ray J, Liu H, Kuehl JV, Melnyk RA, Lamson JS, Suh Y, Carlson HK, Esquivel Z, Sadeeshkumar H, Chakraborty R, Zane GM, Rubin BE, Wall JD, Visel A, Bristow J, Blow MJ, Arkin AP, Deutschbauer AM. 2018. Mutant phenotypes for thousands of bacterial genes of unknown function. Nature 557:503–509. doi: 10.1038/s41586-018-0124-0. [DOI] [PubMed] [Google Scholar]
- 32.McClintock B. 1950. The origin and behavior of mutable loci in maize. Proc Natl Acad Sci U S A 36:344–355. doi: 10.1073/pnas.36.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weil CF, Kunze R. 2000. Transposition of maize Ac/Ds transposable elements in the yeast Saccharomyces cerevisiae. Nat Genet 26:187–190. doi: 10.1038/82827. [DOI] [PubMed] [Google Scholar]
- 34.Michel AH, Hatakeyama R, Kimmig P, Arter M, Peter M, Matos AJ, De Virgilio C, Kornmann B. 2017. Functional mapping of yeast genomes by saturated transposition. Elife 6:e23570. doi: 10.7554/eLife.23570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo Y, Park JM, Cui B, Humes E, Gangadharan S, Hung S, FitzGerald PC, Hoe K-L, Grewal SIS, Craig NL, Levin HL. 2013. Integration profiling of gene function with dense maps of transposon integration. Genetics 195:599–609. doi: 10.1534/genetics.113.152744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carr PD, Tuckwell D, Hey PM, Simon L, d'Enfert C, Birch M, Oliver JD, Bromley MJ. 2010. The transposon impala is activated by low temperatures: use of a controlled transposition system to identify genes critical for viability of Aspergillus fumigatus. Eukaryot Cell 9:438–448. doi: 10.1128/EC.00324-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mielich K, Shtifman-Segal E, Golz JC, Zeng G, Wang Y, Berman J, Kunze R. 2018. Maize transposable elements Ac/Ds as insertion mutagenesis tools in Candida albicans. G3 (Bethesda) 8:1139–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skrzypek MS, Binkley J, Binkley G, Miyasato SR, Simison M, Sherlock G. 2017. The Candida Genome Database (CGD): incorporation of Assembly 22, systematic identifiers and visualization of high throughput sequencing data. Nucleic Acids Res 45:D592–D596. doi: 10.1093/nar/gkw924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lazarow K, Doll ML, Kunze R. 2013. Molecular biology of maize Ac/Ds elements: an overview. Methods Mol Biol 1057:59–82. doi: 10.1007/978-1-62703-568-2_5. [DOI] [PubMed] [Google Scholar]
- 40.Vollbrecht E, Duvick J, Schares JP, Ahern KR, Deewatthanawong P, Xu L, Conrad LJ, Kikuchi K, Kubinec TA, Hall BD, Weeks R, Unger-Wallace E, Muszynski M, Brendel VP, Brutnell TP. 2010. Genome-wide distribution of transposed Dissociation elements in maize. Plant Cell 22:1667–1685. doi: 10.1105/tpc.109.073452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sipiczki M. 2000. Where does fission yeast sit on the tree of life? Genome Biol 1:REVIEWS1011. doi: 10.1186/gb-2000-1-2-reviews1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hirschman JE, Hitz BC, Karra K, Krieger CJ, Miyasato SR, Nash RS, Park J, Skrzypek MS, Simison M, Weng S, Wong ED. 2012. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res 40:D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wood V, Harris MA, McDowall MD, Rutherford K, Vaughan BW, Staines DM, Aslett M, Lock A, Bahler J, Kersey PJ, Oliver SG. 2012. PomBase: a comprehensive online resource for fission yeast. Nucleic Acids Res 40:D695–D699. doi: 10.1093/nar/gkr853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teng X, Dayhoff-Brannigan M, Cheng W-C, Gilbert CE, Sing CN, Diny NL, Wheelan SJ, Dunham MJ, Boeke JD, Pineda FJ, Hardwick JM. 2013. Genome-wide consequences of deleting any single gene. Mol Cell 52:485–494. doi: 10.1016/j.molcel.2013.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dowell RD, Ryan O, Jansen A, Cheung D, Agarwala S, Danford T, Bernstein DA, Rolfe PA, Heisler LE, Chin B, Nislow C, Giaever G, Phillips PC, Fink GR, Gifford DK, Boone C. 2010. Genotype to phenotype: a complex problem. Science 328:469–469. doi: 10.1126/science.1189015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hou J, van Leeuwen J, Andrews BJ, Boone C. 2018. Genetic network complexity shapes background-dependent phenotypic expression. Trends Genet 34:578–586. doi: 10.1016/j.tig.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi Q-M, Wang Y-M, Zheng X-D, Teck Ho Lee R, Wang Y. 2007. Critical role of DNA checkpoints in mediating genotoxic-stress-induced filamentous growth in Candida albicans. Mol Biol Cell 18:815–826. doi: 10.1091/mbc.e06-05-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bruno VM, Wang Z, Marjani SL, Euskirchen GM, Martin J, Sherlock G, Snyder M. 2010. Comprehensive annotation of the transcriptome of the human fungal pathogen Candida albicans using RNA-seq. Genome Res 20:1451–1458. doi: 10.1101/gr.109553.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chandler CH, Chari S, Dworkin I. 2013. Does your gene need a background check? How genetic background impacts the analysis of mutations, genes, and evolution. Trends Genet 29:358–366. doi: 10.1016/j.tig.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fournier T, Schacherer J. 2017. Genetic backgrounds and hidden trait complexity in natural populations. Curr Opin Genet Dev 47:48–53. doi: 10.1016/j.gde.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Costanzo M, VanderSluis B, Koch EN, Baryshnikova A, Pons C, Tan G, Wang W, Usaj M, Hanchard J, Lee SD, Pelechano V, Styles EB, Billmann M, van Leeuwen J, van Dyk N, Lin Z-Y, Kuzmin E, Nelson J, Piotrowski JS, Srikumar T, Bahr S, Chen Y, Deshpande R, Kurat CF, Li1 SC, Li Z, Mattiazzi Usaj M, Okada H, Pascoe N, San Luis B-J, Sharifpoor S, Shuteriqi E, Simpkins SW, Snider J, Garadi Suresh H, Tan Y, Zhu H, Malod-Dognin N, Janjic V, Przulj N, Troyanskaya OG, Stagljar I, Xia T, Ohya Y. 2016. A global genetic interaction network maps a wiring diagram of cellular function. Science 353:aaf1420. doi: 10.1126/science.aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.VanderSluis B, Costanzo M, Billmann M, Ward HN, Myers CL, Andrews BJ, Boone C. 2018. Integrating genetic and protein-protein interaction networks maps a functional wiring diagram of a cell. Curr Opin Microbiol 45:170–179. doi: 10.1016/j.mib.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ihmels J, Collins SR, Schuldiner M, Krogan NJ, Weissman JS. 2007. Backup without redundancy: genetic interactions reveal the cost of duplicate gene loss. Mol Sys Biol 3:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Musso G, Costanzo M, Huangfu M, Smith AM, Paw J, San Luis B-J, Boone C, Giaever G, Nislow C, Emili A, Zhang Z. 2008. The extensive and condition-dependent nature of epistasis among whole-genome duplicates in yeast. Genome Res 18:1092–1099. doi: 10.1101/gr.076174.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matangkasombut O, Buratowski RM, Swilling NW, Buratowski S. 2000. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev 14:951–962. [PMC free article] [PubMed] [Google Scholar]
- 56.Mietton F, Ferri E, Champleboux M, Zala N, Maubon D, Zhou Y, Harbut M, Spittler D, Garnaud C, Courçon M, Chauvel M, d’Enfert C, Kashemirov BA, Hull M, Cornet M, McKenna CE, Govin J, Petosa C. 2017. Selective BET bromodomain inhibition as an antifungal therapeutic strategy. Nat Commun 8:15482. doi: 10.1038/ncomms15482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Byrne KH, Wolfe KP. 2005. The Yeast Gene Order browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res 15:1456–1461. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chua P, Roeder GS. 1995. Bdf1, a yeast chromosomal protein required for sporulation. Mol Cell Biol 15:3685–3696. doi: 10.1128/MCB.15.7.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lygerou Z, Conesa C, Lesage P, Swanson RN, Ruet A, Carlson M, Sentenac A, Séraphin B. 1994. The yeast BDF1 gene encodes a transcription factor involved in the expression of a broad class of genes including snRNAs. Nucleic Acids Res 22:5332–5340. doi: 10.1093/nar/22.24.5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ihmels J, Bergmann S, Gerami-Nejad M, Yanai B, McClellan M, Berman J, Barkai N. 2005. Rewiring of the yeast transcriptional network through the evolution of motif usage. Science 309:938–940. doi: 10.1126/science.1113833. [DOI] [PubMed] [Google Scholar]
- 61.Chapa y Lazo B, Bates S, Sudbery P. 2005. The G1 cyclin Cln3 regulates morphogenesis in Candida albicans. Eukaryot Cell 4:90–94. doi: 10.1128/EC.4.1.90-94.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Davis DA, Bruno VM, Loza L, Filler SG, Mitchell AP. 2002. Candida albicans Mds3p, a conserved regulator of pH responses and virulence identified through insertional mutagenesis. Genetics 162:1573–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suliman HS, Appling DR, Robertus JD. 2007. The gene for cobalamin-independent methionine synthase is essential in Candida albicans: a potential antifungal target. Arch Biochem Biophys 467:218–226. doi: 10.1016/j.abb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Burrack LS, Applen SE, Berman J. 2011. The requirement for the Dam1 complex is dependent upon the number of kinetochore proteins and microtubules. Curr Biol 21:889–896. doi: 10.1016/j.cub.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Padmanabhan S, Thakur J, Siddharthan R, Sanyal K. 2008. Rapid evolution of Cse4p-rich centromeric DNA sequences in closely related pathogenic yeasts, Candida albicans and Candida dubliniensis. Proc Natl Acad Sci U S A 105:19797–19802. doi: 10.1073/pnas.0809770105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Richter G, Fischer M, Krieger C, Eberhardt S, Lüttgen H, Gerstenschläger I, Bacher A. 1997. Biosynthesis of riboflavin: characterization of the bifunctional deaminase-reductase of Escherichia coli and Bacillus subtilis. J Bacteriol 179:2022–2028. doi: 10.1128/jb.179.6.2022-2028.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gangadharan S, Mularoni L, Fain-Thornton J, Wheelan SJ, Craig NL. 2010. DNA transposon Hermes inserts into DNA in nucleosome-free regions in vivo. Proc Natl Acad Sci U S A 107:21966–21972. doi: 10.1073/pnas.1016382107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Meir Y-JJ, Weirauch MT, Yang H-S, Chung P-C, Yu RK, Wu SC-Y. 2011. Genome-wide target profiling of piggyBac and Tol2 in HEK 293: pros and cons for gene discovery and gene therapy. BMC Biotechnol 11:28. doi: 10.1186/1472-6750-11-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lazarow K, Du M-L, Weimer R, Kunze R. 2012. A hyperactive transposase of the maize transposable element Activator (Ac). Genetics 191:747–756. doi: 10.1534/genetics.112.139642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cohen Y, Schuldiner M. 2011. Advanced methods for high-throughput microscopy screening of genetically modified yeast libraries. Methods Mol Biol 781:127–159. doi: 10.1007/978-1-61779-276-2_8. [DOI] [PubMed] [Google Scholar]
- 71.Ryan O, Shapiro RS, Kurat CF, Mayhew D, Baryshnikova A, Chin B, Lin Z-Y, Cox MJ, Vizeacoumar F, Cheung D, Bahr S, Tsui K, Tebbji F, Sellam A, Istel F, Schwarzmuller T, Reynolds TB, Kuchler K, Gifford DK, Whiteway M, Giaever G, Nislow C, Costanzo M, Gingras A-C, Mitra RD, Andrews B, Fink GR, Cowen LE, Boone C. 2012. Global gene deletion analysis exploring yeast filamentous growth. Science 337:1353–1356. doi: 10.1126/science.1224339. [DOI] [PubMed] [Google Scholar]
- 72.Weill U, Yofe I, Sass E, Stynen B, Davidi D, Natarajan J, Ben-Menachem R, Avihou Z, Goldman O, Harpaz N, Chuartzman S, Kniazev K, Knoblach B, Laborenz J, Boos F, Kowarzyk J, Ben-Dor S, Zalckvar E, Herrmann JM, Rachubinski RA, Pines O, Rapaport D, Michnick SW, Levy ED, Schuldiner M. 2018. Genome-wide SWAp-Tag yeast libraries for proteome exploration. Nat Methods 15:617–622. doi: 10.1038/s41592-018-0044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yofe I, Weill U, Meurer M, Chuartzman S, Zalckvar E, Goldman O, Ben-Dor S, Schütze C, Wiedemann N, Knop M, Khmelinskii A, Schuldiner M. 2016. One library to make them all: streamlining the creation of yeast libraries via a SWAp-Tag strategy. Nat Methods 13:371–378. doi: 10.1038/nmeth.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tsankov AM, Thompson DA, Socha A, Regev A, Rando OJ. 2010. The role of nucleosome positioning in the evolution of gene regulation. PLoS Biol 8:e1000414. doi: 10.1371/journal.pbio.1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Griffin JE, Gawronski JD, DeJesus MA, Ioerger TR, Akerley BJ, Sassetti CM. 2011. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog 7:e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mitrovich QM, Guthrie C. 2007. Evolution of small nuclear RNAs in S. cerevisiae, C. albicans, and other hemiascomycetous yeasts. RNA 13:2066–2080. doi: 10.1261/rna.766607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Muzzey D, Schwartz K, Weissman JS, Sherlock G. 2013. Assembly of a phased diploid Candida albicans genome facilitates allele-specific measurements and provides a simple model for repeat and indel structure. Genome Biol 14:R97. doi: 10.1186/gb-2013-14-9-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Becker JM, Kauffman SJ, Hauser M, Huang L, Lin M, Sillaots S, Jiang B, Xu D, Roemer T. 2010. Pathway analysis of Candida albicans survival and virulence determinants in a murine infection model. Proc Natl Acad Sci U S A 107:22044–22049. doi: 10.1073/pnas.1009845107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cabezón V, Llama-Palacios A, Nombela C, Monteoliva L, Gil C. 2009. Analysis of Candida albicans plasma membrane proteome. Proteomics 9:4770–4786. doi: 10.1002/pmic.200800988. [DOI] [PubMed] [Google Scholar]
- 80.Hromatka BS, Noble SM, Johnson AD. 2005. Transcriptional response of Candida albicans to nitric oxide and the role of the YHB1 gene in nitrosative stress and virulence. Mol Biol Cell 16:4814–4826. doi: 10.1091/mbc.e05-05-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meir Z, Osherov N. 2018. Vitamin biosynthesis as an antifungal target. J Fungi [Basel) 4:72. doi: 10.3390/jof4020072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Amberg DC. Burke DJ, Strathern JN. 2005. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 83.Zeng G. 2014. One-step targeted gene deletion in Candida albicans haploids. Nat Protoc 9:464–473. doi: 10.1038/nprot.2014.029. [DOI] [PubMed] [Google Scholar]
- 84.Ausubel FM, Brent R. 1995. Current protocols in molecular biology. Wiley, New York, NY. [Google Scholar]
- 85.Hoffman CS, Winston F. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 86.Gerami-Nejad M, Zacchi LF, McClellan M, Matter K, Berman J. 2013. Shuttle vectors for facile gap repair cloning and integration into a neutral locus in Candida albicans. Microbiology 159:565–579. doi: 10.1099/mic.0.064097-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Allen C, Büttner S, Aragon AD, Thomas JA, Meirelles O, Jaetao JE, Benn D, Ruby SW, Veenhuis M, Madeo F, Werner-Washburne M. 2006. Isolation of quiescent and nonquiescent cells from yeast stationary-phase cultures. J Cell Physiol 174:89–100. doi: 10.1083/jcb.200604072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:3. [Google Scholar]
- 89.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Breiman L. 2001. Random forests. Mach Learn 45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 92.Pedregosa F. 2011. Scikit-learn: machine learning in Python. J Mach Learn Res 12:2825–2830. [Google Scholar]
- 93.Breiman L. 1984. Classification and regression trees. Wadsworth Statistics/Probability series, p 184–185. Wadsworth International Group, Belmont, CA. [Google Scholar]
- 94.NCBI Resource Coordinators. 2018. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 46:D8–D13. doi: 10.1093/nar/gkx1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Engel SR, Dietrich FS, Fisk DG, Binkley G, Balakrishnan R, Costanzo MC, Dwight SS, Hitz BC, Karra K, Nash RS, Weng S, Wong ED, Lloyd P, Skrzypek MS, Miyasato SR, Simison M, Cherry JM. 2014. The reference genome sequence of Saccharomyces cerevisiae: then and now. G3 4:389–398. doi: 10.1534/g3.113.008995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wood V, Gwilliam R, Rajandream M-A, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, Basham D, Bowman S, Brooks K, Brown D, Brown S, Chillingworth T, Churcher C, Collins M, Connor R, Cronin A, Davis P, Feltwell T, Fraser A, Gentles S, Goble A, Hamlin N, Harris D, Hidalgo J, Hodgson G, Holroyd S, Hornsby T, Howarth S, Huckle EJ, Hunt S, Jagels K, James K, Jones L, Jones M, Leather S, McDonald S, McLean J, Mooney P, Moule S, Mungall K, Murphy L, Niblett D, Odell C, Oliver K, O'Neil S, Pearson D, et al. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- 97.Harris MA, Lock A, Bahler J, Oliver SG, Wood V. 2013. FYPO: the fission yeast phenotype ontology. Bioinformatics 29:1671–1678. doi: 10.1093/bioinformatics/btt266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Baryshnikova A, Costanzo M, Kim Y, Ding H, Koh J, Toufighi K, Youn J-Y, Ou J, San Luis B-J, Bandyopadhyay S, Hibbs M, Hess D, Gingras A-C, Bader GD, Troyanskaya OG, Brown GW, Andrews B, Boone C, Myers CL. 2010. Quantitative analysis of fitness and genetic interactions in yeast on a genome scale. Nat Methods 7:1017–1024. doi: 10.1038/nmeth.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Madden T. 2013. The BLAST sequence analysis tool In The NCBI handbook, 2nd ed National Center for Biotechnology Information, Bethesda, MD. [Google Scholar]
- 100.Pearson WR. 2013. An introduction to sequence similarity (“homology”) searching. Curr Protoc Bioinformatics Chapter 3:Unit3.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Virulence of haploid reads, transposon insertion read maps, and machine learning performance. Download FIG S1, TIF file, 0.2 MB (336.4KB, tif) .
Copyright © 2018 Shtifman Segal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Annotated features in C. albicans. Download Table S1, PDF file, 0.2 MB (245.6KB, pdf) .
Copyright © 2018 Shtifman Segal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Number of insertions and reads in regions with low and high likelihood of nucleosome occupancy. Download Table S2, PDF file, 0.02 MB (16.7KB, pdf) .
Copyright © 2018 Shtifman Segal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Hit maps of C. albicans and S. cerevisiae genes with conserved organization of essential domains, examples of misannotated C. albicans ORFs, and alignment of proteins that they predict. Download FIG S2, TIF file, 0.4 MB (760.8KB, tif) .
Copyright © 2018 Shtifman Segal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Specific examples of C. albicans genes and their paralogs in S. cerevisiae and/or S. pombe. Download FIG S4, TIF file, 0.4 MB (697.5KB, tif) .
Copyright © 2018 Shtifman Segal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gene ontology (GO) term analysis of Core5 essential genes and Core5 nonessential genes. Download FIG S3, TIF file, 0.2 MB (155.6KB, tif) .
Copyright © 2018 Shtifman Segal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expanded supplemental figure legends with references. Download Text S1, DOCX file, 0.1 MB (56.7KB, docx) .
Copyright © 2018 Shtifman Segal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Riboflavin biosynthesis pathway analysis. Download Table S3, PDF file, 0.1 MB (67.9KB, pdf) .
Copyright © 2018 Shtifman Segal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Yeast strains and plasmids and primers used. Download Table S4, PDF file, 0.4 MB (360.1KB, pdf) .
Copyright © 2018 Shtifman Segal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
All of the code and required dependencies for analysis of the TnSeq data are available at https://github.com/berman-lab/transposon-pipeline.
Library insertion sequences are available at NCBI under project PRJNA490565 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA490565). Datasets S1 through S9 are available at https://doi.org/10.6084/m9.figshare.c.4251182.