Abstract
BZ junctions, which connect B-DNA to Z-DNA, are necessary for local transformation of B-DNA to Z-DNA in the genome. However, the limited information on the junction-forming sequences and junction structures has led to a lack of understanding of the structural diversity and sequence preferences of BZ junctions. We determined three crystal structures of BZ junctions with diverse sequences followed by spectroscopic validation of DNA conformation. The structural features of the BZ junctions were well conserved regardless of sequences via the continuous base stacking through B-to-Z DNA with A-T base extrusion. However, the sequence-dependent structural heterogeneity of the junctions was also observed in base step parameters that are correlated with steric constraints imposed during Z-DNA formation. Further, circular dichroism and fluorescence-based analysis of BZ junctions revealed that a base extrusion was only found at the A-T base pair present next to a stable dinucleotide Z-DNA unit. Our findings suggest that Z-DNA formation in the genome is influenced by the sequence preference for BZ junctions.
INTRODUCTION
The DNA inside cells usually forms a B-DNA conformation. However, when exposed to certain biochemical and biophysical environments, DNA can adopt a non-B-DNA conformation, which affects various cellular functions such as replication, transcription and genome stability (1–3). Z-DNA, one of the non-B-DNA conformers, adopts a distinct left-handed conformation with a zig-zag arrangement of the phosphate backbone (2) as the result of the alternative stacks of syn and anti conformations of the bases (4). Therefore, Z-DNA preferentially occurs at the purine and pyrimidine repeat sequences, as purine bases can adopt a syn conformation without energy penalty (5). Considering that there are many CG repeat sequences in the genome, it is expected that Z-DNA may be present in numerous genome sites under the appropriate physiological conditions; this has been proven experimentally (6). It is expected that BZ junctions with diverse sequences are formed in each side of the Z-DNA since Z-DNA formation in linear B-DNA is accompanied by the formation of a junction between B-DNA and Z-DNA (i.e. a BZ junction). However, there is limited information on the BZ junction structure and the junction forming site, which has prevented a comprehensive understanding its role in Z-DNA formation in the genome. Therefore, it is necessary to study the structural and sequential diversities of BZ junctions to gain a comprehensive understanding of Z-DNA formation along with BZ junctions and their cellular roles.
Z-DNA has a role in enhancing gene expression by relieving the torsional stress during transcription, as demonstrated by a bioinformatics validation of the co-localization of Z-DNA forming regions with CpG islands in transcription start sites (6) and by confirming the presence of Z-DNA in the transcriptionally active chromosome (7). In addition, Z-DNA formation in the mammalian genome induces large-scale gene deletion through a non-homologous end joining pathway of DNA repair and genomic instability; therefore, it is considered to be a potential cause of several diseases (8). Z-DNA can be stabilized in biological systems in negatively super-coiled conditions as well as in the presence of polyamines (9–11) or Z-DNA-binding proteins (12–15), which specifically recognize phosphate backbones of Z-DNA in a structure-independent manner (13,15–17). These results commonly suggest that Z-DNAs and BZ junctions affect genetic and non-genetic functions of the genome either directly or indirectly.
The previously determined crystal structure of junctions between B-DNA and Z-DNA revealed the distinct structural features of BZ junction in which: (i) an A-T base pair at the junction is broken and extruded out of the base stacking and (ii) B-DNA is connected to Z-DNA with tight base packing (18). The same structural features of BZ junctions have been confirmed in solution by observing the extrusion of the adenine base at the junction site using 2-aminopurine (2AP), a fluorescent analog of adenine, as a probe for junction formation (19). Previous studies conducted by gel migration assays and circular dichroism (CD) suggested that the diverse sequences can form BZ junctions at high salt conditions (20,21). However, an outstanding question in this field is whether the structural features found in the known BZ structure are also commonly present in BZ junctions in other sequences. Since only one BZ junction structure is available, we cannot answer this fundamental question. A more intriguing issue is determining where BZ junctions can be formed in the genome. Considering that many predicted Z-DNA forming sequences occur in the genome, we can easily expect that many Z-DNAs as well as BZ junctions can be formed in the genome. However, it is entirely unknown if all sequences outside of the predicted Z-DNA sites can really form BZ junctions.
In this study, we investigated BZ junctions in diverse sequences to reveal the structural heterogeneity of BZ junctions and the sequence preference for BZ junction formation by determining three crystal structures of oligodeoxynucleotides (ODNs) containing BZ junctions and investigating the base extrusion sites using 2AP modified ODNs with diverse sequences. We describe the sequential preference for the formation of BZ junctions and their structural diversity. Our results help expand our knowledge of the physiological roles of Z-DNA and BZ junctions in the genome.
MATERIALS AND METHODS
Expression and purification of protein
The gene coding for the Zα domain (140–202) of human ADAR1 (hZαADAR1) was subcloned into pET28a+ (Novagen Inc., WI, USA) and transformed into BL21 (DE3) (Novagen Inc., WI, USA). The cells were grown in a Luria-Bertani medium containing 30 μg/ml kanamycin to an optical density of 0.5–0.6 at 600 nm (OD600) at 37°C. They were induced with 0.5 mM isopropyl β-d-1-thiogalactoside at 37°C for 4 h. hZαADAR1 was purified using methods described previously (17). After purification in a His-affinity column (GE Healthcare, NJ, USA) and removal of the N-terminal six histidines with thrombin, the protein in buffer A (20 mM HEPES-NaOH, pH 7.5, and 10 mM NaCl) was further purified using Resource S (GE Healthcare, NJ, USA). The purified protein was dialyzed against buffer containing 5 mM HEPES-NaOH (pH 7.5) and 10 mM NaCl. After dialysis, the protein was concentrated to over 1 mM by ultrafiltration on a Centricon-YM3 device (Millipore, MA, USA). All the buffer change steps were carried out using molecular porous membrane tubing (Spectrum Laboratories, Inc., CA, USA). The protein concentration was measured spectroscopically using an extinction coefficient of 6990 M−1cm−1 at 280 nm, calculated at www.expasy.org.
Circular dichroism (CD) experiments
The conversion of duplex oligonucleotides (Supplementary Table S7) from B-DNA to Z-DNA was monitored by measuring the CD spectra using a Jasco J-810 CD spectrometer at 25°C, with 15 μM double-stranded DNA (dsDNA). All solutions were prepared in a buffer containing 5 mM HEPES-NaOH (pH 7.5) and 10 mM NaCl. hZαADAR1 was added to dsDNA to final concentrations of 15 μM ([P]/[N] = 1), 30 μM ([P]/[N] = 2), 60 μM ([P]/[N] = 4), 90 μM ([P]/[N] = 6), 120 μM ([P]/[N] = 8) and 180 μM ([P]/[N] = 12). [P] and [N] stand for protein concentration and DNA concentration, respectively. The mixtures were equilibrated for 1 h prior to the measurement. Spectra were recorded between 230 nm and 320 nm at 1 nm intervals averaged over 2 s. The maximum volume of the protein added to the sample did not exceed 5% of the total volume. The conversion of duplex oligonucleotides from B-DNA to Z-DNA in NaClO4 was monitored by using the CD spectra as described above. Duplex oligonucleotides (15 μM) (Cosmogenetech, Seoul, South Korea) were dissolved in 100, 500, 750 mM, 1, 2, 3, 5, 7 and 9 M of a NaClO4 solution, followed by annealing after boiling at 97°C. Duplex oligonucleotides were cooled down overnight prior to the experiment. hZαADAR1 was used for the conversion experiment of Z-E1 and Z-low energy penalty (LEP) at the same molar ratio described above. Each type of dsDNA (15 μM) was used for the experiment, and all CD experiments were conducted at 25°C.
Structure determination
The structures of hZαADAR1 in a complex with BZ junction sequences were determined by the molecular replacement method. Initial solutions were obtained by MOLREP (22) using the crystal structure of the BZ junction (PDB code: 2ACJ) (18) as a search model. Rigid body refinement of the initial solution was performed using REFMAC (23). The interactive model was built using COOT (24). Statistics for the final refinements are provided in Supplementary Table S2. The quality of the structure was checked using MolProbity (25). All structural figures were generated using PYMOL (26). A Ramachandran plot analysis of the BZ34/hZαADAR1 complex structure showed 95.0, 5.0 and 0.0% of residues in favored regions, allowed regions and outlier regions, respectively. A Ramachandran plot analysis of the BZ56/hZαADAR1 complex structure showed 98.3, 1.7 and 0.0% of residues in favored regions, allowed regions and outlier regions, respectively. A Ramachandran plot analysis of the BZ78/hZαADAR1 complex structure showed 98.7, 1.3 and 0.0% of residues in favored regions, allowed regions and outlier regions, respectively.
Steady-state fluorescence
Fluorescence measurements were carried out at 25°C using a 3 mm Quartz Fluorometer Cell Microsquare (Starna Cells, Inc., Atascadero, CA, USA). Titrations of duplex oligonucleotides (Supplementary Table S7) in a buffer containing 5 mM HEPES-NaOH (pH 7.5) and 10 mM NaCl at each concentration of hZαADAR1 were performed to reach equilibrium as monitored by the CD spectra, in which the wavelength was fixed at 255 and 280 nm. Fluorescence intensities of 7.5 μM dsDNA were measured with increasing hZαADAR1 concentrations. Duplex oligonucleotides were excited at 320 nm, and emission data were collected between 335 and 500 nm. Fluorescence emission spectra were corrected by subtracting the control spectra containing only buffer because the 2AP fluorescence is highly affected by the position relative to adenines in the oligonucleotide (27). The formation of BZ junctions with 2′-O-methyl guanine and 2AP-labeled LEP and E1 at each concentration of NaClO4 was monitored with CD at wavelengths from 230 to 320 nm at 25°C. The same NaClO4 concentration steps in CD were used in the fluorescence experiment. Both types of labeled DNA were excited at 320 nm, and emission data were collected from 350 to 600 nm for 2′-O-methyl guanine and 335 to 500 nm for 2AP at 25°C. All experiments were conducted with 15 μM labeled dsDNA under ambient conditions.
RESULTS
The common structural features in BZ junctions
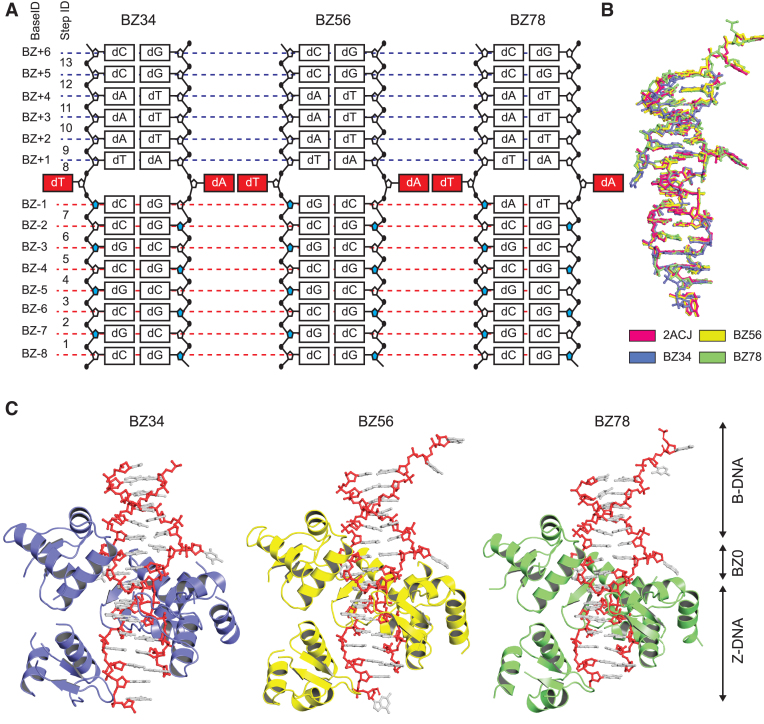
We designed four double-stranded ODNs (BZ12, BZ34, BZ56 and BZ78; Table 1) with different DNA sequences at the BZ-1 to BZ0 positions as compared to the sequences of 2ACJ. The detailed design strategy is provided in the ‘Materials and Methods’ section in supplementary information. To investigate the structural features of BZ junctions with different sequences, we crystallized the junction forming sequences in the presence of hZαADAR1, a Z-DNA-binding domain of human ADAR1, which is necessary for the stabilization of the Z-DNA side of the BZ junctions. Accordingly, crystal structures of BZ34, BZ56 and BZ78 were determined in a complex with hZαADAR1 (Figure 1;Supplementary Figure S1 and Supplementary Table S1). Consistent with the crystal structure of 2ACJ, CG-rich regions (BZ-8 to BZ-1 positions) in the three structures adopted the Z-DNA conformation by binding to four ZαADAR1. BZ+1 to BZ+6 positions with an AT repeat sequence remained as B-form DNA without protein contact (Figure 1C and Supplementary Figure S2). A structural comparison between 2ACJ and the current three BZ junctions revealed that the DNA in 2ACJ had root mean square deviations of 0.70, 0.36 and 0.45 Å, with DNAs in BZ34, BZ56 and BZ78 for 522 atoms, respectively (Figure 1B). These results suggest that the overall structural features observed in 2ACJ are well conserved in the three BZ junctions regardless of the sequence differences.
Table 1.
Oligonucleotide sequence for X-ray crystallography
| Abbreviation | Sequence | Objective |
|---|---|---|
| 2ACJ | 5′-GT CGCGCGCCATAAACC-3′ | BZ junction sequence used in previous study |
| 3′- GCGCGCGGTATTTGG CA-5′ | ||
| BZ12 | 5′-GT CGCGCGCCGTAAACC-3′ | Changed BZ0 position from A-T to G-C |
| 3′- GCGCGCGGCATTTGG CA-5′ | ||
| BZ34 | 5′-GT CGCGCGCCTTAAACC-3′ | Changed BZ0 position from A-T to T-A |
| 3′- GCGCGCGGAATTTGG CA-5′ | ||
| BZ56 | 5′-GT CGCGCGCGATAAACC-3′ | Changed BZ-1 position from C:G to G:C (changed CC steps to CG step |
| 3′- GCGCGCGCTATTTGG CA-5′ | at BZ-2/BZ-1) | |
| BZ78 | 5′-GT CGCGCGCAATAAACC-3′ | Changed BZ-1 position from C:G to A:T (change CC steps to CA step |
| 3′- GCGCGCGTTATTTGG CA-5′ | at BZ-2/BZ-1) |
Figure 1.
Overall structures of BZ junctions. (A) Schematic diagram of DNA composition of BZ34, BZ56 and BZ78. The BZ junction positions are red and extruded from the stacking. The sugar backbones with syn conformations are colored in blue in the Z-DNA part. Base and step IDs are listed on the left side. (B) Structural comparison between the previously reported BZ junction structure (PDB code: 2ACJ) and three crystal structures of BZ junctions in this study. The structures of DNA are represented by ball and stick models. The structures of 2ACJ, BZ34, BZ56 and BZ78 are colored in pink, light blue, yellow and green, respectively. (C) The crystal structures of BZ34, BZ56 and BZ78. The hZαADAR1 domains of BZ34, BZ56 and BZ78 are colored in light blue, yellow and green, respectively. The DNA molecules of BZ34, BZ56 and BZ78 are represented by ball and stick models. The phosphate backbones of DNA are colored in red. The Z-DNA, BZ junction and B-DNA parts are labeled as Z-DNA, BZ0 and B-DNA using arrowed lines, respectively.
The energies required for forming an anti–syn conformation are known to be 2.4, 1.3 and 0.7 kcal/mol for CC, CA and CG dinucleotides, respectively (28), as the syn conformation of pyrimidine is less favorable than that of purine (29). Accordingly, the CG dinucleotide at BZ-2/-1 positions of BZ56 adopted an anti–syn stacking and Z-DNA conformation (Supplementary Figure S3). However, the CC dinucleotide at the BZ-2/-1 positions of 2ACJ and BZ34, and the CA dinucleotide at the same position of BZ78 also formed Z-DNA with an anti–syn conformation (Supplementary Figure S3). A-T base pairs at the BZ0 position in BZ34, BZ56 and BZ78 were extruded from the base stacking in a manner similar to that of 2ACJ. BZ34 consisted of the same sequence composition as 2ACJ (except the sequence at the BZ0 position), and it adopted extruded bases at the junction site with orientations and positions similar to those of 2ACJ (Figure 1 and Supplementary Figure S3).
When the base step parameters of BZ junctions (Supplementary Table S2) were compared with those of typical B- and Z-DNA (Supplementary Table S4), the Z-DNA part (BZ-8 to BZ-3) and the B-DNA part (BZ+3 to BZ+6) of BZ junctions were similar to those of the typical Z- and B-DNAs, except for the rise distances in the B-DNA part (Supplementary Figures S4a and S5) as judged by P-values that represent the statistical significance of the similarity. However, the base step parameters near the BZ junction (BZ-2 to BZ+2) of the BZ junction structures were very different from those of typical B- or Z-DNAs (Supplementary Figures S4a and S5). Large differences near the BZ junction were found in twist, tilt, rise and shift parameters when statistical significance was considered (Supplementary structural analysis; Supplementary Figure S5 and Supplementary Table S5). Similarly, when the average base pair parameters (Supplementary Table S3) of the BZ junction were compared to those of B- and Z-DNAs (Supplementary Figure S4b) with statistical analyses (Supplementary Tables S4 and S5), the near-BZ junction showed a significant difference from the typical B- and Z-DNAs in terms of buckle, propeller and stagger parameters (Supplementary structural analysis; Supplementary Figure S6), while differences in the B- and Z-DNA parts were observed in propeller and stagger parameters, respectively.
Collectively, we propose that BZ junction structures share common structural features with base extrusion of adenine and thymine, and these are independent of their sequence in the dsDNA and are very different from typical B- and Z-DNAs.
Structural heterogeneity of BZ junctions
While the overall structural features were well conserved in four BZ junction structures, local structural heterogeneity of BZ junctions was observed. To investigate the structural differences between the four BZ junctions, the base step parameters of four crystal structures of BZ junctions were plotted along each base step (Supplementary Figure S7a), and their standard error of mean (SEM) was calculated (Supplementary Table S6). The statistical significance of structural heterogeneity was judged by the outlier test at each step (Supplementary Figure S8a). This analysis revealed that SEM outliers of tilt, roll and rise parameters were present near the BZ junction (see the Supplementary structural analysis). When the base pair parameters of four BZ junction structures were compared, structural heterogeneities near the BZ junction were also detected in opening, shear, stretch and stagger parameters (Supplementary Figure S7b, Supplementary structural analysis, Supplementary Table S6 and Supplementary Figure S8b).
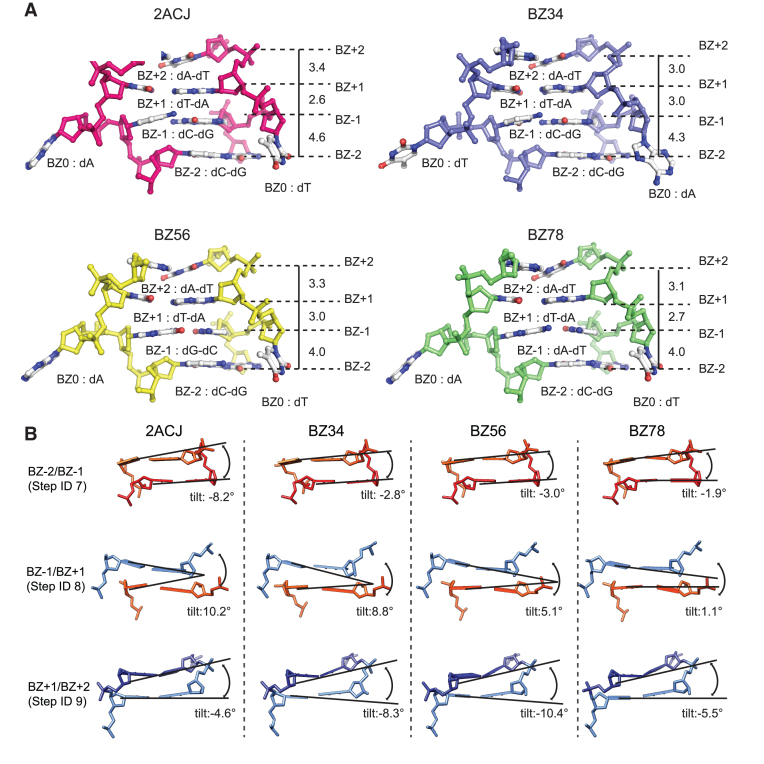
The significant structural heterogeneity in terms of tilt angle and rise distance was found at BZ-1, where sequence differences between BZ junction structures (Table 1) were present. The rise step parameters at the BZ-2/-1 step (step ID 7) from 2ACJ and BZ34, composed of the CC/GG step showed a larger difference (4.6 and 4.3 Å, respectively) than those of BZ56 and BZ78 with CG/GC and CA/GT, respectively (Figure 2A and Supplementary Figure S7a). In addition, tilt angles of 2ACJ and BZ34 were larger than those in other BZ junctions (Figure 2B). This sequence-dependent structural change was also observed in the roll angle at step 7 (Supplementary Figure S7a). Therefore, we anticipated that a large structural change at the BZ-1 position is correlated with the sequence. An upward shift in the cytosine base was observed at the BZ-1 positions in the crystal structures of 2ACJ and BZ34 (Figure 2A and Supplementary Figure S3). Consequently, step rise distance, roll and tilt angles also highly deviated from the values in typical B-DNA, Z-DNA or BZ56. This high structural alteration at 2ACJ and BZ34 can be interpreted by the steric constraints imposed by pyrimidine in the syn conformation at BZ-1 as discussed in a previous study (30). Structural deviations were less significant in BZ56 and BZ78 due to the presence of purine base at the BZ-1 position. However, this interpretation could not be applied to all parameter changes. For example, structural changes in opening, shift and stretch were also observed in BZ56 near the BZ junction although the energy penalty during Z-DNA formation is less in BZ56. Therefore, we also proposed that an intrinsic structural heterogeneity is present near the junction in a sequence independent manner. In addition to structural heterogeneities near the BZ junction, they were also found in the B-DNA side, as indicated by the presence of outliers at the B-DNA base ID in roll, shift and stretch parameters (Supplementary Figure S8). Consistent with this observation, the flexibility of the B-DNA in the BZ junction forming sequences has also been reported (31).
Figure 2.
Structural heterogeneity near the BZ junction. (A) The phosphate backbone, sugar and bases near the BZ junction of 2ACJ, BZ34, BZ56 and BZ78 are represented by ball-and-stick models. 2ACJ, BZ34, BZ56 and BZ78 are shown in magenta, blue, yellow and green colors, respectively, and are labeled. The base positions near the BZ junction are labeled as BZ-2, BZ-1, BZ0, BZ+1 and BZ+2. The bases at each position are labeled by position number. The rise base step parameters are shown in Å scale. (B) The side views of BZ-2/BZ-1 (Step ID 7), BZ-1/BZ+1 (Step ID 8) and BZ+1/BZ+2 (Step ID 9) of 2ACJ, BZ34, BZ56 and BZ78 are represented by stick models. BZ-2, BZ-1, BZ+1 and BZ+2 are colored red, light orange, light blue and dark blue, respectively. The tilt angles at each step are represented by black solid lines and are labeled.
Base extrusion sites
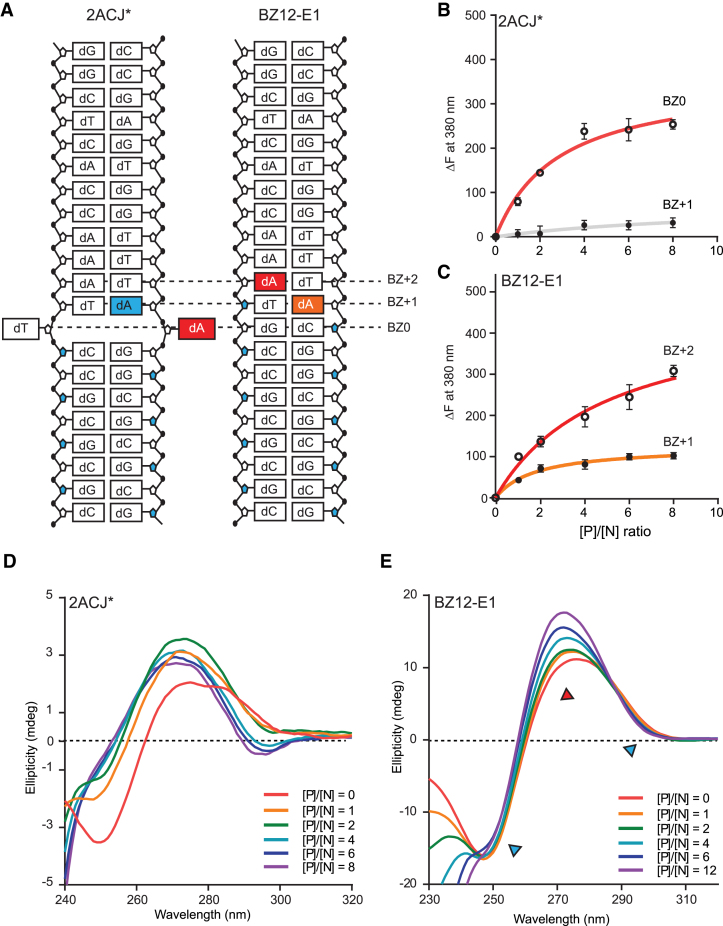
Among the four ODNs used for structural study, BZ12 did not crystallize, regardless of extensive crystallization attempts. We assumed that BZ12 could not form a stable BZ junction with the base extrusion at BZ0. To test this possibility, we designed various ODNs with a modified base 2AP, which were previously used for detecting extruding bases at the junction (19) (Supplementary Table S7). We then monitored the base extrusion as indicated by the fluorescence change (ΔF) upon hZαADAR1-induced Z-DNA formation. In addition, Z-DNA formation was monitored by CD spectra at the same conditions. 2ACJ* (BZ0) and 2ACJ* (BZ+1) that have 2AP at the BZ0 and BZ+1 positions, respectively, were introduced as control ODNs. The large and negligible fluorescence changes of 2ACJ*(BZ0) and 2ACJ*(BZ+1) at 380 nm, respectively, indicated that base at BZ0 can flip out, but the base at BZ+1 cannot (Figure 3B; Supplementary Figure S9a and b). 2ACJ* showed representative ellipticity change during the B-to-Z transition (i.e. a positive ellipticity change at 255 and a negative ellipticity change at 292 nm) (Figure 3D), indicating that the B-DNA conformation of CG repeats (BZ-8 to BZ-1) in 2ACJ* was converted to Z-DNA. However, in the case of BZ12-E1, ΔF from 2AP at both BZ+1 and BZ+2 increased in proportion to the concentration of hZαADAR1 (Figure 3C; Supplementary Figure S9c and d), suggesting that both bases are partially exposed to the solvent. The insignificant ellipticity change at 255 and 292 nm in the CD spectra of BZ12-E1 at various protein/DNA ratios suggests that a typical B-to-Z transition did not occur in BZ12-E1 under Z-DNA-inducing conditions (Figure 3E).
Figure 3.
BZ junction formation and B-to-Z transition of 2ACJ* and BZ12-E1. (A) Schematic drawing of 2AP-modified 2ACJ* and BZ12-E1 ODNs. 2AP at BZ0 and BZ+1 positions of 2ACJ* are represented by red and blue colored boxes, respectively. 2AP at BZ+1 and BZ+2 positions of BZ12-E1 are colored in orange and red, respectively. The BZ junction position of the 2ACJ structure is represented by BZ0. (B) Fluorescence intensity changes (ΔF) at 380 nm of 2AP-labeled 2ACJ* (BZ0) and 2ACJ* (BZ+1) are indicated by open and closed black circles, respectively, and the regressions are represented by red and gray solid lines, respectively. (C) The fluorescent intensity changes (ΔF) at 380 nm of 2AP-labeled BZ12-E1 (BZ+1) and BZ12-E1 (BZ+2) are represented by closed and open circles, respectively. Regressions are represented by orange and red solid lines, respectively. The ratios between protein and DNA ([P]/[N] ratio) were 0, 1, 2, 4, 6 and 8, where [P]/[N] stands for the molar ratio of the protein (hZαADAR1) over DNA. (D) CD spectra of the 2ACJ* sequence in the presence of various amounts of the hZαADAR1 were monitored. The ratios between protein and DNA ([P]/[N] ratio) were 0, 1, 2, 4, 6 and 8. (E) CD spectra of BZ12-E1 sequence by adding hZαADAR1 at [P]/[N] ratios from 0 to 12. The spectral bands, which indicate Z-DNA features, are represented by a blue triangle (255 and 292 nm). The spectrum band showing the most significant changes at 270 nm is represented as a red triangle.
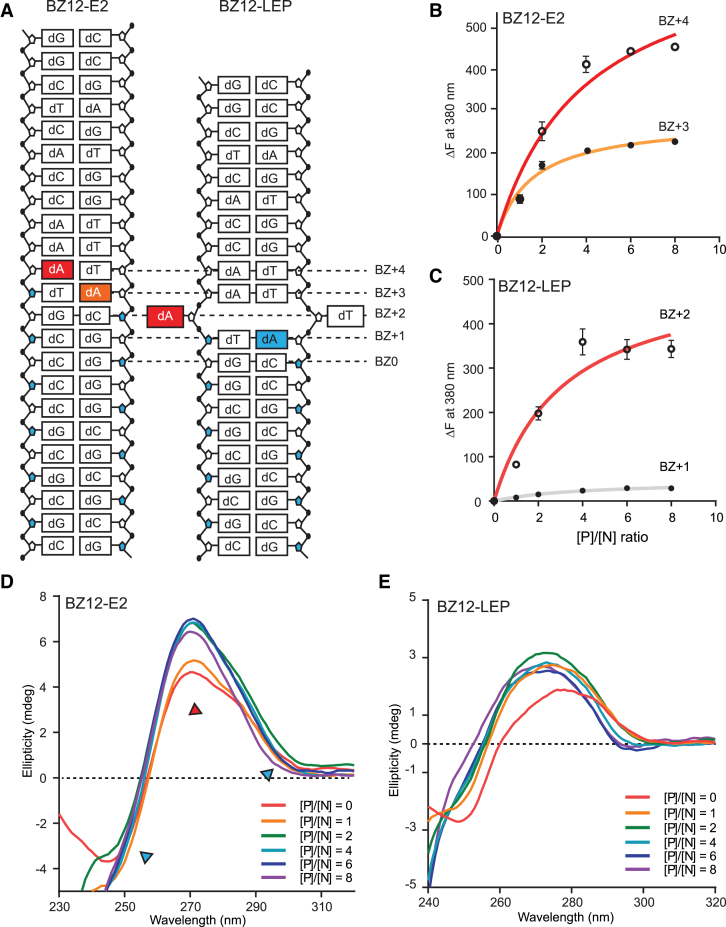
Further fluorescence and CD studies of BZ12-E2 (which has an extra CC step at the BZ0 site) revealed that BZ12-E1 and BZ12-E2 have very similar spectral features (Figure 4B–D; Supplementary Figure S10a and b). Since BZ12-E2 has much less chance of forming Z-DNA due to the large energy penalty during Z-DNA formation, neither BZ12-E1 nor BZ12-E2 were expected to form a typical BZ junction with the B- and Z-DNA on each side. However, the fluorescence and CD spectra of BZ12-LEP (which has G instead of C at the BZ-1 position) were very similar to those of 2ACJ* (Figure 4C; Supplementary Figure S10c and d), revealing that BZ12-LEP had a BZ junction with base extrusion at BZ+2.
Figure 4.
BZ junction formation and B-to-Z transition of BZ12-E2 and BZ12-LEP. (A) Schematic drawing of 2AP-modified BZ-E2 and BZ12-LEP ODNs. 2AP at BZ+3 and BZ+4 positions of BZ12-E2 are colored in orange and red, respectively. The 2AP-substituted adenines at BZ+1 and BZ+2 positions of BZ12-LEP are represented by blue and red colored boxes, respectively. (B) The fluorescent intensity changes (ΔF) at 380 nm of 2AP-labeled BZ12-E2 (BZ+3) and BZ12-E2 (BZ+4) are represented by closed and open circles, respectively, and the regressions are represented by orange and red solid lines, respectively. (C) Fluorescence intensity changes (ΔF) at 380 nm of 2AP-labeled BZ12-LEP (BZ+1) and BZ12-LEP (BZ+2) are indicated by closed black and open circles, respectively, and the regressions are shown by gray and red solid lines, respectively. The ratios between protein and DNA ([P]/[N] ratio) were 0, 1, 2, 4, 6 and 8, where [P]/[N] stands for the molar ratio of protein (hZαADAR1) over DNA. (D) CD spectra of BZ12-E2 upon adding hZαADAR1 at a [P]/[N] ratio from 0 to 8. The spectra bands, which indicate Z-DNA features, are represented by a blue triangle (255 and 292 nm). The spectrum band showing the most significant changes at 270 nm is represented by a red triangle. (E) CD spectra of BZ12-LEP ODN upon adding hZαADAR1 for [P]/[N] ratios of 0–8.
Considering that a flipped base and continuous base stacking are structural features of a typical BZ junction, we concluded that the A-T base pair present next to the Z-DNA dinucleotide unit with anti–syn conformation was broken and extruded in the BZ junction, allowing continuous base packing between Z-DNA and B-DNA. However, the G-C base pair could not be used as a junction site, even though it was next to the anti–syn Z-DNA dinucleotide pair. A theoretical study has suggested that the negative peak at 292 nm in the Z-DNA conformer was related to the strong stacking interaction of CG steps, while a peak increase at ∼270 nm indicates loose base stacking (32). Therefore, the increased positive peak at ∼270 nm and decreased negative peak at 292 nm detected in BZ12-E1 and BZ12-E2 upon hZαADAR1 (Figures 3E and 4D) represented loose stacking and breakage of base pairs instead of the tight base stacking observed in the BZ junction structure. Therefore, the current results suggest that BZ12 cannot form a typical BZ junction structure. However, we cannot rule out the presence of other types of BZ junctions in BZ12, such as the melted junction, which is similar to the known Z–Z junction (33).
The next question was whether BZ junction formation is necessary for Z-DNA formation in ODNs that contain both Z- and B-DNA forming sequences. To answer this question, we synthesized ODNs containing only the Z-DNA side of BZ12-E1 or BZ12-LEP (Z-E1 and Z-LEP, respectively; Supplementary Table S7) and confirmed that both Z-E1 and Z-LEP formed Z-DNA (Supplementary Figure S11). Therefore, the Z-DNA part of BZ12-E1 can form Z-DNA when present in Z-E1. But, it cannot form Z-DNA when present in BZ12-E1 (Figure 3E). These results demonstrated that the formation of the BZ junction structure is required for the B-to-Z transition of the Z-DNA side in ODNs that have a heterologous phase composed of B-DNA and Z-DNA.
To further validate the hypothesis that CG flipping could not occur at the BZ junction site, we synthesized BZ12-E1 and BZ12-LEP with a modified guanine base (2′-O-methyl guanine) at the BZ0 position, BZ12-E1 (BZ0) and BZ12-LEP (BZ0) (Supplementary Table S7), respectively, and we investigated the CD and fluorescence changes under Z-DNA-inducing conditions (Supplementary Figure S12) (34,35,36). Since the fluorescence of 2′-O-methyl guanine can be affected by protein, Z-DNA formation of both BZ12-E1 (BZ0) and BZ12-LEP (BZ0) was induced by salt (9 M NaClO4). The CD spectra of both BZ12-E1 (BZ0) and BZ12-LEP (BZ0) showed an increase in ellipticity at 255 nm under high salt conditions, but a negative peak at 292 nm was only observed in BZ12-LEP (BZ0), indicating the formation of a typical Z-DNA only in BZ12-LEP (BZ0) at high salt conditions (Supplementary Figure S12a and b). To validate the applicability of the salt-induced Z-DNA formation method in the current studies, Z-DNA formation of the junction-forming ODNs was also examined in the presence of NaClO4 in the same way as the protein-induced Z-DNA formation was examined (Supplementary Figure S13). From these experiments, it was confirmed that the sequence-dependency of Z-DNA formation was consistently observed in both cases, suggesting that both methods can be used for the study of Z-DNA and junction formation.
The modified guanine showed a fluorescence change when exposed to the solvent (34,35,36), as confirmed by the enhanced fluorescence signal from the single-stranded BZ12-E1 (BZ0) and BZ12-LEP (BZ0) (Supplementary Figure S12). No significant fluorescence change was observed in BZ12-E1 (BZ0) and BZ12-LEP (BZ0) (Supplementary Figure S12c and d) under Z-DNA-inducing conditions. This result suggests that 2′-O-methyl guanine at the BZ0 position was not exposed to the solvent. However, when the transition was tested using the BZ12-E1 (BZ+2) and BZ12-LEP (BZ+2) containing 2AP at the BZ+2 position under the same salt conditions, a 2AP fluorescence signal and CD transition were only detected in BZ12-LEP (BZ+2) (Supplementary Figure S14). To collaborate this analysis, we calculated the midpoint NaClO4 concentration in the B-to-Z transition of the junction forming ODNs using the CD spectra (Supplementary Figure S13) in the same way as used in the previous study (37). Consistent to our analysis, BZ12-E1 retained to be B-DNA at high salt concentration, since midpoint was not determined as there was no change in the ellipticity at 255 nm, while BZ12-LEP flipped to Z-DNA with a midpoint at 5.02 M NaClO4 (Supplementary Table S8). Furthermore, such midpoint analyses also revealed that BZ12, which does not form the BZ junction, had a higher midpoint salt concentration while the midpoints concentrations of BZ56 and BZ78 were relatively lower. This suggests that midpoint concentration of the B-to-Z transition has the correlation with the junction formation.
Taking these results together, we concluded that base flipping occurred not at BZ0 but at BZ+2 in the case of BZ12-LEP, and no base flipping was observed in the case of BZ12-E1. The current results also suggest a way to predict a sequence that forms BZ junction. When one pair of energetically unfavorable dinucleotide repeat units for adopting Z-DNA is present prior to the junction site, a junction can be formed (for example, CC in the case of 2ACJ and BZ34, and CA in the case of BZ78), but no BZ junction is formed when two pairs are present (for example, CCGT in the case of BZ12). However, this principle is likely to vary depending on the length of the Z-DNA or B-DNA, which affects the overall stability of the BZ junction. Furthermore, in terms of the BZ junction structure, the presence of an energetically unfavorable dinucleotide repeat prior to the junction site is likely to cause the largest structural changes in the BZ junction structure.
DISCUSSION
Structural analyses of the three BZ junctions in this study and a previously known BZ junction revealed that BZ junction structures adopted common structural features found in the previously known BZ junction structure independent of the sequence. These features are base extrusion and continuous base stacking through B-to-Z-DNA. Our structural comparison of four BZ junction structures to the typical B- and Z-DNAs showed that many DNA steps and pair parameters highly deviated from the typical B- and Z-DNAs near the BZ junction (BZ-2 to BZ+2). Moreover, we also found that base extrusion at the junction site affects the structural changes in B- and Z-DNA sides as well. A structural comparison of BZ junction structures also revealed the presence of a sequence-dependent structural heterogeneity near the BZ junction since the non-CG repeat sequence in the Z-DNA side of the BZ junction resulted in larger structural alternations. Although a previous study has suggested that BZ junctions could be formed in any sequence with the same probability (38), we found that BZ junctions with base flipping occurred not at a G-C pair but at an A-T base pair. In addition, we also provided clues to understand sequence preference for junction formation, and we demonstrated that BZ junction formation is required for Z-DNA formation. In other words, ODNs containing Z- and B-DNA-forming sequences cannot be induced to Z-DNA when a BZ junction is not formed.
Increasing experimental evidence proves Z-DNA formation in the genome. Therefore, identification of the Z-DNA-forming sites and the mechanism of Z-DNA formation is necessary for a comprehensive understanding of many Z-DNA driven genetic events in the genome. So far, Z-DNA forming sequences have been predicted or identified based on the energy preference or a pattern of sequences (6,39). However, our results strongly suggest that the BZ junction formation ability should be considered during the prediction of Z-DNA-forming sites, since our results demonstrate that Z-DNA cannot be induced when a BZ junction is not formed. In summary, we provide a more generalized concept of where BZ junctions and Z-DNA are present in the genome. In addition, we elucidated common structural features of BZ junctions as well as the sequence-dependent structural heterogeneities of BZ junctions. Therefore, our study is a milestone in the understanding of Z-DNA in the genome.
DATA AVAILABILITY
The coordinates and structure factors have been deposited under PDB accession codes 5ZU1 (BZ34), 5ZUO (BZ56) and 5ZUP (BZ78).
Supplementary Material
ACKNOWLEDGEMENTS
Author contributions: K.K.K. conceived of and supervised the work. K.K.K., D.K., J.H., S.C.H., D.S., S.L., S.P. and H.S. contributed to data interpretation. J.H.H. and S.P. synthesized the modified DNA oligomers. D.K. determined the crystal structure. D.K. and J.H. produced and purified proteins and performed CD and fluorescent experiments. K.K.K., D.K. and J.H. wrote the manuscript. All authors reviewed the final manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Samsung Science & Technology Foundation [SSTF-BA1301-01 to K.K.K.]. Funding for open access charge: Samsung Science & Technology Foundation [SSTF-BA1301-01 to K.K.K.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Bacolla A., Wells R.D.. Non-B DNA conformations, genomic rearrangements, and human disease. J. Biol. Chem. 2004; 279:47411–47414. [DOI] [PubMed] [Google Scholar]
- 2. Rich A. DNA comes in many forms. Gene. 1993; 135:99–109. [DOI] [PubMed] [Google Scholar]
- 3. Wang G., Vasquez K.M.. Non-B DNA structure-induced genetic instability. Mutat. Res. 2006; 598:103–119. [DOI] [PubMed] [Google Scholar]
- 4. Wang A.H., Quigley G.J., Kolpak F.J., Crawford J.L., van Boom J.H., van der Marel G., Rich A.. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979; 282:680–686. [DOI] [PubMed] [Google Scholar]
- 5. Rich A., Nordheim A., Wang A.H.. The chemistry and biology of left-handed Z-DNA. Annu. Rev. Biochem. 1984; 53:791–846. [DOI] [PubMed] [Google Scholar]
- 6. Khuu P., Sandor M., DeYoung J., Ho P.S.. Phylogenomic analysis of the emergence of GC-rich transcription elements. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:16528–16533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nordheim A., Pardue M.L., Lafer E.M., Moller A., Stollar B.D., Rich A.. Antibodies to left-handed Z-DNA bind to interband regions of Drosophila polytene chromosomes. Nature. 1981; 294:417–422. [DOI] [PubMed] [Google Scholar]
- 8. Bacolla A., Wells R.D.. Non-B DNA conformations as determinants of mutagenesis and human disease. Mol. Carcinog. 2009; 48:273–285. [DOI] [PubMed] [Google Scholar]
- 9. Lafer E.M., Valle R.P., Moller A., Nordheim A., Schur P.H., Rich A., Stollar B.D.. Z-DNA-specific antibodies in human systemic lupus erythematosus. J. Clin. Invest. 1983; 71:314–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Singleton C.K., Klysik J., Stirdivant S.M., Wells R.D.. Left-handed Z-DNA is induced by supercoiling in physiological ionic conditions. Nature. 1982; 299:312–316. [DOI] [PubMed] [Google Scholar]
- 11. Kim D., Lee Y.H., Hwang H.Y., Kim K.K., Park H.J.. Z-DNA binding proteins as targets for structure-based virtual screening. Curr. Drug Targets. 2010; 11:335–344. [DOI] [PubMed] [Google Scholar]
- 12. Herbert A., Alfken J., Kim Y.G., Mian I.S., Nishikura K., Rich A.. A Z-DNA binding domain present in the human editing enzyme, double-stranded RNA adenosine deaminase. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:8421–8426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schwartz T., Behlke J., Lowenhaupt K., Heinemann U., Rich A.. Structure of the DLM-1-Z-DNA complex reveals a conserved family of Z-DNA-binding proteins. Nat. Struct. Biol. 2001; 8:761–765. [DOI] [PubMed] [Google Scholar]
- 14. Rothenburg S., Deigendesch N., Dittmar K., Koch-Nolte F., Haag F., Lowenhaupt K., Rich A.. A PKR-like eukaryotic initiation factor 2alpha kinase from zebrafish contains Z-DNA binding domains instead of dsRNA binding domains. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:1602–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ha S.C., Lokanath N.K., Van Quyen D., Wu C.A., Lowenhaupt K., Rich A., Kim Y.G., Kim K.K.. A poxvirus protein forms a complex with left-handed Z-DNA: crystal structure of a Yatapoxvirus Zalpha bound to DNA. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:14367–14372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim D., Hur J., Park K., Bae S., Shin D., Ha S.C., Hwang H.Y., Hohng S., Lee J.H., Lee S. et al. . Distinct Z-DNA binding mode of a PKR-like protein kinase containing a Z-DNA binding domain (PKZ). Nucleic Acids Res. 2014; 42:5937–5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schwartz T., Rould M.A., Lowenhaupt K., Herbert A., Rich A.. Crystal structure of the Zalpha domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science. 1999; 284:1841–1845. [DOI] [PubMed] [Google Scholar]
- 18. Ha S.C., Lowenhaupt K., Rich A., Kim Y.G., Kim K.K.. Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases. Nature. 2005; 437:1183–1186. [DOI] [PubMed] [Google Scholar]
- 19. Kim D., Reddy S., Kim D.Y., Rich A., Lee S., Kim K.K., Kim Y.G.. Base extrusion is found at helical junctions between right- and left-handed forms of DNA and RNA. Nucleic Acids Res. 2009; 37:4353–4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lu M., Guo Q., Kallenbach N.R., Sheardy R.D.. Conformational properties of B-Z junctions in DNA. Biochemistry. 1992; 31:4712–4719. [DOI] [PubMed] [Google Scholar]
- 21. Otokiti E.O., Sheardy R.D.. Sequence effects on the relative thermodynamic stabilities of B-Z junction-forming DNA oligomeric duplexes. Biophys. J. 1997; 73:3135–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vagin A., Teplyakov A.. Molecular replacement with MOLREP. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:22–25. [DOI] [PubMed] [Google Scholar]
- 23. Murshudov G.N., Vagin A.A., Dodson E.J.. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 1997; 53:240–255. [DOI] [PubMed] [Google Scholar]
- 24. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen V.B., Arendall W.B. 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C.. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schrodinger, LLC. The PyMol Molecular Graphics System, Version 1.3r1. 2010; https://pymol.org/2.
- 27. Davis S., Matsumura M., Williams A., Nordlund T.. Position dependence of 2-aminopurine spectra in adenosine pentadeoxynucleotides. J. Fluoresc. 2003; 13:249–259. [Google Scholar]
- 28. Ho P.S., Ellison M.J., Quigley G.J., Rich A.. A computer aided thermodynamic approach for predicting the formation of Z-DNA in naturally occurring sequences. EMBO J. 1986; 5:2737–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Haschemeyer A.E., Rich A.. Nucleoside conformations: an analysis of steric barriers to rotation about the glycosidic bond. J. Mol. Biol. 1967; 27:369–384. [DOI] [PubMed] [Google Scholar]
- 30. Eichman B.F., Schroth G.P., Basham B.E., Ho P.S.. The intrinsic structure and stability of out-of-alternation base pairs in Z-DNA. Nucleic Acids Res. 1999; 27:543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bothe J.R., Lowenhaupt K., Al-Hashimi H.M.. Sequence-specific B-DNA flexibility modulates Z-DNA formation. J. Am. Chem. Soc. 2011; 133:2016–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miyahara T., Nakatsuji H.. Indicator of the stacking interaction in the DNA Double-Helical Structure: ChiraSac study. J. Phys. Chem. A. 2015; 119:8269–8278. [DOI] [PubMed] [Google Scholar]
- 33. de Rosa M., de Sanctis D., Rosario A.L., Archer M., Rich A., Athanasiadis A., Carrondo M.A.. Crystal structure of a junction between two Z-DNA helices. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:9088–9092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Park S., Otomo H., Zheng L., Sugiyama H.. Highly emissive deoxyguanosine analogue capable of direct visualization of B-Z transition. Chem. Commun. (Camb.). 2014; 50:1573–1575. [DOI] [PubMed] [Google Scholar]
- 35. Yamamoto S., Park S., Sugiyama H.. Development of a visible nanothermometer with a highly emissive 2′-O-methylated guanosine analogue. RSC advance. 2015; 5:104601–104605. [Google Scholar]
- 36. Han J.H., Yamamoto S., Park S., Sugiyama H.. Development of a Vivid FRET System Based on a Highly Emissive dG-dC Analogue Pair. Chemistry. 2017; 23:7607–7613. [DOI] [PubMed] [Google Scholar]
- 37. Sugiyama H., Kawai K., Matsunaga A., Fujimoto K., Saito I., Robinson H., Wang A.H.. Synthesis, structure and thermodynamic properties of 8-methylguanine-containing oligonucleotides: Z-DNA under physiological salt conditions. Nucleic Acids Res. 1996; 24:1272–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee J., Kim Y.G., Kim K.K., Seok C.. Transition between B-DNA and Z-DNA: free energy landscape for the B-Z junction propagation. J. Phys. Chem. B. 2010; 114:9872–9881. [DOI] [PubMed] [Google Scholar]
- 39. Li H., Xiao J., Li J., Lu L., Feng S., Droge P.. Human genomic Z-DNA segments probed by the Z alpha domain of ADAR1. Nucleic Acids Res. 2009; 37:2737–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates and structure factors have been deposited under PDB accession codes 5ZU1 (BZ34), 5ZUO (BZ56) and 5ZUP (BZ78).