Abstract
WRKY transcription factors constitute one of the largest transcription factor families in plants, and play crucial roles in plant growth and development, defense regulation and stress responses. However, knowledge about this family in maize is limited. In the present study, we identified a drought-induced WRKY gene, ZmWRKY106, based on the maize drought de novo transcriptome sequencing data. ZmWRKY106 was identified as part of the WRKYII group, and a phylogenetic tree analysis showed that ZmWRKY106 was closer to OsWRKY13. The subcellular localization of ZmWRKY106 was only observed in the nucleus. The promoter region of ZmWRKY106 included the C-repeat/dehydration responsive element (DRE), low-temperature responsive element (LTR), MBS, and TCA-elements, which possibly participate in drought, cold, and salicylic acid (SA) stress responses. The expression of ZmWRKY106 was induced significantly by drought, high temperature, and exogenous abscisic acid (ABA), but was weakly induced by salt. Overexpression of ZmWRKY106 improved the tolerance to drought and heat in transgenic Arabidopsis by regulating stress-related genes through the ABA-signaling pathway, and the reactive oxygen species (ROS) content in transgenic lines was reduced by enhancing the activities of superoxide dismutase (SOD), peroxide dismutase (POD), and catalase (CAT) under drought stress. This suggested that ZmWRKY106 was involved in multiple abiotic stress response pathways and acted as a positive factor under drought and heat stress.
Keywords: WRKY, ZmWRKY106, drought tolerance, thermotolerance, maize
1. Introduction
Changing environmental factors, such as abiotic stresses, influence plant growth and development [1]. Among them, drought and heat stresses seriously threaten crop productivity and quality. Plants must respond appropriately to changing environmental challenges to survive. Thus, it is important to explore the stress response mechanisms of plants and to enhance their tolerance to drought and heat to increase crop productivity without expanding cultivated land [2].
Environmental stresses initiate transcription factor (TF)-mediated expression of a variety of genes in plants, including bZIP, AP2/EREBP, MYB/MYC, NAC and WRKY [1,3,4,5,6]. WRKY TFs are identified by their conserved DNA-binding WRKY domains (WRKYGQK) in N-termini, and a zinc -finger motif (C-X4-5-C-X22-23-H-X1-H or C-X7-C-X23-H-X1-C) in C-termini [7,8]. It has been reported that WRKYs participated in defense responses by binding to the W-box located in the promoters of plant defense-related genes [9,10,11]. Another study found that 15 WRKY rice genes were induced by infection with the pathogen Magnaporthe grisea [12]. Statistically, 13 rice WRKY genes have regulated resistance against pathogens [13,14,15,16,17].
Recently, studies have revealed the involvement of the WRKY family in plant responses to abiotic stresses [18,19]. For example, ABO3/WRKY63 took part in responses to abscisic acid (ABA) and drought stress in Arabidopsis. AtWRKY57-overexpressing Arabidopsis exhibited improved tolerance to drought by combining the promoter sequences of NCED3 through the ABA pathway [20]. Overexpression of OsWRKY30 enhanced resistance to drought stress in rice by the phosphorylation process of mitogen-activated protein kinases (MAPKs) [11]. TaWRKY2 is a nuclear-located protein, and overexpression of TaWRKY2 in Arabidopsis led to enhanced tolerance to drought and salt stresses by improving the expressions of STZ and RD29B; moreover, the exogenous expression of TaWRKY19 in Arabidopsis not only conferred resistance to salt and drought, but also improved freezing tolerance [21]. In addition, CmWRKY10-overexpression in chrysanthemum revealed enhanced resistance to drought stress by regulating stress-related genes [19].
Maize (Zea mays) is a major food and economic crop. A few studies on the genome-wide analysis of WRKYs in maize have been reported in recent years. Wei et al. (2012) identified 136 WRKY proteins encoded by 119 ZmWRKY genes in maize, and Zhang et al. (2017) identified three additional new ZmWRKY genes and analyzed the gene expression profiles of ZmWRKYs using data from microarray, three RNA-seq studies, and the results of RT-PCR, which improved knowledge of WRKYs in maize [22,23]. In this paper, we performed drought-treated de novo transcriptome sequencing of maize (SRP144573) to investigate potential drought-tolerant WRKY genes in the maize genome. We identified a drought-responsive WRKY gene, ZmWRKY106, (Gene ID: GRMZM2G013391), which was named by Wei et al. (2012) and Zhang et al. (2017) [22,23]. The exogenous expression of ZmWRKY106 in Arabidopsis led to enhanced tolerance of drought and heat.
2. Results
2.1. De Novo Transcriptome Sequencing Analysis
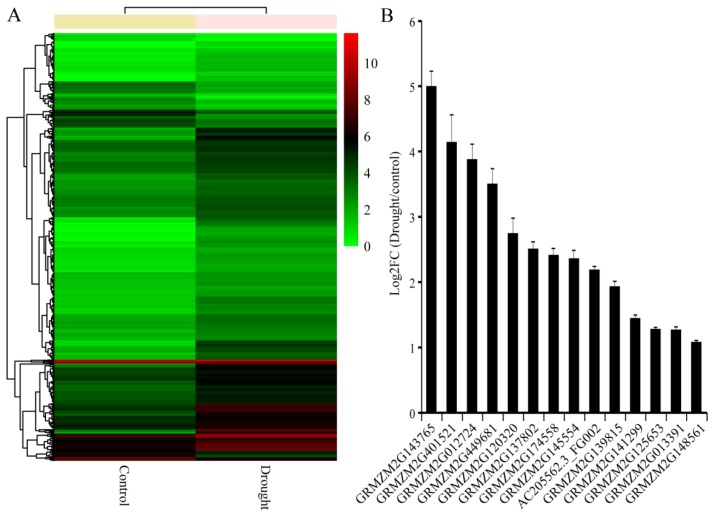
To find maize stress-responsive genes under drought stress, three-leaf seedlings were dehydrated on filter paper for 4 h, and then were collected for transcriptome sequencing analysis. The results showed that the transcription levels of many genes had changed after drought treatment (Figure 1A). Gene ontology (GO) analyses were used to classify the differentially expressed genes (DEGs) into functional groups. Almost 30 functionally enriched GO terms were identified for DEGs, and the results are shown in Supplementary Figure S1A. Among the predominantly enriched GO terms, signaling process was the most enriched term related to biological process. To further understand which pathways the stress-responsive genes may be involved in, the DEGs were analyzed against the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database. The top 20 enriched pathways were identified, and the “plant hormone signal transduction” pathway enriched the most DEGs under drought treatment (Supplementary Figure S1B). DEGs including many transcription factors that play vital roles in plant growth, development, morphogenesis, and abiotic stress responses through regulating the expression of downstream genes [1,4,18]. Among these transcription factors, WRKYs play important roles in response to biotic and abiotic stresses [10,19]. We searched for ZmWRKYs among the DEGs, and found 14 ZmWRKYs induced by drought treatment (Figure 1B). We chose the gene GRMZM2G013391 named ZmWRKY106 for further study.
Figure 1.
De novo transcriptome sequencing analysis of maize under drought stress. (A) Cluster analysis of the differentially expressed genes (DEGs) under drought treatment. (B) Transcription levels of the 14 differentially expressed ZmWRKYs under drought treatment. Error bar represent standard deviations (SD). The data represent means ± SD of three biological replications.
2.2. Phylogenetic Analysis of Maize ZmWRKY106
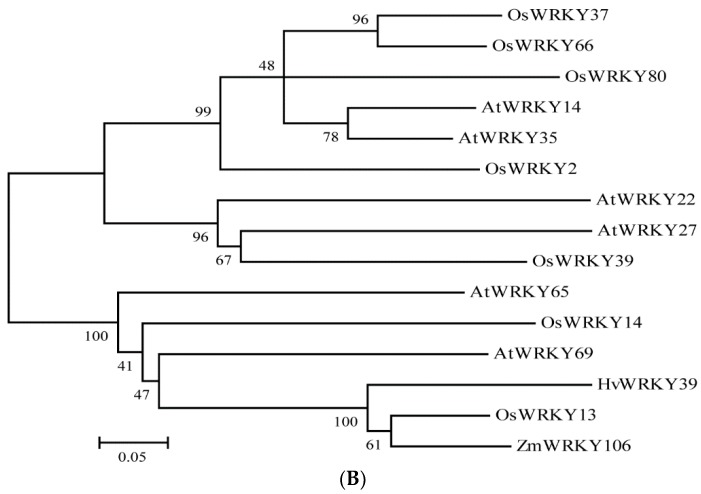
After selecting from the drought-treated maize de novo transcriptome data, we got a putative WRKY gene ZmWRKY106 encoding 277 amino acids. The BLASTp online tool was used to search for the homologous amino acid sequences of ZmWRKY106 in rice and Arabidopsis. The amino acid sequence alignment and phylogeny analysis of ZmWRKY106 orthologs are shown in Figure 2. ZmWRKY106 shared a mean identity of 28.47% with its rice, Arabidopsis, and barley orthologs and had a conserved signature WRKYGQK at the N-terminus followed by a C2H2 zinc-finger motif (C-X5-C-X23-H-X1-H), which characterized group II (Figure 2A). The sequences outside the conserved domain/motif were very different. The results of phylogenesis showed that ZmWRKY106 was closer to OsWRKY13, with a 61% bootstrap rate, followed by HvWRKY39, with a frequency of 100% (Figure 2B). However, the identity of ZmWRKY106 with other orthologs was lower than that with OsWRKY13, which indicated that ZmWRKY106 may have an extensive difference from other members.
Figure 2.
Multiple alignment and phylogenetic relationships of ZmWRKY106 with other orthologs in rice, Arabidopsis, and barley. The phylogenetic tree was produced using the aligned file with 1000 bootstraps in the MEGA 5.0 program. (A) Multiple alignment of ZmWRKY106 homologous proteins in rice, Arabidopsis, and barley. The different background colors represent the similar degree of amino acid sequences. (B) Phylogenetic relationship of ZmWRKY106 and other orthologs in different species. The first red box indicates the WRKYGQK motif, and the second indicates the conserved C2H2 zinc-finger motif.
2.3. ZmWRKY106 Was Localized in the Nucleus
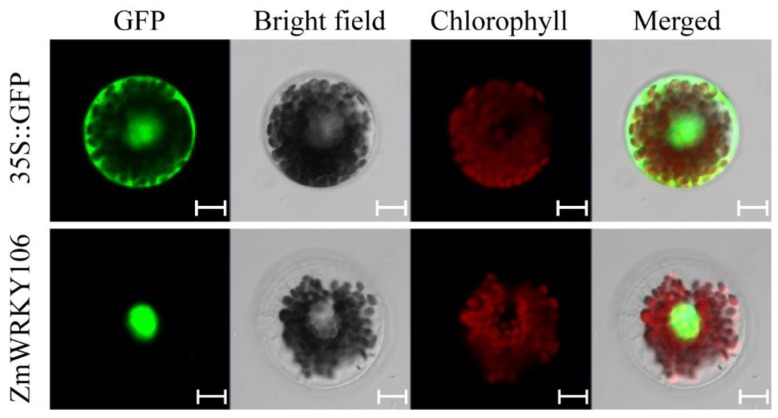
The transient expression vector p16318h-ZmWRKY106 was transformed to maize mesophyll protoplasts by the PEG-mediated method to determine the cell localization. After incubation in darkness for 18 h, the fluorescence signals were monitored by a confocal laser scanning microscope. As shown in Figure 3, relative to the control distributed throughout the cell, the p16318h-ZmWRKY106 fusion protein was specifically detected in the nucleus.
Figure 3.
Subcellular localization of ZmWRKY106. The p16318hGFP and p16318hGFP-ZmWRKY106 constructs were transiently expressed in maize protoplasts. The green indicates green fluorescent, and the red indicates chloroplast autofluorescence. Results were observed after transformation for 18 h with confocal microscopy. Scale bars = 10 μm.
2.4. ZmWRKY106 Promoter Domain Contained Various Stress-Related Cis-Elements
To further understand the regulation mechanism of ZmWRKY106, we isolated the promoter region upstream of the ZmWRKY106 ATG start codon. Types of cis-elements correlated to stress were present in the promoter region, including the C-repeat/DRE element referred to cold and dehydration response, low-temperature responsive element LTR and the drought-induced element MBS. In addition, there was another TCA-element that participated in salicylic acid (SA) response in the promoter region of ZmWRKY106 (Table 1). This analysis suggested that ZmWRKY106 may function in abiotic stress response.
Table 1.
Putative cis-elements in the ZmWRKY106 promoter.
| Elements | Sequence | Function |
|---|---|---|
| C-repeat/DRE | TGGCCGAC | involved in cold- and dehydration-responsiveness |
| LTR | CCGAAA | involved in low-temperature responsiveness |
| MBS | TAACTG | MYB binding site involved in drought-inducibility |
| TCA-element | TCAGAAGAGG | involved in SA responsiveness |
DRE—dehydration responsive element; LTR—low-temperature responsive; SA—salicylic acid.
2.5. ZmWRKY106 Was Involved in Abiotic Stress Responses
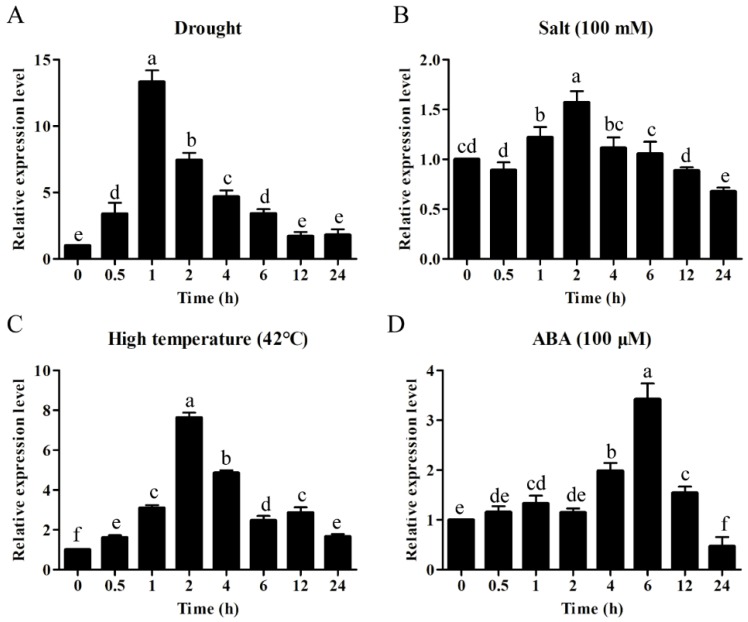
To explore the possible signal pathways which ZmWRKY106 may be involved in, we performed qRT-PCR to investigate the expression patterns of ZmWRKY106 in maize treated with drought, high-salt, high-temperature, and ABA treatments. ZmWRKY106 was remarkably induced by drought, high temperature and ABA, but was weakly induced by salt (Figure 4). For dehydration treatment, the transcript of ZmWRKY106 was rapidly up-regulated more than 10-fold after 1 h of dehydration stress (Figure 4A). ZmWRKY106 was slightly induced by salt at a maximum level of about 1.5-fold (Figure 4B). High temperature also significantly affected the expression of ZmWRKY106. Under high-temperature stress, the transcription level of ZmWRKY106 increased gradually, peaked at 7.6-fold after 2 h of stress, and then rapidly declined to a constitutive level. With exogenous ABA treatment, the transcription level of ZmWRKY106 was increased more than three-fold at 6 h after treatment.
Figure 4.
Expression patterns of ZmWRKY106 under (A) drought, (B) high-salt, (C) high-temperature, and (D) exogenous abscisic acid (ABA) stresses. The ordinates are the relative expression level (fold) of ZmWRKY106 compared to the non-stressed control. The horizontal ordinate is treatment time for 0, 0.5, 1, 2, 4, 6, 12 and 24 h. All experiments were repeated three times. Error bars represent standard deviations (SDs). All the data represent the means ± SDs of three independent biological replicates. The different letters in the bar graphs indicate significant differences at p < 0.05.
2.6. ZmWRKY106 Enhanced Drought Tolerance in Transgenic Arabidopsis
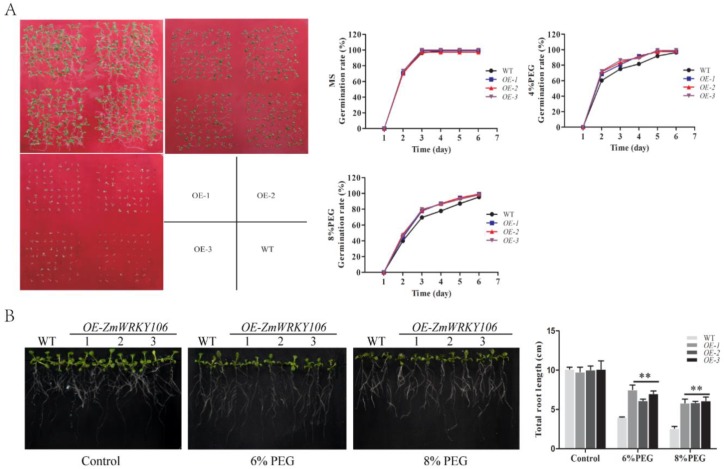
To investigate the function of ZmWRKY106, the pBI121-ZmWRKY106 recombinant was transformed into wild-type (WT) Arabidopsis (Columbia-0). T3 generation transgenic lines with relatively high expressions were selected by qRT-PCR for further analysis. The expression levels of transgenic lines are exhibited in Supplementary Figure S2. On MS medium, no significant differences in seed germination rates were observed between transgenic and WT plants. In the presence of 4% PEG6000, the germination rate of transgenic seeds was nearly 9% higher than WT after four days. Moreover, the germination was suppressed under 8% PEG6000, but transgenic seeds showed a higher germination rate than WT seeds (Figure 5A). For root growth assays, as shown in Figure 5B, ZmWRKY106 transgenic lines had similar phenotypes to WT on MS medium. When supplemented with PEG6000, the growth of all transgenic and WT plants was repressed; however, transgenic plants showed clear differences compared to WT ones, with significantly longer total root lengths than those of WT under both PEG treatments. These results showed that ZmWRKY106 transgenic lines had a stronger capacity to resist drought.
Figure 5.
Phenotypes of ZmWRKY106 transgenic Arabidopsis under drought treatment. (A) Seed germinations of wild-type (WT) and ZmWRKY106-overexpressing lines. (B) Root lengths of WT and ZmWRKY106 transgenic plants. Five-day-old seedlings were transferred to MS medium supplemented with or without PEG6000 for seven days, and then root lengths were measured. All the data represent the means ± SDs of three independent biological replicates and asterisks (**) represent the significant differences at p < 0.01 (Student’s t-test).
2.7. ZmWRKY106 Enhanced Heat Tolerance in Transgenic Arabidopsis
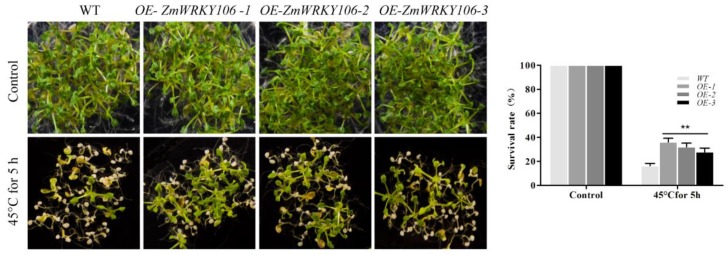
Under high temperature, the expression of ZmWRKY106 was up-regulated. Following this result, we observed the phenotypes among WT and transgenic lines under 45 °C (Figure 6). The survival rates of transgenic and WT plants were 100% under normal conditions, while higher a survival rate was exhibited in OE lines than WT after heat treatment for 5 h. ZmWRKY106-overexpressing lines had a survival rate of more than 30%, compared to less than 20% for WT plants after heat treatment. This suggested that ZmWRKY106 may improve thermotolerance of transgenic plants.
Figure 6.
Survival rates of WT and ZmWRKY106 transgenic lines under heat stress. Five-day-old seedlings were placed at 45 °C for 5 h and then resumed growth at 22 °C. The data represent the means ± SDs of three independent biological replicates. Asterisks (**) represent the significant differences (p < 0.01) compared with the control (Student’s t-test).
2.8. ZmWRKY106 Regulated the Expression of Stress-Related Genes
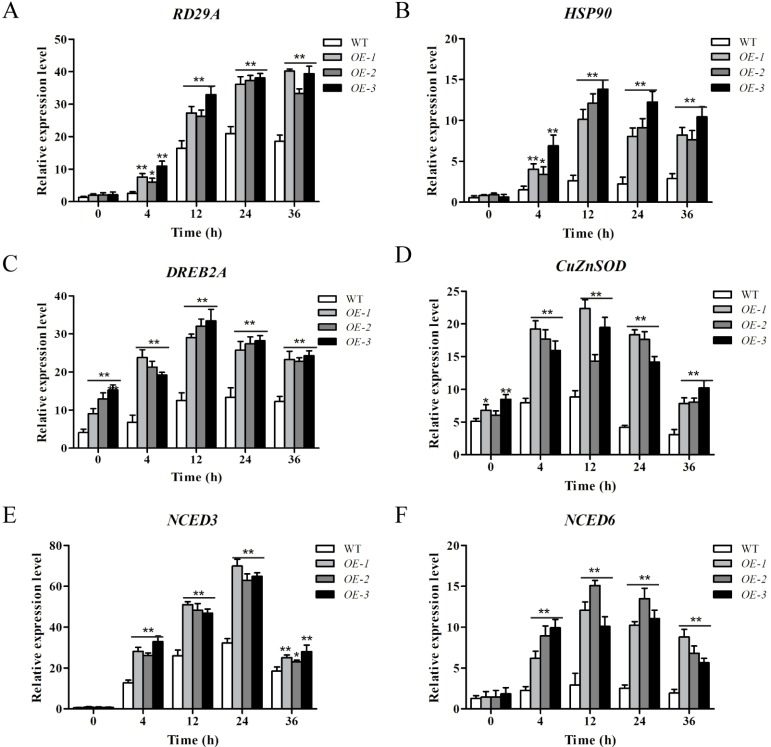
To understand the molecular mechanisms of ZmWRKY106 in stress responses, expression of stress-responsive genes, including RD29A, HSP90, DREB2A, CuZnSOD, NCED3, and NCED6, was examined using qRT-PCR under normal and drought conditions. The results showed that the expression levels of HSP90 and NCED3 were low in both WT and OE lines under normal conditions, while the expression levels in OE lines remained higher than WT plants after treatment (Figure 7B,E). Meanwhile, CuZnSOD and NCED6 in OE lines were up-regulated after 4 h of stress treatment, and sharply increased to the maximum (Figure 7D,F). The expression levels of RD29A and DREB2A in OE lines were remarkably higher following all treatments (Figure 7A,C). Because the expressions of ABA and stress-related genes were altered in transgenic lines, we conjectured that ZmWRKY106 may play a role in the abiotic stress response by regulating stress-related genes through the ABA-signaling pathway.
Figure 7.
The relative expression of stress-related genes. (A) RD29A, (B) HSP90, (C) DREB2A, (D) CuZnSOD, (E) NCED3, and (F) NCED6 were examined under control and drought conditions for various time points (4, 12, 24 and 36 h). Values are means ± SDs of three replicates, and asterisks (* or **) represent the significant differences at p < 0.05 or p < 0.01, respectively (Student’s t-test).
2.9. Overexpression of ZmWRKY106 Reduced Reactive Oxygen Species (ROS) Content and Enhanced the Activities of Superoxide Dismutase (SOD), Peroxide Dismutase (POD), and Catalase (CAT) under Drought Treatment
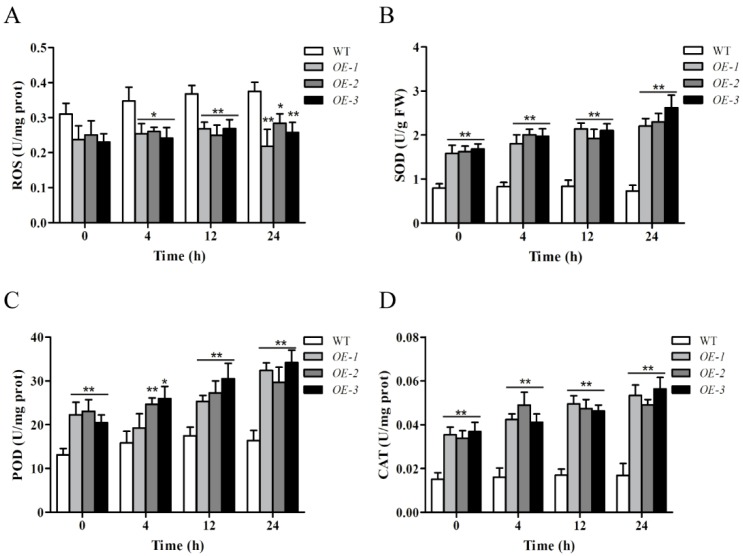
The ROS content and the enzyme activities were assessed in transgenic lines and WT plants at 0, 4, 12 and 24 h after drought treatment (Figure 8). As shown in Figure 8A, the ROS accumulation in transgenic lines was less than that in WT plants at all times, while the ROS content was increased in WT plants and remained at a higher level during the whole experiment. The activities of SOD, POD and CAT were increased in OE lines compared to WT lines (Figure 8B–D). The activity of SOD was almost unchanged in WT before and after drought treatment, whereas in OE lines the activity of SOD was greater, and reached a maximum at 24 h after drought treatment (Figure 8B). Increases of POD activity were observed in both WT and transgenic lines, but the increases in WT were smaller, and there was consistently higher POD activity in transgenic lines than in WT lines (Figure 8C). In the case of CAT, the CAT activity of WT lines remained consistent at 0.016 U and had almost no significant change during the stress treatment; however, the CAT concentration in the OE lines remained significantly higher compared to that in WT lines under drought stress (Figure 8D). In a word, overexpression of ZmWRKY106 reduced ROS content by enhancing the activities of SOD, POD and CAT to resist drought stress.
Figure 8.
(A) The reactive oxygen species (ROS) content and the activities of (B) superoxide dismutase (SOD), (C) peroxide dismutase (POD), and (D) catalase (CAT) under different conditions at different time points (0, 4, 12, and 24 h). Values are means ± SDs of three replicates, and asterisks (* or **) represent the significant differences at p < 0.05 or p < 0.01, respectively (Student’s t-test).
3. Discussion
Biotic and abiotic stresses seriously affect plant growth and development. Under adverse environments, transcriptome changes are the earliest responses, and transcriptional regulation plays a crucial role in plant defense responses [15]. Thus far, many TFs have been identified as participating in plant defense responses, including MYB, bZIP, and WRKY proteins. There are many more biotic stress-related genes in WRKYs than in other TFs, and an increasing number of studies have revealed that WRKY TFs play positive or negative roles in plants’ disease prevention [24]. For example, AtWRKY46, coordinated with AtWRKY70 and AtWRKY53, positively regulated basal resistance to Pseudomonas syringae [25]; OsWRKY6 played a positive role in plant defense response by activating the expression of defense-related genes [26]; GhWRKY44 was induced by pathogen injection, and overexpression of GhWRKY44 led to enhanced resistance against bacterial and fungal pathogens [27]. These results all suggest that the WRKY family plays an important role in responding to biotic stresses [28].
However, knowledge about the role of WRKYs in abiotic stresses is limited [29,30]. Maize is a major food and economic crop and plays an important role in basic and applied biological research. So far, known research about WRKYs has been mostly related to defense response in dicotyledon plants such as Arabidopsis, tomato, and tobacco, but little information about the role of maize WRKYs has been reported [31,32,33]. It is rather crucial to elucidate the functional maize WRKY protein in abiotic stress response. In maize, Wei et al. [22] have identified 136 WRKY proteins encoded by 119 WRKY genes, numbered them, and performed a phylogenetic tree analysis of the maize WRKYs with orthologs in Arabidopsis, rice, and barley, which improved knowledge of WRKYs in maize. In addition, Zhang et al. [23] identified three new additional ZmWRKY genes, analyzed the gene expression profiles of ZmWRKYs using data from various studies, and found that ten genes, including ZmWRKY9, ZmWRKY25, ZmWRKY47, ZmWRKY97, ZmWRKY80, ZmWRKY39, ZmWRKY106, ZmWRKY53, ZmWRKY36 and ZmWRKY113, were responsive under drought treatment in at least in three studies, which provided the basis for cloning functional ZmWRKY genes. In this study, we revealed the function of ZmWRKY106 in abiotic stress responses. Our study showed that ZmWRKY106 belongs to group II, shares a mean identity with its rice, Arabidopsis and barley orthologs, and is closer to OsWRKY13 (Figure 2).
Increasing evidence has indicated that WRKYs play an important role in abiotic stress response, for example, GmWRKY21 improved freezing tolerance in transgenic Arabidopsis, and GmWRKY54 played a positive role in response to salt and drought stresses, whereas GmWRKY13 markedly increased sensitivity to salt and mannitol [34]. Overexpression of AtWRKY25 and AtWRKY33 in Arabidopsis led to enhanced resistance to salt and hypersensitivity to ABA [35]. In rice, OsWRKY11 enhanced heat and drought tolerance [29]. In barley, Hv-WRKY38 played key roles in the response to cold and drought stresses, and enhanced drought tolerance in turf and forage grass [36,37]. In this study, expression profiles analysis revealed that ZmWRKY106 was induced significantly by drought, high temperature and ABA (Figure 4), possibly related to various stress-related cis-elements in its promoter region (Table 1). Under drought treatment, the transgenic seeds of ZmWRKY106 germinated faster than WT seeds, and roots of OE lines were remarkably longer than those of WT lines (Figure 5). Meanwhile, overexpression of ZmWRKY106 reduced ROS content and enhanced the activities of SOD, POD and CAT under drought treatment (Figure 8). Furthermore, the survival rates of OE lines were higher than those of WT lines (Figure 6). These results all showed that ZmWRKY106 exhibited drought tolerance and thermotolerance.
ABA is a major phytohormone referred to plant response under drought stress, and there exist ABA-dependent and ABA-independent pathways in drought stress response. In our study, the expression levels of six stress-related genes were assessed under normal and drought conditions (Figure 7). DREB2A is a well-known marker gene in ABA-independent stress responses [38]. ABRE and DRE/CRT motifs were found in the promoters of many stress-inducible genes, such as RD29A, which contained several DREs and one ABRE in the promoter domain, and was strongly induced by cold, drought and salt stresses [39,40,41]. HSP90 played a major role in stress signal transduction, and overexpression of HSP90 affected the phenotype of transgenic plants [42,43,44,45]. In our study, the expressions of RD29A, HSP90, and DREB2A genes were all up-regulated in ZmWRKY106 transgenic lines (Figure 7A–C), suggesting that ZmWRKY106 may play a positive role in drought and heat response. CmWRKY10 acted as a positive factor in response to drought stress by regulating the expression of DREB1A, DREB2A, CuZnSOD, NCED3A, and NCED3B, which proved that CmWRKY10 enhanced the drought tolerance through the ABA-dependent pathway [19]. These genes could play key roles in the physiological process of abiotic stress response [46,47]. We found that the expressions of ABA-related genes were higher in transgenic lines, which indicated that overexpression of ZmWRKY106 led to enhanced tolerance of drought stress through the ABA-dependent pathway (Figure 7D–F). These results all indicated that ZmWRKY106 may play a role in the abiotic stress response by regulating stress-related genes through the ABA-signaling pathway (Figure 7). Nevertheless, the role and regulation mechanisms of ZmWRKY106 in maize still need further research.
4. Materials and Methods
4.1. De Novo Transcriptome Sequencing
Three-leaf stage untreated maize seedlings and seedlings dehydrated on filter paper for 4 h were collected for RNA-seq analysis. The detailed process of RNA-seq was undertaken as previously described [18]. The transcriptome data are available in the National Center for Biotechnology Information (NCBI) under accession number SRP144573.
4.2. Plant Materials and Stress Treatments
The seeds of maize (X178) used in this study were provided by Zhuan-Fang Hao (Institute of Crop Science, Chinese Academy of Agricultural Sciences, Beijing, China). The maize seeds were sown as previously described [48]. Three-leaf stage maize seedlings were exposed to drought, salt, high-temperature and ABA treatments. For dehydration treatment, seedlings were quickly cleaned and then transferred on to filter paper to rapidly dry in air as previously described [49]. Seedlings were placed in 42 °C chambers for high-temperature treatment. For salt and ABA treatments, seedlings were exposed to water solutions supplemented with 100 mM NaCl, and 100 μM ABA, respectively. The samples were collected at 0, 0.5, 1, 2, 4, 6, 12 and 24 h after treatment. Harvested seedlings were dropped immediately into liquid nitrogen and stored at −80 °C for RNA extraction.
4.3. RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
Total RNAs were extracted from maize tissue using an RNAprep plant kit (Tiangen, Beijing, China), and cDNA was synthesized as previously described [18]. The qRT-PCR was performed with SuperReal PreMix Plus (Tiangen, Beijing, China) by an ABI Prism 7500 system (Applied Biosystems, Foster City, CA, USA). The specific primers of ZmWRKY106 are listed in Supplementary Table S1. Each PCR was repeated three times and data were analyzed, as previously described [50].
4.4. Gene Isolation and Sequence Analysis
The full length of the ZmWRKY106 gene was amplified by PCR with specific primers from maize cDNA. The primers of ZmWRKY106-F and ZmWRKY106-R are listed in Supplementary Table S1. The PCR products were cloned into pLB vector (Tiangen, China) and sequenced. The homologs of ZmWRKY106 in different species were searched for in the NCBI database. Sequence alignments of ZmWRKY106 orthologs were performed by ClustalX software. The phylogenetic tree was constructed using the neighbor-joining method by the MEGA 5.0 program with bootstrap analysis of 1000 replicates [51].
4.5. Subcellular Localization
The coding region of ZmWRKY106 was fused to the subcellular localization vector p16318h with green fluorescent protein (GFP) tags containing the CaMV35S promoter. The specific primers are listed in Supplementary Table S1. For transient expression assays, the p16318h-ZmWRKY106 reconstruction plasmid was transformed to maize mesophyll protoplasts by the PEG-mediated method, while the p16318hGFP vector was transformed as control [52]. The fluorescence signals were observed by a confocal laser scanning microscope (LSM700; CarlZeiss, Oberkochen, Germany) after incubation in darkness at 22 °C for 18 h.
4.6. Cis-Acting Elements in Promoter
The 2.0 kb promoter region upstream of ZmWRKY106 was obtained from maize genomic DNA on the EnsemblPlants website (available online: http://plants.ensembl.org/index.html). Putative cis-acting elements in the promoter region were analyzed using the PLACE database [53].
4.7. Generation of Transgenic Arabidopsis and Its Phenotype under Stress Treatment
Plant expression vector pBI121-ZmWRKY106 was constructed as previously described [54], and was transformed to wild-type (WT) Arabidopsis using the Agrobacterium-mediated floral dip method. Columbia-0 (WT) was used for exogenous expression of ZmWRKY106. The transformed seeds were selected on MS medium containing 50 mM Kanamycin at 22 °C with a photoperiod of 16 h light/8 h dark (60% humidity) to obtain the positive plants. Three T3 generation overexpression lines (OE-ZmWRKY106-1, OE-ZmWRKY106-2, OE-ZmWRKY106-3) with higher expression levels of ZmWRKY106 were selected by qRT-PCR for further analysis. Arabidopsis seeds were grown as described previously [50]. Four-week-old seedlings of transgenic and WT Arabidopsis were collected at 0, 4, 12, 24 and 36 h after drought treatment (dried on filter paper) to examine the expression of stress-related genes by qRT-PCR. The specific primers of stress-related genes are listed in Supplementary Table S1. Three biological replicates were performed for qRT-PCR.
For the germination assay, WT and transgenic Arabidopsis seeds were placed on MS medium and MS medium supplemented with 4% (w/v) or 8% PEG6000. When the radicle had emerged from the seed coat, we considered the seed germinated. Seed germination was followed for five days, and the germination rate was analyzed. For the root growth assay, five-day-old seedlings were transferred to MS medium with or without 6% and 8% PEG6000 for seven days, and then root lengths were measured. Each treatment contained three independent replicates.
For high-temperature stress assay, five-day-old seedlings were placed at 45 °C for 5 h and then resumed growth at 22 °C as described previously [18]. After growing under normal conditions for seven days, we took photos and analyzed the survival rate. Each treatment contained three independent replicates. Values are means ±SD and statistically significant differences were based on the Student’s test.
4.8. Measurements of Reactive Oxygen Species (ROS) Content and Enzyme Activity
To better understand the function of ZmWRKY106 under drought treatment, we assessed the activities of superoxide dismutase (SOD), peroxide dismutase (POD), catalase (CAT) and the ROS content in WT and transgenic lines at 0, 4, 12 and 24 h after drought stress. The ROS content and the activities of SOD, POD, and CAT were measured as previously described [19]. To obtain reproducible results, each experiment was repeated three times. Values are means ±SD and statistically significant differences were based on the Student’s test.
5. Conclusions
We identified a drought-induced WRKYII gene ZmWRKY106 based on maize drought de novo transcriptome sequencing data (SRP144573). ZmWRKY106 was only observed in the nucleus. The expression of ZmWRKY106 was induced significantly by drought, high-temperature, and exogenous abscisic acid (ABA) treatments, but was induced weakly by salt. Further research revealed that overexpression of ZmWRKY106 could improve tolerance to drought and heat in transgenic Arabidopsis by regulating stress-related genes through the ABA-signaling pathway, and could reduce the ROS content in transgenic lines by enhancing the activities of SOD, POD and CAT under drought stress. These results may provide a basis for understanding the functions of ZmWRKY106 in abiotic stress response in maize.
Acknowledgments
We thank Li-Na Ning for critically reading the manuscript.
Abbreviations
| ABA | abscisic acid |
| ABRE | ABA-responsive element |
| DRE | dehydration responsive element |
| GFP | green fluorescent protein |
| LTR | low-temperature responsive |
| MAPKs | mitogen-activated protein kinases |
| qRT-PCR | quantitative real-time PCR |
| RT-PCR | reverse transcription PCR |
| SA | salicylic acid |
| TF | transcription factor |
| WT | wild type |
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/19/10/3046/s1.
Author Contributions
Conceptualization, C.-T.W., J.-N.R., and J.-F.Y.; methodology, C.-T.W., J.-N.R., and Y.-W.L.; validation, C.-T.W., J.-N.R., and Y.-W.L.; formal analysis, C.-T.W. and J.-N.R.; investigation, C.-T.W. and J.-N.R.; data, C.-T.W. and J.-N.R.; writing—original draft preparation, C.-.T.W. and J.-N.R.; writing—review and editing, Z.-S.X. and J.-D.F.; supervision, J.-F.Y. and Y.-W.L.; project administration, Z.-S.X. and J.-D.F.; funding acquisition, M.L. and D.Z.
Funding
This study was financially supported by the Funding Project for Beijing Advanced Innovation Center for Food Nutrition and Human Health, and the Open Research Fund Program of Beijing Key Lab of Plant Resource Research and Development, Beijing Technology and Business University.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Xu Z.S., Chen M., Li L.C., Ma Y.Z. Functions of the ERF transcription factor family in plants. Botany. 2008;86:969–977. doi: 10.1139/B08-041. [DOI] [Google Scholar]
- 2.Mehta R.H., Ponnuchamy M., Kumar J., Reddy N.R. Exploring drought stress-regulated genes in senna (Cassia angustifolia Vahl.): A transcriptomic approach. Funct. Integr. Genom. 2017;17:1–25. doi: 10.1007/s10142-016-0523-y. [DOI] [PubMed] [Google Scholar]
- 3.Rushton P.J., Somssich I.E. Transcriptional control of plant genes responsive to pathogens. Curr. Biol. 1998;1:311–315. doi: 10.1016/1369-5266(88)80052-9. [DOI] [PubMed] [Google Scholar]
- 4.Tuteja N. Abscisic acid and abiotic stress signaling. Plant Signal Behav. 2007;2:135–138. doi: 10.4161/psb.2.3.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh D., Laxmi A. Transcriptional regulation of drought response: A tortuous network of transcriptional factors. Front. Plant Sci. 2015;6:895. doi: 10.3389/fpls.2015.00895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gahlaut V., Jaiswal V., Kumar A., Gupta P.K. Transcription factors involved in drought tolerance and their possible role in developing drought tolerant cultivars with emphasis on wheat (Triticum aestivum L.) Theor. Appl. Genet. 2016;129:2019–2042. doi: 10.1007/s00122-016-2794-z. [DOI] [PubMed] [Google Scholar]
- 7.Eulgem T., Rushton P.J., Robatzek S., Somssich I.E. The WRKY superfamily of plant transcription factor. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 8.Jiang W., Wu J., Zhang Y., Yin L., Lu J. Isolation of a WRKY30 gene from Muscadinia rotundifolia (Michx) and validation of its function under biotic and abiotic stresses. Protoplasma. 2015;252:1361–1374. doi: 10.1007/s00709-015-0769-6. [DOI] [PubMed] [Google Scholar]
- 9.Ciolkowski I., Wanke D., Birkenbihl R.P., Somssich I.E. Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 2008;68:81–92. doi: 10.1007/s11103-008-9353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verk M.C.V., Pappaioannou D., Neeleman L., Bol J.F., Linthorst H.J.M. A novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiol. 2008;146:1983–1995. doi: 10.1104/pp.107.112789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen H., Liu C., Zhang Y., Meng X., Zhou X., Chu C., Wang X.P. OsWRKY30 is activated by MAP kinases to confer drought tolerance in rice. Plant Mol. Biol. 2012;80:241–253. doi: 10.1007/s11103-012-9941-y. [DOI] [PubMed] [Google Scholar]
- 12.Ryu H.S., Han M., Lee S.K., Cho J.I., Ryoo N., Heu S., Lee Y.H., Bhoo S.H., Wang G.L., Hahn T.R., et al. A comprehensive expression analysis of the WRKY gene superfamily in rice plants during defense response. Plant Cell Rep. 2006;25:836–847. doi: 10.1007/s00299-006-0138-1. [DOI] [PubMed] [Google Scholar]
- 13.Cheng H.T., Li H.B., Deng Y., Xiao J.H., Li X.H., Wang S.P. The WRKY45-2-WRKY13-WRKY42 transcriptional regulatory cascade is required for rice resistance to fungal pathogen. Plant Physiol. 2015;167:1087–1099. doi: 10.1104/pp.114.256016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi C., Hwang S.H., Fang I.R., Kwon S.I., Park S.R., Ahn I., Kim J.B., Hwang D.J. Molecular characterization of Oryza sativa WRKY6, which binds to W-box-like element 1 of the Oryza sativa pathogenesis–related (PR) 10a promoter and confers reduced susceptibility to pathogens. New Phytol. 2015;208:846–859. doi: 10.1111/nph.13516. [DOI] [PubMed] [Google Scholar]
- 15.Wang H.H., Meng J.M., Peng X.X., Tang X.K., Zhou P.L., Xiang J.H., Deng X.B. Rice WRKY4 acts as a transcriptional activator mediating defense responses toward Rhizoctonia solani, the causing agent of rice sheath blight. Plant Mol. Biol. 2015;89:157–171. doi: 10.1007/s11103-015-0360-8. [DOI] [PubMed] [Google Scholar]
- 16.Hwang S.H., Kwon S.I., Jang J.Y., Fang I.L., Lee H., Choi C., Park S., Ahn I., Bae S.C., Hwang D.J. OsWRKY51, a rice transcription factor, functions as a positive regulator in defense response against Xanthomonas oryzae pv. oryzae. Plant Cell Rep. 2016;35:1975–1985. doi: 10.1007/s00299-016-2012-0. [DOI] [PubMed] [Google Scholar]
- 17.Peng X., Wang H., Jang J.C., Xiao T., He H., Jiang D., Tang X. OsWRKY80-OsWRKY4 module as a positive regulatory circuit in rice resistance against Rhizoctonia solani. Rice. 2016;9:63–76. doi: 10.1186/s12284-016-0137-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He G.H., Xu J.Y., Wang Y.X., Liu J.M., Li P.S., Chen M., Ma Y.Z., Xu Z.S. Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC Plant Biol. 2016;16:116. doi: 10.1186/s12870-016-0806-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaffar M.A., Song A.P., Faheem M., Chen S.M., Jiang J.F., Liu C., Fan Q.Q., Chen F.D. Involvement of CmWRKY10 in drought tolerance of chrysanthemum through the ABA–Signaling pathway. Int. J. Mol. Sci. 2016;17:693. doi: 10.3390/ijms17050693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang Y.J., Liang G., Yu D.Q. Activated expression of WRKY57 confers drought tolerance in Arabidopsis. Mol. Plant. 2012;5:1375–1388. doi: 10.1093/mp/sss080. [DOI] [PubMed] [Google Scholar]
- 21.Niu C.F., Wei W., Zhou Q.Y., Tian A.G., Hao Y.J., Zhang W.K., Ma B., Lin Q., Zhang Z.B., Zhang J.S., et al. Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ. 2012;35:1156–1170. doi: 10.1111/j.1365-3040.2012.02480.x. [DOI] [PubMed] [Google Scholar]
- 22.Wei K.F., Chen J., Chen Y.F., Wu L.J., Xie D.X. Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in Maize. DNA Res. 2012;19:153–164. doi: 10.1093/dnares/dsr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang T., Tan D.F., Zhang L., Zhang X.Y., Han Z.X. Phylogenetic analysis and drought-responsive expression profiles of the WRKY transcription factor family in maize. Agri Gene. 2017;3:99–108. doi: 10.1016/j.aggene.2017.01.001. [DOI] [Google Scholar]
- 24.Pandey S.P., Somssich I.E. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009;150:1648–1655. doi: 10.1104/pp.109.138990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu Y.R., Dong Q.Y., Yu D.Q. Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae. Plant Sci. 2012;185–186:288–297. doi: 10.1016/j.plantsci.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 26.Huang S.H., Yie S.W., Hwang D.J. Heterologous expression of OsWRKY6 gene in Arabidopsis activates the expression of defense related genes and enhances resistance to pathogens. Plant Sci. 2011;181:316–323. doi: 10.1016/j.plantsci.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 27.Li J., Wang J., Wang N.X., Guo X.Q., Gao Z. GhWRKY44, a WRKY transcription factor of cotton, mediates defense responses to pathogen infection in transgenic Nicotiana benthamiana. Plant Cell Tissue Organ Cult. 2015;121:127–140. doi: 10.1007/s11240-014-0688-9. [DOI] [Google Scholar]
- 28.Ulker B., Somssich I.E. WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 2004;7:491–498. doi: 10.1016/j.pbi.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 29.Wu X.L., Shiroto Y., Kishitani S., Ito Y., Toriyama K. Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 2009;28:21–30. doi: 10.1007/s00299-008-0614-x. [DOI] [PubMed] [Google Scholar]
- 30.Yan Y., Jia H.H., Wang F., Wang C., Liu S.C., Guo X.Q. Overexpression of GhWRKY27a reduces tolerance to drought stress and resistance to Rhizoctonia solani infection in transgenic Nicotiana benthamiana. Front. Physiol. 2015;6:265. doi: 10.3389/fphys.2015.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rizhsky L., Liang H., Mittler R. The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol. 2002;130:1143–1151. doi: 10.1104/pp.006858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li S.J., Fu Q.T., Chen L.G., Huang W.D., Yu D.Q. Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta. 2011;233:1237–1252. doi: 10.1007/s00425-011-1375-2. [DOI] [PubMed] [Google Scholar]
- 33.Atamian H.S., Eulgem T., Kaloshian I. SlWRKY70 is required for Mi-1-mediated resistance to aphids and nematodes in tomato. Planta. 2012;235:299–309. doi: 10.1007/s00425-011-1509-6. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Q.Y., Tian A.G., Zou H.F., Xie Z.M., Lei G., Huang J., Wang C.M., Wang H.W., Zhang J.S., Chen S.Y. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 2008;6:486–503. doi: 10.1111/j.1467-7652.2008.00336.x. [DOI] [PubMed] [Google Scholar]
- 35.Jiang Y., Deyholos M.K. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 2009;69:91–105. doi: 10.1007/s11103-008-9408-3. [DOI] [PubMed] [Google Scholar]
- 36.Marè C., Mazzucotelli E., Crosatti C., Francia E., Stanca A.M., Cattivelli L. Hv-WRKY38: A new transcription factor involved in cold- and drought-response in barley. Plant Mol. Biol. 2004;55:399–416. doi: 10.1007/s11103-004-0906-7. [DOI] [PubMed] [Google Scholar]
- 37.Xiong X., James V.A., Zhang H.N., Altpeter F. Constitutive expression of the barley HvWRKY38 transcription factor enhances drought tolerance in turf and forage grass (Paspalum notatum Flugge) Mol. Breed. 2010;25:419–432. doi: 10.1007/s11032-009-9341-4. [DOI] [Google Scholar]
- 38.Shinozaki K., Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007;58:221–227. doi: 10.1093/jxb/erl164. [DOI] [PubMed] [Google Scholar]
- 39.Qin Y.X., Wang M.C., Tian Y.C., He W.X., Han L., Xia G.M. Over-expression of TaMYB33 encoding a novel wheat MYB transcription factor increases salt and drought tolerance in Arabidopsis. Mol. Biol. Rep. 2012;39:7183–7192. doi: 10.1007/s11033-012-1550-y. [DOI] [PubMed] [Google Scholar]
- 40.Xiong L., Schumaker K.S., Zhu J.K. Cell signaling during cold, drought, and salt stress. Plant Cell. 2000;14:S165. doi: 10.1105/tpc.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Msanne J., Lin J., Stone J.M., Awada T. Characterization of abiotic stress-responsive Arabidopsis thaliana RD29A and RD29B genes and evaluation of transgenes. Planta. 2011;234:97–107. doi: 10.1007/s00425-011-1387-y. [DOI] [PubMed] [Google Scholar]
- 42.Pratt W.B., Galigniana M.D., Harrell J.M., Defranco D.B. Role of hsp90 and the hsp90-binding immunophilins in signalling protein movement. Cell. Signal. 2004;16:857–872. doi: 10.1016/j.cellsig.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 43.Yamada K., Fukao Y., Hayashi M., Fukazawa M., Suzuki I., Nishimura M. Cytosolic HSP90 regulates the heat shock response that is responsible for heat acclimation in Arabidopsis thaliana. J. Biol. Chem. 2007;282:37794–37804. doi: 10.1074/jbc.M707168200. [DOI] [PubMed] [Google Scholar]
- 44.Queitsch C., Sangster T.A., Lindquist S. Hsp90 as a capacitor of phenotypic variation. Nature. 2000;417:618–624. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- 45.Sangster T.A., Bahrami A., Wilczek A., Watanabe E., Schellenberg K., Mclellan C., Kelley A., Kong S.W., Queitsch C., Lindquist S. Phenotypic diversity and altered environmental plasticity in Arabidopsis thaliana with reduced Hsp90 levels. PLoS ONE. 2007;2:e648. doi: 10.1371/journal.pone.0000648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seki M., Kamei A., Yamaguchi-Shinozaki K., Shinozaki K. Molecular responses to drought, salinity and frost: Common and different paths for plant protection. Curr. Opin. Biotechnol. 2003;14:194–199. doi: 10.1016/S0958-1669(03)00030-2. [DOI] [PubMed] [Google Scholar]
- 47.Shinozaki K., Yamaguchi-shinozaki K., Seki M. Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Biol. 2003;6:410–417. doi: 10.1016/S1369-5266(03)00092-X. [DOI] [PubMed] [Google Scholar]
- 48.Song W., Zhao H., Zhang X., Lei L., Lai J. Genome-Wide identification of VQ motif-containing proteins and their expression profiles under abiotic stresses in Maize. Front. Plant Sci. 2016;6:1177. doi: 10.3389/fpls.2015.01177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu W.X., Zhang F.C., Zhang W.Z., Song L.F., Wu W.H., Chen Y.F. Arabidopsis Di19 functions as a transcription factor and modulates PR1, PR2, and PR5 expression in response to drought stress. Mol. Plant. 2013;6:1487–1502. doi: 10.1093/mp/sst031. [DOI] [PubMed] [Google Scholar]
- 50.Feng Z.J., Cui X.Y., Cui X.Y., Chen M., Yang G.X., Ma Y.Z., He G.Y., Xu Z.S. The soybean GmDi19-5 interacts with GmLEA3.1 and increases sensitivity of transgenic plants to abiotic stresses. Front. Plant Sci. 2015;6:179. doi: 10.3389/fpls.2015.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toppo S., Vanin S., Bosello V., Tosatto S.C. Evolutionary and structural insights into the multifaceted glutathione peroxidase (Gpx) superfamily. Antioxid. Redox Signal. 2008;10:1501–1514. doi: 10.1089/ars.2008.2057. [DOI] [PubMed] [Google Scholar]
- 53.Higo K., Ugawa Y., Iwamoto M., Korenaga T. Plant cis–acting regulatory DNA elements (PLACE) database. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu Z.S., Xia L.Q., Chen M., Cheng X.G., Zhang R.Y., Li L.C., Li Y.X., Zhao Y.X., Lu Y., Ni Z.Y., et al. Isolation and molecular characterization of the Triticum aestivum L. ethylene-responsive factor 1 (TaERF1) that increases multiple stress tolerance. Plant Mol. Biol. 2007;65:719–732. doi: 10.1007/s11103-007-9237-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.