Abstract
A novel pathway of vitamin D activation by CYP11A has previously been elucidated. To define the mechanism of action of its major dihydroxy-products, we tested the divergence and overlap between the gene expression profiles of human epidermal keratinocytes treated with either CYP11A1-derived 20,23(OH)2D3 or classical 1,25(OH)2D3. Both secosteroids have significant chemical similarity with the only differences being the positions of the hydroxyl groups. mRNA was isolated and examined by microarray analysis using Illumina’s HumanWG-6 chip/arrays and subsequent bioinformatics analyses. Marked differences in the up- and downregulated genes were observed between 1,25(OH)2D3- and 20,23(OH)2D3-treated cells. Hierarchical clustering identified both distinct, opposite and common (overlapping) gene expression patterns. CYP24A1 was a common gene strongly activated by both compounds, a finding confirmed by qPCR. Ingenuity pathway analysis identified VDR/RXR signaling as the top canonical pathway induced by 1,25(OH)2D3. In contrast, the top canonical pathway induced by 20,23(OH)2D3 was AhR, with VDR/RXR being the second nuclear receptor signaling pathway identified. QPCR analyses validated the former finding by revealing that 20,23(OH)2D3 stimulated CYP1A1 and CYP1B1 gene expression, effects located downstream of AhR. Similar stimulation was observed with 20(OH)D3, the precursor to 20,23(OH)2D3, as well as with its downstream metabolite, 17,20,23(OH)3D3. Using a Human AhR Reporter Assay System we showed marked activation of AhR activity by 20,23(OH)2D3, with weaker stimulation by 20(OH)D3. Finally, molecular modeling using an AhR LBD model predicted vitamin D3 hydroxyderivatives to be good ligands for this receptor. Thus, our microarray, qPCR, functional studies and molecular modeling indicate that AhR is the major receptor target for 20,23(OH)2D3, opening an exciting area of investigation on the interaction of different vitamin D3-hydroxyderivatives with AhR and the subsequent downstream activation of signal transduction pathways in a cell-type-dependent manner.
Keywords: vitamin D, dihydroxyvitamin D, epidermal keratinocytes, nuclear receptor signaling, microarray
1. Introduction
Vitamin D3 (D3) is formed by ultraviolet B radiation (UVB)-mediated breaking of the B ring of 7-dehydrocholesterol (7DHC) followed by thermal isomerization of the resulting pre-vitamin D3 to D3 [1,2]. The vast majority of circulating D3 is generated in epidermal keratinocytes [3]. D3 is a prohormone that is activated by sequential hydroxylations at C25 (by CYP2R1 or CYP27A1) and C1α (by CYP27B1) to 1,25-dihydroxyvitamin D3 (1,25(OH)2D3), the hormonally active form, referred to as the canonical pathway [4,5,6,7]. At the systemic level, C25 hydroxylation takes place in the liver with the resulting 25(OH)D3 being hydroxylated at C1α in kidneys [3,4,5,6,7]. The same pathway operates in peripheral tissues including epidermal keratinocytes, the major site of D3 formation [1].
In addition to the canonical pathway of vitamin D activation described above, novel CYP11A-mediated pathways have been discovered (reviewed in [8]). Specifically, CYP11A1, the first enzyme of steroidogenesis that hydroxylates and then cleaves the side chain of cholesterol to produce pregnenolone (reviewed in [9,10]), can also hydroxylate and cleave the side chain of 7DHC, and hydroxylate the side chain of D3 and D2 without cleavage [11,12,13,14,15,16]. The two major products of CYP11A1 action on D3, with defined stereochemistry, are 20S-hydroxyvitamin D3 (20(OH)D3) and 20S,23S-dihydroxyvitamin D3 (20,23(OH)2D3) [17]. These pathways operate in cultured epidermal, human and pig keratinocytes, dermal fibroblasts, colon cancer cells, and have also been described ex vivo for placenta and adrenal glands [18,19,20,21,22,23]. Importantly, the major products of these pathways are detectable in vivo in human serum, epidermis and adrenal glands [24].
The classical, hormonally-active dihydroxy form of vitamin D3, 1,25(OH)2D3, in addition to playing a fundamental role in body calcium and phosphorous homeostasis and in the proper functioning of the skeletomuscular system, has pleiotropic effects on different organs and cell functions (reviewed in [3,6,25,26,27,28,29]). These studies show that 1,25(OH)2D3 has immunomodulatory properties, is involved in the regulation of reproduction, pregnancy, child development, neurodevelopment, regulation of global metabolic and endocrine homeostasis and functions of the cardiovascular system, and has anticancer activities (reviewed in [2,6,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50]). At the cellular level, it regulates proliferation, differentiation, apoptosis, senescence, metabolism, migration, secretory activities, and protective and reparative mechanisms against oxidative stress and radiation. It is widely accepted that these functions are regulated by different signal transduction pathways initiated by 1,25(OH)2D3 binding to the vitamin D receptor (VDR) at the genomic binding site, and to some degree at a nongenomic binding site, in a cell-type dependent manner (reviewed [6,45,46,47,51,52,53,54]). In the skin, 1,25(OH)2D3 regulates the epidermal barrier and hair cycling and has radioprotective, anti-cancer and anti-inflammatory properties [1,3,52,53,55,56,57,58,59].
The novel secosteroids, produced by the non-canonical activation pathways initiated by CYP11A1, inhibit the proliferation of epidermal keratinocytes, melanocytes and dermal fibroblasts and promote the differentiation of keratinocytes. Furthermore, they inhibit fibrotic activities of fibroblasts and have immunomodulatory properties (reviewed in [19,49,60]). Importantly, 20(OH)D3 and 20,23(OH)2D3 are non-calcemic at pharmacological doses [61,62,63] which is in contrast to the highly calcemic effects of 1,25(OH)2D3 and 25(OH)D3. 20(OH)D3 and 20,23(OH)2D3 also attenuate the symptoms of skin fibrosis, rheumatoid arthritis and have photoprotective properties [8,19,23,64,65]. The CYP11A1-derived secosteroids have pleiotropic phenotypic effects that are cell-type–dependent [19,23,60,61,62,65,66,67,68,69,70,71,72,73,74,75]. They can act as biased agonists of the VDR [19,60,76,77] and can act as inverse agonists on retinoic acid orphan receptors (ROR) α and γ [60,78].
To better define the signaling pathways and mechanisms underlying the similarities and differences between phenotypic activities of classical 1,25(OH)2D3 and the major dihydroxy product of CYP11A1 action on vitamin D3, 20,23(OH)2D3, we examined and compared the gene expression profiles of human keratinocytes exposed to these secosteroids. Bioinformatics analysis was performed and differences and similarities in the activities of these structurally similar but distinct dihydroxy-D3 species were compared.
2. Results and Discussion
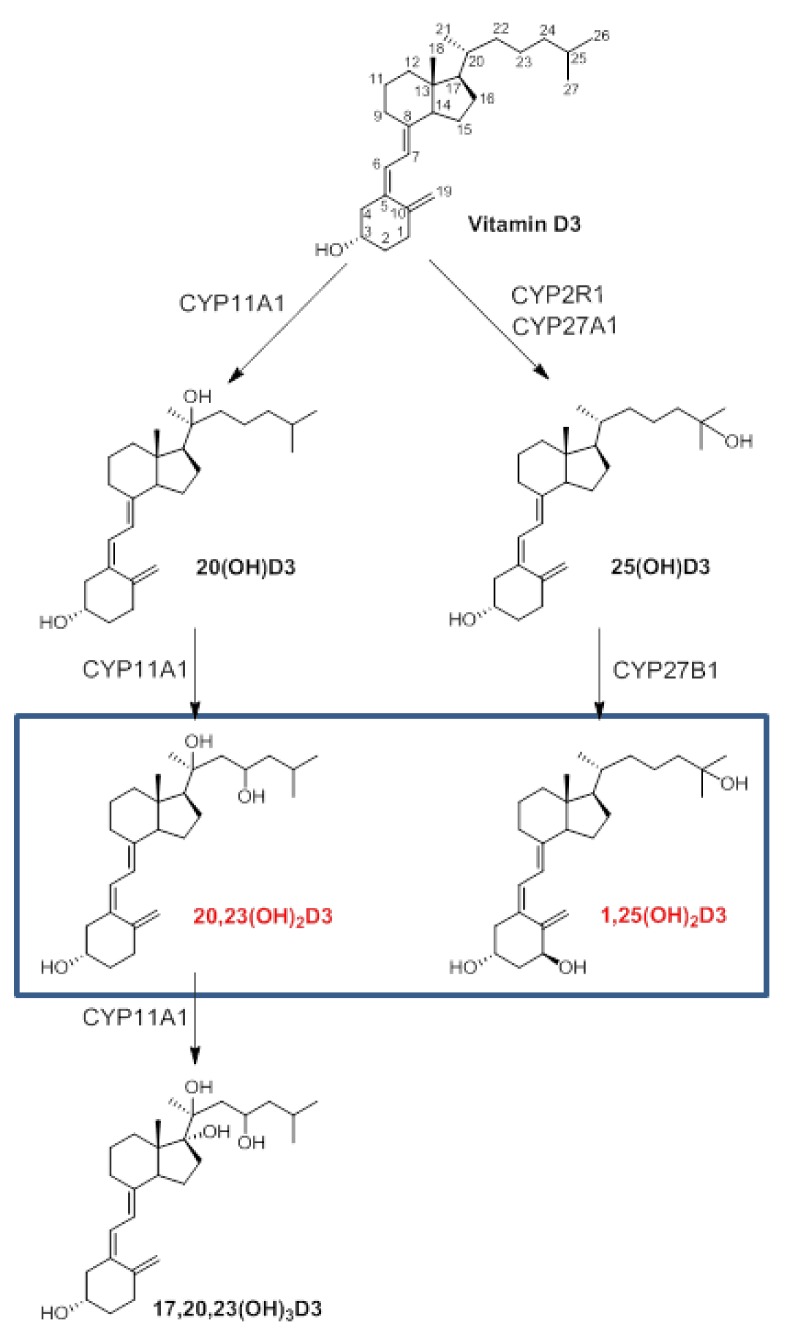
The structures and sequences of the reactions producing 1,25(OH)2D3 and 20,23(OH)2D3 in the epidermis are shown in Figure 1.
Figure 1.
Epidermal pathways of vitamin D3 activation to produce 1,25(OH)2D3 and 20,23(OH)2D3 and the downstream metabolite 17,20,23(OH)3D3. The rectangle marks the secosteroids used for microarray analyses.
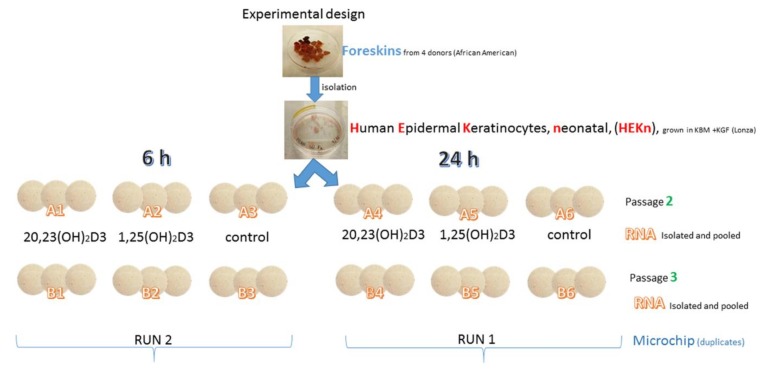
The schematic outline of the experimental design is presented in Figure 2. Briefly, to test the divergence and overlap between the gene expression patterns, human neonatal epidermal keratinocytes combined from four African-American [79] donors were treated with 1,25(OH)2D3 or 20,23(OH)2D3 for 6 or 24 h. Microarray assays were performed using Illumina’s HumanWG-6_V2 (Platform GPL13376) chip/array as described in Materials and Methods and the raw data has been deposited at the NCBI GEO (GSE117351).
Figure 2.
Outline of the experimental design.
The relative changes in gene expression (average of two independent experiments that used triplicate cell cultures), were normalized vs. vehicle control (0.1% ethanol). Table 1 shows marked differences in the number of genes up- or downregulated by either 1,25(OH)2D3 or 20,23(OH)2D3 using 1.5-, 2- and 4-fold cut-off values (FC). Average signal values for filtered gene clusters with FC ≥ ±1.5 are shown in Supplemental excel file #1. Briefly, treatment with 1,25(OH)2D3 for 6 h leads to changes in the expression of 148 vs. 37 genes for 20,23(OH)2D3 when using 1.5-FC, and 38 vs. 21 and 3 vs. 0 when using 2- and 4-FC, respectively. After 24 h, this trend changed to 410 and 4079 genes regulated, respectively, by 1,25(OH)2D3 and 20,23(OH)2D3 with 1.5-FC value, and 119 and 1611 for 2-FC value and 12 and 199 genes for 4-FC value, respectively (Table 1).
Table 1.
Number of genes up or downregulated in keratinocytes by 1,25(OH)2D3 or 20,23(OH)2D3 in comparison to vehicle control using 1.5-, 2- and 4-fold cut-off values.
| Time | Genes | 1,25(OH)2D3 | 20,23(OH)2D3 | ||||
|---|---|---|---|---|---|---|---|
| >1.5-Fold | >2-Fold | >4-Fold | >1.5-Fold | >2-Fold | >4-Fold | ||
| 6 h | Upregulated | 116 | 35 | 3 | 33 | 21 | 0 |
| Downregulated | 32 | 3 | 0 | 4 | 0 | 0 | |
| 24 h | Upregulated | 266 | 98 | 12 | 2013 | 763 | 98 |
| Downregulated | 144 | 21 | 0 | 2066 | 848 | 101 | |
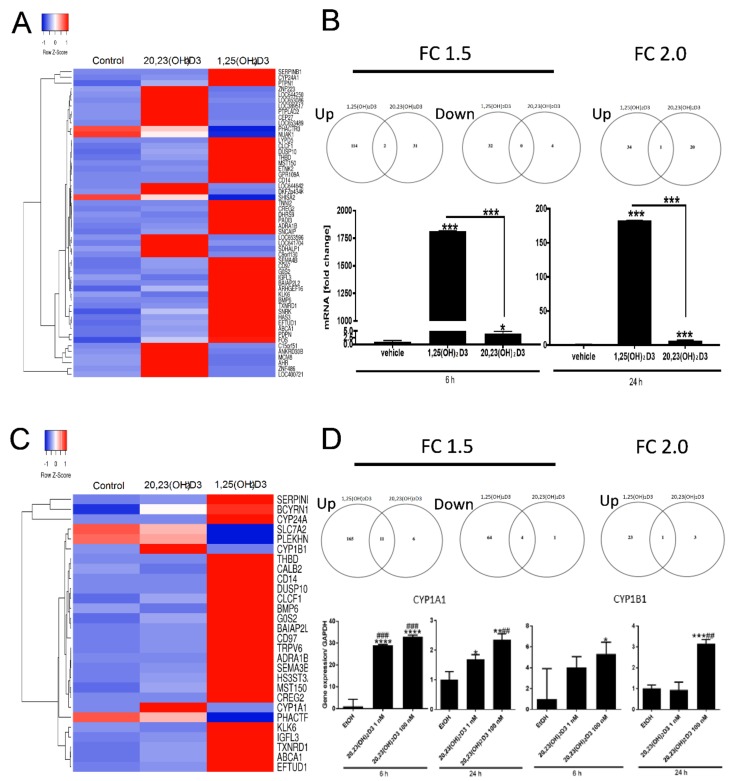
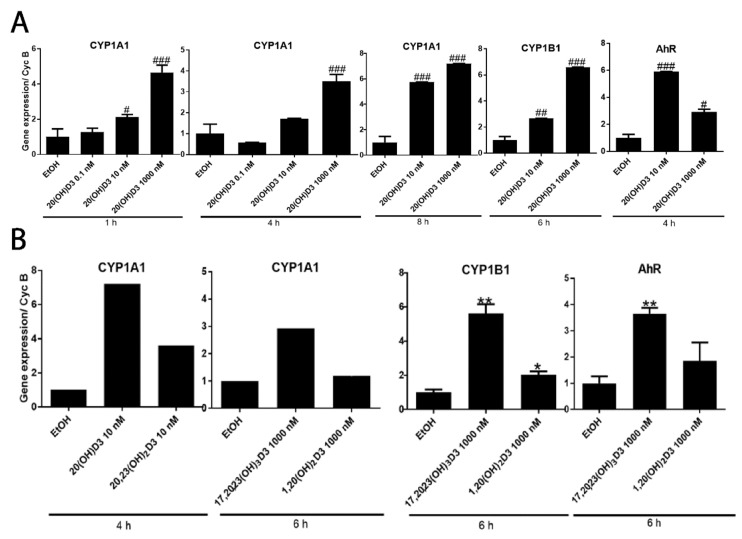
Hierarchical clustering identified patterns of genes responding to either 1,25(OH)2D3 or 20,23(OH)2D3, or to both. Selected gene clusters, representing the altered expression after 6 h of incubation as well as Venn diagrams are shown in Figure 3A. The heat maps corresponding to relative gene expression levels displayed both distinct or opposite, or common (overlapping) gene expression. For 2-FC there was only 1 common gene (CYP24A1) stimulated by both 1,25(OH)2D3 (82 fold) and 20,23(OH)2D3 (3.4 fold). This differential stimulation of CYP24A1 was further confirmed by qPCR (Figure 3B) and is consistent with the literature on 1,25(OH)2D3 [1,3,6,27,51] and 20,23(OH)2D3 [69,80]. For 1.5-FC there were two common genes, CYP24A1 and the gene with a target id ILMN_131812 (identified as small ILF3/NF90-associated RNA A1 (SNAR-A1)), for which expression was stimulated.
Figure 3.
Changes in gene expression in human epidermal keratinocytes treated with 1,25(OH)2D3 or 20,23(OH)2D3 for 6 h. (A) Heat map of the gene expression pattern for experiment #1 with corresponding Venn diagrams for FC ≥ 2 and 1.5. (B) Effect of 10−7 M of 1,25(OH)2D3 or 20,23(OH)2D3 on CYP24A1 expression in keratinocytes after 6 and 24 h treatment. (C) Heat map of the gene expression pattern for experiment #2 with corresponding Venn diagrams for FC-2 and 1.5. (D) Effect of 10−9 and 10−7 M of 20,23(OH)2D3 on CYP1A1 and CYP1B1 expression in keratinocytes after 6 or 24 h treatment. Data represent means ± SD (n = 3) where * p < 0.05, ** p < 0.01, *** p < 0.001 and **** p < 0.0001 at student t-test; ## p < 0.01 and ### p < 0.001 at one-way ANOVA test.
Ingenuity pathway analysis using FC ≥ ±1.5 was performed. The top canonical pathway induced by 1,25(OH)2D3 was VDR/RXR signaling (Table 2) (Supplemental Figure S1A), which was expected [3,6,51,81,82]. This was followed by the roles of osteoblasts, osteoclasts and chondrocytes in rheumatoid arthritis; the role of macrophages, fibroblasts and endothelial cells in rheumatoid arthritis; and Toll-like receptor signaling (Table 2), which is consistent with previously reported functions of 1,25(OH)2D3 [3,6,30,31,45,51,54]. Interestingly, the next top nuclear receptor signaling pathway activated by 1,25(OH)2D3 was linked to the glucocorticoid receptor (GR) followed by the aryl hydrocarbon receptor [74], PPAR, PPARα/RXRα, LXR/RXR, and RAR (Table 2). The inclusion of these additional pathways could be secondary to the use of the same dimeric partner, RXR, and communication between receptors, or alternatively by activation by signaling pathways downstream of VDR. For example, it is already known that 1,25(OH)2D3 can selectively activate local elements of hypothalamo-pituitary adrenal axis in keratinocytes [71]. The significance of additional nuclear receptor signaling is out of the scope of this paper and is a goal of our future research.
Table 2.
Canonical pathways activated by 1,25(OH)2D3 in human epidermal keratinocytes after 6 h of treatment. Nuclear receptors are marked in bold.
| Ingenuity Canonical Pathways | p-Value | Overlap (%) | Downregulated | No Change | Upregulated | No Overlap with Dataset |
|---|---|---|---|---|---|---|
| VDR/RXR Activation | 1.0 ×10−10 | 12.8 | 34/78 (44%) | 0/78 (0%) | 43/78 (55%) | 1/78 (1%) |
| Role of Osteoblasts, Osteoclasts and Chondrocytes in Rheumatoid Arthritis | 2.2 × 10−7 | 5.02 | 100/219 (46%) | 0/219 (0%) | 114/219 (52%) | 5/219 (2%) |
| Role of Macrophages, Fibroblasts and Endothelial Cells in Rheumatoid Arthritis | 4.4 × 10−6 | 3.72 | 144/296 (49%) | 0/296 (0%) | 138/296 (47%) | 14/296 (5%) |
| Toll-like Receptor Signaling | 9.3 × 10−6 | 8.11 | 39/74 (53%) | 0/74 (0%) | 33/74 (45%) | 2/74 (3%) |
| Hepatic Cholestasis | 1.2 × 10−5 | 4.94 | 83/162 (51%) | 0/162 (0%) | 75/162 (46%) | 4/162 (2%) |
| Glucocorticoid Receptor Signaling | 1.5 × 10−5 | 3.64 | 152/275 (55%) | 0/275 (0%) | 117/275 (43%) | 6/275 (2%) |
| Role of Cytokines in Mediating Communication between Immune Cells | 3.1 × 10−5 | 9.09 | 32/55 (58%) | 0/55 (0%) | 20/55 (36%) | 3/55 (5%) |
| IL-10 Signaling | 8.5 × 10−5 | 7.35 | 33/68 (49%) | 0/68 (0%) | 34/68 (50%) | 1/68 (1%) |
| IL-6 Signaling | 0.00012 | 5.17 | 57/116 (49%) | 0/116 (0%) | 59/116 (51%) | 0/116 (0%) |
| p38 MAPK Signaling | 0.000126 | 5.13 | 62/117 (53%) | 0/117 (0%) | 52/117 (44%) | 3/117 (3%) |
| MIF Regulation of Innate Immunity | 0.000151 | 9.76 | 19/41 (46%) | 0/41 (0%) | 21/41 (51%) | 1/41 (2%) |
| Molecular Mechanisms of Cancer | 0.000155 | 2.74 | 201/365 (55%) | 0/365 (0%) | 157/365 (43%) | 7/365 (2%) |
| iNOS Signaling | 0.0002 | 9.09 | 23/44 (52%) | 0/44 (0%) | 20/44 (45%) | 1/44 (2%) |
| Aryl Hydrocarbon Receptor Signaling | 0.000331 | 4.29 | 73/140 (52%) | 0/140 (0%) | 61/140 (44%) | 6/140 (4%) |
| PPAR Signaling | 0.000398 | 5.32 | 52/94 (55%) | 0/94 (0%) | 38/94 (40%) | 4/94 (4%) |
| LPS/IL-1 Mediated Inhibition of RXR Function | 0.000631 | 3.2 | 119/219 (54%) | 0/219 (0%) | 88/219 (40%) | 12/219 (5%) |
| TNFR2 Signaling | 0.000891 | 10.3 | 14/29 (48%) | 0/29 (0%) | 14/29 (48%) | 1/29 (3%) |
| HMGB1 Signaling | 0.001202 | 4.17 | 54/120 (45%) | 0/120 (0%) | 63/120 (53%) | 3/120 (3%) |
| MIF-mediated Glucocorticoid Regulation | 0.001318 | 9.09 | 16/33 (48%) | 0/33 (0%) | 16/33 (48%) | 1/33 (3%) |
| ILK Signaling | 0.001479 | 3.23 | 102/186 (55%) | 0/186 (0%) | 77/186 (41%) | 7/186 (4%) |
| IL-17A Signaling in Fibroblasts | 0.001549 | 8.57 | 20/35 (57%) | 0/35 (0%) | 15/35 (43%) | 0/35 (0%) |
| Role of JAK2 in Hormone-like Cytokine Signaling | 0.001549 | 8.57 | 18/35 (51%) | 0/35 (0%) | 14/35 (40%) | 3/35 (9%) |
| PI3K Signaling in B Lymphocytes | 0.001585 | 3.91 | 66/128 (52%) | 0/128 (0%) | 57/128 (45%) | 5/128 (4%) |
| Factors Promoting Cardiogenesis in Vertebrates | 0.003236 | 4.35 | 52/92 (57%) | 0/92 (0%) | 37/92 (40%) | 3/92 (3%) |
| TNFR1 Signaling | 0.004169 | 6.12 | 27/49 (55%) | 0/49 (0%) | 20/49 (41%) | 2/49 (4%) |
| Antioxidant Action of Vitamin C | 0.004169 | 4.04 | 51/99 (52%) | 0/99 (0%) | 42/99 (42%) | 6/99 (6%) |
| Acute Phase Response Signaling | 0.005248 | 2.96 | 88/169 (52%) | 0/169 (0%) | 79/169 (47%) | 2/169 (1%) |
| Differential Regulation of Cytokine Production in Macrophages and T Helper Cells by IL-17A and IL-17F | 0.006166 | 11.1 | 7/18 (39%) | 0/18 (0%) | 11/18 (61%) | 0/18 (0%) |
| PPARα/RXRα Activation | 0.006607 | 2.79 | 98/179 (55%) | 0/179 (0%) | 67/179 (37%) | 14/179 (8%) |
| Hepatic Fibrosis/Hepatic Stellate Cell Activation | 0.007244 | 2.73 | 96/183 (52%) | 0/183 (0%) | 84/183 (46%) | 3/183 (2%) |
| Type II Diabetes Mellitus Signaling | 0.007586 | 3.42 | 50/117 (43%) | 0/117 (0%) | 64/117 (55%) | 3/117 (3%) |
| Estrogen-Dependent Breast Cancer Signaling | 0.008318 | 4.76 | 30/63 (48%) | 0/63 (0%) | 33/63 (52%) | 0/63 (0%) |
| LXR/RXR Activation | 0.008511 | 3.31 | 68/121 (56%) | 0/121 (0%) | 53/121 (44%) | 0/121 (0%) |
| RAR Activation | 0.008511 | 2.63 | 86/190 (45%) | 0/190 (0%) | 100/190 (53%) | 4/190 (2%) |
The top canonical nuclear receptor pathway induced by 20,23(OH)2D3 was AhR signaling (Supplemental Figure S2A) with VDR/RXR being next (Supplemental Figure S3A) (Table 3). While the identification of the VDR/RXR as the target for 20,23(OH)2D3 is consistent with previously reported functional data and molecular modeling [60,65,69,83], identification of the AhR as its primary target was unexpected and hence it was further analyzed in detail as described below. Table 4 shows that for 1,25(OH)2D3 the nuclear signaling pathways VDR/RXR, followed by AhR, PPARα/RXRα, RAR and LXR/RXR, were among the top toxicity-related pathways identified. The top signaling pathways for 20,23(OH)2D3 were linked to the activation of AhR and VDR/RXR (Table 5). Table 6 and Table 7 show certain functional similarities between top diseases and bifunctions affected by both molecules. For example, cancer, and organismal injury and abnormalities, are the top two diseases affected by both molecules. These phenotypic similarities are consistent with previously reported studies comparing the biological effects of 1,25(OH)2D3 and CYP11A1-derived D3-hydroxyderivatives, including 20,23(OH)2D3, and indicate similarities between the effects on cell proliferation and differentiation, as well as similar anti-inflammatory, photoprotective and anti-cancer actions [23,60,61,62,64,72,80,84].
Table 3.
Canonical pathways activated by 20,23(OH)2D3 in human epidermal keratinocytes after 6 h of treatment. Nuclear receptors are marked in bold.
| Ingenuity Canonical Pathways | p-Value | Overlap (%) | Downregulated | No change | Upregulated | No Overlap with Dataset |
|---|---|---|---|---|---|---|
| 2-ketoglutarate Dehydrogenase Complex | 0.004898 | 25 | 2/4 (50%) | 0/4 (0%) | 2/4 (50%) | 0/4 (0%) |
| Aryl Hydrocarbon Receptor Signaling | 0.012589 | 1.43 | 64/140 (46%) | 0/140 (0%) | 70/140 (50%) | 6/140 (4%) |
| Aldosterone Signaling in Epithelial Cells | 0.014791 | 1.32 | 80/152 (53%) | 0/152 (0%) | 69/152 (45%) | 3/152 (2%) |
| TCA Cycle II (Eukaryotic) | 0.027542 | 4.35 | 14/23 (61%) | 0/23 (0%) | 9/23 (39%) | 0/23 (0%) |
| Bupropion Degradation | 0.0302 | 4 | 12/25 (48%) | 0/25 (0%) | 12/25 (48%) | 1/25 (4%) |
| D-myo-inositol (1,4,5)-Trisphosphate Biosynthesis | 0.032359 | 3.7 | 13/27 (48%) | 0/27 (0%) | 13/27 (48%) | 1/27 (4%) |
| Acetone Degradation I (to Methylglyoxal) | 0.032359 | 3.7 | 13/27 (48%) | 0/27 (0%) | 13/27 (48%) | 1/27 (4%) |
| Xenobiotic Metabolism Signaling | 0.042658 | 0.738 | 133/271 (49%) | 0/271 (0%) | 122/271 (45%) | 16/271 (6%) |
| Estrogen Biosynthesis | 0.045709 | 2.63 | 17/38 (45%) | 0/38 (0%) | 20/38 (53%) | 1/38 (3%) |
| Nicotine Degradation III | 0.064565 | 1.85 | 23/54 (43%) | 0/54 (0%) | 22/54 (41%) | 9/54 (17%) |
| Melatonin Degradation I | 0.067608 | 1.75 | 25/57 (44%) | 0/57 (0%) | 24/57 (42%) | 8/57 (14%) |
| Superpathway of Melatonin Degradation | 0.072444 | 1.61 | 27/62 (44%) | 0/62 (0%) | 27/62 (44%) | 8/62 (13%) |
| GM-CSF Signaling | 0.072444 | 1.61 | 26/62 (42%) | 0/62 (0%) | 36/62 (58%) | 0/62 (0%) |
| Nicotine Degradation II | 0.074131 | 1.59 | 26/63 (41%) | 0/63 (0%) | 25/63 (40%) | 12/63 (19%) |
| VDR/RXR Activation | 0.091201 | 1.28 | 34/78 (44%) | 0/78 (0%) | 43/78 (55%) | 1/78 (1%) |
| Acute Myeloid Leukemia Signaling | 0.091201 | 1.27 | 39/79 (49%) | 0/79 (0%) | 38/79 (48%) | 2/79 (3%) |
| TR/RXR Activation | 0.097724 | 1.18 | 39/85 (46%) | 0/85 (0%) | 46/85 (54%) | 0/85 (0%) |
| Regulation of Actin-based Motility by Rho | 0.105196 | 1.1 | 45/91 (49%) | 0/91 (0%) | 39/91 (43%) | 7/91 (8%) |
| Antioxidant Action of Vitamin C | 0.114025 | 1.01 | 53/99 (54%) | 0/99 (0%) | 40/99 (40%) | 6/99 (6%) |
| Rac Signaling | 0.119399 | 0.962 | 47/104 (45%) | 0/104 (0%) | 55/104 (53%) | 2/104 (2%) |
| Type I Diabetes Mellitus Signaling | 0.125893 | 0.909 | 54/110 (49%) | 1/110 (1%) | 46/110 (42%) | 9/110 (8%) |
| RhoA Signaling | 0.138676 | 0.82 | 56/122 (46%) | 0/122 (0%) | 61/122 (50%) | 5/122 (4%) |
| 3-phosphoinositide Biosynthesis | 0.175792 | 0.633 | 72/158 (46%) | 0/158 (0%) | 76/158 (48%) | 10/158 (6%) |
| RhoGDI Signaling | 0.190985 | 0.578 | 75/173 (43%) | 0/173 (0%) | 94/173 (54%) | 4/173 (2%) |
| Superpathway of Inositol Phosphate Compounds | 0.212814 | 0.513 | 87/195 (45%) | 0/195 (0%) | 96/195 (49%) | 12/195 (6%) |
| Actin Cytoskeleton Signaling | 0.233884 | 0.461 | 93/217 (43%) | 0/217 (0%) | 115/217 (53%) | 9/217 (4%) |
| Signaling by Rho Family GTPases | 0.249459 | 0.427 | 106/234 (45%) | 0/234 (0%) | 124/234 (53%) | 4/234 (2%) |
| Protein Ubiquitination Pathway | 0.268534 | 0.392 | 121/255 (47%) | 0/255 (0%) | 126/255 (49%) | 8/255 (3%) |
Table 4.
Ingenuity toxicity list secondary to keratinocytes treatment with 1,25(OH)2D3 in humans for 6 h. Nuclear receptors are marked in bold.
| Ingenuity Toxicity Lists | p-Value | Overlap (%) | Downregulated | No Change | Upregulated | No Overlap with Dataset |
|---|---|---|---|---|---|---|
| VDR/RXR Activation | 1.0 × 10−10 | 12.8 | 34/78 (44%) | 0/78 (0%) | 43/78 (55%) | 1/78 (1%) |
| Hepatic Cholestasis | 1.4 × 10−5 | 4.85 | 85/165 (52%) | 0/165 (0%) | 76/165 (46%) | 4/165 (2%) |
| Renal Necrosis/Cell Death | 2.8 × 10−5 | 2.59 | 241/501 (48%) | 0/501 (0%) | 242/501 (48%) | 18/501 (4%) |
| Aryl Hydrocarbon Receptor Signaling | 9.6 × 10−5 | 4.35 | 79/161 (49%) | 0/161 (0%) | 67/161 (42%) | 15/161 (9%) |
| Liver Necrosis/Cell Death | 0.000525 | 2.87 | 131/279 (47%) | 0/279 (0%) | 136/279 (49%) | 12/279 (4%) |
| Liver Proliferation | 0.000776 | 3.08 | 110/227 (48%) | 0/227 (0%) | 106/227 (47%) | 11/227 (5%) |
| Cardiac Hypertrophy | 0.00138 | 2.24 | 207/401 (52%) | 0/401 (0%) | 171/401 (43%) | 23/401 (6%) |
| LPS/IL-1 Mediated Inhibition of RXR Function | 0.00138 | 2.79 | 124/251 (49%) | 0/251 (0%) | 93/251 (37%) | 34/251 (14%) |
| Hepatic Stellate Cell Activation | 0.001549 | 8.57 | 17/35 (49%) | 0/35 (0%) | 18/35 (51%) | 0/35 (0%) |
| Increases Liver Steatosis | 0.002188 | 4.82 | 43/83 (52%) | 0/83 (0%) | 37/83 (45%) | 3/83 (4%) |
| Mechanism of Gene Regulation by Peroxisome Proliferators via PPARα | 0.003631 | 4.21 | 54/95 (57%) | 0/95 (0%) | 40/95 (42%) | 1/95 (1%) |
| Hepatic Fibrosis | 0.004169 | 4.04 | 42/99 (42%) | 0/99 (0%) | 54/99 (55%) | 3/99 (3%) |
| Increases Liver Damage | 0.005495 | 3.74 | 48/107 (45%) | 0/107 (0%) | 56/107 (52%) | 3/107 (3%) |
| PPARα/RXRα Activation | 0.007244 | 2.73 | 100/183 (55%) | 0/183 (0%) | 69/183 (38%) | 14/183 (8%) |
| Acute Renal Failure Panel (Rat) | 0.007943 | 4.84 | 31/62 (50%) | 0/62 (0%) | 24/62 (39%) | 7/62 (11%) |
| RAR Activation | 0.008511 | 2.63 | 86/190 (45%) | 0/190 (0%) | 100/190 (53%) | 4/190 (2%) |
| Cardiac Necrosis/Cell Death | 0.008913 | 2.23 | 141/269 (52%) | 0/269 (0%) | 114/269 (42%) | 14/269 (5%) |
| Cardiac Fibrosis | 0.008913 | 2.6 | 104/192 (54%) | 0/192 (0%) | 72/192 (38%) | 16/192 (8%) |
| LXR/RXR Activation | 0.008913 | 3.25 | 69/123 (56%) | 0/123 (0%) | 54/123 (44%) | 0/123 (0%) |
| Nongenotoxic Hepatocarcinogenicity Biomarker Panel | 0.00912 | 9.09 | 11/22 (50%) | 0/22 (0%) | 10/22 (45%) | 1/22 (5%) |
| Increases Renal Damage | 0.016218 | 3.7 | 38/81 (47%) | 0/81 (0%) | 36/81 (44%) | 7/81 (9%) |
| NRF2-mediated Oxidative Stress Response | 0.019498 | 2.14 | 99/234 (42%) | 0/234 (0%) | 103/234 (44%) | 32/234 (14%) |
| TGF-β Signaling | 0.021878 | 3.33 | 54/90 (60%) | 0/90 (0%) | 35/90 (39%) | 1/90 (1%) |
Table 5.
Ingenuity toxicity list secondary to keratinocytes treatment with 20,23(OH)2D3 in humans for 6 h. Nuclear receptors are marked in bold.
| Ingenuity Toxicity Lists | p-Value | Overlap (%) | Downregulated | No Change | Upregulated | No Overlap with Dataset |
|---|---|---|---|---|---|---|
| Cytochrome P450 Panel—Substrate is a Vitamin (Human) | 0.007244 | 16.7 | 2/6 (33%) | 0/6 (0%) | 4/6 (67%) | 0/6 (0%) |
| Aryl Hydrocarbon Receptor Signaling | 0.016218 | 1.24 | 71/161 (44%) | 0/161 (0%) | 75/161 (47%) | 15/161 (9%) |
| Cytochrome P450 Panel—Substrate is a Sterol (Human) | 0.016982 | 7.14 | 5/14 (36%) | 0/14 (0%) | 9/14 (64%) | 0/14 (0%) |
| Cytochrome P450 Panel—Substrate is a Xenobiotic (Human) | 0.021878 | 5.56 | 7/18 (39%) | 0/18 (0%) | 9/18 (50%) | 2/18 (11%) |
| Nongenotoxic Hepatocarcinogenicity Biomarker Panel | 0.026303 | 4.55 | 12/22 (55%) | 0/22 (0%) | 9/22 (41%) | 1/22 (5%) |
| Cytochrome P450 Panel—Substrate is a Xenobiotic (Mouse) | 0.0302 | 4 | 5/25 (20%) | 0/25 (0%) | 7/25 (28%) | 13/25 (52%) |
| Cytochrome P450 Panel—Substrate is a Xenobiotic (Rat) | 0.030903 | 3.85 | 5/26 (19%) | 0/26 (0%) | 7/26 (27%) | 14/26 (54%) |
| Xenobiotic Metabolism Signaling | 0.063096 | 0.595 | 155/336 (46%) | 0/336 (0%) | 139/336 (41%) | 42/336 (13%) |
| VDR/RXR Activation | 0.091201 | 1.28 | 34/78 (44%) | 0/78 (0%) | 43/78 (55%) | 1/78 (1%) |
| TR/RXR Activation | 0.097724 | 1.18 | 39/85 (46%) | 0/85 (0%) | 46/85 (54%) | 0/85 (0%) |
| Hepatic Fibrosis | 0.114025 | 1.01 | 49/99 (49%) | 0/99 (0%) | 47/99 (47%) | 3/99 (3%) |
| Increases Liver Hyperplasia/ Hyperproliferation |
0.116145 | 0.99 | 39/101 (39%) | 0/101 (0%) | 52/101 (51%) | 10/101 (10%) |
| Renal Necrosis/Cell Death | 0.12388 | 0.399 | 224/501 (45%) | 1/501 (0%) | 258/501 (51%) | 18/501 (4%) |
| Fatty Acid Metabolism | 0.133352 | 0.855 | 57/117 (49%) | 0/117 (0%) | 39/117 (33%) | 21/117 (18%) |
| Cardiac Fibrosis | 0.209894 | 0.521 | 89/192 (46%) | 0/192 (0%) | 87/192 (45%) | 16/192 (8%) |
| Liver Proliferation | 0.24322 | 0.441 | 98/227 (43%) | 0/227 (0%) | 118/227 (52%) | 11/227 (5%) |
Table 6.
Categories of biological functions with diseases or function annotation activated by 1,25(OH)2D3 in human epidermal keratinocytes after 6 h of treatment.
| Categories of Biological Function | Diseases or Functions Annotation | p-Value | Activation z-Score | # of Genes |
|---|---|---|---|---|
| Cancer, Organismal Injury and Abnormalities | growth of tumor | 1.5 × 10−12 | 0.033 | 31 |
| Cellular Movement | cell movement | 4.5 × 10−12 | 1.845 | 52 |
| Cancer, Cellular Development, Cellular Growth and Proliferation, Organismal Injury and Abnormalities, Tumor Morphology | proliferation of tumor cells | 7.5 × 10−12 | −0.27 | 23 |
| Cellular Movement | migration of cells | 1.3 × 10−11 | 1.801 | 48 |
| Cellular Growth and Proliferation | proliferation of cells | 4.2 × 10−11 | 0.347 | 70 |
| Carbohydrate Metabolism | metabolism of polysaccharide | 4.4 × 10−11 | 0.755 | 16 |
| Cell Death and Survival | apoptosis of tumor cell lines | 8.3 × 10−11 | 1.089 | 36 |
| Cellular Movement | invasion of cells | 8.7 × 10−11 | 0.571 | 30 |
| Inflammatory Response | inflammatory response | 1.4 × 10−10 | 0.766 | 28 |
| Cellular Development | differentiation of cells | 1.5 × 10−10 | 2.774 | 51 |
| Cell Death and Survival | cell survival | 3.1 × 10−10 | 2.346 | 38 |
| Cell Death and Survival | apoptosis | 5.7 × 10−10 | 1.307 | 55 |
| Cell Death and Survival | necrosis | 6.3 × 10−10 | 1.7 | 54 |
| Cell Death and Survival | cell viability | 6.4 × 10−10 | 2.547 | 36 |
| Tissue Morphology | quantity of cells | 7.1 × 10−10 | 1.065 | 43 |
| Cardiovascular System Development and Function, Organismal Development | vasculogenesis | 9.4 × 10−10 | 0.844 | 26 |
| Carbohydrate Metabolism | synthesis of polysaccharide | 9.6 × 10−10 | 0.297 | 13 |
| Cellular Growth and Proliferation, Tissue Development | proliferation of connective tissue cells | 1.2 × 10−9 | 1.232 | 23 |
| Cellular Movement | cell movement of tumor cell lines | 1.2 × 10−9 | 0.955 | 28 |
| Cell Death and Survival | cell death of tumor cell lines | 1.2 × 10−9 | 0.81 | 39 |
| Cellular Development, Cellular Growth and Proliferation | proliferation of tumor cell lines | 1.9 × 10−9 | 0.324 | 39 |
| Organismal Survival | morbidity or mortality | 2.3 × 10−9 | −1.006 | 51 |
| Embryonic Development, Organismal Development | development of body trunk | 2.7 × 10−9 | 0.06 | 32 |
| Cell Death and Survival | cell death | 3.1 × 10−9 | 1.556 | 62 |
| Cancer, Organismal Injury and Abnormalities | growth of malignant tumor | 3.3 × 10−9 | 0.518 | 19 |
| Cellular Development, Cellular Growth and Proliferation, Connective Tissue Development and Function, Tissue Development | proliferation of fibroblasts | 4.0 × 10−9 | 0.604 | 17 |
| Cellular Function and Maintenance, Hematological System Development and Function | function of myeloid cells | 4.5 × 10−9 | 14 | |
| Cell Signaling, Small Molecule Biochemistry | synthesis of nitric oxide | 4.7 × 10−9 | 0.89 | 15 |
| Dermatological Diseases and Conditions | psoriasis | 5.0 × 10−9 | 22 |
Table 7.
Categories of biological functions with diseases or function annotation activated by 20,23(OH)2D3 in human epidermal keratinocytes after 6 h of treatment.
| Categories of Biological Function | Diseases or Functions Annotation | p-Value | # of Genes |
|---|---|---|---|
| Cancer, Organismal Injury and Abnormalities, Respiratory Disease | carcinoma in lung | 6.5 × 10−5 | 8 |
| Vitamin and Mineral Metabolism | metabolism of vitamin | 0.000254 | 3 |
| Developmental Disorder, Skeletal and Muscular Disorders | hypertrophy of smooth muscle | 0.000325 | 2 |
| Cancer, Organismal Injury and Abnormalities, Respiratory Disease | non-small cell lung cancer | 0.000334 | 7 |
| Infectious Diseases | internalization of virus | 0.000355 | 2 |
| Cancer, Organismal Injury and Abnormalities | adenocarcinoma | 0.000603 | 20 |
| Cancer, Organismal Injury and Abnormalities, Reproductive System Disease | prostate cancer | 0.000618 | 7 |
| Cancer, Gastrointestinal Disease, Hepatic System Disease, Organismal Injury and Abnormalities | liver adenoma | 0.000692 | 2 |
| Cancer, Endocrine System Disorders, Organismal Injury and Abnormalities | endocrine gland tumor | 0.000718 | 9 |
| Cell Death and Survival | apoptosis of germ cells | 0.000799 | 3 |
| Lipid Metabolism, Small Molecule Biochemistry, Vitamin and Mineral Metabolism | catabolism of terpenoid | 0.000828 | 2 |
| Cancer, Organismal Injury and Abnormalities | epithelial cancer | 0.000989 | 23 |
| Endocrine System Development and Function, Lipid Metabolism, Small Molecule Biochemistry, Vitamin and Mineral Metabolism | synthesis of estrogen | 0.00114 | 2 |
| Ophthalmic Disease, Organismal Injury and Abnormalities | age-related macular degeneration type 6 | 0.00122 | 1 |
| Cell Cycle | arrest in sub-G1 phase of endometrial cancer cell lines | 0.00122 | 1 |
| Cell Morphology, Connective Tissue Development and Function | blebbing of pulmonary fibroblasts | 0.00122 | 1 |
| Organismal Injury and Abnormalities | calcification of uterus | 0.00122 | 1 |
| Cell Cycle, Cell Death and Survival | chromosome condensation of pulmonary fibroblasts | 0.00122 | 1 |
| Cell-To-Cell Signaling and Interaction, Inflammatory Response | cytotoxic reaction of bone marrow cells | 0.00122 | 1 |
| Tissue Morphology | deficiency of mast cells | 0.00122 | 1 |
| Cell Cycle | delay in G1/S phase transition of hepatoma cell lines | 0.00122 | 1 |
| Embryonic Development, Organ Development, Organismal Development, Tissue Development, Visual System Development and Function | development of outflow pathway | 0.00122 | 1 |
| Embryonic Development, Organ Development, Organismal Development, Reproductive System Development and Function, Tissue Development | development of placenta decidua | 0.00122 | 1 |
| Embryonic Development, Organ Development, Organismal Development, Reproductive System Development and Function, Tissue Development | development of placental spongiotrophoblast layer | 0.00122 | 1 |
| Cardiovascular System Development and Function, Tissue Morphology | diameter of portal vein | 0.00122 | 1 |
| Cardiovascular System Development and Function, Tissue Morphology | diameter of umbilical vein | 0.00122 | 1 |
| Hereditary Disorder, Ophthalmic Disease, Organismal Injury and Abnormalities | digenic early-onset glaucoma | 0.00122 | 1 |
| Cancer, Organismal Injury and Abnormalities, Reproductive System Disease | estrogen receptor positive endometrial cancer | 0.00122 | 1 |
| Connective Tissue Disorders, Organismal Injury and Abnormalities | fibrosis of submucosa | 0.00122 | 1 |
| Digestive System Development and Function, Embryonic Development, Organ Development, Organismal Development, Tissue Development | formation of salivary duct | 0.00122 | 1 |
| Cardiovascular System Development and Function, Embryonic Development, Lymphoid Tissue Structure and Development, Organ Development, Organismal Development, Respiratory System Development and Function, Tissue Development | formation of tracheal duct | 0.00122 | 1 |
| Cancer, Gastrointestinal Disease, Organismal Injury and Abnormalities | hyperplasia of pylorus | 0.00122 | 1 |
| Cancer, Cardiovascular Disease, Organismal Injury and Abnormalities | hyperplasia of vasculature | 0.00122 | 1 |
| Developmental Disorder, Gastrointestinal Disease | hypertrophy of gastric epithelium | 0.00122 | 1 |
| Dermatological Diseases and Conditions, Developmental Disorder | hypertrophy of skin | 0.00122 | 1 |
Because of the unexpected differences between 1,25(OH)2D3 and 20,23(OH)2D3, the 6 h incubation experiment was repeated in a similar manner as shown in Figure 2 and microarray analyses were performed using Illumina’s HumanWG-6_V2 (Platform GPL13376) chip/array. Average signal values for filtered gene clusters with FC ≥ ±1.5 are shown in Supplemental excel file #2. The heat maps corresponding to relative gene expression and Venn diagrams are shown in Figure 3C. Again, for a 2-fold cut-off value there was only one common gene (CYP24A1) whose expression was stimulated by both 1,25(OH)2D3 (80-fold) and 20,23(OH)2D3 (2.9-fold). For FC ≥ ±1.5 there were 11 common genes upregulated and 4 downregulated. Again, ingenuity pathway analysis showed that VDR/RXR was the top canonical pathway induced by 1,25(OH)2D3, followed by the role of osteoblasts, osteoclasts and chondrocytes in rheumatoid arthritis. As before, other nuclear receptor signaling pathways included LXR/RXR, GR and AhR. For 20,23(OH)2D3 the top nuclear receptor signaling pathways were again AhR and VDR/RXR. Of note, this microarray showed that 20,23(OH)2D3 upregulated two genes downstream of AhR signaling, CYP1A1 and CYP1B1, by factors of 2.4 and 2.6, respectively. This stimulation was confirmed by qPCR (Figure 3D). VDR/RXR was identified as the top toxicity pathway for 1,25(OH)2D3 and AhR for 20,23(OH)2D3.
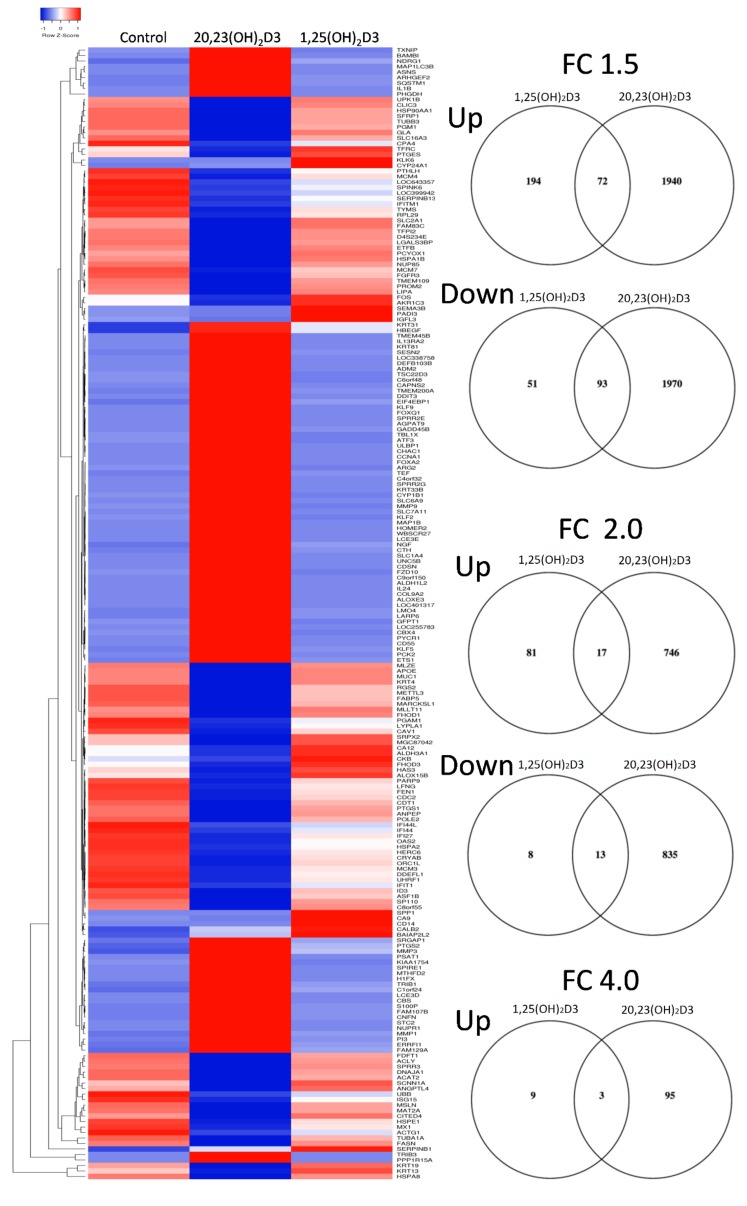
More robust data were obtained with 24 h of treatment for which the average signal values for filtered gene clusters with FC ≥ ±1.5 are shown in Supplemental excel file #3. Because of the large number of genes affected (Table 1), the heat map of differentially expressed genes and Venn diagrams were generated using the 4-FC value which show three overlapping genes (CYP24A1, MMP3 and SERPINB1) as well as distinct gene expression patterns (Figure 4). For FC ≥ ±1.5, 93 and 72 common genes were up- and downregulated, respectively. Ingenuity pathway analysis using FC ≥ ±2.0 was consistent with results obtained after 6 h of treatment. Again, the top canonical pathway for 1,25(OH)2D3 was VDR/RXR (Supplemental Figure S1B) followed by MIF-related glucocorticoid regulation and regulation of the innate immunity system (Table 8). AhR signaling was also listed. The top canonical pathways induced by 20,23(OH)2D3 were AhR signaling (Supplemental Figure S2B) and the cholesterol biosynthesis pathway (Table 9). Interestingly, the involvement of a second nuclear receptor complex was emphasized by VDR/RXR activation (Supplemental Figure S3B), with p53 signaling also being listed. The latter is consistent with the photoprotective properties of 20,23(OH)2D3 and activation of p53 by its direct precursor, 20(OH)D3 [23]. The top affected toxicity pathways for 1,25(OH)2D3 included VDR/RXR, xenobiotic metabolism, cardiac fibrosis and cytochrome P450s (Table 10). AhR signaling was also listed. For 20,23(OH)2D3, AhR signaling was again listed as the top toxicity pathway followed by cholesterol synthesis, p53 signaling and again VDR/RXR (Table 11). The top upstream gene regulation pathways for 1,25(OH)2D3 included vitamin D3-VDR-RXR, calcitriol, dexamethasone, progesterone and β-estradiol, while for 20,23(OH)2D3, included TP53 (p53 tumor suppressor), β-estradiol, lipopolysaccharide, TNF (tumor necrosis factor) and TGF β1 (transforming growth factor-β1) (not shown).
Figure 4.
Heat map of gene expression pattern in human epidermal keratinocytes treated with 10−7 M of 1,25(OH)2D3 or 20,23(OH)2D3 for 24 h. On the right are the corresponding Venn diagrams for FC ≥ 4, 2 and 1.5.
Table 8.
Canonical pathways activated by 1,25(OH)2D3 in human epidermal keratinocytes after 24 h of treatment. Nuclear receptors are marked in bold.
| Ingenuity Canonical Pathways | p-Value | Overlap (%) | Downregulated | No Change | Upregulated | No Overlap with Dataset |
|---|---|---|---|---|---|---|
| VDR/RXR Activation | 7.9 × 10−15 | 15.4 | 23/78 (29%) | 0/78 (0%) | 54/78 (69%) | 1/78 (1%) |
| MIF-mediated Glucocorticoid Regulation | 2.6 × 10−5 | 12.1 | 12/33 (36%) | 0/33 (0%) | 20/33 (61%) | 1/33 (3%) |
| MIF Regulation of Innate Immunity | 6.3 × 10−5 | 9.76 | 19/41 (46%) | 0/41 (0%) | 21/41 (51%) | 1/41 (2%) |
| α-tocopherol Degradation | 0.000162181 | 50 | 0/4 (0%) | 0/4 (0%) | 4/4 (100%) | 0/4 (0%) |
| Antioxidant Action of Vitamin C | 0.000177828 | 5.05 | 43/99 (43%) | 0/99 (0%) | 50/99 (51%) | 6/99 (6%) |
| Retinoate Biosynthesis I | 0.000691831 | 9.09 | 9/33 (27%) | 0/33 (0%) | 20/33 (61%) | 4/33 (12%) |
| Coagulation System | 0.000831764 | 8.57 | 17/35 (49%) | 0/35 (0%) | 18/35 (51%) | 0/35 (0%) |
| Estrogen Biosynthesis | 0.001047129 | 7.89 | 18/38 (47%) | 0/38 (0%) | 19/38 (50%) | 1/38 (3%) |
| iNOS Signaling | 0.00162181 | 6.82 | 17/44 (39%) | 0/44 (0%) | 26/44 (59%) | 1/44 (2%) |
| Role of IL-17A in Arthritis | 0.002884032 | 5.56 | 27/54 (50%) | 0/54 (0%) | 27/54 (50%) | 0/54 (0%) |
| Parkinson’s Signaling | 0.003162278 | 12.5 | 9/16 (56%) | 0/16 (0%) | 7/16 (44%) | 0/16 (0%) |
| LXR/RXR Activation | 0.003890451 | 3.31 | 61/121 (50%) | 0/121 (0%) | 60/121 (50%) | 0/121 (0%) |
| CD40 Signaling | 0.004897788 | 4.62 | 31/65 (48%) | 0/65 (0%) | 33/65 (51%) | 1/65 (2%) |
| IL-10 Signaling | 0.005495409 | 4.41 | 32/68 (47%) | 0/68 (0%) | 35/68 (51%) | 1/68 (1%) |
| Role of MAPK Signaling in the Pathogenesis of Influenza | 0.005754399 | 4.35 | 27/69 (39%) | 0/69 (0%) | 39/69 (57%) | 3/69 (4%) |
| LPS/IL-1 Mediated Inhibition of RXR Function | 0.006025596 | 2.28 | 102/219 (47%) | 0/219 (0%) | 105/219 (48%) | 12/219 (5%) |
| Role of Osteoblasts, Osteoclasts and Chondrocytes in Rheumatoid Arthritis | 0.006025596 | 2.28 | 111/219 (51%) | 0/219 (0%) | 103/219 (47%) | 5/219 (2%) |
| IL-17 Signaling | 0.006456542 | 4.17 | 33/72 (46%) | 0/72 (0%) | 39/72 (54%) | 0/72 (0%) |
| LPS-stimulated MAPK Signaling | 0.00676083 | 4.11 | 33/73 (45%) | 0/73 (0%) | 40/73 (55%) | 0/73 (0%) |
| Toll-like Receptor Signaling | 0.007079458 | 4.05 | 32/74 (43%) | 0/74 (0%) | 40/74 (54%) | 2/74 (3%) |
| BMP signaling pathway | 0.007585776 | 3.95 | 43/76 (57%) | 0/76 (0%) | 31/76 (41%) | 2/76 (3%) |
| Intrinsic Prothrombin Activation Pathway | 0.01023293 | 6.9 | 12/29 (41%) | 0/29 (0%) | 16/29 (55%) | 1/29 (3%) |
| 4-1BB Signaling in T Lymphocytes | 0.011481536 | 6.45 | 19/31 (61%) | 0/31 (0%) | 12/31 (39%) | 0/31 (0%) |
| Acute Phase Response Signaling | 0.012302688 | 2.37 | 76/169 (45%) | 0/169 (0%) | 91/169 (54%) | 2/169 (1%) |
| Endothelin-1 Signaling | 0.013182567 | 2.33 | 89/172 (52%) | 0/172 (0%) | 77/172 (45%) | 6/172 (3%) |
| Inhibition of Angiogenesis by TSP1 | 0.013803843 | 5.88 | 21/34 (62%) | 0/34 (0%) | 11/34 (32%) | 2/34 (6%) |
| Xenobiotic Metabolism Signaling | 0.014454398 | 1.85 | 127/271 (47%) | 0/271 (0%) | 128/271 (47%) | 16/271 (6%) |
| IL-17A Signaling in Fibroblasts | 0.014454398 | 5.71 | 15/35 (43%) | 0/35 (0%) | 20/35 (57%) | 0/35 (0%) |
| Interferon Signaling | 0.015488166 | 5.56 | 28/36 (78%) | 0/36 (0%) | 8/36 (22%) | 0/36 (0%) |
| Thyroid Hormone Biosynthesis | 0.015848932 | 33.3 | 1/3 (33%) | 0/3 (0%) | 2/3 (67%) | 0/3 (0%) |
| April Mediated Signaling | 0.016982437 | 5.26 | 18/38 (47%) | 0/38 (0%) | 20/38 (53%) | 0/38 (0%) |
| Inhibition of Matrix Metalloproteases | 0.017782794 | 5.13 | 17/39 (44%) | 0/39 (0%) | 21/39 (54%) | 1/39 (3%) |
| RAR Activation | 0.018197009 | 2.11 | 87/190 (46%) | 0/190 (0%) | 99/190 (52%) | 4/190 (2%) |
| Thrombin Signaling | 0.018620871 | 2.09 | 90/191 (47%) | 0/191 (0%) | 96/191 (50%) | 5/191 (3%) |
| B Cell Activating Factor Signaling | 0.018620871 | 5 | 21/40 (53%) | 0/40 (0%) | 19/40 (48%) | 0/40 (0%) |
| Dermatan Sulfate Biosynthesis (Late Stages) | 0.022387211 | 4.55 | 16/44 (36%) | 0/44 (0%) | 26/44 (59%) | 2/44 (5%) |
| IL-6 Signaling | 0.023442288 | 2.59 | 52/116 (45%) | 0/116 (0%) | 64/116 (55%) | 0/116 (0%) |
| p38 MAPK Signaling | 0.023988329 | 2.56 | 52/117 (44%) | 0/117 (0%) | 62/117 (53%) | 3/117 (3%) |
Table 9.
Canonical pathways activated by 20,23(OH)2D3 in human epidermal keratinocytes after 24 h of treatment. Nuclear receptors are marked in bold.
| Ingenuity Canonical Pathways | p-Value | Overlap (%) |
Downregulated | No Change | Upregulated | No Overlap with Dataset |
|---|---|---|---|---|---|---|
| Aryl Hydrocarbon Receptor Signaling | 9.5 × 10−10 | 22.9 | 80/140 (57%) | 0/140 (0%) | 54/140 (39%) | 6/140 (4%) |
| Superpathway of Cholesterol Biosynthesis | 9.1 × 10−9 | 46.4 | 22/28 (79%) | 0/28 (0%) | 4/28 (14%) | 2/28 (7%) |
| Cell Cycle Control of Chromosomal Replication | 6.3 × 10−8 | 44.4 | 21/27 (78%) | 0/27 (0%) | 6/27 (22%) | 0/27 (0%) |
| Mismatch Repair in Eukaryotes | 2.3 × 10−7 | 56.2 | 15/16 (94%) | 0/16 (0%) | 1/16 (6%) | 0/16 (0%) |
| Unfolded protein response | 3.5 × 10−7 | 29.6 | 24/54 (44%) | 0/54 (0%) | 29/54 (54%) | 1/54 (2%) |
| Fatty Acid α-oxidation | 1.5 × 10−6 | 47.4 | 10/19 (53%) | 0/19 (0%) | 6/19 (32%) | 3/19 (16%) |
| Ethanol Degradation IV | 6.8 × 10−6 | 40.9 | 17/22 (77%) | 0/22 (0%) | 2/22 (9%) | 3/22 (14%) |
| Cholesterol Biosynthesis I | 7.9 × 10−6 | 53.8 | 12/13 (92%) | 0/13 (0%) | 1/13 (8%) | 0/13 (0%) |
| p53 Signaling | 8.3 × 10−6 | 20.4 | 53/98 (54%) | 0/98 (0%) | 45/98 (46%) | 0/98 (0%) |
| VDR/RXR Activation | 1.5 × 10−5 | 21.8 | 26/78 (33%) | 0/78 (0%) | 51/78 (65%) | 1/78 (1%) |
| GADD45 Signaling | 1.7 × 10−5 | 42.1 | 14/19 (74%) | 0/19 (0%) | 5/19 (26%) | 0/19 (0%) |
| Putrescine Degradation III | 2.8 × 10−5 | 40 | 12/20 (60%) | 0/20 (0%) | 5/20 (25%) | 3/20 (15%) |
| Histamine Degradation | 4.5 × 10−5 | 43.8 | 10/16 (63%) | 0/16 (0%) | 3/16 (19%) | 3/16 (19%) |
| Dopamine Degradation | 4.7 × 10−5 | 33.3 | 16/27 (59%) | 0/27 (0%) | 6/27 (22%) | 5/27 (19%) |
| Tryptophan Degradation X (Mammalian, via Tryptamine) | 6.2 × 10−5 | 36.4 | 13/22 (59%) | 0/22 (0%) | 5/22 (23%) | 4/22 (18%) |
| Xenobiotic Metabolism Signaling | 9.3 × 10−5 | 13.3 | 131/271 (48%) | 0/271 (0%) | 124/271 (46%) | 16/271 (6%) |
| Oxidative Ethanol Degradation III | 0.000109648 | 38.9 | 13/18 (72%) | 0/18 (0%) | 2/18 (11%) | 3/18 (17%) |
| Mevalonate Pathway I | 0.000112202 | 46.2 | 8/13 (62%) | 0/13 (0%) | 3/13 (23%) | 2/13 (15%) |
| Estrogen-mediated S-phase Entry | 0.000125893 | 33.3 | 18/24 (75%) | 0/24 (0%) | 6/24 (25%) | 0/24 (0%) |
| Hereditary Breast Cancer Signaling | 0.000165959 | 16.3 | 77/129 (60%) | 0/129 (0%) | 45/129 (35%) | 7/129 (5%) |
| Glucocorticoid Receptor Signaling | 0.000269153 | 12.7 | 122/275 (44%) | 0/275 (0%) | 147/275 (53%) | 6/275 (2%) |
| Adipogenesis pathway | 0.000371535 | 15.7 | 66/127 (52%) | 0/127 (0%) | 55/127 (43%) | 6/127 (5%) |
| Interferon Signaling | 0.000537032 | 25 | 24/36 (67%) | 0/36 (0%) | 12/36 (33%) | 0/36 (0%) |
| Superpathway of Serine and Glycine Biosynthesis I | 0.000630957 | 57.1 | 3/7 (43%) | 0/7 (0%) | 4/7 (57%) | 0/7 (0%) |
| Superpathway of Geranylgeranyldiphosphate Biosynthesis I (via Mevalonate) | 0.000630957 | 35.3 | 11/17 (65%) | 0/17 (0%) | 4/17 (24%) | 2/17 (12%) |
| Semaphorin Signaling in Neurons | 0.000758578 | 20.8 | 27/53 (51%) | 0/53 (0%) | 24/53 (45%) | 2/53 (4%) |
| Glutaryl-CoA Degradation | 0.000776247 | 41.7 | 7/12 (58%) | 0/12 (0%) | 4/12 (33%) | 1/12 (8%) |
| Pancreatic Adenocarcinoma Signaling | 0.000794328 | 16 | 53/106 (50%) | 0/106 (0%) | 53/106 (50%) | 0/106 (0%) |
| Role of CHK Proteins in Cell Cycle Checkpoint Control | 0.001047129 | 20 | 35/55 (64%) | 0/55 (0%) | 20/55 (36%) | 0/55 (0%) |
| Glycolysis I | 0.001096478 | 28 | 20/25 (80%) | 0/25 (0%) | 4/25 (16%) | 1/25 (4%) |
| NRF2-mediated Oxidative Stress Response | 0.001230269 | 13.3 | 89/180 (49%) | 0/180 (0%) | 87/180 (48%) | 4/180 (2%) |
| HIF1α Signaling | 0.001412538 | 15.7 | 52/102 (51%) | 0/102 (0%) | 48/102 (47%) | 2/102 (2%) |
| Aldosterone Signaling in Epithelial Cells | 0.001548817 | 13.8 | 82/152 (54%) | 0/152 (0%) | 67/152 (44%) | 3/152 (2%) |
| Serotonin Degradation | 0.001737801 | 17.9 | 38/67 (57%) | 0/67 (0%) | 16/67 (24%) | 13/67 (19%) |
Table 10.
Toxicity-related pathways identified by Ingenuity in human keratinocytes treated with 1,25(OH)2D3 for 24 h. Nuclear receptors are marked in bold.
| Ingenuity Toxicity Lists | p-Value | Overlap (%) | Downregulated | No Change | Upregulated | No Overlap with Dataset |
|---|---|---|---|---|---|---|
| VDR/RXR Activation | 7.9 × 10−15 | 15.4 | 23/78 (29%) | 0/78 (0%) | 54/78 (69%) | 1/78 (1%) |
| Xenobiotic Metabolism Signaling | 0.00040738 | 2.38 | 144/336 (43%) | 0/336 (0%) | 150/336 (45%) | 42/336 (13%) |
| Cardiac Fibrosis | 0.000537032 | 3.12 | 85/192 (44%) | 0/192 (0%) | 91/192 (47%) | 16/192 (8%) |
| Cytochrome P450 Panel—Substrate is an Eicosanoid (Human) | 0.000562341 | 28.6 | 1/7 (14%) | 0/7 (0%) | 6/7 (86%) | 0/7 (0%) |
| Cytochrome P450 Panel—Substrate is a Fatty Acid (Human) | 0.001202264 | 20 | 3/10 (30%) | 0/10 (0%) | 7/10 (70%) | 0/10 (0%) |
| Cardiac Hypertrophy | 0.00128825 | 2 | 193/401 (48%) | 0/401 (0%) | 185/401 (46%) | 23/401 (6%) |
| Liver Proliferation | 0.00128825 | 2.64 | 103/227 (45%) | 0/227 (0%) | 113/227 (50%) | 11/227 (5%) |
| Hepatic Fibrosis | 0.001862087 | 4.04 | 49/99 (49%) | 1/99 (1%) | 46/99 (46%) | 3/99 (3%) |
| LXR/RXR Activation | 0.004073803 | 3.25 | 62/123 (50%) | 0/123 (0%) | 61/123 (50%) | 0/123 (0%) |
| Renal Necrosis/Cell Death | 0.005011872 | 1.6 | 238/501 (48%) | 0/501 (0%) | 245/501 (49%) | 18/501 (4%) |
| LPS/IL-1 Mediated Inhibition of RXR Function | 0.010715193 | 1.99 | 107/251 (43%) | 0/251 (0%) | 110/251 (44%) | 34/251 (14%) |
| Positive Acute Phase Response Proteins | 0.010964782 | 6.67 | 11/30 (37%) | 0/30 (0%) | 19/30 (63%) | 0/30 (0%) |
| Liver Necrosis/Cell Death | 0.016218101 | 1.79 | 118/279 (42%) | 0/279 (0%) | 149/279 (53%) | 12/279 (4%) |
| RAR Activation | 0.018197009 | 2.11 | 87/190 (46%) | 0/190 (0%) | 99/190 (52%) | 4/190 (2%) |
| Increases Liver Damage | 0.019054607 | 2.8 | 46/107 (43%) | 0/107 (0%) | 58/107 (54%) | 3/107 (3%) |
| Fatty Acid Metabolism | 0.023988329 | 2.56 | 48/117 (41%) | 0/117 (0%) | 48/117 (41%) | 21/117 (18%) |
| Increases Liver Hepatitis | 0.030902954 | 3.85 | 22/52 (42%) | 0/52 (0%) | 29/52 (56%) | 1/52 (2%) |
Table 11.
Toxicity-related pathways identified by Ingenuity in human keratinocytes treated with 20,23(OH)2D3 for 24 h. Nuclear receptors are marked in bold.
| Ingenuity Toxicity Lists | p-Value | Overlap (%) | Downregulated | No Change | Upregulated | No Overlap with Dataset |
|---|---|---|---|---|---|---|
| Aryl Hydrocarbon Receptor Signaling | 2.6 × 10−9 | 21.1 | 90/161 (56%) | 0/161 (0%) | 56/161 (35%) | 15/161 (9%) |
| Cholesterol Biosynthesis | 1.1 × 10−8 | 62.5 | 13/16 (81%) | 0/16 (0%) | 3/16 (19%) | 0/16 (0%) |
| Renal Necrosis/Cell Death | 1.9 × 10−7 | 13.2 | 228/501 (46%) | 0/501 (0%) | 255/501 (51%) | 18/501 (4%) |
| Primary Glomerulonephritis Biomarker Panel (Human) | 1.7 × 10−6 | 63.6 | 5/11 (45%) | 0/11 (0%) | 6/11 (55%) | 0/11 (0%) |
| p53 Signaling | 2.6 × 10−6 | 21.2 | 54/99 (55%) | 0/99 (0%) | 45/99 (45%) | 0/99 (0%) |
| VDR/RXR Activation | 1.5 × 10−5 | 21.8 | 26/78 (33%) | 0/78 (0%) | 51/78 (65%) | 1/78 (1%) |
| Liver Proliferation | 3.0 × 10−5 | 14.5 | 96/227 (42%) | 0/227 (0%) | 120/227 (53%) | 11/227 (5%) |
| Cardiac Hypertrophy | 5.1 × 10−5 | 12.2 | 176/401 (44%) | 0/401 (0%) | 202/401 (50%) | 23/401 (6%) |
| Liver Necrosis/Cell Death | 7.8 × 10−5 | 13.3 | 117/279 (42%) | 0/279 (0%) | 150/279 (54%) | 12/279 (4%) |
| Oxidative Stress | 0.000380189 | 21.1 | 39/57 (68%) | 0/57 (0%) | 17/57 (30%) | 1/57 (2%) |
| Mechanism of Gene Regulation by Peroxisome Proliferators via PPARα | 0.000645654 | 16.8 | 38/95 (40%) | 0/95 (0%) | 56/95 (59%) | 1/95 (1%) |
| Xenobiotic Metabolism Signaling | 0.000794328 | 11.6 | 152/336 (45%) | 0/336 (0%) | 142/336 (42%) | 42/336 (13%) |
| Increases Renal Proliferation | 0.002398833 | 13.9 | 68/137 (50%) | 0/137 (0%) | 62/137 (45%) | 7/137 (5%) |
| Fatty Acid Metabolism | 0.002398833 | 14.5 | 65/117 (56%) | 0/117 (0%) | 31/117 (26%) | 21/117 (18%) |
| Increases Liver Steatosis | 0.003890451 | 15.7 | 32/83 (39%) | 0/83 (0%) | 48/83 (58%) | 3/83 (4%) |
| Decreases Depolarization of Mitochondria and Mitochondrial Membrane | 0.004570882 | 25 | 18/24 (75%) | 0/24 (0%) | 5/24 (21%) | 1/24 (4%) |
| Cell Cycle: G1/S Checkpoint Regulation | 0.004677351 | 16.7 | 35/66 (53%) | 0/66 (0%) | 28/66 (42%) | 3/66 (5%) |
Table 12 and Table 13 show some similarities and differences with dermatological diseases and conditions; with cancer, organismal injury and abnormalities being the main diseases affected by 20,23(OH)2D3 and 1,25(OH)2D3. With regard to molecular and cellular functions, cellular growth and proliferation, cell death and survival, cellular movement and cell cycle were the major functions for 20,23(OH)2D3, and cellular movement, cell signaling, small molecule biochemistry, lipid metabolism and cellular development for 1,25(OH)2D3. Among the 25 networks activated by 20,23(OH)2D3, the top five included: (1) connective tissue disorders, neurological diseases, organismal injuries and abnormalities, (2) RNA post-transcriptional modification, carbohydrate metabolism and lipid metabolism, (3) connective tissue, developmental, skeletal and muscular disorders, (4) cellular movement, endocrine system disorders, gastrointestinal diseases and (5) nucleic acid metabolism, small molecules biochemistry and dermatological diseases and conditions. Among the 15 networks activated by 1,25(OH)2D3, the top five included: (1) cancer, organismal functions, organismal injuries and abnormalities, (2) cell-to-cell signaling and interaction, cellular assembly and organization, cellular development, (3) cellular growth and proliferation, tissue development and cancer, (4) molecular transport, carbohydrate and lipid metabolism and (5) protein degradation, protein synthesis, cellular assembly and organization.
Table 12.
Categories of biological functions with diseases or function annotation activated by 1,25(OH)2D3 in human epidermal keratinocytes after 24 of treatment.
| Categories of Biological Function | Diseases or Functions Annotation | p-Value | Activation z-Score | # of Genes |
|---|---|---|---|---|
| Cancer, Organismal Injury and Abnormalities | benign neoplasia | 1.0 × 10−9 | 0.927 | 26 |
| Dermatological Diseases and Conditions | psoriasis | 2.6 × 10−9 | 20 | |
| Cardiovascular System Development and Function, Cellular Movement | cell movement of endothelial cells | 8.8 × 10−9 | 1.37 | 15 |
| Cancer, Cellular Movement, Organismal Injury and Abnormalities, Tumor Morphology | invasion of tumor cells | 1.2 × 10−8 | 1.596 | 11 |
| Cell Signaling, Small Molecule Biochemistry | synthesis of nitric oxide | 1.9 × 10−8 | −0.217 | 13 |
| Cardiovascular System Development and Function, Cellular Movement | homing of endothelial cells | 3.2 × 10−8 | 1.597 | 7 |
| Cancer, Organismal Injury and Abnormalities, Tumor Morphology | invasion of tumor | 3.4 × 10−8 | 1.63 | 12 |
| Lipid Metabolism, Small Molecule Biochemistry | metabolism of eicosanoid | 3.4 × 10−8 | 2.747 | 12 |
| Cardiovascular Disease, Hematological Disease | Thrombosis | 5.9 × 10−8 | −0.946 | 10 |
| Organismal Injury and Abnormalities | Fibrosis | 1.1 × 10−7 | −1.401 | 17 |
| Lipid Metabolism, Small Molecule Biochemistry | metabolism of prostaglandin | 1.2 × 10−7 | 2.589 | 10 |
| Immunological Disease | hypersensitive reaction | 1.5 × 10−7 | 0.914 | 15 |
| Cardiovascular System Development and Function, Organismal Development | vasculogenesis | 1.6 × 10−7 | 1.825 | 20 |
| Inflammatory Response | inflammation of organ | 1.8 × 10−7 | −0.022 | 26 |
| Dermatological Diseases and Conditions, Inflammatory Disease, Inflammatory Response | Dermatitis | 1.8 × 10−7 | −0.355 | 15 |
| Cardiovascular System Development and Function, Organismal Development | vascularization of hindlimb | 2.3 X 10−7 | 1.994 | 4 |
| Cellular Movement | homing | 3.4 × 10−7 | 1.468 | 17 |
| Cancer, Cellular Development, Cellular Growth and Proliferation, Organismal Injury and Abnormalities, Tumor Morphology | proliferation of tumor cells | 5.4 × 10−7 | −0.189 | 15 |
| Cancer, Organismal Injury and Abnormalities | growth of tumor | 6.0 × 10−7 | 0.402 | 20 |
| Cellular Movement | invasion of cells | 6.3 × 10−7 | 1.731 | 21 |
Table 13.
Categories of biological functions with diseases or function annotation activated by 20,23(OH)2D3 in human epidermal keratinocytes after 24 h of treatment.
| Categories of Biological Function | Diseases or Functions Annotation | p-Value | Activation z-Score | # of Genes |
|---|---|---|---|---|
| Cellular Growth and Proliferation | proliferation of cells | 7.0 × 10−33 | −3.052 | 551 |
| Dermatological Diseases and Conditions | psoriasis | 4.6 × 10−29 | 142 | |
| Cell Death and Survival | cell death | 1.2 × 10−27 | 1.116 | 493 |
| Cell Death and Survival | necrosis | 1.9 × 10−26 | 1.15 | 400 |
| Cell Death and Survival | apoptosis | 1.9 × 10−25 | 0.619 | 405 |
| Cell Death and Survival | cell death of tumor cell lines | 7.2 × 10−25 | 0.971 | 266 |
| Cell Death and Survival | apoptosis of tumor cell lines | 7.6 × 10−23 | 0.672 | 219 |
| Cellular Movement | cell movement | 1.3 × 10−22 | −0.187 | 334 |
| Cellular Movement | migration of cells | 6.7 × 10−21 | −0.554 | 301 |
| Cancer, Organismal Injury and Abnormalities | abdominal neoplasm | 8.7 × 10−20 | −1.733 | 1030 |
| Infectious Diseases | Viral Infection | 1.2 × 10−19 | 0.737 | 261 |
| Cancer, Organismal Injury and Abnormalities | tumorigenesis of tissue | 6.5 × 10−19 | −0.349 | 1047 |
| Cancer, Organismal Injury and Abnormalities | abdominal cancer | 2.3 × 10−18 | −1.938 | 1014 |
| Cancer, Organismal Injury and Abnormalities | cancer | 4.2 × 10−18 | 1.528 | 1215 |
| Cancer, Organismal Injury and Abnormalities | neoplasia of epithelial tissue | 2.3 × 10−17 | −0.365 | 1026 |
| Cellular Development, Cellular Growth and Proliferation | proliferation of tumor cell lines | 3.0 × 10−17 | −2.431 | 245 |
| Cancer, Organismal Injury and Abnormalities | benign neoplasia | 3.5 × 10−17 | −0.029 | 165 |
| Cell Death and Survival | cell survival | 4.7 × 10−17 | −0.537 | 225 |
| Cancer, Organismal Injury and Abnormalities | advanced stage solid tumor | 7.1 × 10−17 | −0.397 | 106 |
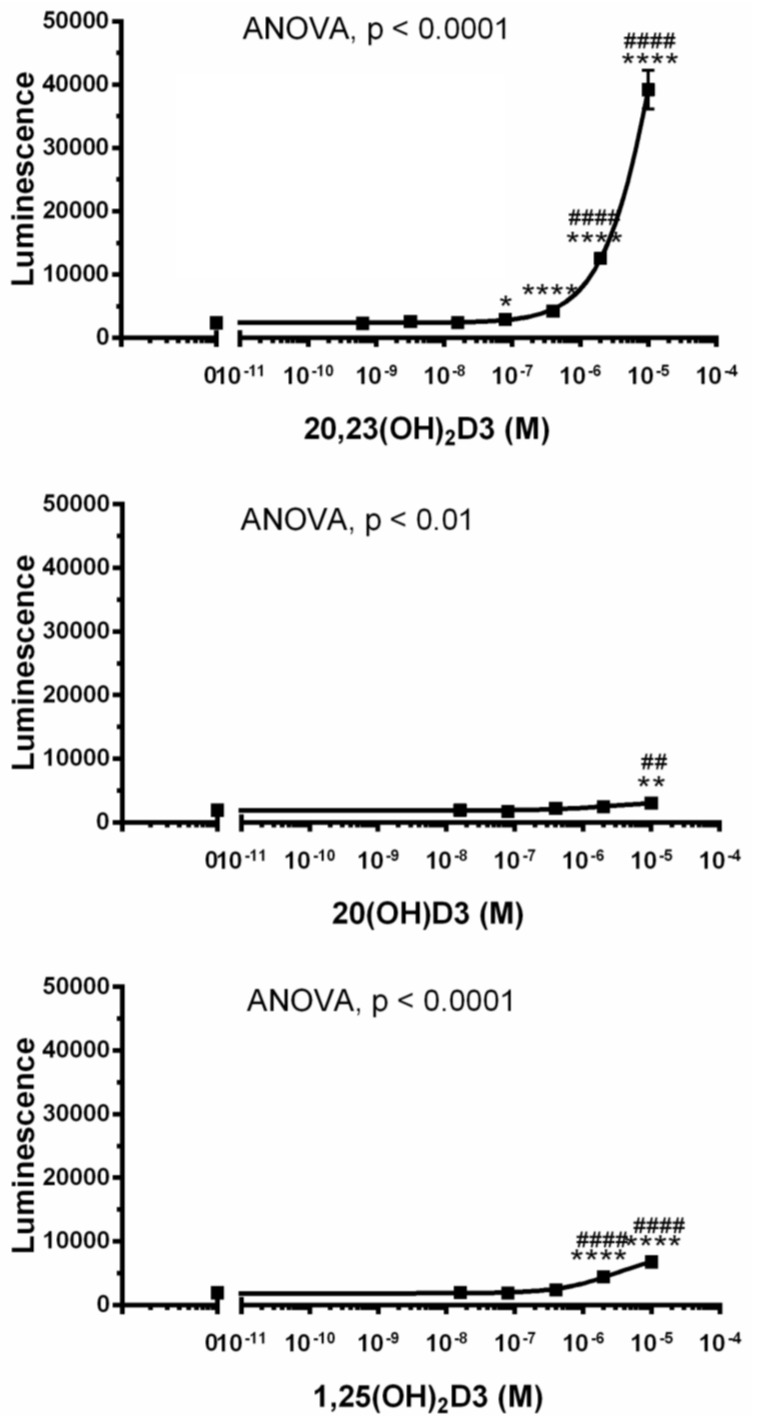
Because of the unexpected finding that AhR signaling represented the top regulatory pathway activated by 20,23(OH)2D3, and is validated by qPCR analysis of CYP1A1 and CYP1B1 genes expression (Figure 3D), we examined whether 20(OH)D3, which is the precursor to 20,23(OH)2D3, and 17,20,23(OH)2D3 and 1,20(OH)2D3, which are downstream metabolites (see Figure 1), also affected the expression of genes linked to AhR in HaCaT keratinocytes. Figure 5A shows that 20(OH)D3 stimulated the expression of CYP1A1 and CYP1B1 in a dose-dependent fashion, with a stimulatory effect also seen for the AhR gene. 17,20,23(OH)3D3 (1 µM) could also stimulate CYPA1, CYP1B1 and AhR expression, while 1,20(OH)2D3 had only a small effect on CYPB1 and no effect on CYP1A1 and AhR. Finally, we used a Human AhR Reporter Assay System (INDIGO, Biosciences) to analyze the effect of several D3-hydroxyderivatives on AhR-mediated transactivation. The kit contains AhR Reporter Cells that contain the luciferase reporter gene functionally linked to an AhR-responsive promoter, which provides a sensitive surrogate measure of the changes in AhR-mediated activation of luciferase reporter. Figure 6 shows that there was marked activation of AhR activity by 20,23(OH)2D3 with weaker but significant activation by 20(OH)D3 or 1,25(OH)2D3. Thus, the functional studies support the microarray analysis indicating that hydroxyderivatives of D3 can act on AhR. This finding can be explained by the promiscuous nature of AhR and its activity [85].
Figure 5.
Changes in CYP1A1, CYP1B1 and AhR gene expression in HaCaT keratinocytes treated with vitamin D3 hydroxyderivatives as a function of the time of treatment. A. Dose-dependent effect of 20(OH)D3 on the gene expression. Data represent means ± SD (n = 3) where # p < 0.05, ## p < 0.01 and ### p < 0.001 at one-way ANOVA test. B. Effect of 1,25(OH)2D3 and 20,23(OH)2D3 on the gene expression as indicated. Data represent means (n = 2) for CYP1A1, or means ± SD (n = 3) for CYP1B1 and AhR where * p < 0.05, ** p < 0.01, at student t-test.
Figure 6.
Stimulation of AhR activity by 20(OH)D3, 22,23(OH)2D3, and 1,25(OH)2D3. The assays for 22,23(OH)2D3 were performed in quadruplicate, while for 20(OH)D3 and 1,25(OH)2D3 in triplicate. Data represent means ± SD where * p < 0.05, ** p < 0.01 and **** p < 0.0001 at student t-test; ## p < 0.01 and #### p < 0.0001 at one-way ANOVA test.
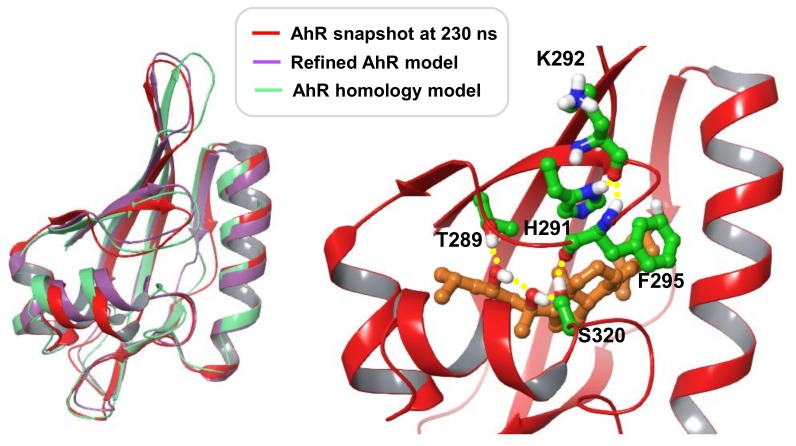
An additional mechanistic insight into the above interactions was provided by modeling using the crystal structure of the ligand-binding domain (LBD) of human AhR. The presently available crystal structure of the human AhR (PDB: 5NJ8) is missing the LBD region. A model of the human AhR LBD with bound 20S,23S(OH)2D3 was developed as described under Methods. Briefly, the final model was based on the homology modelling template of C-terminal Per-ARNT-Sim domain of Hypoxia-Inducible Factor-2α, PDB entry code 3H82. The sequence identity between human AhR and the modelled sequence is 27%; the alignment is shown in Supplemental Figure S4. Short molecular dynamic simulation runs were performed on selected docked poses of 20S,23R/S(OH)2D3 epimers in order to identify binding modes most favorably accommodated in the binding site of the homology model. The selected complex with 20S,23S(OH)2D3 was simulated for 100 ns to allow for local structural adjustments of flexible regions to the presence of the vitamin D3 scaffold. The final conformer obtained is referred to as the ‘refined AhR model’. Further ligand-induced effects were explored through a 250 ns simulation production run starting with this model. Over the first 130 ns the ligand-induced conformational changes were in the vicinity of F295 and S320. The latter is in a flexible region with two adjacent glycine residues while F295 is part of a loop structure ‘covering’ the binding pocket. The conformation adopted by 130 ns in these regions were maintained for the rest of the simulation time, likely stabilized by a hydrogen bonding network that formed, involving ligand hydroxyl groups, T289, S320 side chains and the backbone of F295 as shown in a representative simulation snapshot at 230 ns in Figure 7. Interactions of this network link the more rigid beta-sheet structure of the pocket containing T289 with two loop regions. The flexible ‘belt’ between G309-H326 includes a short helical segment near S320 that also shifted due to the presence of the ligand. This binding mode also changes the preferred orientation of H291 which by 130 ns simulation time forms a stable hydrogen bond with the backbone carbonyl of K292, an interaction not present in the initial or refined AhR models. Alanine mutation of T281, H285 of mouse AhR corresponding to the human residues T289 and H291 was shown to dramatically decrease Hsp90 binding [86]. Figure 7 illustrates that differences in the structural fold of AhR between the homology model, the refined AhR model and the simulation conformer are mainly within loops and the flexible ‘belt’ region.
Figure 7.
Structural fold of AhR models. To the left: Superimposed are the initial homology model, the refined AhR model and a molecular dynamic simulation snapshot at 230 ns. To the right: Close-up view of the simulation snapshot at 230 ns, displaying the ligand and AhR residues involved in an interaction network, as discussed in the text. 20S,23S(OH)2D3 is shown with carbon atoms colored light brown, AhR residue carbons colored green; all other atoms are colored by atom type (O: red, N: blue, S: yellow). Hydrogen bonding interactions are indicated with yellow spheres.
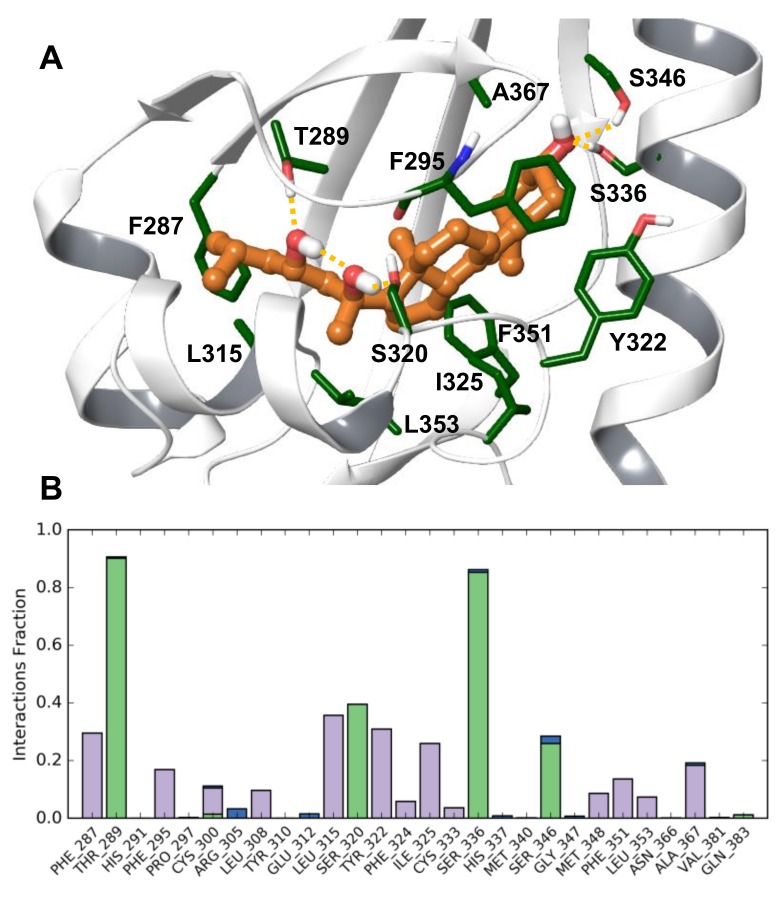
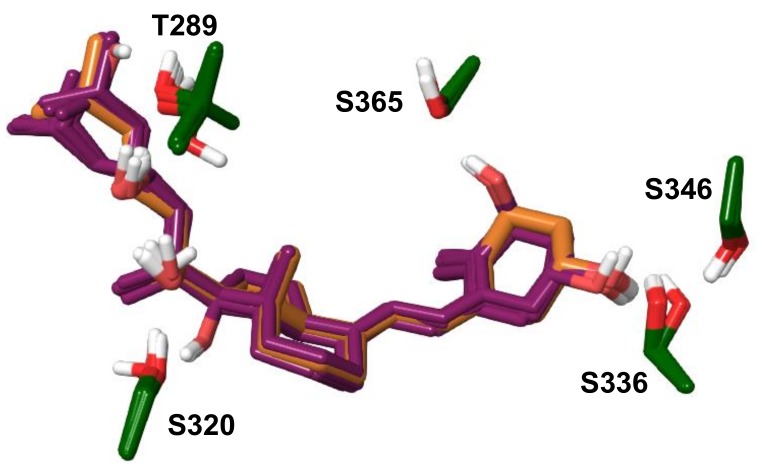
The proposed binding model of 20S,23S(OH)2D3 is shown in Figure 8A through a representative simulation conformer at 230 ns. Figure 8B shows the fraction of simulation time during which interactions are present with each AhR residue, as averaged over 130—250 ns. The most stable polar interactions are hydrogen bonding between 23-OH and T289 at 90% and between 3-OH and S336 at 85%, with S346 also contributing 26% of the simulation time. These interactions anchor the two end regions of the scaffold in the pocket. 20-OH is hydrogen bonding with S320 for 39% of the stimulation time. Due to intra-molecular hydrogen bonding between the ligand hydroxyls, the 20-OH group is positioned to act as a hydrogen bond donor to S320, which allows S320 to interact with F295. This interaction is likely important for these loop conformational changes and their effect on H291. Ligand–protein contacts versus simulation time are shown in Supplemental Figure S5.
Figure 8.
(A) The proposed binding model 20S,23S(OH)2D3 at human AhR. (A) Representative simulation snapshot at 230 ns. Shown residues contribute to the binding of the ligand over simulation time. (B) Fraction of simulation time during which interactions are present with each AhR residue, averaged over 130–250 ns of the simulation production run.
Loop conformational changes induced by 20S,23S(OH)2D3 are likely specific to this ligand. Therefore, for docking other vitamin D3 analogs the refined AhR model was utilized, applying the Induced Fit method. As shown in Table 14, Glide XP docking scores of three analogs are notably lower compared to other compounds within this set, 20(OH)D3, 1,25(OH)2D3 and 1,20(OH)2D3. Docked poses of all analogues are very similar, and also closely overlap with 20S,23S(OH)2D3 in the refined AhR model. Docked poses for all analogs are displayed in Figure 9, along with the binding mode of 20S,23S(OH)2D3 for comparison. Residues from Induced Fit structures contributing to polar interactions with ligands are shown only; all residues in proximity of docked ligands are included in Supplemental Figure S6. Docked Vitamin D3 analogs share similar hydrogen bonding interactions through hydroxyl groups: 1-OH interacts with S365, 3-OH with S336 and possibly S346, 17-OH and 20-OH with S320, 23-OH with T289. Docking results predict that 25-OH interacts with T289. A short, 20 ns molecular dynamic simulation was performed on 20(OH)D3, 1,25(OH)2D3, 17,20,23S(OH)3D3, starting with docked poses. The ligands maintained the binding mode and predicted interactions during simulation except for 1,25(OH)2D3. Therefore, simulation of the latter was extended another 50 ns, during which the pose of 1,25(OH)2D3 shifted, disrupting hydrogen bonding between 3-OH and S336 that was only present for 35% of simulation time. In comparison, in the case of 20(OH)D3 and 17,20,23S(OH)3D3 the same interaction was present 85% and 66% of time, respectively. Due to mobility of the aliphatic chain in the binding site, 25-OH formed contacts with T289 30% and Y310 37% of the simulation time. 1,25(OH)2D3 may have a distinct binding mode and interaction with AhR than the other analogs. Interactions of 17,20,23S(OH)3D3 are analogous to those of 20S,23S(OH)2D3. However, hydrogen bonding between 17-OH with S320 may interfere with structural changes such as those induced by 20S,23S(OH)2D3 during the 250 ns simulation production run. While 20(OH)D3 is also predicted to form analogous contacts, the absence of 23(OH) interactions is likely significant.
Table 14.
Glide XP scores of vitamin D3 analogs docked into the refined human AhR LBD model.
| Compound | Score | Compound | Score |
|---|---|---|---|
| 20SOHD3 | −13.3 | 1,20S,23S(OH)3D3 | −16.1 |
| 1,25(OH)2D3 | −13.1 | 1,20S,23R(OH)3D3 | −16.4 |
| 20S,23S(OH)2D3 | −15.1 | 17,20S,23S(OH)3D3 | −14.9 |
| 20S,23R(OH)2D3 | −15.7 | 17,20S,23R(OH)3D3 | −15.4 |
| 1,20S(OH)2D3 | −13.0 |
Figure 9.
Induced Fit docked vitamin D3 analogs displayed simultaneously. The pose of 20S,23S(OH)2D3 from the refined AhR model is shown for comparison, with carbon atoms colored light brown. Only AhR residues involved in polar interactions are shown.
Modelling Conclusions
Molecular dynamic simulation of the developed AhR-20S,23S(OH)2D3 model predicts strong hydrogen bonding interactions between this ligand and T289, S336. A hydrogen bond formed with S320 is also well maintained during simulation. A number of AhR residues have favorable non-polar contacts with the ligand (Figure 8). The simulation trajectory predicts that ligand-specific interactions induce a conformational change in the region in the vicinity of S320 and F295, also leading to a distinct position and interaction of H291. The interaction network that forms during simulation due to the ligand links the beta-sheet structure of the pocket with two loops, restraining the conformation of flexible regions in the binding site. The presented model is also consistent with the observed effect of 20S,23S(OH)2D3 on AhR since, in particular, T289 and H291 are essential residues for Hsp90 binding.
Docking of a set of D3 analogs predicts ligand binding modes close to that of 20S,23S(OH)2D3, as well as analogous interactions with AhR. Short simulation runs of docked poses of 20(OH)D3 and 17,20,23S(OH)3D3 predict stability of the starting ligand poses. While forming interactions analogous to those of 20S,23S(OH)2D3, these two analogs lack features that contribute to the induced effects of 20S,23S(OH)2D3 during simulation. The 20(OH)D3 analog lacks hydrogen bonding through 23-OH and in the case of 17,20,23S(OH)3D3 the 17-OH group may interfere with the interactions between S320 and the F295 backbone. Stability of the docked pose of 1,25(OH)2D3 was also explored through molecular dynamic simulation. Shifting and fluctuations of 1,25(OH)2D3 over simulation time suggests that this ligand would not adopt a binding mode close to that of 20S,23S(OH)2D3 in the AhR binding site. Thus, modelling predictions are consistent with the distinct effects of these D3 analogs on AhR.
3. Materials and Methods
3.1. Materials
Vitamin D3 (D3) and 1,25(OH)2D3 were purchased from Sigma-Aldrich (St. Louis, MO, USA). 20,23(OH)2D3 was produced by hydroxylation of D3 by CYP11A, extracted with dichloromethane and purified as described in References [14,66]. 20S-Hydroxyvitamin D3 (20(OH)D3, 1α,20S-dihydroxyvitamin D3 (1,20(OH)2D3) and 17,20S,23S-trihydroxyvitamin D3 (17,20,23(OH)3D3) were also synthesized using CYP11A1 as described before [66,87]. An extinction coefficient of 18,000 M−1 cm−1 at 263 nm was used to quantify concentrations of 20,23(OH)2D3 [88] and the secosteroids were divided, dried and stored at −80 °C until use. Secosteroids were dissolved in ethanol prior to experiments to obtain stock solutions of 10−4 M.
The structures of the secosteroids tested and the routes of enzymatic synthesis that include C25 and C1 hydroxylation for 1,25(H)2D3 [5], and the sequential hydroxylation of the D3 side chain by CYP11A1 producing 20,23(OH)2D3 and 17,20.23(OH)3D3 [14,66], are shown in Figure 1.
3.2. Cell Culture
Neonatal foreskins of African American [79] donors were used to isolate neonatal human epidermal keratinocytes (HEK) following standard protocols described previously [69,89]. The use of human tissues were approved both by the IRB at the UTHSC as an exempt protocol #4 and by the IRB at the University of Alabama Birmingham, as they are not subject to FDA regulation and not Human Subject Research. Cells were grown in keratinocyte basal medium (KBM) supplemented with keratinocyte growth factors (KGF) (Lonza, Walkersville, MD, USA) on collagen coated plates [68] and second and third passages were used for the experiments [69]. Human epidermal HaCaT keratinocytes were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with glucose, l-glutamine, pyridoxine hydrochloride (Cell Grow), 5% fetal bovine serum (FBS) (Atlanta Biologicals, Flowery Branch, GA, USA) and 1% penicillin/streptomycin/amphotericin antibiotic solution (Thermo Fisher Scientific, Waltham, MA, USA). Human cells were cultured at 37 °C, with a CO2 concentration of 5%, 100% humidity, and media were changed every second and/or third day.
Prior to treatment with secosteroids, HaCaT cells were serum deprived for 24 h and the medium was changed to DMEM medium containing 5% charcoal-treated FBS (ctFBS) (Atlanta Biologicals, Flowery Branch, GA, USA) to which D3 hydroxymetabolites from the stock solutions were added. For epidermal neonatal keratinocytes, the KBM with KGF was supplemented with 0.5% bovine serum albumin (BSA) prior to the addition of D3 derivatives.
3.3. Microarray Assays
Petri dishes (100 mm in diameter) were seeded with human neonatal keratinocytes that were combined from five different black donors at either passage 2 or 3. After reaching 70–80% of confluence, cells were treated with 10−7 M of either 20,23(OH)2D3 or 1,25(OH)2D3, or with 0.1% ethanol (EtOH) as a solvent control for 6 or 24 h. After, these cells were isolated from three plates per each experimental condition and combined for passage 2 and 3, separately (Figure 2).
The RNA from HEK treated with either 20,23(OH)2D3 or 1,25(OH)2D3, or 0.1% ethanol control, was isolated using the Absolutely RNA Miniprep Kit (Qiagen, Germantown, MD, USA). High purity RNA samples were subjected to microarray analysis at the Molecular Resources Center at the UTHSC. Expression profiling was accomplished using whole-genome gene expression direct hybridization assay using Illumina’s HumanWG-6_V2 (Platform GPL13376) chip/array (Illumina, San Diego, CA, USA). Each array contains full-length 50-mer probes representing more than 22,000 well-annotated RefSeq transcripts, including up-to-date genes derived from the National Center for Biotechnology Information Reference Sequence (NCBI RefSeq) database. Initially, 250 ng total RNA was converted to cDNA, followed by an in vitro transcription step to generate labeled cRNA following the manufacturer’s recommendations (Applied Biosystems, Foster City, CA, USA).
The labeled probes were then mixed with hybridization reagents and hybridized overnight to the Human BeadChips. Following washing and staining, the BeadChips were imaged using the Illumina BeadArray Reader to measure fluorescence intensity at each probe. The intensity of the signal corresponds to the quantity of the respective mRNA in the original sample.
3.4. Bioinformatics Analysis
For generating networks, a data set containing gene identifiers and corresponding expression values was uploaded into the application. Each identifier was mapped to its corresponding object in Ingenuity’s Knowledge Base. A FC of ±2 or ±1.5, where indicated, was set to identify molecules whose expression was significantly differentially regulated. These molecules, called Network Eligible molecules, were overlaid onto a global molecular network developed from information contained in Ingenuity’s Knowledge Base. Networks of Network Eligible Molecules were then algorithmically generated based on their connectivity. The Functional Analysis identified the biological functions and/or diseases that were most significant to the entire data set. Molecules from the dataset that met the FC cutoff of ±2 or ±1.5 and were associated with biological functions and/or diseases in Ingenuity’s Knowledge Base were considered for the analysis. Right-tailed Fisher’s exact test was used to calculate a p-value determining the probability that each biological function and/or disease assigned to that data set is due to chance alone.
3.5. Real-Time Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
Semiconfluent cultures of human neonatal keratinocytes or HaCaT cells were treated for 6 h, 24 h or as indicated in the figure legends with vitamin D3 hydroxyderivatives or ethanol, and RNA isolated as described above. Reverse transcription was done using the Transcriptor First Strand cDNA Synthesis Kit (Applied Biosystems, Foster City, CA, USA) with 100 ng RNA per reaction. qRT-PCR was performed using cDNA diluted 10-fold in sterile water and a TaqMan PCR Master Mix. Reactions (in triplicate) were performed at 50 °C for 2 min, 95 °C for 10 min and then 50 cycles of 95 °C for 15 s and 60 °C for 1 min. The primers and probes were designed with the universal probe library (Roche). Data were collected on a Roche Light Cycler 480. The amount of amplified product for each gene was compared to that of Cyclophilin B or GAPDH using a comparative CT (ΔΔCT) method. Supplemental Table S1 lists the primers used for qRT-PCR amplifications.
3.6. Interaction of Hydroxyvitamin D Derivatives with AhR
Interaction of 20(OH)D3, 20,23(OH)2D3 and 1,25(OH)2D3 with AhR was evaluated using the Human AhR Reporter Assay System (INDIGO Biosciences, State College, PA, USA) according to the manufacture’s protocol. Briefly, AhR reporter cells were recovered on a 96-well plate frame using the cell recovery medium for 5 h, followed by treatment with vitamin D3 hydroxyderivatives in the compound screening medium for 22 h. After removing the media from the wells, luciferase detection reagent was added to the wells and luminescence was measured using a Cytation 5 Cell Imaging Multi-Mode Reader (Winooski, VT, USA).
3.7. Statistical Analyses
Data are presented as means ± SD (n = 3–4), and were analyzed with a Student’s t-test (for two groups) or ANOVA using Prism 4.00 (GraphPad Software, San Diego, CA, USA). Statistically significant differences are denoted with asterisks for t-tests or for one way ANOVA with # as indicated in the figure legends.
3.8. Data Deposition
The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GEO: GSE117351).
3.9. Development of a Human AhR LBD Model Complexed with 20S,23S(OH)2D3
The strategy applied utilized tools implemented in the Schrödinger software package, version 2017-4 (Schrödinger, LLC, New York, NY, USA). Homology modelling of the human AhR ligand-binding domain was based on crystal structures of the C-terminal Per-ARNT-Sim domain of Hypoxia-Inducible Factor-2α (HIF-2α PAS-B), PDB entry codes 3H82 and 4XT2. Based on the two templates, two homology models were built using the energy-based homology model building method in Schrödinger. The sequence identity is 27% between human AhR and template sequences in the modelled LBD region. The sequence alignment is shown in Supplemental Figure S4. Residues were numbered according to the human AhR sequence (Uniprot ID P35869). Co-crystallized ligands were included. Modeled loops that contained gaps in the sequence alignment were refined through default loop refinement options. The models were relaxed through restrained energy minimization in Protein Preparation Wizard (OPLS3 force field).
Initial binding mode hypotheses were generated through docking 20S,23R/S(OH)2D3 into the two obtained AhR models. Out of the top scoring poses at both AhR models, eight were selected for protein-ligand complex refinement (Prime tool in Schrödinger software), followed by a 10 ns molecular dynamic simulation run for each complex using Desmond. Four poses induced distortions in rigid, AhR beta-sheet/helical backbone structures within 10 ns simulation and were not considered further. Out of the remaining poses, the most favorable contacts were formed by two similar poses of the epimers: 20S,23S/R(OH)2D3. Simulation of these poses was extended to 20 ns, which suggested that 20S,23S(OH)2D3 is more favorably accommodated than its R epimer. The 20S,23S(OH)2D3 complex conformer at 20 ns showed an overall RMSD (root-mean-square deviation) of 1.48 from its homology modelling template (PDB: 3H82). In order to allow flexible regions to adjust to the presence of the bound vitamin D3 scaffold, the model was further simulated for 100 ns or for 230 or 25 ns as indicated. The final conformer was relaxed through restrained energy minimization and is referred to as the ‘refined AhR model’. The overall RMSD of this model from its homology modelling template (PDB: 3H82) is 1.58, suggesting stability of the AhR structure over simulation time. The model structure contains only two residues with backbone dihedrals in disallowed regions, both of which are glycines: Gly309 and Gly374.
3.10. Docking Method
The induced Fit docking method was used as implemented in Schrödinger, version 2017-4 (Schrödinger, LLC, New York, NY, USA). This method combines Prime tools and Glide docking, taking into account the flexibility of residues in proximity to the ligand. 20,23(OH)2D3 was Induced Fit docked into the two human AhR homology models using default parameters except optimization of side chains was extended to 6 Å around the ligand and Glide re-docking was done in extra-precision mode. Docking of vitamin D3 analogs into the refined AhR model also utilized Induced Fit with default options, except that van der Waals scaling parameters for docking were set to 1.0 for both protein and ligand (no scaling), and re-docking was done in extra-precision mode.
3.11. Molecular Dynamic Simulation Method
Molecular dynamic simulations were performed using Desmond (Schrödinger, LLC, New York, NY, USA) with the OPLS3 force field. Structures were solved in TIP3P explicit waters with boundary conditions in a 10 Å buffered orthorhombic system. Counter-ions were added. The NPT ensemble was employed with temperature fixed at 300 K and pressure at 1.01 bar. The cutoff radius for Coulombic interactions was set to 10 Å. The trajectory was recorded at 10 ps intervals.
4. Conclusions
Gene expression profile analysis demonstrated that 20,23(OH)2D3 and 1,25(OH)2D3 induce distinct and overlapping gene expression patterns in keratinocytes linked to the activation of common (VDR-dependent) and distinct (involving other nuclear receptors) signal transduction pathways. Taking into consideration the strong chemical similarity between 20,23(OH)2D3 and 1,25(OH)2D3 (Figure 1), the marked differences in gene expression panels (Table 1, Figure 3 and Figure 4) were unexpected. This is because our previous studies predominantly showed phenotypic similarities between the effects of both dihydroxy-vitamin D3 species, such as regulation of cell proliferation and differentiation, and anti-inflammatory, photoprotective and anticancer functions, with only a couple of notable differences where 20,23(OH)2D3 displayed no calcemic effects and poor activation of CYP24A1. These predominantly overlapping effects are most likely secondary to the redundancy of downstream phenotypic regulators and intercommunication between distinct transduction pathways at the cell or organ levels. The similarities are likely related to the activation of the VDR. The most significant and unexpected finding is the identification of AhR as the major receptor for 20,23(OH)2D3, which also appears to be activated by other CYP11A1-derived vitamin D3 derivatives, and possibly by 1,25(OH)2D3 to some extent, as predicted by molecular modeling. The future challenge is to precisely define the interaction of different vitamin D3 hydroxyderivatives with the ligand binding domain of AhR and how it is affected by the location of the OH-group on the side chain or at C1α, and how the activation of downstream signal transduction pathways occurs.
Acknowledgments
We thank Quynh Tran for initial normalization of the microarray data.
Abbreviations
| 1,20(OH)2D3 | 1,20-dihyhydroxyvitamin D3 |
| 17,20,23(OH)2D3 | 17,20,23-trihyhydroxyvitamin D3 |
| 1α,25(OH)2D3 | 1α,25-dihydroxyvitamin D3 |
| 20(OH)D3 | 20-hydroxyvitamin D3 |
| 20,23(OH)2D3 | 20,23-dihyhydroxyvitamin D3 |
| 25(OH)D3 | 25-hydroxyvitamin D3 |
| 7-DHC | 7-dehydrocholesterol |
| AA | African-American |
| AhR | aryl hydrocarbon receptor |
| GR | glucocorticoid receptor |
| LBD | ligand binding domain |
| LXR | liver X receptor |
| PPAR | peroxisome proliferator-activated receptor |
| RAR | retinoic acid receptor |
| RXR | retinoid X receptor |
| VDR | vitamin D receptor |
| UVB | ultraviolet B radiation |
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/19/10/3072/s1.
Author Contributions
A.T.S. designed experiments, analyzed data and wrote the manuscript. T.-K.K. performed the main experiments, took part in data analysis and preparation of the manuscript. Z.J. performed the main experiments, took part in data analysis and preparation of the manuscript. A.A.B. performed experiments, took part in data analysis and preparation of the manuscript. M.A.Ż. performed the experiments, took part in data analysis and preparation of the manuscript. H.X. took part in data analysis and preparation of the manuscript. T.R.S. performed bioinformatics analysis and took part in preparation of the manuscript. R.C.T. took part in data analysis and preparation of the manuscript. A.M.J. took part in data analysis and preparation of the manuscript. D.K.C. performed bioinformatics analysis and took part in preparation of the manuscript.
Funding
The studies were supported by grants R21AR066505, 1R01AR073004-01A1 and 1 RO1 AR071189-01A1 from NIH and VA Merit award 1I01BX004293-01A1 to ATS, grant R01ES017014 to TRS, N402 662840 from the Polish Ministry of Science and Higher Education to MAZ and internal funds from the University of Western Australia, Australia and the University of Memphis Feinstone Center for Genomic Research.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Holick M.F. Vitamin D: A millenium perspective. J. Cell. Biochem. 2003;88:296–307. doi: 10.1002/jcb.10338. [DOI] [PubMed] [Google Scholar]
- 2.Wacker M., Holick M.F. Sunlight and vitamin D: A global perspective for health. Dermato-Endocrinology. 2013;5:51–108. doi: 10.4161/derm.24494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holick M.F. Vitamin D deficiency. N. Engl. J. Med. 2007;357:266–281. doi: 10.1056/NEJMra070553. [DOI] [PubMed] [Google Scholar]
- 4.Zhu J., DeLuca H.F. Vitamin d 25-hydroxylase—Four decades of searching, are we there yet? Arch. Biochem. Biophys. 2012;523:30–36. doi: 10.1016/j.abb.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 5.Jones G., Prosser D.E., Kaufmann M. Cytochrome P450-mediated metabolism of vitamin D. J. Lipid Res. 2014;55:13–31. doi: 10.1194/jlr.R031534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christakos S., Dhawan P., Verstuyf A., Verlinden L., Carmeliet G. Vitamin D: Metabolism, molecular mechanism of action, and pleiotropic effects. Physiol. Rev. 2016;96:365–408. doi: 10.1152/physrev.00014.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adams J.S., Hewison M. Extrarenal expression of the 25-hydroxyvitamin D-1-hydroxylase. Arch. Biochem. Biophys. 2012;523:95–102. doi: 10.1016/j.abb.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slominski A.T., Li W., Kim T.K., Semak I., Wang J., Zjawiony J.K., Tuckey R.C. Novel activities of CYP11A1 and their potential physiological significance. J. Steroid Biochem. Mol. Biol. 2015;151:25–37. doi: 10.1016/j.jsbmb.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tuckey R.C. Progesterone synthesis by the human placenta. Placenta. 2005;26:273–281. doi: 10.1016/j.placenta.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 10.Miller W.L., Auchus R.J. The molecular biology, biochemistry, and physiology of human steroidogenesis and its disorders. Endocr. Rev. 2011;32:81–151. doi: 10.1210/er.2010-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guryev O., Carvalho R.A., Usanov S., Gilep A., Estabrook R.W. A pathway for the metabolism of vitamin D3: Unique hydroxylated metabolites formed during catalysis with cytochrome P450scc (CYP11A1) Proc. Natl. Acad. Sci. USA. 2003;100:14754–14759. doi: 10.1073/pnas.2336107100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slominski A., Zjawiony J., Wortsman J., Semak I., Stewart J., Pisarchik A., Sweatman T., Marcos J., Dunbar C., R C.T. A novel pathway for sequential transformation of 7-dehydrocholesterol and expression of the P450scc system in mammalian skin. Eur. J. Biochem. 2004;271:4178–4188. doi: 10.1111/j.1432-1033.2004.04356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slominski A., Semak I., Zjawiony J., Wortsman J., Li W., Szczesniewski A., Tuckey R.C. The cytochrome P450scc system opens an alternate pathway of vitamin D3 metabolism. FEBS J. 2005;272:4080–4090. doi: 10.1111/j.1742-4658.2005.04819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tuckey R.C., Li W., Zjawiony J.K., Zmijewski M.A., Nguyen M.N., Sweatman T., Miller D., Slominski A. Pathways and products for the metabolism of vitamin D3 by cytochrome P450scc. FEBS J. 2008;275:2585–2596. doi: 10.1111/j.1742-4658.2008.06406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slominski A., Semak I., Wortsman J., Zjawiony J., Li W., Zbytek B., Tuckey R.C. An alternative pathway of vitamin D metabolism. Cytochrome P450scc (CYP11A1)-mediated conversion to 20-hydroxyvitamin D2 and 17,20-dihydroxyvitamin D2. FEBS J. 2006;273:2891–2901. doi: 10.1111/j.1742-4658.2006.05302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen M.N., Slominski A., Li W., Ng Y.R., Tuckey R.C. Metabolism of vitamin D2 to 17,20,24-trihydroxyvitamin D2 by cytochrome P450scc (CYP11A1) Drug Metab. Dispos. 2009;37:761–767. doi: 10.1124/dmd.108.025619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin Z., Marepally S.R., Ma D., Myers L.K., Postlethwaite A.E., Tuckey R.C., Cheng C.Y., Kim T.K., Yue J., Slominski A.T., et al. Chemical synthesis and biological activities of 20s,24s/r-dihydroxyvitamin D3 epimers and their 1alpha-hydroxyl derivatives. J. Med. Chem. 2015;58:7881–7887. doi: 10.1021/acs.jmedchem.5b00881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slominski A.T., Kim T.K., Shehabi H.Z., Semak I., Tang E.K., Nguyen M.N., Benson H.A., Korik E., Janjetovic Z., Chen J., et al. In vivo evidence for a novel pathway of vitamin D(3) metabolism initiated by P450scc and modified by CYP27B1. FASEB J. 2012;26:3901–3915. doi: 10.1096/fj.12-208975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slominski A.T., Kim T.K., Shehabi H.Z., Tang E.K., Benson H.A., Semak I., Lin Z., Yates C.R., Wang J., Li W., et al. In vivo production of novel vitamin D2 hydroxy-derivatives by human placentas, epidermal keratinocytes, Caco-2 colon cells and the adrenal gland. Mol. Cell. Endocrinol. 2014;383:181–192. doi: 10.1016/j.mce.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slominski A.T., Kim T.K., Li W., Tuckey R.C. Classical and non-classical metabolic transformation of vitamin D in dermal fibroblasts. Exp. Dermatol. 2016;25:231–232. doi: 10.1111/exd.12872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Slominski A.T., Zmijewski M.A., Semak I., Sweatman T., Janjetovic Z., Li W., Zjawiony J.K., Tuckey R.C. Sequential metabolism of 7-dehydrocholesterol to steroidal 5,7-dienes in adrenal glands and its biological implication in the skin. PLoS ONE. 2009;4:e4309. doi: 10.1371/journal.pone.0004309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slominski A.T., Kim T.K., Chen J., Nguyen M.N., Li W., Yates C.R., Sweatman T., Janjetovic Z., Tuckey R.C. Cytochrome P450scc-dependent metabolism of 7-dehydrocholesterol in placenta and epidermal keratinocytes. Int. J. Biochem. Cell Biol. 2012;44:2003–2018. doi: 10.1016/j.biocel.2012.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slominski A.T., Janjetovic Z., Kim T.K., Wasilewski P., Rosas S., Hanna S., Sayre R.M., Dowdy J.C., Li W., Tuckey R.C. Novel non-calcemic secosteroids that are produced by human epidermal keratinocytes protect against solar radiation. J. Steroid Biochem. Mol. Biol. 2015;148:52–63. doi: 10.1016/j.jsbmb.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slominski A.T., Kim T.K., Li W., Postlethwaite A., Tieu E.W., Tang E.K., Tuckey R.C. Detection of novel CYP11A1-derived secosteroids in the human epidermis and serum and pig adrenal gland. Sci. Rep. 2015;5:14875. doi: 10.1038/srep14875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeLuca H.F. Overview of general physiologic features and functions of vitamin D. Am. J. Clin. Nutr. 2004;80:1689S–1696S. doi: 10.1093/ajcn/80.6.1689S. [DOI] [PubMed] [Google Scholar]
- 26.Holick M.F. Vitamin D and bone health. J. Nutr. 1996;126:1159S–1164S. doi: 10.1093/jn/126.suppl_4.1159S. [DOI] [PubMed] [Google Scholar]
- 27.Bikle D.D. Vitamin D: An ancient hormone. Exp. Dermatol. 2011;20:7–13. doi: 10.1111/j.1600-0625.2010.01202.x. [DOI] [PubMed] [Google Scholar]
- 28.Pike J.W., Christakos S. Biology and mechanisms of action of the vitamin D hormone. Endocrinol. Metab. Clin. N. Am. 2017;46:815–843. doi: 10.1016/j.ecl.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Driel M., van Leeuwen J. Vitamin D endocrinology of bone mineralization. Mol. Cell. Endocrinol. 2017;453:46–51. doi: 10.1016/j.mce.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 30.Hewison M. An update on vitamin D and human immunity. Clin. Endocrinol. 2012;76:315–325. doi: 10.1111/j.1365-2265.2011.04261.x. [DOI] [PubMed] [Google Scholar]
- 31.Vanherwegen A.S., Gysemans C., Mathieu C. Regulation of immune function by vitamin d and its use in diseases of immunity. Endocrinol. Metab. Clin. N. Am. 2017;46:1061–1094. doi: 10.1016/j.ecl.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 32.Berridge M.J. Vitamin D deficiency: Infertility and neurodevelopmental diseases (attention deficit hyperactivity disorder, autism, and schizophrenia) Am. J. Physiol. Cell Physiol. 2018;314:C135–C151. doi: 10.1152/ajpcell.00188.2017. [DOI] [PubMed] [Google Scholar]
- 33.Liu N.Q., Hewison M. Vitamin d, the placenta and pregnancy. Arch. Biochem. Biophys. 2012;523:37–47. doi: 10.1016/j.abb.2011.11.018. [DOI] [PubMed] [Google Scholar]
- 34.Ganguly A., Tamblyn J.A., Finn-Sell S., Chan S.Y., Westwood M., Gupta J., Kilby M.D., Gross S.R., Hewison M. Vitamin D, the placenta and early pregnancy: Effects on trophoblast function. J. Endocrinol. 2018;236:R93–R103. doi: 10.1530/JOE-17-0491. [DOI] [PubMed] [Google Scholar]
- 35.Shin J.S., Choi M.Y., Longtine M.S., Nelson D.M. Vitamin d effects on pregnancy and the placenta. Placenta. 2010;31:1027–1034. doi: 10.1016/j.placenta.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knabl J., Vattai A., Ye Y., Jueckstock J., Hutter S., Kainer F., Mahner S., Jeschke U. Role of placental VDR expression and function in common late pregnancy disorders. Int. J. Mol. Sci. 2017;18:2340. doi: 10.3390/ijms18112340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lorenzen M., Boisen I.M., Mortensen L.J., Lanske B., Juul A., Blomberg Jensen M. Reproductive endocrinology of vitamin D. Mol. Cell. Endocrinol. 2017;453:103–112. doi: 10.1016/j.mce.2017.03.023. [DOI] [PubMed] [Google Scholar]
- 38.Cui X., Gooch H., Petty A., McGrath J.J., Eyles D. Vitamin D and the brain: Genomic and non-genomic actions. Mol. Cell. Endocrinol. 2017;453:131–143. doi: 10.1016/j.mce.2017.05.035. [DOI] [PubMed] [Google Scholar]
- 39.Di Somma C., Scarano E., Barrea L., Zhukouskaya V.V., Savastano S., Mele C., Scacchi M., Aimaretti G., Colao A., Marzullo P. Vitamin D and neurological diseases: An endocrine view. Int. J. Mol. Sci. 2017;18:2482. doi: 10.3390/ijms18112482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Welsh J. Function of the vitamin D endocrine system in mammary gland and breast cancer. Mol. Cell. Endocrinol. 2017;453:88–95. doi: 10.1016/j.mce.2017.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Angellotti E., Pittas A.G. The role of vitamin d in the prevention of type 2 diabetes: To D or not to D? Endocrinology. 2017;158:2013–2021. doi: 10.1210/en.2017-00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dimova R., Tankova T., Chakarova N. Vitamin d in the spectrum of prediabetes and cardiovascular autonomic dysfunction. J. Nutr. 2017;147:1607–1615. doi: 10.3945/jn.117.250209. [DOI] [PubMed] [Google Scholar]
- 43.Berridge M.J. Vitamin D deficiency and diabetes. Biochem. J. 2017;474:1321–1332. doi: 10.1042/BCJ20170042. [DOI] [PubMed] [Google Scholar]
- 44.Kassi E., Nasiri-Ansari N., Papavassiliou A.G. Vitamin d affects glucocorticoid action in target cells. Oncotarget. 2017;8:7220–7221. doi: 10.18632/oncotarget.13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plum L.A., DeLuca H.F. Vitamin D, disease and therapeutic opportunities. Nat. Rev. Drug Discov. 2010;9:941–955. doi: 10.1038/nrd3318. [DOI] [PubMed] [Google Scholar]
- 46.Feldman D., Krishnan A.V., Swami S., Giovannucci E., Feldman B.J. The role of vitamin d in reducing cancer risk and progression. Nat. Rev. Cancer. 2014;14:342–357. doi: 10.1038/nrc3691. [DOI] [PubMed] [Google Scholar]
- 47.Fleet J.C., DeSmet M., Johnson R., Li Y. Vitamin d and cancer: A review of molecular mechanisms. Biochem. J. 2012;441:61–76. doi: 10.1042/BJ20110744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Masuda S., Jones G. Promise of vitamin d analogues in the treatment of hyperproliferative conditions. Mol. Cancer Ther. 2006;5:797–808. doi: 10.1158/1535-7163.MCT-05-0539. [DOI] [PubMed] [Google Scholar]
- 49.Slominski A.T., Brozyna A.A., Skobowiat C., Zmijewski M.A., Kim T.K., Janjetovic Z., Oak A.S., Jozwicki W., Jetten A.M., Mason R.S., et al. On the role of classical and novel forms of vitamin d in melanoma progression and management. J. Steroid Biochem. Mol. Biol. 2018;177:159–170. doi: 10.1016/j.jsbmb.2017.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slominski A.T., Brozyna A.A., Zmijewski M.A., Jozwicki W., Jetten A.M., Mason R.S., Tuckey R.C., Elmets C.A. Vitamin D signaling and melanoma: Role of vitamin d and its receptors in melanoma progression and management. Lab. Investig. 2017;97:706–724. doi: 10.1038/labinvest.2017.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Norman A.W. Minireview: Vitamin d receptor: New assignments for an already busy receptor. Endocrinology. 2006;147:5542–5548. doi: 10.1210/en.2006-0946. [DOI] [PubMed] [Google Scholar]
- 52.Mason R.S., Reichrath J. Sunlight vitamin d and skin cancer. Anticancer Agents Med. Chem. 2013;13:83–97. doi: 10.2174/187152013804487272. [DOI] [PubMed] [Google Scholar]
- 53.Bikle D.D., Elalieh H., Welsh J., Oh D., Cleaver J., Teichert A. Protective role of vitamin D signaling in skin cancer formation. J. Steroid Biochem. Mol. Biol. 2013;136:271–279. doi: 10.1016/j.jsbmb.2012.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neme A., Seuter S., Carlberg C. Selective regulation of biological processes by vitamin d based on the spatio-temporal cistrome of its receptor. Biochim. Biophys. Acta. 2017;1860:952–961. doi: 10.1016/j.bbagrm.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 55.Bikle D.D. Vitamin D receptor, UVR, and skin cancer: A potential protective mechanism. J. Investig. Dermatol. 2008;128:2357–2361. doi: 10.1038/jid.2008.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Elias P.M. Structure and function of the stratum corneum extracellular matrix. J. Investig. Dermatol. 2012;132:2131–2133. doi: 10.1038/jid.2012.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dixon K.M., Norman A.W., Sequeira V.B., Mohan R., Rybchyn M.S., Reeve V.E., Halliday G.M., Mason R.S. 1alpha,25(OH)(2)-vitamin D and a nongenomic vitamin d analogue inhibit ultraviolet radiation-induced skin carcinogenesis. Cancer Prev. Res. (Phila) 2011;4:1485–1494. doi: 10.1158/1940-6207.CAPR-11-0165. [DOI] [PubMed] [Google Scholar]
- 58.Makarova A., Wang G., Dolorito J.A., Kc S., Libove E., Epstein E.H., Jr. Vitamin D3 produced by skin exposure to UVR inhibits murine basal cell carcinoma carcinogenesis. J. Investig. Dermatol. 2017;137:2613–2619. doi: 10.1016/j.jid.2017.05.037. [DOI] [PubMed] [Google Scholar]
- 59.Slominski A.T., Zmijewski M.A., Semak I., Zbytek B., Pisarchik A., Li W., Zjawiony J., Tuckey R.C. Cytochromes P450 and skin cancer: Role of local endocrine pathways. Anticancer Agents Med. Chem. 2014;14:77–96. doi: 10.2174/18715206113139990308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Slominski A.T., Kim T.K., Hobrath J.V., Oak A.S.W., Tang E.K.Y., Tieu E.W., Li W., Tuckey R.C., Jetten A.M. Endogenously produced nonclassical vitamin D hydroxy-metabolites act as “Biased” Agonists on VDR and inverse agonists on RORalpha and RORgamma. J. Steroid Biochem. Mol. Biol. 2017;173:42–56. doi: 10.1016/j.jsbmb.2016.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang J., Slominski A., Tuckey R.C., Janjetovic Z., Kulkarni A., Chen J., Postlethwaite A.E., Miller D., Li W. 20-Hydroxyvitamin D(3) inhibits proliferation of cancer cells with high efficacy while being non-toxic. Anticancer Res. 2012;32:739–746. [PMC free article] [PubMed] [Google Scholar]
- 62.Slominski A.T., Janjetovic Z., Fuller B.E., Zmijewski M.A., Tuckey R.C., Nguyen M.N., Sweatman T., Li W., Zjawiony J., Miller D., et al. Products of vitamin D3 or 7-dehydrocholesterol metabolism by cytochrome P450scc show anti-leukemia effects, having low or absent calcemic activity. PLoS ONE. 2010;5:e9907. doi: 10.1371/journal.pone.0009907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen J., Wang J., Kim T.K., Tieu E.W., Tang E.K., Lin Z., Kovacic D., Miller D.D., Postlethwaite A., Tuckey R.C., et al. Novel vitamin d analogs as potential therapeutics: Metabolism, toxicity profiling, and antiproliferative activity. Anticancer Res. 2014;34:2153–2163. [PMC free article] [PubMed] [Google Scholar]
- 64.Tongkao-On W., Carter S., Reeve V.E., Dixon K.M., Gordon-Thomson C., Halliday G.M., Tuckey R.C., Mason R.S. Cyp11a1 in skin: An alternative route to photoprotection by vitamin D compounds. J. Steroid Biochem. Mol. Biol. 2015;148:72–78. doi: 10.1016/j.jsbmb.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 65.Slominski A., Janjetovic Z., Tuckey R.C., Nguyen M.N., Bhattacharya K.G., Wang J., Li W., Jiao Y., Gu W., Brown M., et al. 20s-hydroxyvitamin D3, noncalcemic product of CYP11A1 action on vitamin D3, exhibits potent antifibrogenic activity in vivo. J. Clin. Endocrinol. Metab. 2013;98:E298–E303. doi: 10.1210/jc.2012-3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tuckey R.C., Li W., Shehabi H.Z., Janjetovic Z., Nguyen M.N., Kim T.K., Chen J., Howell D.E., Benson H.A., Sweatman T., et al. Production of 22-hydroxy metabolites of vitamin D3 by cytochrome P450scc (CYP11A1) and analysis of their biological activities on skin cells. Drug Metab. Dispos. 2011;39:1577–1588. doi: 10.1124/dmd.111.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zbytek B., Janjetovic Z., Tuckey R.C., Zmijewski M.A., Sweatman T.W., Jones E., Nguyen M.N., Slominski A.T. 20-Hydroxyvitamin D3, a product of vitamin D3 hydroxylation by cytochrome P450scc, stimulates keratinocyte differentiation. J. Investig. Dermatol. 2008;128:2271–2280. doi: 10.1038/jid.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Janjetovic Z., Zmijewski M.A., Tuckey R.C., DeLeon D.A., Nguyen M.N., Pfeffer L.M., Slominski A.T. 20-Hydroxycholecalciferol, product of vitamin D3 hydroxylation by P450scc, decreases NF-kappaB activity by increasing IkappaB alpha levels in human keratinocytes. PLoS ONE. 2009;4:e5988. doi: 10.1371/journal.pone.0005988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Janjetovic Z., Tuckey R.C., Nguyen M.N., Thorpe E.M., Jr., Slominski A.T. 20,23-dihydroxyvitamin D3, novel P450scc product, stimulates differentiation and inhibits proliferation and NF-kappaB activity in human keratinocytes. J. Cell. Physiol. 2010;223:36–48. doi: 10.1002/jcp.21992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wasiewicz T., Szyszka P., Cichorek M., Janjetovic Z., Tuckey R.C., Slominski A.T., Zmijewski M.A. Antitumor effects of vitamin D analogs on hamster and mouse melanoma cell lines in relation to melanin pigmentation. Int. J. Mol. Sci. 2015;16:6645–6667. doi: 10.3390/ijms16046645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wierzbicka J.M., Zmijewski M.A., Piotrowska A., Nedoszytko B., Lange M., Tuckey R.C., Slominski A.T. Bioactive forms of vitamin d selectively stimulate the skin analog of the hypothalamus-pituitary-adrenal axis in human epidermal keratinocytes. Mol. Cell. Endocrinol. 2016;437:312–322. doi: 10.1016/j.mce.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Skobowiat C., Oak A.S., Kim T.K., Yang C.H., Pfeffer L.M., Tuckey R.C., Slominski A.T. Noncalcemic 20-hydroxyvitamin D3 inhibits human melanoma growth in in vitro and in vivo models. Oncotarget. 2017;8:9823–9834. doi: 10.18632/oncotarget.14193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Janjetovic Z., Brozyna A.A., Tuckey R.C., Kim T.K., Nguyen M.N., Jozwicki W., Pfeffer S.R., Pfeffer L.M., Slominski A.T. High basal NF-kappaB activity in nonpigmented melanoma cells is associated with an enhanced sensitivity to vitamin D3 derivatives. Br. J. Cancer. 2011;105:1874–1884. doi: 10.1038/bjc.2011.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wierzbicka J.M., Binek A., Ahrends T., Nowacka J.D., Szydlowska A., Turczyk L., Wasiewicz T., Wierzbicki P.M., Sadej R., Tuckey R.C., et al. Differential antitumor effects of vitamin d analogues on colorectal carcinoma in culture. Int. J. Oncol. 2015;47:1084–1096. doi: 10.3892/ijo.2015.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Piotrowska A., Wierzbicka J., Slebioda T., Wozniak M., Tuckey R.C., Slominski A.T., Zmijewski M.A. Vitamin D derivatives enhance cytotoxic effects of H2O2 or cisplatin on human keratinocytes. Steroids. 2016;110:49–61. doi: 10.1016/j.steroids.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lin Z., Marepally S.R., Goh E.S.Y., Cheng C.Y.S., Janjetovic Z., Kim T.K., Miller D.D., Postlethwaite A.E., Slominski A.T., Tuckey R.C., et al. Investigation of 20s-hydroxyvitamin D3 analogs and their 1alpha-OH derivatives as potent vitamin d receptor agonists with anti-inflammatory activities. Sci. Rep. 2018;8:1478. doi: 10.1038/s41598-018-19183-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lin Z., Chen H., Belorusova A.Y., Bollinger J.C., Tang E.K.Y., Janjetovic Z., Kim T.K., Wu Z., Miller D.D., Slominski A.T., et al. 1alpha,20S-dihydroxyvitamin D3 interacts with vitamin D receptor: Crystal structure and route of chemical synthesis. Sci. Rep. 2017;7:10193. doi: 10.1038/s41598-017-10917-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Slominski A.T., Kim T.K., Takeda Y., Janjetovic Z., Brozyna A.A., Skobowiat C., Wang J., Postlethwaite A., Li W., Tuckey R.C., et al. RORalpha and ROR gamma are expressed in human skin and serve as receptors for endogenously produced noncalcemic 20-hydroxy- and 20,23-dihydroxyvitamin D. FASEB J. 2014;28:2775–2789. doi: 10.1096/fj.13-242040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Le Poole I.C., van den Berg F.M., van den Wijngaard R.M., Galloway D.A., van Amstel P.J., Buffing A.A., Smits H.L., Westerhof W., Das P.K. Generation of a human melanocyte cell line by introduction of HPV16 E6 and E7 genes. In Vitro Cell. Dev. Biol. Anim. 1997;33:42–49. doi: 10.1007/s11626-997-0021-6. [DOI] [PubMed] [Google Scholar]
- 80.Slominski A.T., Kim T.K., Li W., Yi A.K., Postlethwaite A., Tuckey R.C. The role of CYP11A1 in the production of vitamin D metabolites and their role in the regulation of epidermal functions. J. Steroid Biochem. Mol. Biol. 2014;144PA:28–39. doi: 10.1016/j.jsbmb.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bikle D.D., Oda Y., Tu C.L., Jiang Y. Novel mechanisms for the vitamin d receptor (VDR) in the skin and in skin cancer. J. Steroid Biochem. Mol. Biol. 2015;148:47–51. doi: 10.1016/j.jsbmb.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Carlberg C. What do we learn from the genome-wide perspective on vitamin D3? Anticancer Res. 2015;35:1143–1151. [PubMed] [Google Scholar]
- 83.Kim T.K., Wang J., Janjetovic Z., Chen J., Tuckey R.C., Nguyen M.N., Tang E.K., Miller D., Li W., Slominski A.T. Correlation between secosteroid-induced vitamin D receptor activity in melanoma cells and computer-modeled receptor binding strength. Mol. Cell. Endocrinol. 2012;361:143–152. doi: 10.1016/j.mce.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Slominski A.T., Janjetovic Z., Kim T.K., Wright A.C., Grese L.N., Riney S.J., Nguyen M.N., Tuckey R.C. Novel vitamin D hydroxyderivatives inhibit melanoma growth and show differential effects on normal melanocytes. Anticancer Res. 2012;32:3733–3742. [PMC free article] [PubMed] [Google Scholar]
- 85.Soshilov A.A., Denison M.S. Ligand promiscuity of aryl hydrocarbon receptor agonists and antagonists revealed by site-directed mutagenesis. Mol. Cell. Biol. 2014;34:1707–1719. doi: 10.1128/MCB.01183-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Soshilov A., Denison M.S. Ligand displaces heat shock protein 90 from overlapping binding sites within the aryl hydrocarbon receptor ligand-binding domain. J. Biol. Chem. 2011;286:35275–35282. doi: 10.1074/jbc.M111.246439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tuckey R.C., Janjetovic Z., Li W., Nguyen M.N., Zmijewski M.A., Zjawiony J., Slominski A. Metabolism of 1alpha-hydroxyvitamin D3 by cytochrome P450scc to biologically active 1alpha,20-dihydroxyvitamin D3. J. Steroid Biochem. Mol. Biol. 2008;112:213–219. doi: 10.1016/j.jsbmb.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hiwatashi A., Nishii Y., Ichikawa Y. Purification of cytochrome P-450d1 alpha (25-hydroxyvitamin D3-1 alpha-hydroxylase) of bovine kidney mitochondria. Biochem. Biophys. Res. Commun. 1982;105:320–327. doi: 10.1016/S0006-291X(82)80047-8. [DOI] [PubMed] [Google Scholar]
- 89.Janjetovic Z., Nahmias Z.P., Hanna S., Jarrett S.G., Kim T.K., Reiter R.J., Slominski A.T. Melatonin and its metabolites ameliorate ultraviolet b-induced damage in human epidermal keratinocytes. J. Pineal Res. 2014;57:90–102. doi: 10.1111/jpi.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.