To the Editor: Abdominal aortic aneurysm (AAA), a disease defined as a 50% increase in the aortic diameter or the diameter larger than 30 mm compared to the normal abdominal aortic, is a severe and chronic disorder occurring mostly among men older than 65 years of age.[1] Understanding the risk factors and the biomarkers for AAA formation is imperative for creating targeted interventions to prevent, screen, or slow the disease progression.
Inflammatory processes are important in the formation of AAA. C-reactive protein (CRP) is an acute phase reactant produced by the liver that has long been used as a marker of systemic inflammation. Compelling epidemiological studies have suggested CRP is associated with the development of AAA. However, it is unclear whether this association is causal or because of confounding and reverse causality existing in observational studies.[2,3] Moreover, limited randomized trials have provided data that can precisely demonstrate the underlying causal relationships between CRP and AAA.[4]
Mendelian randomization (MR) analysis, a natural randomized trial, has been widely used to study the causal relevance of emerging risk factors with diseases.[5] Genetic variants which are randomly allocated at birth, are used as instrumental variables in MR. In this study, using established genetic variants that determine CRP level, we performed an MR to investigate the causal role of CRP in AAA.
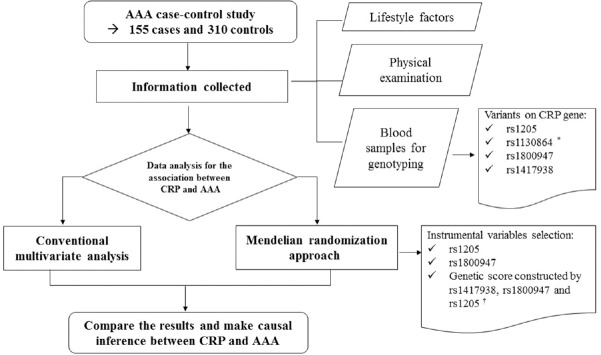
The study included 465 Chinese participants aged 40 years and above from an age- and gender-matched case–control study and details of the design were described previously.[6] AAA patients (n = 155) were diagnosed at the Vascular and Endovascular Surgery Department of the Chinese PLA General Hospital. The patients were diagnosed by abdominal Doppler ultrasound or computed tomography. Patients with inflammatory AAA, mental disorders, or pregnant were excluded from the study. The control group came from two sources, control Group 1 (n = 155) were selected from the same hospital and control Group 2 (n = 155) were selected from communities in Fangshan District in Beijing. The control participants were age- and gender-matched with AAA patients but were negative for AAA by abdominal Doppler ultrasound [Figure 1].
Figure 1.
The flow chart of the study design. *rs1130864 was excluded from the data analysis for its high linkage disequilibrium with rs1417938; †rs1417938 was not used as an instrumental variable individually because its extremely low F-statistic in the first stage of MR analysis, but it was included into the genetic score. MR: Mendelian randomization.
All participants were interviewed in person using a standardized questionnaire to collect data related to sociodemographics, history of chronic diseases and lifestyle factors, including smoking status, alcohol consumption, and physical activity. Participants underwent a standard physical examination including height, weight, systolic, and diastolic blood pressure and abdominal ultrasound or computed tomography. Peripheral venous fasting blood samples were obtained to detect biochemical parameters. Biochemical parameters were analyzed at the clinical laboratory of the Chinese PLA Hospital.[6]
Four common single-nucleotide polymorphisms (SNPs) (rs1205, rs1130864, rs1800947, and rs1417938) on this gene were selected from the International HapMap Project collection of Han Chinese data and because they were reported to be significantly related to CRP levels in a Chinese population.[7] Genomic DNA was extracted from the peripheral blood using the salt fractionation method. Genotypes were analyzed with Sequenom's MassARRAY® genotyping platform.[8] All SNPs passed the quality control with genotyping call rate of >95%. We obtained the linkage disequilibrium estimates using data from SNP Annotation and Proxy Search, and rs1130864 was excluded from the analysis for its high linkage disequilibrium with rs1417938 (r2= 0.962) [Table S1].
Supplementary Table S1.
Pairwise linkage disequilibrium matrix for SNPs in the CRP gene
| Items | rs1417938 | rs1800947 | rs1130864 | rs1205 |
|---|---|---|---|---|
| rs1417938 | 1.000 | 0.020 | 0.962 | 0.191 |
| rs1800947 | 0.020 | 1.000 | 0.021 | 0.106 |
| rs1130864 | 0.962 | 0.021 | 1.000 | 0.198 |
| rs1205 | 0.191 | 0.106 | 0.198 | 1.000 |
Data on estimates of pairwise linkage disequilibrium metrics were obtained from SNAP. http://www.broadinstitute.org/mpg/snap/index.php. Values represent r2 values, a measure of linkage disequilibrium. r2 values range from 0 to 1; with 0 representing complete equilibrium and 1 representing complete disequilibrium. SNAP: SNP annotation and proxy search; SNP: Single nucleotides polymorphisms; CRP: C-reactive protein.
Both separate SNPs and the constructed genetic score were used as the genetic instruments in our study. Instrumental variables were selected according to its relationship with CRP level and the F-statistics in the first stage regression of MR analysis; therefore, rs1800947, rs1205, and genetic score were involved for their F-statistics larger than 10 or approximated to 10. rs1417938 was not used as an instrumental variable individually because its F-statistic was extremely low, but it was included into the genetic score to maximize power and avoid the weak instrument bias in MR analysis [Figure 1 and Table S2].
Supplementary Table S2.
F-statistics for first stage regression of MR analysis using different instrumental variables
| Optional instrumental variables | F-statistic without adjustment |
|---|---|
| rs1417938 | 0.21 |
| rs1800947 | 7.07 |
| rs1205 | 14.15 |
| Genetic score (binary variable) | 19.30 |
| Genetic score (continuous variable) | 22.33 |
MR: Mendelian randomization.
Before calculating the genetic score, we first defined the exposure allele (or the risk allele) as the allele related to elevate circulating CRP levels according to our data and previous report, that is, T allele in rs1417938, G allele in rs1800947, and C allele in rs1205.[9] We then calculated an unweighted CRP genetic score by adding the number of CRP-increasing alleles that the person had inherited at each variant. We treated genetic score as instrumental variable in two ways – one way was to do the median split to obtain a binary variable (below or above the median genetic score), and the other way was to use the original values of the genetic score.
All statistical analysis was undertaken using STATA software package (version 13.0, StataCorp, College Station, Texas, USA). Power analysis was undertaken using PASS software (version 14.0, NCSS, LLC. Kaysville, Utah, USA, ncss.com/software/pass). Hardy–Weinberg Equilibrium was tested in control group using the Chi-square test. Concentrations of CRP were natural log transformed for its skewed distribution (LnCRP). Variables are expressed as numbers and percentages (categorical variables), mean ± standard deviation (continuous variables with normal distribution), median and interquartile range for continuous variables with abnormal distribution. First, we compared the basic characteristics of the participants as divided by genetic variants and genetic score, respectively, to assess whether the potential confounding factors differed across the instrumental variables. Differences between the groups were analyzed using t-test and Kruskal–Wallis rank test for continuous variables and Chi-square tests for categorical variables. Second, we measured the difference of CRP level for each instrumental variable using linear regression. Third, we used two-stage least squares regression for the instrumental analysis. The first stage was to do the linear regression of Ln-transferred CRP levels on the genetic instruments, obtaining the predicted values for CRP levels. The second stage was to do the logistic regression of AAA on the predicted CRP levels. The standard errors were corrected by bootstrapping both first and second stage together 1000 times. Fourth, we compared the associations derived from the instrumental variable analysis to the observational analysis by conducting multivariable analysis. In this step, the original genetic score was used as instrumental variable. Potential confounding factors were adjusted gradually, including age and gender, lifestyle factors including smoking, drinking, and physical activity, history of diseases, and body mass index (BMI). P = 0.05 was considered as the statistical significance threshold.
Table 1 shows the basic characteristics of the study participants. There were significant differences for smoking, drinking, history of hypertension and hyperlipidemia, and the concentration of plasma CRP between cases and controls, and no differences were found for age, gender, physical activity, history of diabetes, BMI, and genotypes of rs1417938, rs1800947, and rs1205.
Table 1.
Basic characteristics of the study participants
| Characteristics | Case group (n = 155) | Control groups (N = 310) | P* | |
|---|---|---|---|---|
| Control group from hospital (n = 155) | Control group from community (n = 155) | |||
| Age (years), mean ± SD | 69.2 ± 10.0 | 69.6 ± 10.9 | 69.5 ± 9.0 | 0.727 |
| Female, n (%) | 17 (11.0) | 17 (11.0) | 17 (11.0) | 1.000 |
| Smoker, n (%) | 132 (85.2) | 69 (44.5) | 98 (63.2) | <0.001 |
| Drinker, n (%) | 79 (51.0) | 58 (37.4) | 67 (43.2) | 0.029 |
| Physical activity, n (%) | 94 (60.7) | 87 (56.1) | 108 (69.7) | 0.530 |
| History of chronic diseases, n (%) | ||||
| Hypertension | 108 (70.0) | 64 (41.3) | 79 (51.6) | <0.001 |
| Type 2 diabetes | 18 (11.6) | 27 (17.4) | 19 (12.4) | 0.341 |
| Hyperlipidemia | 76 (49.0) | 34 (21.9) | 27 (17.4) | <0.001 |
| BMI (kg/m2)†, mean ± SD | 24.4 ± 3.4 | 25.3 ± 3.2 | 24.2 ± 3.5 | 0.338 |
| LnCRP (mg/L), median, (Q1,Q3) | −1.3 (−2.3, 0.02) | −2.2 (−2.8, −1.0) | −1.5 (−1.6, −1.3) | 0.014 |
| rs1417638, n (%) | ||||
| AT | 12 (8.0) | 20 (13.1) | 26 (17.2) | 0.030 |
| TT | 139 (92.1) | 133 (87.0) | 125 (82.8) | |
| rs1800947, n (%) | ||||
| CC | 0 (0) | 1 (0.7) | 1 (0.7) | 0.607 |
| CG | 8 (5.3) | 8 (5.2) | 8 (5.3) | |
| GG | 143 (94.7) | 144 (94.1) | 142 (94.3) | |
| rs1205, n (%) | ||||
| CC | 23 (15.3) | 31 (20.4) | 22 (14.7) | 0.801 |
| CT | 76 (50.7) | 73 (48.0) | 80 (53.3) | |
| TT | 51 (34.0) | 48 (31.6) | 48 (32.0) | |
*P values were calculated by comparing characteristics between AAA and all controls; †The BMI is the weight in kilograms divided by the square of the height in meters. BMI: Body mass index; SD: Standard deviation; CRP: C-reactive protein; IQR: Interquartile range; AAA: Abdominal aortic aneurysm; LnCRP: Consentrations of CRP that were natural log transformed.
All the basic characteristics were comparable when we grouped participants based on the genotypes of rs1800947 and rs1205, and the genetic score, respectively, thus indicating that these genetic variants are reliable instrumental variables. Only in rs1417938, we found that age and BMI were a significant difference in AT and TT groups, but did not find the significant difference in CRP [Tables S3–S6]. All the SNPs were in Hardy–Weinberg equilibrium (P > 0.01), and the allele frequencies in the control group from the community were shown in Table S7.
Supplementary Table S3.
Basic characteristics of the participants, according to genotype of rs1417938
| Items | AT (n = 58) | TT (n = 397) | P |
|---|---|---|---|
| Age (years), mean ± SD | 66.8 ± 10.5 | 69.7 ± 9.9 | 0.035 |
| Female sex (%) | 10.3 | 10.6 | 0.957 |
| Han nation (%) | 1.7 | 2.0 | 0.882 |
| Smoker (%) | 63.8 | 64.0 | 0.978 |
| Drinker (%) | 46.6 | 44.3 | 0.751 |
| Physical activity (%) | 63.8 | 67.3 | 0.601 |
| History of chronic disease (%) | |||
| Hypertension | 46.6 | 55.2 | 0.219 |
| Diabetes | 19.0 | 13.4 | 0.251 |
| Hyperlipidemia | 20.7 | 31.0 | 0.109 |
| BMI (kg/m2), mean ± SD* | 25.8 ± 2.9 | 24.5 ± 3.4 | 0.005 |
| LnCRP (mg/L), median (Q1,Q3) | −1.6 (−1.9, −1.2) | −1.5 (−2.2, −0.9) | 0.545 |
*The BMI is the weight in kilograms divided by the square of the height in meters. BMI: Body mass index; SD: Standard deviation; IQR: Interquartile range; CRP: C-reactive protein.
Supplementary Table S6.
Basic characteristics of the participants, according to the genetic risk score
| Characterostics | Genetic score below median* (n = 192) | Genetic score above median* (n = 260) | P |
|---|---|---|---|
| Age (years), mean ± SD | 68.8 ± 10.4 | 69.8 ± 9.6 | 0.315 |
| Female sex (%) | 8.9 | 11.9 | 0.295 |
| Han nation (%) | 3.1 | 1.2 | 0.138 |
| Smoker (%) | 65.6 | 62.7 | 0.521 |
| Drinker (%) | 45.3 | 44.2 | 0.819 |
| Physical activity (%) | 66.7 | 66.9 | 0.954 |
| History of chronic disease (%) | |||
| Hypertension | 50.0 | 56.9 | 0.144 |
| Diabetes | 13.0 | 14.6 | 0.629 |
| Hyperlipidemia | 26.0 | 32.3 | 0.149 |
| BMI (kg/m2), mean ± SD† | 25.0 ± 3.3 | 24.4 ± 3.4 | 0.078 |
| LnCRP (mg/L), median (Q1,Q3) | −1.6 (−2.4, −1.3) | −1.5 (−1.9, −0.7) | <0.001 |
*The median of genetic score was 5. Genetic score below median: Genetic score <5; genetic score above the median: genetic score ≥5. Of all the participants, 13 had missing data for SNPs and were excluded from the analysis for the need to construct the genetic score, †The BMI is the weight in kilograms divided by the square of the height in meters. BMI: Body mass index; SD: Standard deviation; CRP: C-reactive protein; SNP: Single nucleotides polymorphisms.
Supplementary Table S7.
Minor allelic frequencies for CRP SNPs in different groups of the study population
| Major homozygote | Heterozygote | Minor homozygote | Minor allele frequency | |
|---|---|---|---|---|
| Control group from community | ||||
| rs1417938 | 125 (TT, 83.5%) | 26 (AT, 15.7%) | 0 (AA, 0.7%) | 0.086 |
| rs1800947 | 142 (GG, 93.5%) | 8 (CG, 6.4%) | 1 (CC, 0.1%) | 0.033 |
| rs1205 | 48 (TT, 34.4%) | 80 (CT, 48.5%) | 22 (CC, 17.1%) | 0.413 |
| Control group from community | ||||
| rs1417938 | 133 (TT, 87.4%) | 20 (AT, 12.2%) | 0 (AA, 0.4%) | 0.065 |
| rs1800947 | 144 (GG, 93.6%) | 8 (CG, 6.3%) | 1 (CC, 0.1%) | 0.033 |
| rs1205 | 48 (TT, 30.9%) | 73 (CT, 49.4%) | 31 (CC, 19.7%) | 0.444 |
| Cases group | ||||
| rs1417938 | 139 (TT, 92.2%) | 12 (AT, 7.6%) | 0 (AA, 0.2%) | 0.040 |
| rs1800947 | 143 (GG, 94.8%) | 8 (CG, 5.2%) | 0 (CC, 0.1%) | 0.026 |
| rs1205 | 51 (TT, 35.2%) | 76 (CT, 48.3%) | 23 (CC, 16.5%) | 0.407 |
CRP: C-reactive protein; SNP: Single nucleotides polymorphisms.
Supplementary Table S4.
Basic characteristics of the participants, according to genotype of rs1800947
| Characteristics | CC/CG (n = 26) | GG (n = 429) | P |
|---|---|---|---|
| Age (years), mean ± SD | 69.0 ± 10.4 | 69.4 ± 10.0 | 0.830 |
| Female sex (%) | 11.5 | 10.5 | 0.866 |
| Han nation (%) | 3.9 | 1.9 | 0.481 |
| Smoker (%) | 61.5 | 64.1 | 0.791 |
| Drinker (%) | 26.9 | 45.7 | 0.062 |
| Physical activity (%) | 65.4 | 66.9 | 0.873 |
| History of chronic disease (%) | |||
| Hypertension | 57.8 | 53.9 | 0.702 |
| Diabetes | 19.2 | 13.8 | 0.435 |
| Hyperlipidemia | 30.8 | 29.6 | 0.899 |
| BMI (kg/m2), mean ± SD* | 24.9 ± 2.8 | 24.6 ± 3.4 | 0.718 |
| LnCRP (mg/L), median (Q1,Q3) | −2.3 (−3.0, −1.6) | −1.5 (−2.2, −1.0) | 0.004 |
*The BMI is the weight in kilograms divided by the square of the height in meters. BMI: Body mass index; SD: Standard deviation; CRP: C-reactive protein.
Supplementary Table S5.
Basic characteristics of the participants, according to genotype of rs1205
| Characteristics | TT (n = 147) | CT (n = 229) | CC (n = 76) | P |
|---|---|---|---|---|
| Age (years), mean ± SD | 69.4 ± 9.8 | 69.6 ± 9.8 | 70.8 ± 7.7 | 0.918 |
| Female sex (%) | 7.5 | 12.7 | 10.5 | 0.282 |
| Han nation (%) | 2.7 | 1.3 | 2.6 | 0.575 |
| Smoker (%) | 68.0 | 62.0 | 61.8 | 0.454 |
| Drinker (%) | 44.9 | 47.6 | 35.5 | 0.186 |
| Physical activity (%) | 67.4 | 66.8 | 65.8 | 0.973 |
| History of chronic disease (%) | ||||
| Hypertension | 50.3 | 56.3 | 54.0 | 0.524 |
| Diabetes | 10.9 | 14.9 | 17.1 | 0.380 |
| Hyperlipidemia | 27.2 | 32.3 | 26.3 | 0.448 |
| BMI (kg/m2), mean ± SD* | 24.6 ± 3.4 | 25.1 ± 3.6 | 23.1 ± 3.4 | 0.532 |
| LnCRP (mg/L), median (Q1,Q3) | −1.6 (−2.4, −1.2) | −1.5 (−2.1, −1.1) | −1.2 (−1.7, −0.3) | 0.0007 |
*The BMI is the weight in kilograms divided by the square of the height in meters. BMI: Body mass index; SD: Standard deviation; CRP: C-reactive protein.
Figure 2 shows the genetic association with LnCRP and estimated causal relationship between CRP and AAA. We found that G allele of rs1800947 (0.782 unit per G allele with 95% confidence interval [CI], 0.204–1.360, P = 0.008), C allele of rs1205 (0.358 unit per C allele with 95% CI, 0.171–0.545, P < 0.001), higher genetic score (0.423 unit per allele with 95% CI: 0.247–0.599, P < 0.001) were significantly associated with elevated CRP concentrations. MR analyses showed that genetically elevated CRP level were not significantly associated with higher risk of AAA (odds ratio [OR] = 1.163 for rs1800947, 95% CI, 0.174–7.748, P = 0.876; OR = 0.767 for rs1205, 95% CI, 0.249–2.367, P = 0.645; OR = 1.130 for binary genetic score, 95% CI, 0.524–2.439, P = 0.755; OR = 0.196 for continuous genetic score, 95% CI, 0.605–2.364, P = 0.606).
Figure 2.

Effect of genetically elevated CRP levels on abdominal aortic aneurysm using MR approach in a Chinese population. *Mean difference in LnCRP was assessed using regression model with genetic variants and genetic score as repressors, respectively; †OR was for the association and was estimated from the instrumental variable analysis by using genetic variants and genetic score as instrumental variables, respectively. No other covariates were adjusted in the MR model. CRP: C-reactive protein; MR: Mendelian randomization; OR: Odds ratio.
Table 2 compares the conventional observational multivariable results and the MR estimates for the association between CRP and AAA in three models. In model 1, age and gender were adjusted, and we observed that observational analysis obtained significant directionally association of CRP with risk of AAA (OR = 1.357, 95% CI: 1.163–1.583, P < 0.001), but instrumental variable analysis found nonsignificant association (OR = 1.203, 95% CI, 0.574–2.519, P = 0.625). When further adjusted for smoking, drinking, and physical activity in model 2, we observed significant associations in observational analysis for CRP and AAA, but not in instrumental variable analysis (OR in observational analysis, 1.276, 95% CI, 1.085–1.502, P = 0.003; OR in instrumental variable analysis, 1.291, 95% CI, 0.606–2.754, P = 0.508). In model 3, BMI and history of chronic diseases were additionally adjusted, and similar results were observed for the association between CRP and AAA as in model 2.
Table 2.
The effect size between LnCRP and AAA estimated from the conventional multivariate model and instrumental variable analysis
| Items | Instrumental variable analysis | Observational multivariable | ||||
|---|---|---|---|---|---|---|
| OR | 95% CI | P | OR | 95% CI | P | |
| Model 1* | 1.203 | 0.574–2.519 | 0.625 | 1.357 | 1.163–1.583 | <0.001 |
| Model 2* | 1.291 | 0.606–2.754 | 0.508 | 1.276 | 1.085–1.502 | 0.003 |
| Model 3* | 1.132 | 0.480–2.667 | 0.777 | 1.249 | 1.049–1.489 | 0.013 |
*Model 1: Adjusted for age and gender; Model 2: Adjusted for ever smoked, ever drunk, and physical activity based on Model 1; Model 3: Adjusted for BMI and history of hypertension; diabetes, and hyperlipidemia based on Model 2. OR: Odds ratio; CI: Confidence interval; CRP: C-reactive protein; AAA: Abdominal aortic aneurysm.
Our findings showed that the genetically elevated CRP concentrations were not associated with AAA, indicating that this association was not causal.
By using genetic variants as the instrumental variables, this approach could provide evidence of causality between exposure and outcome. To be effective and strong instrumental variables, the genetic variants must satisfy these assumptions: (1) the genetic variants should be related to the exposure we studied; (2) the instrument should not be associated with confounding factors of the exposure and outcome association; and (3) the genetic variants must affect the outcome only through the exposure pathway, and no other pathways.[5] Both separate CRP genetic variants (except rs1417938) and genetic score were found to be related significantly to the plasma CRP levels, and the F-statistics were reasonably large for these variants and genetic score, which satisfied the first assumption of the instrumental variables. Our results showed that the basic characteristics were equally distributed across each instrumental variable so that the second assumption was satisfied according to the current data although they could not represent all the confounders. For the third assumption, the functions of the genetic variants we chose located in CRP gene and their relationships with CRP level have been widely studied.[10] In addition, the genetic score, we constructed could combine information from multiple variants that associated with the CRP, and could maximize power and avoid the weak instrument bias.[11]
Our finding did not find plasma CRP was a determinant of the AAA. This was inconsistent with the results of most observational studies, including one study based on the same population used in the present study.[2,12] Atherosclerosis and inflammation, with increased levels of related markers, such as CRP, are considered the common pathophysiologic process for vascular-related disease.[13] However, some studies in vivo did not find the significant association between plasma CRP and cardiovascular events, which agreed with our finding.[14] In the pathway of AAA, we suppose that plasma CRP may be an early predictor of AAA, but not a causal factor of the AAA development; and therefore, the observed association may derive from reverse causality. Irrespective of the causal effect of plasma CRP on AAA, however, there are considerable evidence persisting that inflammatory activation and degradation of the elastic media are the important pathophysiologic process in the development of AAA. More evidence is needed to identify specific genetic, environmental, and biochemical factors for AAA development.
There were several limitations in our study. One limitation is the low power of the study mainly led by small sample size and the lack of enough common genetic variants in CRP genes to construct a complete genetic score. The concentration of CRP in the circulation is influenced by many polymorphisms in CRP gene and other genes, more power would be obtained if more genetic variants could be involved.[15] Therefore, larger sample size and more genotyping data are expected to confirm the CRP and AAA through MR studies. Moreover, our study was conducted in Chinese population; therefore, the results cannot be generalized to other ethnic populations.
Supplementary information is linked to the online version of the paper on the Chinese Medical Journal website.
Financial support and sponsorship
The study was supported by the National Natural Science Foundation of China (81230066, 81502874) and The Eleventh Five-year Plan in Health Care Foundation of PLA (09BJZ04).
Conflicts of interest
There are no conflicts of interest.
Acknowledgments
We would like to thank Professor Tao Wu from School of Public Health in Peking University for her advices and thank all participants in the study.
Footnotes
Edited by: Yi Cui
REFERENCES
- 1.Bird AN, Davis AM. Screening for abdominal aortic aneurysm. JAMA. 2015;313:1156–7. doi: 10.1001/jama.2015.0996. doi: 10.1001/jama.2015.0996. [DOI] [PubMed] [Google Scholar]
- 2.Folsom AR, Yao L, Alonso A, Lutsey PL, Missov E, Lederle FA, et al. Circulating biomarkers and abdominal aortic aneurysm incidence: The atherosclerosis risk in communities (ARIC) study. Circulation. 2015;132:578–85. doi: 10.1161/CIRCULATIONAHA.115.016537. doi: 10.1161/circulationaha.115.016537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stather PW, Sidloff DA, Dattani N, Gokani VJ, Choke E, Sayers RD, et al. Meta-analysis and meta-regression analysis of biomarkers for abdominal aortic aneurysm. Br J Surg. 2014;101:1358–72. doi: 10.1002/bjs.9593. doi: 10.1002/bjs.9593. [DOI] [PubMed] [Google Scholar]
- 4.Mojtahedzadeh M, Chelkeba L, Ranjvar-Shahrivar M, Najafi A, Moini M, Najmeddin F, et al. Randomized trial of the effect of magnesium sulfate continuous infusion on IL-6 and CRP serum levels following abdominal aortic aneurysm surgery. Iran J Pharm Res. 2016;15:951–6. [PMC free article] [PubMed] [Google Scholar]
- 5.Burgess S, Timpson NJ, Ebrahim S, Davey Smith G. Mendelian randomization: Where are we now and where are we going? Int J Epidemiol. 2015;44:379–88. doi: 10.1093/ije/dyv108. doi: 10.1093/ije/dyv108. [DOI] [PubMed] [Google Scholar]
- 6.Wei Y, Xiong J, Zuo S, Chen F, Chen D, Wu T, et al. Association of polymorphisms on chromosome 9p21.3 region with increased susceptibility of abdominal aortic aneurysm in a Chinese Han population. J Vasc Surg. 2014;59:879–85. doi: 10.1016/j.jvs.2013.10.095. doi: 10.1016/j.jvs. 2013.10.095. [DOI] [PubMed] [Google Scholar]
- 7.Kong H, Qian YS, Tang XF, Zhang J, Gao PJ, Zhang Y, et al. C-reactive protein (CRP) gene polymorphisms, CRP levels and risk of incident essential hypertension: Findings from an observational cohort of Han Chinese. Hypertens Res. 2012;35:1019–23. doi: 10.1038/hr.2012.89. doi: 10.1038/hr.2012.89. [DOI] [PubMed] [Google Scholar]
- 8.Bradić M, Costa J, Chelo IM. Genotyping with Sequenom. Methods Mol Biol. 2011;772:193–210. doi: 10.1007/978-1-61779-228-1_11. doi: 10.1007/978-1-61779-228-1_11. [DOI] [PubMed] [Google Scholar]
- 9.Lee CC, You NC, Song Y, Hsu YH, Manson J, Nathan L, et al. Relation of genetic variation in the gene coding for C-reactive protein with its plasma protein concentrations: Findings from the women's health initiative observational cohort. Clin Chem. 2009;55:351–60. doi: 10.1373/clinchem.2008.117176. doi: 10.1373/clinchem.2008.117176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crawford DC, Sanders CL, Qin X, Smith JD, Shephard C, Wong M, et al. Genetic variation is associated with C-reactive protein levels in the third national health and nutrition examination survey. Circulation. 2006;114:2458–65. doi: 10.1161/CIRCULATIONAHA.106.615740. doi: 10.1161/circulationaha.106.615740. [DOI] [PubMed] [Google Scholar]
- 11.Hopewell JC, Clarke R. Emerging risk factors for stroke: What have we learned from Mendelian randomization studies? Stroke. 2016;47:1673–8. doi: 10.1161/STROKEAHA.115.010646. doi: 10.1161/strokeaha.115.010646. [DOI] [PubMed] [Google Scholar]
- 12.Shangwei Z, Yingqi W, Jiang X, Zhongyin W, Juan J, Dafang C, et al. Serum high-sensitive C-reactive protein level and CRP genetic polymorphisms are associated with abdominal aortic aneurysm. Ann Vasc Surg. 2017;45:186–92. doi: 10.1016/j.avsg.2017.05.024. doi: 10.1016/j.avsg.2017.05.024. [DOI] [PubMed] [Google Scholar]
- 13.Pai JK, Pischon T, Ma J, Manson JE, Hankinson SE, Joshipura K, et al. Inflammatory markers and the risk of coronary heart disease in men and women. N Engl J Med. 2004;351:2599–610. doi: 10.1056/NEJMoa040967. doi: 10.1056/NEJMoa040967. [DOI] [PubMed] [Google Scholar]
- 14.Koike T, Kitajima S, Yu Y, Nishijima K, Zhang J, Ozaki Y, et al. Human C-reactive protein does not promote atherosclerosis in transgenic rabbits. Circulation. 2009;120:2088–94. doi: 10.1161/CIRCULATIONAHA.109.872796. doi: 10.1161/circulationaha.109.872796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dehghan A, Dupuis J, Barbalic M, Bis JC, Eiriksdottir G, Lu C, et al. Meta-analysis of genome-wide association studies in >80 000 subjects identifies multiple loci for C-reactive protein levels. Circulation. 2011;123:731–8. doi: 10.1161/CIRCULATIONAHA.110.948570. doi: 10.1161/circulationaha.110.948570. [DOI] [PMC free article] [PubMed] [Google Scholar]