Well diffracting crystals of the ectodomain of rat ectonucleotide phosphodiesterase/pyrophosphatase-3 were obtained for a construct lacking the SMB domains, while the structure of the full-length ectodomain reveals the orientation of the SMB domains but has a partially disordered active-site structure, probably owing to the absence of the active-site zinc ions.
Keywords: ectonucleotide phosphodiesterase/pyrophosphatase, NPP3, ectonucleotidase, ATP hydrolysis, allergic response
Abstract
Ectonucleotide phosphodiesterase/pyrophosphatase-3 (NPP3, ENPP3) is an ATP-hydrolyzing glycoprotein that is located in the extracellular space. The full-length ectodomain of rat NPP3 was expressed in HEK293S GntI− cells, purified using two chromatographic steps and crystallized. Its structure at 2.77 Å resolution reveals that the active-site zinc ions are missing and a large part of the active site and the surrounding residues are flexible. The SMB-like domains have the same orientation in all four molecules in the asymmetric unit. The SMB2 domain is oriented as in NPP2, but the SMB1 domain does not interact with the PDE domain but extends further away from the PDE domain. Deletion of the SMB domains resulted in crystals that diffracted to 2.4 Å resolution and are suitable for substrate-binding studies.
1. Introduction
Ectonucleotide phosphodiesterase/pyrophosphatase-3 (NPP3) is an extracellular membrane-associated glycoprotein that hydrolyzes ATP, producing AMP and pyrophosphate (PPi). NPP3 has been identified on the cell surface of basophils and mast cells, serving as a marker protein for the estimation of hypersensitivity to allergens (Bühring et al., 2004 ▸; Hauswirth et al., 2007 ▸). Recent studies have shown that NPP3 negatively regulates chronic allergic responses by hydrolyzing extracellular ATP, which enhances allergic inflammation (Tsai et al., 2015 ▸). Hence, NPP3 could be a novel therapeutic target for allergic diseases. NPP3 may also have an intracellular function in the modulation of the level of intracellular nucleotide sugars in the endoplasmic reticulum and Golgi (Korekane et al., 2013 ▸). The NPP3-mediated hydrolysis of UDP-N-acetylglucosamine (UDP-GlcNAc) produces UMP, which is a potent competitive inhibitor of N-acetylglucosaminyltransferase (GnT-IX). NPP3 affects the cellular glycosylation profile.
NPP3 is a member of the NPP protein family, which comprises seven glycoproteins (Stefan et al., 2005 ▸; Bollen et al., 2000 ▸; Zimmermann et al., 2012 ▸). All family members share pyrophosphate/phosphodiesterase activity for the hydrolysis of nucleotides and phospholipids. This activity is mediated by the catalytic phosphodiesterase (PDE) domain. NPP1, NPP2 and NPP3 contain an additional nuclease-like domain and two somatomedin B (SMB)-like domains. NPP1 and NPP3 hydrolyze different extracellular nucleotides such as nucleotide triphosphates or dinucleotides. In contrast, NPP2 hydrolyzes lysophosphatidylcholine to produce lysophosphatidic acid, while NPP4 is known for the conversion of dinucleotides (Albright et al., 2012 ▸; Tokumura et al., 2002 ▸). Further identified substrates include adenine dinucleotide (NPP5), glycerophosphocholine (NPP6) and sphingomyelin (NPP7) (Gorelik, Randriamihaja et al., 2017 ▸; Morita et al., 2016 ▸; Duan et al., 2003 ▸).
Crystal structures have been determined for NPP1 (Kato et al., 2012 ▸; Jansen et al., 2012 ▸), NPP2 (Nishimasu et al., 2011 ▸; Hausmann et al., 2011 ▸), NPP4 (Albright et al., 2014 ▸), NPP5 (Gorelik, Liu et al., 2017 ▸), NPP6 (Morita et al., 2016 ▸) and NPP7 (Gorelik, Liu et al., 2017 ▸). These structures revealed the conserved arrangement of the nuclease and PDE domains of NPP1 and NPP2 and the significant differences in the architecture of the substrate-binding pockets for the binding of nucleotide and phospholipid substrates, while the core catalytic center comprising two zinc ions and an asparagine residue binding to the zinc-coordinated phosphate group is absolutely conserved. The PDE domain of NPPs is related to alkaline phosphatase (AP) and the two enzyme families share the catalytic dizinc center with all metal ligands.
To characterize the substrate specificity and obtain insight into the structure of NPP3, we have overexpressed rat NPP3 in HEK293 cells using a construct comprising the complete ectodomain. These crystals were not suitable for substrate-binding studies owing to a partly disordered active-site structure and loss of the two zinc ions. However, the orientation of the SMB domains could be analyzed in comparison to NPP1 and NPP2. A new construct lacking the SMB domains resulted in a new crystal form with superior diffraction properties and a well defined active-site structure.
2. Materials and methods
2.1. Construct design
The cDNA coding for the NPP3 ectodomain from Rattus norvegicus (UniProtKB P97675) was cloned with (Glu49–Ile875) and without (Trp140–Ile875) the SMB-like domains into the pHLsec vector (Aricescu et al., 2006 ▸; Table 1 ▸). This results in a fusion with an N-terminal signal sequence, leading to secretion of the expressed target protein with a C-terminal His6 tag. The sequences were verified by DNA sequencing.
Table 1. Macromolecule-production information.
Additional amino acids of NPP349–875 are underlined.
| Source organism | R. norvegicus |
| DNA source | RnNPP3 cDNA |
| Expression vector | pHLsec (Aricescu et al., 2006 ▸) |
| Expression host | HEK293S GntI− (Reeves et al., 2002 ▸) |
| Complete amino-acid sequence of construct product | AETGEEHIGSCRKKCFDSSHRGLEGCRCDSGCTDRGDCCWDFEDTCVKSTQIWTCNSFRCGETRLEAALCSCADDCLQRKDCCTDYKAVCQGEVPWVTEACASSQEPQCPEGFDQPPVILFSMDGFRAEYLQTWSTLLPNINKLKTCGLHSKYMRAVYPTKTFPNHYTIVTGLYPESHGIIDNNMYDVYLNKNFSLSSVEKSNPAWWSGQPIWLTAMYQGLKAASYYWPGSDVAVNGSFPNIYRNYSNSVPYESRIATLLQWLDLPKAERPSFYTIYVEEPDSAGHKSGPVSAGVIKALQLVDDAFGMLMEGLKQRNLHNCVNIIVLADHGMDQTSCDRVEYMTDYFPEINFYMYQGPAPRIRTRNIPQDFFTFNSEEIVRDLSCRKSDQHFKPYLTPDLPKRLHYAKNVRIDKVHLMVDRQWLAYRNKGSSNCEGGTHGYNNEFKSMEAIFLAHGPSFKEKTVIEPFENIEVYNLLCDLLHIQPAPNNGSHGSLNHLLKAPFYQPSHAEELSKSAGCGFTTPLPKDSLNCSCLALQTSGQEEQVNQRLNLNRGEVSATEKTNLPFGRPRVIQKNKDHCLLYHREYVSGFGKAMKMPMWSSYTVPKPGDTSSLPPTVPDCLRADVRVDPSESQKCSFYLADQNIDHGFLYPPAIKGNNESQYDALITSNLVPMYKEFKKMWDYFHKVLLIKYAIERNGVNVVSGPIFDYNYDGHFDAPDEITNYVAGTDVPVPTHYFVVLTSCKNKTHTPDSCPGWLDVLPFVVPHRPTNVESCPENKAEDLWVEERFKAHIARVRDVELLTGLDFYQEKTQPVSEILQLKTYLPTFETIIGTKHHHHHH |
2.2. Expression
For small-scale expression tests, HEK293S GntI− cells were grown in adherent culture in 24-well TPP plates at 310 K in a humidified atmosphere containing 5% CO2. Cell viability was supported by Dulbecco’s Modified Eagle’s Medium (Gibco) supplemented with 10%(v/v) fetal bovine serum (Gibco), 1% Glutmax (Gibco) and 1% non-essential amino acids (Gibco). The cells were transfected with the expression plasmid after growth to 90% confluency using polyethylenimine (Sigma–Aldrich) as a transfection reagent in a 1.5-fold excess (w/w) with respect to plasmid DNA. For preparative expression, cells from one confluent T175 bottle (Greiner) were cultured in roller bottles (Greiner) with a surface of 1700 cm2 per bottle for 2 d before transfection. The roller bottles were incubated at 310 K with a rotation speed of 0.9 rev min−1 and filled with 250 ml medium per bottle. 2 h after transfection, which followed a similar protocol as stated above, valproic acid (Sigma–Aldrich) was added to a final concentration of 4 mM to enhance expression yields.
2.3. Purification
Both constructs were purified using the same procedure as follows. The culture supernatant (1–2 l) was collected 5 d after transfection and separated from detached cells by centrifugation and filtration using a 0.22 µm bottle-top filter (TPP). To prepare the supernatant for purification by immobilized metal-affinity chromatography, the buffer was exchanged via an ultrafiltration unit (10 kDa molecular-weight cutoff; GE Healthcare) using 5 l buffer A (50 mM Tris, 500 mM NaCl pH 8.0). The resulting protein solution (volume of ∼250 ml) was adjusted to 30 mM imidazole by adding elution buffer B (50 mM Tris, 500 mM NaCl, 500 mM imidazole pH 8.0). After this, the protein solution was loaded onto a HisTrap HP column (5 ml volume; GE Healthcare), washed with 30 ml 30 mM imidazole in buffer A and eluted with a linear gradient from buffer A to buffer B over 5 ml. Fractions containing NPP3 were pooled and concentrated to 2 ml using Amicon Ultra centrifugal filter units (10 kDa molecular-weight cutoff; Merck Millipore) and further purified on a Superdex 200 16/60 gel-filtration column (120 ml volume; GE Healthcare) using a buffer consisting of 20 mM Tris pH 8.0. Pure protein fractions were pooled, concentrated to 5–10 mg ml−1 and stored at 277 K for further use.
2.4. Crystallization
Initial screening for crystallization conditions was performed with commercially available crystallization screens (Hampton Research and Jena Bioscience) at 292 K using the sitting-drop vapor-diffusion technique in 96-well CrystalQuick plates with round wells (Greiner) and a reservoir volume of 90 µl. The crystallization droplets set up by a MicroSys 4000-XL crystallization robot (Zinsser Analytic) consisted of equal volumes (0.1 µl) of reservoir solution and protein solution (10 mg ml−1 NPP3 in 20 mM Tris pH 8.0). Optimizations of the initial hits were performed by changing the pH value and the precipitant and protein concentrations using the hanging-drop vapor-diffusion method. Equal volumes of protein and reservoir solution (1 µl) were mixed and equilibrated against 500 ml reservoir solution in 24-well trays (Nelipak). Crystallization information is given in Table 2 ▸.
Table 2. Crystallization information.
| Construct | NPP349–875 |
| Method | Vapor diffusion |
| Plate type, screening | Three-drop 96-well plates (Greiner) |
| Plate type, optimization | 24-well PVC trays (Nelipak) |
| Temperature (K) | 292 |
| Protein concentration (mg ml−1) | 10.5 |
| Composition of protein solution | 20 mM Tris–HCl pH 8.0 |
| Composition of reservoir solution | 0.2 M triammonium citrate pH 7.0, 20%(w/v) PEG 3350 |
| Volume and ration of drop | 2 µl, 1:1 |
| Volume of reservoir (µl) | 500 |
2.5. Data-collection and crystal structure analysis
Crystals of NPP3140–875 were cryoprotected in reservoir solution by adding glycerol (Sigma–Aldrich) stepwise to the drop until a final concentration of 12%(v/v) was reached. The crystals were flash-cooled in liquid nitrogen. The crystal of NPP349–875 was cooled without adding cryoprotectant since only one crystal was available. Data collection was performed at HZB (BESSY II beamline 14.1, Berlin, Germany; Mueller et al., 2015 ▸; Table 3 ▸). Data processing was performed with DIALS (NPP349–875; Winter et al., 2018 ▸) or XDS (NPP3140–875; Kabsch, 2010 ▸). The NPP349–875 crystal showed strongly anisotropic diffraction, with a CC1/2 limit of >0.5 at ∼3.4 Å resolution in directions best corresponding to a* and b* and much better diffraction approximately along c* to ∼2.8 Å resolution (CC1/2 > 0.5). Anisotropic resolution cutoff and scaling were therefore applied to this data set using the STARANISO server (http://staraniso.globalphasing.org). As a result of this procedure, the overall completeness and the completeness in the highest resolution shell are low. However, the completeness is >96% in all shells up to a resolution of 3.6 Å. For the NPP3140–875 data set the reflections were scaled with AIMLESS (Evans & Murshudov, 2013 ▸). For both structures, molecular replacement was performed with Phaser (McCoy et al., 2007 ▸) with an NPP3 search model generated by MODELLER 9.7 (Šali & Blundell, 1993 ▸) based on NPP2 (PDB entry 2xr9; Hausmann et al., 2011 ▸). However, owing to significant disorder and the low resolution of the data, model building and refinement of the NPP349–875 structure reported here was only possible using the superimposed PDE and nuclease domains of the refined NPP3140–875 structure as a starting point. The structures were refined using REFMAC5 (Murshudov et al., 2011 ▸) and BUSTER (v.2.10.2; Bricogne et al., 2016 ▸). Structure-refinement statistics are given in Table 4 ▸.
Table 3. Data collection and processing.
| Construct | NPP349–875 |
| Beamline | BESSY MX14.1 |
| Detector | Rayonix MX225 |
| Wavelength (Å) | 0.91841 |
| Temperature (K) | 100 |
| Rotation range per image (°) | 1 |
| Total rotation range (°) | 210 |
| Exposure time per image (s) | 7 |
| Space group | P1 |
| a, b, c (Å) | 76.4, 116.3, 124.2 |
| α, β, γ (°) | 86.8, 87.8, 88.2 |
| Mosaicity (°) | 0.21 |
| Resolution range (Å) | 47.2–2.8 (3.04–2.80) |
| Total No. of reflections | 159100 |
| No. of unique reflections | 68486 (3424) |
| Completeness (%) | 65.2 (14.8) |
| Multiplicity | 2.4 (2.6) |
| 〈I/σ(I)〉 | 9.2 (1.3) |
| R p.i.m. (%) | 6.8 (60.3) |
| CC1/2 | 0.996 (0.425) |
| Overall B factor from Wilson plot (Å2) | 67.4 |
| No. of molecules in asymmetric unit | 4 |
| Matthews coefficient (Å3 Da−1) | 2.92 |
| Solvent content (%) | 57.8 |
Table 4. Structure solution and refinement of NPP349–875 .
Values in parentheses are for the outer shell.
| Resolution range (Å) | 47.2–2.8 (2.87–2.80) |
| Completeness (%) | 65.3 (10.0) |
| σ Cutoff | 0.0 |
| No. of reflections, working set | 68479 |
| No. of reflections, test set | 3360 |
| Final R cryst | 0.194 |
| Final R free | 0.238 |
| Cruickshank DPI | 0.438 |
| No. of non-H atoms | |
| Protein | 24770 |
| Ion | 0 |
| Ligand | 0 |
| Water | 0 |
| Total | 24770 |
| R.m.s. deviations | |
| Bonds (Å) | 0.01 |
| Angles (°) | 1.19 |
| Average B factors (Å2) | |
| Protein | 78.2 |
| Ramachandran plot | |
| Most favored (%) | 92.0 |
| Allowed (%) | 6.9 |
| Outliers (%) | 1.1 |
| PDB code | 6g4g |
2.6. Enzyme-activity assays
The nucleotide phosphodiesterase activity of NPP3 was determined from the hydrolysis of p-nitrophenyl thymidine 5′-monophosphate (pNPT-TMP). The absorbance of the released product p-nitrophenolate was detected at 405 nm. To determine the enzymatic activity and catalytic parameters, purified NPP3 was incubated at 298 K in a buffer consisting of 50 mM Tris–HCl, 25 mM NaCl, 0.1 mM ZnCl2 pH 9.5 with the substrate in a total volume of 150 µl. The reaction was stopped after 5–20 min by the addition of 150 µl 0.2 M NaOH. The amount of p-nitrophenolate was quantified colorimetrically at 405 nm and the data were fitted to the Michaelis–Menten equation using OriginPro 8.0 (OriginLab).
3. Results and discussion
3.1. Expression and characterization of NPP349–875
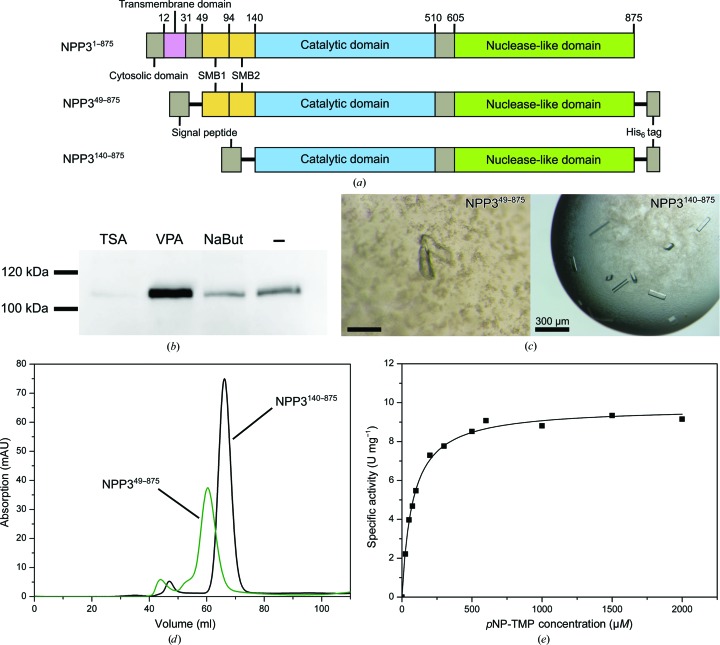
To express NPP3 in a soluble form, the full-length ectodomain was expressed in HEK cells and purified (construct NPP349–875; Fig. 1 ▸ a). Secretion of the expressed protein was enabled by fusion to a secretion signal provided by the expression vector. To enhance the chance of crystallization, HEK293S cells with an N-acetylglucosaminyltransferase deficiency were used to yield a homogeneous glycosylation pattern (Reeves et al., 2002 ▸). NPP3 contains seven putative glycosylation sites. To optimize the expression yield in transient transfection, we tested histone deacetylase inhibitors (Backliwal et al., 2008 ▸; Aricescu et al., 2006 ▸). A significant increase in expressed protein was detected using valproic acid (Fig. 1 ▸ b). The protein was purified to homogeneity by His-tag affinity chromatography and gel-filtration chromatography. The elution volume of the size-exclusion step indicates a monomeric form of the protein for both constructs (Fig. 1 ▸ d). Overall, about 1.0 mg of pure and active protein was obtained from 1 l cell-culture medium. With an enzymatic activity of A max = 10.3 µmol min−1 mg−1 and catalytic parameters of K m = 86.3 µM and k cat = 21 s−1 for the substrate pNP-TMP, NPP3 (Fig. 1 ▸ e) shows a similar catalytic behavior to those described previously for the NPP1 ectodomain (K m = 46 µM and k cat = 16 s−1; Kato et al., 2012 ▸) and for NPP3 membrane fractions (K m = 15 µM; Raza et al., 2011 ▸).
Figure 1.
Expression and purification of NPP3. (a) Domain organization of the full-length protein and the constructs NPP349–875 and NPP3140–875 used for crystallization experiments. (b) Western blot analysis (4 d post-transfection) for the expression level of NPP349–875 after the addition of histone deacetylase inhibitors. 5 µM trichostatin, 4 mM valproic acid and 5 mM sodium butyrate were added 2 h after transfection. (c) Crystals of NPP349–875 and NPP3140–875. (d) Elution profile of the size-exclusion chromatography step (Superdex 200 16/60; 120 ml) of NPP349–875 (green) and NPP3140–875 (black). The elution volume was approximately 60 ml for NPP349–875 (115 kDa) and 69 ml for NPP3140–875 (105 kDa). (e) Plot of the specific activity of NPP349–875 against pNP-TMP concentration.
3.2. Crystallization of NPP349–875 and design of the new construct NPP3140–875
In the initial screening of the construct NPP349–875 crystal growth was observed in a few conditions. Small intergrown crystals were obtained in condition No. 88 of the Index screen [0.2 M triammonium citrate pH 7.0, 20%(w/v) PEG 3350]. In the optimization process, only one single crystal was obtained after ∼3 months using 0.2 M triammonium citrate pH 6.5, 18%(w/v) PEG 3350 (Fig. 1 ▸ c). This crystal diffracted anisotropically to 2.77–3.4 Å resolution and belonged to space group P1 (Table 3 ▸). A molecular-replacement solution was obtained with NPP2 as a search model and partially refined. However, owing to the low resolution of the data and significant disorder in the structure, in particular in the active-site region, we were not able to progress with model building to obtain a sufficiently complete model. We therefore designed a new construct NPP3140–875, omitting the SMB domains (Fig. 1 ▸ a). This construct was expressed and purified using a very similar procedure to the construct NPP349–875 (Fig. 1 ▸ d). Crystallization screening led to several hits, and well shaped single crystals, which were obtained after two months of growth, diffracted to 2.4 Å resolution (Fig. 1 ▸ c). A molecular-replacement solution was obtained with the search model based on the NPP2 structure. These crystals contained an intact active site with bound zinc ions and are well suited for structural analysis of substrate binding, which will be described elsewhere (Döhler et al., 2018 ▸).
3.3. Orientation of the SMB domains in NPP3
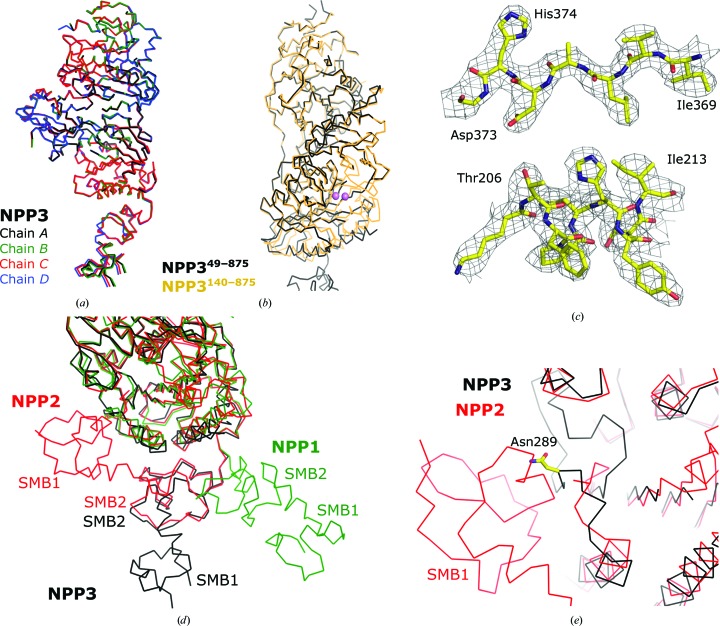
To proceed with the refinement and model building of the NPP349–875 structure, we superimposed NPP3140–875 (PDB entry 6f2t, analyzed to 2.4 Å resolution with R free = 22.9%; Döhler et al., 2018 ▸). Based on the improved starting model, it was possible to rebuild and refine the nuclease-like and SMB domains and to determine the structure of the SMB domains (Table 4 ▸). Chain D (average B factor of 41.6 Å2 for Cα atoms) of the four molecules in the asymmetric unit of NPP349–875 is best defined in the electron-density maps, followed by chains A (47.5 Å2), C (62.1 Å2) and B (71.3 Å2). In all four chains of the NPP349–875 structure the following regions of the PDE domain have no density owing to flexibility: 168–181, 324–336 and 473–492 (Fig. 2 ▸ b). These regions, which are part of or close to the active-site region, are structured in the NPP3140–875 crystals. In addition, no bound metal ions could be identified in the active site, and the Zn2-coordinating ligands Thr206, Asp373 and His374 of an intact NPP active site are well defined in the electron-density map but are not positioned for metal-ion coordination (Fig. 2 ▸ c). Zinc ions were not added during the purification and crystallization of NPP349–875. However, the purified ectodomain exhibited an activity of 10.6 ± 0.3 U mg−1 in the presence of 0.5 mM Zn2+, compared with 10.5 ± 0.1 U mg−1 without any addition of metal ions. This strongly indicates that the metal ions were lost during the relatively long crystallization time, which was perhaps favored by the presence of 0.2 M citrate as a chelating agent in the crystallization buffer and the removal of the zinc-free enzyme from solution by incorporation into the growing crystal. We assume that the loss of zinc ions causes the flexibility of nearby regions of the PDE domain.
Figure 2.
Crystal structure of NPP349–875. (a) Superposition of the four chains in the asymmetric unit. (b) Superposition of NPP349–875 onto NPP3140–875. The zinc ions are shown in purple. (c) Feature-enhanced electron-density map (contoured at 1.3σr.m.s.; Afonine et al., 2015 ▸) of residues 369–373 and 205–213 of the active-site region. (d) Positions of the SMB domains in a superposition of NPP349–875 with NPP1 (PDB entry 4b56; Jansen et al., 2012 ▸) and NPP2 (PDB entry 5lia; Shah et al., 2016 ▸). (e) Close-up view of the SMB1 domain of NPP2 and its interaction with the PDE domain. NPP349–875 is superimposed and the glycosylated Asn289 is shown.
The four molecules in the asymmetric unit have a similar structure, including the SMB domains (Fig. 2 ▸ a). For NPP1, only one structure is available with the SMB domains (PDB code 4b56; Jansen et al., 2012 ▸). The SMB domains are only visible in one of the two chains in the asymmetric unit, whereas they are apparently mobile in the other chain (Jansen et al., 2012 ▸). The SMB2 domain has little interaction with the PDE domain. In contrast, many independent structures (in different space groups) are available for NPP2 with its SMB domains in a similar orientation (PDB entries 5lqq, 5kxa, 5lia, 5l0b, 5ijq, 5hrt, 2xrg and 3nkm; Miller et al., 2017 ▸; Bain et al., 2017 ▸; Shah et al., 2016 ▸; Jones et al., 2016 ▸; Hausmann et al., 2011 ▸, 2016 ▸; Kato et al., 2016 ▸; Nishimasu et al., 2011 ▸). Both SMB domains interact with the PDE domain (Fig. 2 ▸ d). The SMB2 domain of NPP3 has the same orientation as the corresponding domain in NPP2, whereas the SMB1 domain does not interact with the PDB domain but extends further away from the PDE domain (Fig. 2 ▸ d). A superposition of NPP2 and NPP3 at the SMB1–PDE interface shows that the PDE domain of NPP3 has a significantly different fold at this interface, and a similar position of the SMB1 domain of NPP3 would cause clashes with the PDE domain, including the glycosylated Asn289. These regions have the same fold in the NPP3140–875 structure and are thus not affected by the active-site disorder of NPP349–875. The differences between NPP2 and NPP3 in the PDE domain and the observation of identical SMB-domain orientations in all four NPP3 molecules of the asymmetric unit (molecules A and B as well as molecules C and D have similar packing interactions, but molecules A and C have a different crystal packing) indicate that the SMB-domain orientations in the NPP3 structure are physiological. The recently published structure of human NPP3 supports this notion, as both SMB domains are similarly oriented in the two structures (Gorelik et al., 2018 ▸). The difference between the SMB1-domain orientations of NPP2 and NPP3 probably results from the different functional states of the enzymes. Whereas NPP2 is a secreted soluble extracellular enzyme, NPP3 is attached to the membrane via a transmembrane helix formed by residues 12–30. The position of the SMB1 domain extending away from the PDE domain may position the catalytic PDE domain further away from the cell membrane, as would the SMB1-domain orientation observed in NPP2.
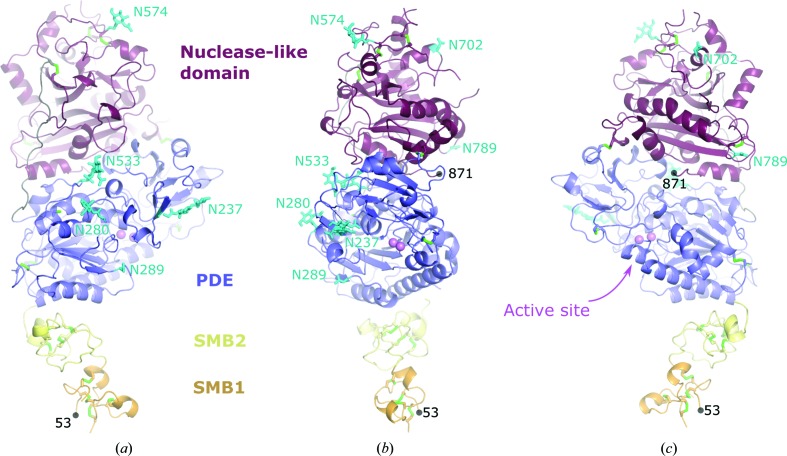
In conclusion, we have developed procedures for the expression and purification of two constructs of the ectodomain of rat NPP3. Construct NPP349–875 yields reasonably well diffracting crystals and is suitable for structural studies of the substrate specificity and catalytic mechanism of NPP3. From construct NPP3140–875 we obtained a low-resolution structure with a partially flexible active-site structure in the absence of bound zinc ions. However, this structure allowed a characterization of the orientation of the two SMB domains, which are likely to be important for stable membrane anchoring, and together with the NPP3140–875 structure a complete model for the ectodomain of NPP3 can be constructed (Fig. 3 ▸).
Figure 3.
Model of the full-length ectodomain of NPP3. (b) View from the front side, from which substrates have access to the active site (marked by the two zinc ions shown in magenta). Additional views are (a) from the left side [relative to the view in (b)] and (c) from the right side. This model was constructed by replacement of the PDE and nuclease-like domains of the NPP349–875 construct investigated in this work with the crystallographic coordinates of NPP3140–875 consisting of only the PDE and nuclease-like domains. Disulfide bridges are depicted in green and glycosylation sites in cyan. The N-terminal (53) and C-terminal (871) residues are marked.
Supplementary Material
PDB reference: ectonucleotide phosphodiesterase/pyrophosphatase-3, 6g4g
Acknowledgments
We acknowledge Antje Keim for technical assistance and support. We gratefully acknowledge the Helmholtz Zentrum Berlin (HZB) for the allocation of synchrotron beam time, for support during data collection and for travel support.
Funding Statement
This work was funded by Deutsche Forschungsgemeinschaft grant Str 477/13-2.
References
- Afonine, P. V., Moriarty, N. W., Mustyakimov, M., Sobolev, O. V., Terwilliger, T. C., Turk, D., Urzhumtsev, A. & Adams, P. D. (2015). Acta Cryst. D71, 646–666. [DOI] [PMC free article] [PubMed]
- Albright, R. A., Chang, W. C., Robert, D., Ornstein, D. L., Cao, W., Liu, L., Redick, M. E., Young, J. I., De La Cruz, E. M. & Braddock, D. T. (2012). Blood, 120, 4432–4440. [DOI] [PMC free article] [PubMed]
- Albright, R. A., Ornstein, D. L., Cao, W., Chang, W. C., Robert, D., Tehan, M., Hoyer, D., Liu, L., Stabach, P., Yang, G., De La Cruz, E. M. & Braddock, D. T. (2014). J. Biol. Chem. 289, 3294–3306. [DOI] [PMC free article] [PubMed]
- Aricescu, A. R., Lu, W. & Jones, E. Y. (2006). Acta Cryst. D62, 1243–1250. [DOI] [PubMed]
- Backliwal, G., Hildinger, M., Kuettel, I., Delegrange, F., Hacker, D. L. & Wurm, F. M. (2008). Biotechnol. Bioeng. 101, 182–189. [DOI] [PubMed]
- Bain, G., Shannon, K. E., Huang, F., Darlington, J., Goulet, L., Prodanovich, P., Ma, G. L., Santini, A. M., Stein, A. J., Lonergan, D., King, C. D., Calderon, I., Lai, A., Hutchinson, J. H. & Evans, J. F. (2017). J. Pharmacol. Exp. Ther. 360, 1–13. [DOI] [PubMed]
- Bollen, M., Gijsbers, R., Ceulemans, H., Stalmans, W. & Stefan, C. (2000). Crit. Rev. Biochem. Mol. Biol. 35, 393–432. [DOI] [PubMed]
- Bricogne, G., Blanc, E., Brandl, M., Flensburg, C., Keller, P., Paciorek, W., Roversi, P., Sharff, A., Smart, O. S., Vonrhein, C. & Womack, T. O. (2016). BUSTER. Cambridge: Global Phasing Ltd.
- Bühring, H.-J., Streble, A. & Valent, P. (2004). Int. Arch. Allergy Immunol. 133, 317–329. [DOI] [PubMed]
- Döhler, C., Zebisch, M. & Sträter, N. (2018). Sci. Rep. 8, 10874. [DOI] [PMC free article] [PubMed]
- Duan, R.-D., Bergman, T., Xu, N., Wu, J., Cheng, Y., Duan, J., Nelander, S., Palmberg, C. & Nilsson, A. (2003). J. Biol. Chem. 278, 38528–38536. [DOI] [PubMed]
- Evans, P. R. & Murshudov, G. N. (2013). Acta Cryst. D69, 1204–1214. [DOI] [PMC free article] [PubMed]
- Gorelik, A., Liu, F., Illes, K. & Nagar, B. (2017). J. Biol. Chem. 292, 7087–7094. [DOI] [PMC free article] [PubMed]
- Gorelik, A., Randriamihaja, A., Illes, K. & Nagar, B. (2017). FEBS J. 284, 3718–3726. [DOI] [PubMed]
- Gorelik, A., Randriamihaja, A., Illes, K. & Nagar, B. (2018). FEBS J. 285, 2481–2494. [DOI] [PubMed]
- Hausmann, J. et al. (2011). Nature Struct. Mol. Biol. 18, 198–204. [DOI] [PMC free article] [PubMed]
- Hausmann, J., Keune, W.-J., Hipgrave Ederveen, A. L., van Zeijl, L., Joosten, R. P. & Perrakis, A. (2016). J. Struct. Biol. 195, 199–206. [DOI] [PubMed]
- Hauswirth, A. W., Sonneck, K., Florian, S., Krauth, M.-T., Böhm, A., Sperr, W. R., Valenta, R., Schernthaner, G.-H., Printz, D., Fritsch, G., Bühring, H.-J. & Valent, P. (2007). Int. J. Immunopathol. Pharmacol. 20, 267–278. [DOI] [PubMed]
- Jansen, S., Perrakis, A., Ulens, C., Winkler, C., Andries, M., Joosten, R. P., van Acker, M., Luyten, F. P., Moolenaar, W. H. & Bollen, M. (2012). Structure, 20, 1948–1959. [DOI] [PubMed]
- Jones, S. B. et al. (2016). ACS Med. Chem. Lett. 7, 857–861. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kato, K., Ikeda, H., Miyakawa, S., Futakawa, S., Nonaka, Y., Fujiwara, M., Okudaira, S., Kano, K., Aoki, J., Morita, J., Ishitani, R., Nishimasu, H., Nakamura, Y. & Nureki, O. (2016). Nature Struct. Mol. Biol. 23, 395–401. [DOI] [PubMed]
- Kato, K., Nishimasu, H., Okudaira, S., Mihara, E., Ishitani, R., Takagi, J., Aoki, J. & Nureki, O. (2012). Proc. Natl Acad. Sci. USA, 109, 16876–16881. [DOI] [PMC free article] [PubMed]
- Korekane, H., Park, J. Y., Matsumoto, A., Nakajima, K., Takamatsu, S., Ohtsubo, K., Miyamoto, Y., Hanashima, S., Kanekiyo, K., Kitazume, S., Yamaguchi, Y., Matsuo, I. & Taniguchi, N. (2013). J. Biol. Chem. 288, 27912–27926. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Miller, L. M., Keune, W.-J., Castagna, D., Young, L. C., Duffy, E. L., Potjewyd, F., Salgado-Polo, F., Engel García, P., Semaan, D., Pritchard, J. M., Perrakis, A., Macdonald, S. J. F., Jamieson, C. & Watson, A. J. B. (2017). J. Med. Chem. 60, 722–748. [DOI] [PubMed]
- Morita, J. et al. (2016). Sci. Rep. 6, 20995. [DOI] [PMC free article] [PubMed]
- Mueller, U., Förster, R., Hellmig, M., Huschmann, F. U., Kastner, A., Malecki, P., Pühringer, S., Röwer, M., Sparta, K., Steffien, M., Ühlein, M., Wilk, P. & Weiss, M. S. (2015). Eur. Phys. J. Plus, 130, 662.
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Nishimasu, H., Okudaira, S., Hama, K., Mihara, E., Dohmae, N., Inoue, A., Ishitani, R., Takagi, J., Aoki, J. & Nureki, O. (2011). Nature Struct. Mol. Biol. 18, 205–212. [DOI] [PubMed]
- Raza, R., Akhtar, T., Hameed, S., Lecka, J., Iqbal, J. & Sevigny, J. (2011). Open Enzym. Inhib. J. 4, 17–22.
- Reeves, P. J., Callewaert, N., Contreras, R. & Khorana, H. G. (2002). Proc. Natl Acad. Sci. USA, 99, 13419–13424. [DOI] [PMC free article] [PubMed]
- Šali, A. & Blundell, T. L. (1993). J. Mol. Biol. 234, 779–815. [DOI] [PubMed]
- Shah, P. et al. (2016). Bioorg. Med. Chem. Lett. 26, 5403–5410.
- Stefan, C., Jansen, S. & Bollen, M. (2005). Trends Biochem. Sci. 30, 542–550. [DOI] [PubMed]
- Tokumura, A., Majima, E., Kariya, Y., Tominaga, K., Kogure, K., Yasuda, K. & Fukuzawa, K. (2002). J. Biol. Chem. 277, 39436–39442. [DOI] [PubMed]
- Tsai, S. H. et al. (2015). Immunity, 42, 279–293. [DOI] [PubMed]
- Winter, G., Waterman, D. G., Parkhurst, J. M., Brewster, A. S., Gildea, R. J., Gerstel, M., Fuentes-Montero, L., Vollmar, M., Michels-Clark, T., Young, I. D., Sauter, N. K. & Evans, G. (2018). Acta Cryst. D74, 85–97. [DOI] [PMC free article] [PubMed]
- Zimmermann, H., Zebisch, M. & Sträter, N. (2012). Purinergic Signal. 8, 437–502. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: ectonucleotide phosphodiesterase/pyrophosphatase-3, 6g4g