Structures of N-acetyl-d-neuraminic acid lyase in the ligand-free form and of a covalent Schiff base adduct are reported. The structure and kinetic data reveal the conserved nature of these proteins among Gram-negative bacteria.
Keywords: N-acetylneuraminate lyase, sialic acid catabolism, enzyme kinetics, Fusobacterium nucleatum
Abstract
N-Acetyl-d-neuraminic acid lyase (NanA) catalyzes the breakdown of sialic acid (Neu5Ac) to N-acetyl-d-mannosamine (ManNAc) and pyruvate. NanA plays a key role in Neu5Ac catabolism in many pathogenic and bacterial commensals where sialic acid is available as a carbon and nitrogen source. Several pathogens or commensals decorate their surfaces with sialic acids as a strategy to escape host innate immunity. Catabolism of sialic acid is key to a range of host–pathogen interactions. In this study, atomic resolution structures of NanA from Fusobacterium nucleatum (FnNanA) in ligand-free and ligand-bound forms are reported at 2.32 and 1.76 Å resolution, respectively. F. nucleatum is a Gram-negative pathogen that causes gingival and periodontal diseases in human hosts. Like other bacterial N-acetylneuraminate lyases, FnNanA also shares the triosephosphate isomerase (TIM)-barrel fold. As observed in other homologous enzymes, FnNanA forms a tetramer. In order to characterize the structure–function relationship, the steady-state kinetic parameters of the enzyme are also reported.
1. Introduction
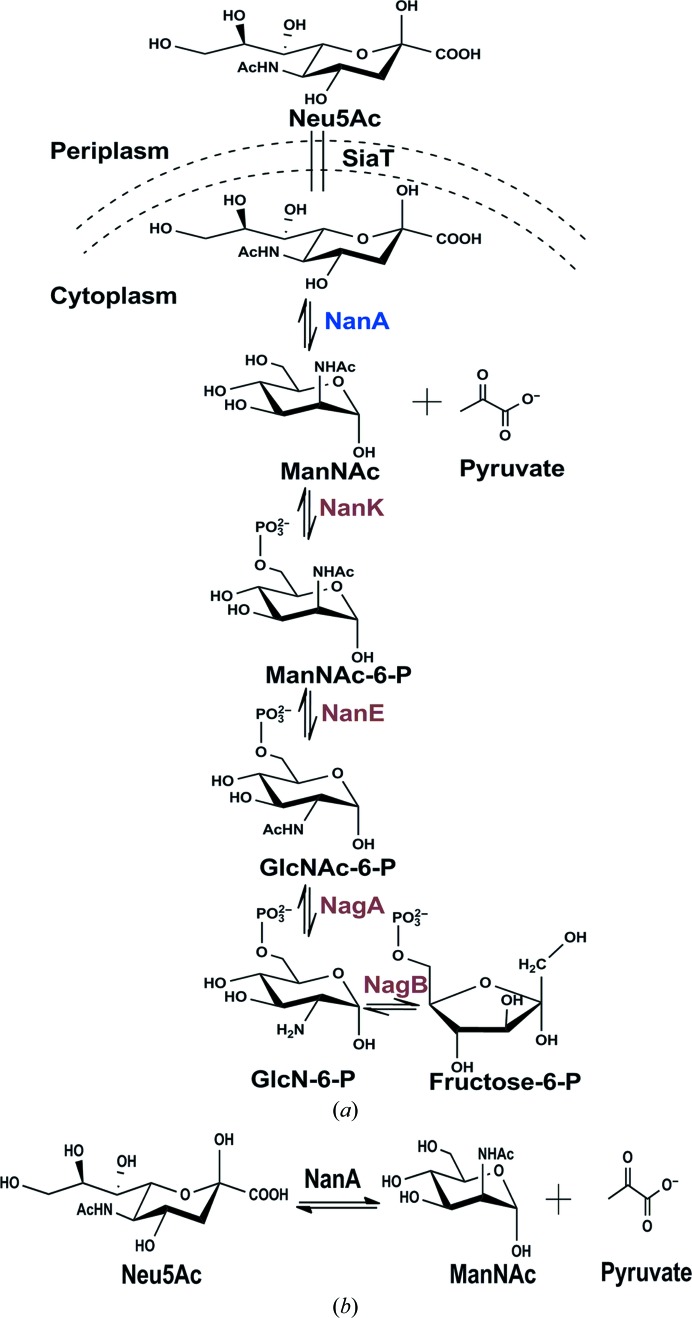
Sialic acids are a family of related nine-carbon, acidic, α-keto sugars which are used by bacteria for molecular mimicry. The most widely recognized type of sialic acid is N-acetylneuraminic acid (Neu5Ac; Varki, 1992 ▸; Maru et al., 2002 ▸). The large structural diversity and broad distribution of sialic acids in nature makes them very important in cell biology (Varki & Schauer, 2009 ▸). Sialic acids are usually found as the terminal nonreducing sugars of glycan chains on the cell surface of higher eukaryotes (Huynh et al., 2013 ▸). While some pathogens have evolved de novo biosynthesis pathways for Neu5Ac (Vimr et al., 2004 ▸), many bacteria solely rely on the acquisition of Neu5Ac from their environment with the help of specific membrane transporters (Severi et al., 2007 ▸; Bouchet et al., 2003 ▸). Once inside, the sialic acid can either enter the catabolic pathway or the surface-incorporation pathway, depending upon the requirements of the bacteria at the time (North et al., 2016 ▸). The surface-exposed sugar is recognized by the complement system in the serum as ‘self’. Studying the enzymes that are involved in the catabolism of sialic acids is important owing to the biological significance of sialic acids in bacteria. There are five enzymes that are responsible for the catabolism of sialic acids (Fig. 1 ▸ a). N-Acetylneuraminate lyase (NanA), the first committed enzyme, cleaves Neu5Ac to N-acetyl-d-mannosamine (ManNAc) and pyruvate (Fig. 1 ▸ b). The second enzyme, N-acetylmannosamine kinase (NanK), phosphorylates ManNAc at the C6 position, which produces N-acetylmannosamine 6-phosphate (ManNAc-6-P). The third enzyme, N-acetylmannosamine-6-phosphate epimerase (NanE), epimerizes ManNAc-6-P to N-acetylglucosamine 6-phosphate (GLcNAc-6-P). The fourth enzyme, N-acetylglucosamine-6-phosphate deacetylase (NagA), eliminates the acetyl group from GlcNAc-6-P to yield glucosamine 6-phosphate (GlcN-6-P), which is finally converted to fructose 6-phosphate by the fifth enzyme, glucosamine-6-phosphate deaminase (NagB) (Martinez et al., 1995 ▸; Ringenberg et al., 2003 ▸; Plumbridge & Vimr, 1999 ▸; Vimrt & Troy, 1985 ▸; Fig. 1 ▸ a). N-Acetyl-d-neuraminic acid lyase (NanA; EC 4.1.3.3) is also known as sialic acid aldolase. NanA is present in both pathogenic and nonpathogenic bacteria and in various mammalian tissues (Izard et al., 1994 ▸; Helmer & Meyer, 1956 ▸; Aisaka et al., 1991 ▸). On the basis of the reaction mechanism, aldolases can be subdivided into classes I and II. NanA belongs to the class I aldolase family, which is characterized by a triosephosphate isomerase (TIM)-barrel fold and by aldol condensation that proceeds through the formation of a Schiff base intermediate between a conserved lysine residue and the substrate pyruvate (Campeotto et al., 2009 ▸). In contrast, class II aldolases differ in the reaction mechanism, in which intermediates are stabilized by a metal cofactor (for example Zn2+; Plater et al., 1999 ▸). Multiple class I aldolase structures have been reported, and all of the structures share the same TIM-barrel fold (Wymer et al., 2001 ▸; Theodossis et al., 2004 ▸; Pauluhn et al., 2008 ▸). Various reports have demonstrated that the enzyme is a tetramer in solution (Huynh et al., 2013 ▸; Aisaka et al., 1991 ▸; Krüger et al., 2001 ▸; Barbosa et al., 2000 ▸). Our study enriches the understanding of N-acetyl-d-neuraminic acid lyase in members of the oral microbiome such as Fusobacterium nucleatum. F. nucleatum is a Gram-negative, nonmotile, anaerobic, spindle-shaped and non-spore-forming bacterium (Kolenbrander et al., 2006 ▸). It plays a vital role in the formation of dental plaque biofilms in humans. Here, we report 2.32 and 1.76 Å resolution structures of N-acetylneuraminate lyase from F. nucleatum in two forms: ligand-free and with ligand bound as a pyruvate Schiff base. The high-resolution structure of the enzyme is compared with those of other known structures of sialic acid aldolases. The atomic resolution structure will provide a pathway for the improvement of prospective antimicrobials. We also report the steady-state enzyme kinetics of this enzyme from F. nucleatum.
Figure 1.
Sialic acid catabolism. (a) SiaT, sialic acid transporter; NanA, N-acetylneuraminate lyase; NanK, N-acetylmannosamine kinase; NanE, N-acetylmannosamine-6-phosphate 2-epimerase; NagA, N-acetylglucosamine-6-phosphate deacetylase; NagB, glucosamine-6-phosphate deaminase. (b) The reaction catalysed by N-acetylneuraminate lyase.
2. Materials and methods
2.1. Production of F. nucleatum NanA
The gene encoding F. nucleatum NanA (FnNanA) was cloned into a pET300/NT-DEST vector as described previously (Bairy et al., 2018 ▸) and transformed into Escherichia coli BL21 (DE3) cells (Novagen) for protein expression (Table 1 ▸). The cell cultures were grown to an OD600 nm of 0.6 and induced with 100 µM isopropyl β-d-1-thiogalactopyranoside (IPTG). The cells were grown at 289 K for 16 h post-induction. The cultures were harvested by centrifugation at 6000g for 15 min at 277 K. The cell pellets were resuspended in lysis buffer [50 mM Tris–HCl pH 8.0, 500 mM NaCl, 20 mM imidazole and one tablet of cOmplete EDTA-free Protease Inhibitor Cocktail (Roche)]. The cells were lysed by passage through a cell disruptor (Constant Systems) three times at 138 MPa. The cell lysate was centrifuged at 18 000g for 35 min to remove cell debris and unlysed cells.
Table 1. F. nucleatum NanA production information.
| Source organism | F. nucleatum strain ATCC 25586 |
| DNA source | Synthetic DNA |
| Forward primer | CAAAAAAGCAGGCTTCATGAAAGGGATATATTCAG |
| Reverse primer | CAAGAAAGCTGGGTTTTAATTTTTTAAAAATTTTTTATG |
| Cloning vector | pMK vector |
| Expression vector | pET300/NT-DEST with an N-terminal His tag |
| Expression host | E. coli BL21 (DE3) |
| Sequence of the recombinant protein produced† | MHHHHHHITSLYKKAGFMKGIYSALMVPYNEDGSINEKGLREIIRYNIDKMKVDGLYVGGSTGENFMISTEEKKRVFEIAIDEAKDSVNLIAQVGSINLNEAVELGKYVTKLGYKCLSAVTPFYYKFDFSEIKDYYETIVRETGNYMIIYSIPFLTGVNMSLSQFGELFENEKIIGVKFTAGDFYLLERVRKAFPDKLIFAGFDEMLLPATVLGVDGAIGSTYNINGIRAKQIFELAKNSKIDEALKIQHTTNDLIEGILSNGLYQTIKEILKLEGVDAGYCRKPMKKISQKQIEFAKELHKKFLKN |
Non-native amino-acid residues originating from the vector are underlined.
The supernatant was loaded onto a 5 ml HisTrap FF column (GE Healthcare) equilibrated with buffer A (50 mM HEPES pH 7.4, 20 mM imidazole, 500 mM NaCl, 6% glycerol, 10 mM β-mercaptoethanol) using an ÄKTA FPLC system (GE Healthcare). Unbound bacterial proteins were eluted with a step gradient of 4 and 6% buffer B (50 mM HEPES pH 7.4, 500 mM imidazole, 500 mM NaCl, 6% glycerol, 10 mM β-mercaptoethanol). The desired protein was eluted using a linear gradient of 6–100% buffer B. The fractions were analyzed by SDS–PAGE to check their purity. Prior to cation-exchange chromatography, the protein was buffer-exchanged with 50 mM HEPES pH 6.8, 10 mM NaCl, 6% glycerol, 10 mM β-mercaptoethanol using Amicon Ultra Centrifugal Filters (10 000 molecular-weight cutoff; Millipore). The protein was loaded onto a cation exchanger (5 ml HiTrap SP FF column; GE Healthcare) equilibrated with the same buffer.
The protein was further purified on a Superdex 200 10/300 GL analytical column in 50 mM HEPES pH 7.4, 50 mM NaCl, 10 mM β-mercaptoethanol. The purity of FnNanA was analyzed by 12% SDS–PAGE. The peak fraction with the highest purity was pooled and then concentrated using Amicon Ultra Centrifugal Filters (10 000 molecular-weight cutoff; Millipore) for crystallization trials. All protein-purification steps were performed at 277 K. The protein concentration was determined using the Bradford assay (Bio-Rad) with bovine serum albumin as a protein standard. The absorbance of each sample was measured at 595 nm using an Ultrospec 2100 pro UV–visible spectrophotometer (GE Healthcare). The purified protein exhibited a molecular mass of ∼35 kDa using mass spectrometry, matching the calculated mass of the translated His6-tagged protein (34.9 kDa; data not shown).
2.2. Crystallization
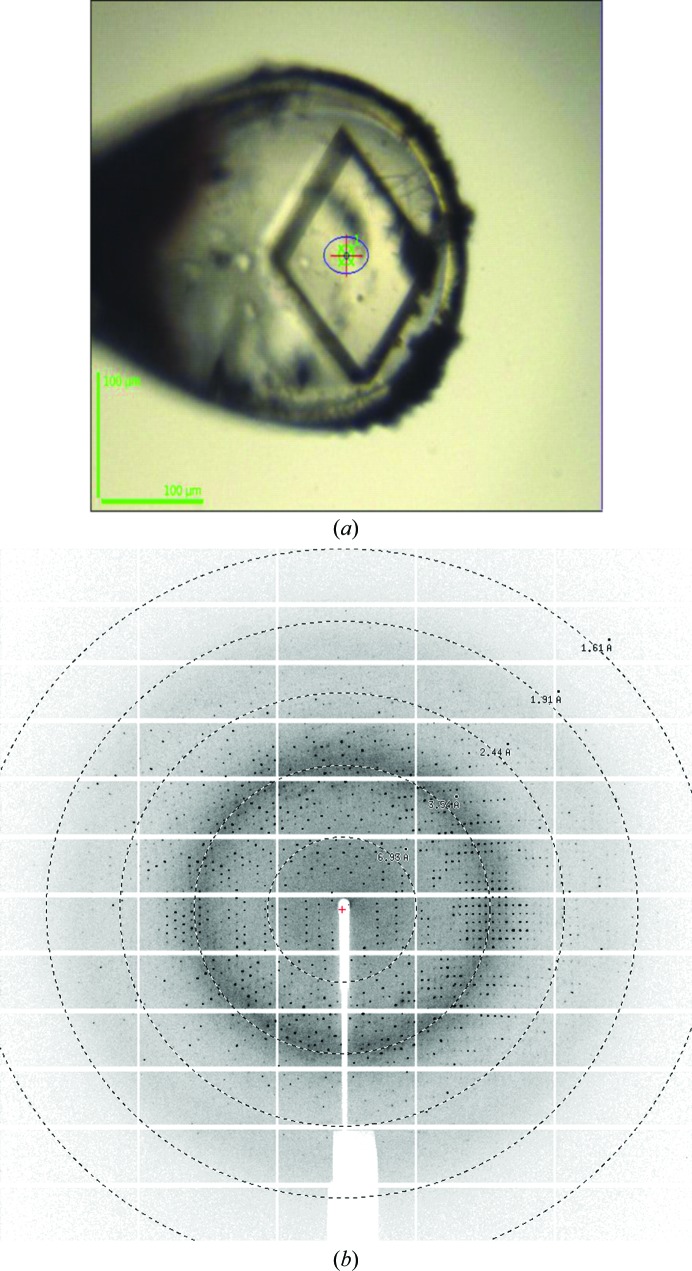
Purified FnNanA was concentrated to 10 mg ml−1 in 50 mM HEPES pH 7.4, 50 mM NaCl, 10 mM β-mercaptoethanol for crystallization setup. Initial hanging-drop crystallization trials were performed using commercially available sparse-matrix crystallization screens from Rigaku Reagents (Wizard 1 and 2), Hampton Research (PEG/Ion, PEG/Ion 2, Crystal Screen and Crystal Screen 2) and Qiagen (The Classics and Classics II Suites) using a Mosquito nanolitre-dispensing robot (TTP Labtech). The crystallization drops consisted of 350 nl protein solution and 350 nl reservoir solution. Several of the commercial screen conditions produced small crystals at 277 and 291 K. These initial hits were optimized to obtain diffraction-quality crystals. For the crystallization of FnNanA with sodium pyruvate, a tenfold molar excess of sodium pyruvate was mixed with 10 mg ml−1 (0.28 mM) protein and incubated for 30 min before crystallization setup. Crystals were obtained after 7–8 d (Fig. 2 ▸ a). All crystals were cryoprotected using 20%(v/v) ethylene glycol in reservoir solution. The crystals were flash-cooled in liquid nitrogen prior to diffraction data collection. Crystallization information is summarized in Table 2 ▸.
Figure 2.
Crystal and diffraction pattern. (a) A typical crystal of ligand-free FnNanA. (b) X-ray diffraction image of a ligand-free FnNanA crystal showing diffraction to 2.32 Å resolution.
Table 2. Crystallization of F. nucleatum NanA.
| Ligand-free FnNanA | Ligand-bound FnNanA with a pyruvate Schiff base | |
|---|---|---|
| Method | Vapour diffusion, hanging drop | Vapour diffusion, hanging drop |
| Plate type | 96-well | 96-well |
| Temperature (K) | 277 | 277 |
| Protein concentration (mg ml−1) | 10 | 10 |
| Buffer composition of protein solution | 50 mM HEPES pH 6.8, 50 mM NaCl, 10 mM β-mercaptoethanol | 50 mM HEPES pH 6.8, 50 mM NaCl, 10 mM β-mercaptoethanol |
| Buffer composition of reservoir solution | 0.1 M CHES pH 9.5, 10%(w/v) PEG 3000, 5 mM adenosine 5′-triphosphate disodium salt hydrate (ATP) | 0.1 M CHES pH 9.5, 10%(w/v) PEG 3000, 2.85 mM sodium pyruvate |
| Volume of drop (nl) | 700 | 700 |
| Volume of reservoir (µl) | 100 | 100 |
2.3. Data collection and processing
X-ray diffraction data were collected from a single crystal of ligand-free FnNanA on beamline ID23 at the European Synchrotron Radiation Facility (ESRF), Grenoble, France at a wavelength of 0.9789 Å. X-ray data for ligand-bound FnNanA with a pyruvate Schiff base were also collected on beamline ID30 at the ESRF at a wavelength of 0.9762 Å. Diffraction data collection was performed at 100 K.
The diffraction data were indexed and integrated using XDS (Kabsch, 2010 ▸). Data reduction and scaling were achieved with AIMLESS (Evans & Murshudov, 2013 ▸) from the CCP4 suite (Winn et al., 2011 ▸). The resulting intensity data were analyzed using phenix.xtriage from the PHENIX suite (Zwart et al., 2005 ▸; Adams et al., 2010 ▸). An estimate of the number of molecules in the asymmetric unit was obtained using MATTHEWS_COEF from the CCP4 suite (Matthews, 1968 ▸; Kantardjieff & Rupp, 2003 ▸). A summary of the data-collection and processing statistics is given in Table 3 ▸.
Table 3. Data collection and processing for F. nucleatum NanA.
Values in parentheses are for the outer shell.
| Ligand-free FnNanA | Ligand-bound FnNanA with a pyruvate Schiff base | |
|---|---|---|
| Diffraction source | Beamline ID23-1, ESRF | Beamline ID30B, ESRF |
| Wavelength (Å) | 0.9789 | 0.9762 |
| Temperature (K) | 100 | 100 |
| Detector | Dectris PILATUS 6M-F | Dectris PILATUS3 6M |
| Crystal-to-detector distance (mm) | 405.721 | 318.80 |
| Rotation range per image (°) | 0.1 | 0.05 |
| Total rotation range (°) | 180 | 230 |
| Exposure time per image (s) | 0.1 | 0.01 |
| Space group | C2 | P21212 |
| a, b, c (Å) | 105.17, 108.98, 141.14 | 82.68, 86.57, 89.99 |
| α, β, γ (°) | 90, 96.9, 90 | 90, 90, 90 |
| Mosaicity (°) | 0.14 | 0.13 |
| Resolution range (Å) | 49.24–2.32 (2.38–2.32) | 86.57–1.76 (1.79–1.76) |
| Total No. of reflections | 150647 (9709) | 213363 (12153) |
| No. of unique reflections | 64567 (4304) | 63236 (3548) |
| Completeness (%) | 94.900 (94.300) | 98.000 (98.200) |
| Multiplicity | 2.300 (2.300) | 3.400 (3.400) |
| 〈I/σ(I)〉 | 10.300 (1.7)† | 10.900 (1.6)‡ |
| R r.i.m. | 0.051 (0.600) | 0.064 (0.999) |
| R p.i.m. | 0.032 (0.371) | 0.034 (0.520) |
| Overall B factor from Wilson plot (Å2) | 59.610 | 29.190 |
We decided to cut the data at 〈I/σ(I)〉 = 2.32 Å as the CC1/2 and multiplicity of the data in the outer shell were 0.88 and 2.3, respectively. 〈I/σ(I)〉 falls below 2.0 in the outer shell at 2.38 Å resolution.
We decided to cut the data at 〈I/σ(I)〉 = 1.76 Å as the CC1/2 and multiplicity of data in the outer shell were 0.65 and 3.4, respectively. 〈I/σ(I)〉 falls below 2.0 in the outer shell at 1.82 Å resolution.
2.4. Structure solution and refinement
A monomer of Pasteurella multocida NanA (PDB entry 4imc; Huynh et al., 2013 ▸), which has 73% sequence identity to FnNanA, was used as a search model for molecular replacement with Phaser (McCoy et al., 2007 ▸) in the PHENIX suite for both ligand-free FnNanA and ligand-bound FnNanA with a pyruvate Schiff base. The phenix.autobuild program (Terwilliger et al., 2008 ▸) was used for initial model building. Structure refinement was performed using phenix.refine (Afonine et al., 2012 ▸). Iterative improvement of the map and the model was performed using alternate cycles of refinement and residue-by-residue analysis in Coot (Emsley et al., 2010 ▸). Water and ligand molecules were added via Coot and modelled into the electron density manually. All structure-refinement statistics are summarized in Table 4 ▸.
Table 4. Structure solution and refinement of F. nucleatum NanA.
Values in parentheses are for the outer shell.
| Ligand-free FnNanA | Ligand-bound FnNanA with a pyruvate Schiff base | |
|---|---|---|
| Resolution range (Å) | 47.10–2.32 (2.35–2.32) | 60.88–1.76 (1.80–1.76) |
| Completeness (%) | 94.5 | 97.7 |
| σ Cutoff | 1.3 | 1.3 |
| No. of reflections | ||
| Working set | 61188 (2602) | 61155 (4313) |
| Test set | 3195 (141) | 2028 (147) |
| Final R cryst | 0.205 (0.4388) | 0.187 (0.3258) |
| Final R free | 0.243 (0.4569) | 0.216 (0.3769) |
| No. of non-H atoms | ||
| Protein | 8895 | 4618 |
| Ligand | 12 | 30 |
| Solvent | 14 | 166 |
| Total | 8921 | 4814 |
| R.m.s. deviations | ||
| Bonds (Å) | 0.004 | 0.007 |
| Angles (°) | 1.01 | 1.12 |
| Average B factors (Å2) | ||
| Overall | 68.03 | 38.71 |
| Protein | 68.02 | 38.55 |
| Ligand | 82.75 | 60.89 |
| Ramachandran plot | ||
| Most favoured (%) | 97.57 | 97.23 |
| Allowed (%) | 2.08 | 2.6 |
| No. of TLS groups | 4 | 2 |
2.5. Sequence comparison based on the structural alignment
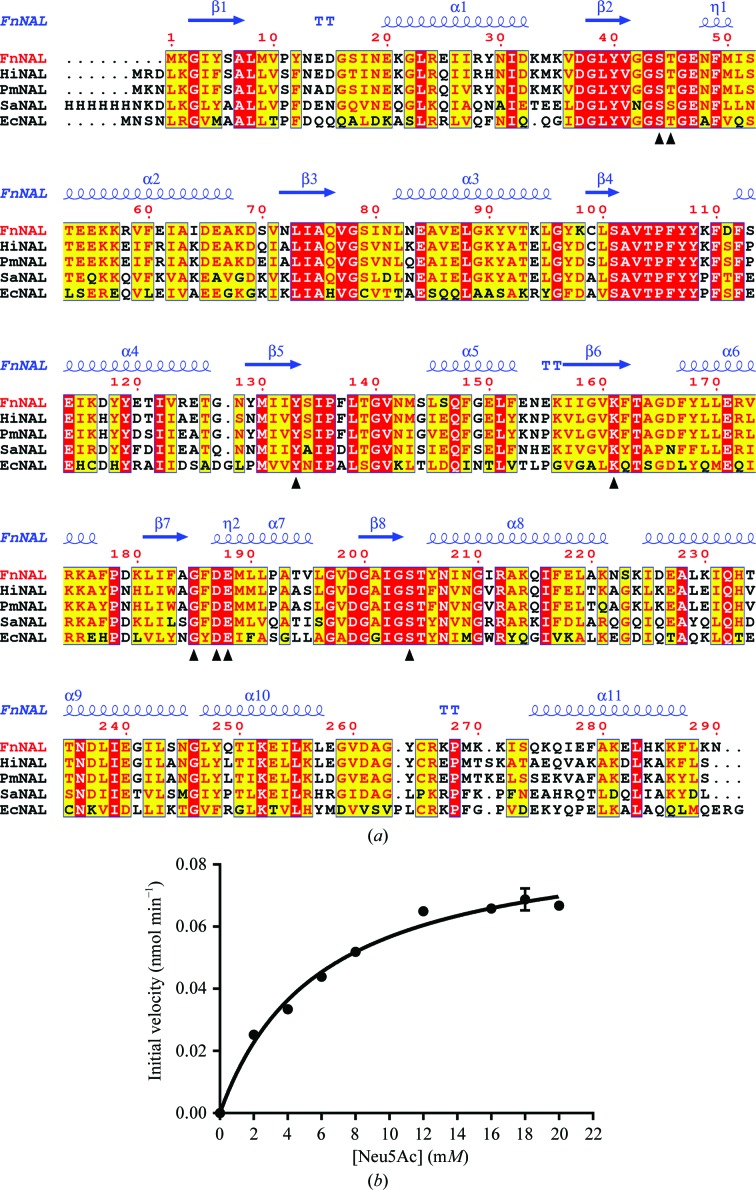
The T-Coffee Expresso server (Armougom et al., 2006 ▸) was used to produce a structure-based sequence alignment of the four known N-acetylneuraminate lyase structures, and a figure was generated with ESPript 3 (Robert & Gouet, 2014 ▸). The sequence identities of the N-acetylneuraminate lyases from Haemophilus influenzae (PDB entry 1f7b; Barbosa et al., 2000 ▸), P. multocida (PDB entry 4imd; Huynh et al., 2013 ▸), Staphylococcus aureus (PDB entry 5a8g; Stockwell et al., 2016 ▸) and E. coli (PDB entry 1nal; Izard et al., 1994 ▸) are 74, 72, 57 and 36%, respectively.
2.6. Enzyme assay
N-Acetylneuraminate lyase activity was assessed by quantifying the amount of pyruvate produced using a standard coupled assay (Zhu et al., 2010 ▸). The pyruvate released by the cleavage of N-acetylneuraminic acid is oxidized by pyruvate oxidase in the presence of phosphate (Pi) and oxygen to give acetylphosphate, CO2 and H2O2. Horseradish peroxide (HRP) was used to catalyze the reaction between the fluorometric probe and H2O2. The resulting H2O2 was then detected using a fluorogenic substrate (Sugahara et al., 1980 ▸).
3. Results and discussion
3.1. Structure determination
The structure of ligand-free FnNanA was refined to 2.32 Å resolution. The structure has 0.35% Ramachandran outliers. The conserved Ser79 residue is in the disallowed region of the Ramachandran plot in each monomer of ligand-free FnNanA. However, electron density is well defined for Ser79.
The crystals of ligand-bound FnNanA with a pyruvate Schiff base intermediate belonged to space group P21212 and diffracted to 1.76 Å resolution. The two monomers that were identified appear to form a tetramer with a similar monomer arrangement to those in sialic acid aldolases from other species. The structure has 0.17% Ramachandran outliers. One residue, Lys267 of chain B, is present in the disallowed region; the electron density for this residue is not well defined. All residues were modelled into electron density, except for Tyr108 and Lys109 of chain B in ligand-bound FnNanA with a pyruvate Schiff base and the residues at the N-termini of all monomers.
3.2. The overall structure of N-acetylneuraminate lyase
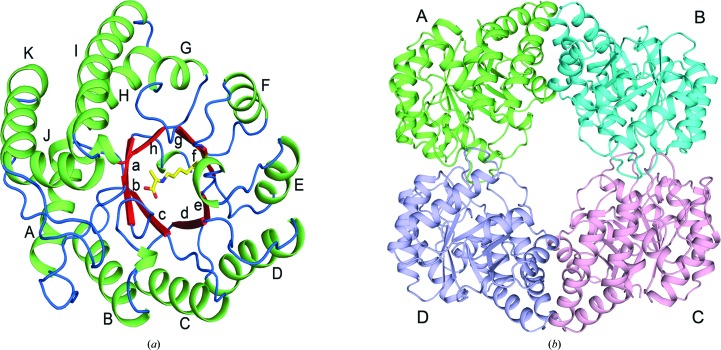
The monomeric structure of FnNanA shows that the enzyme adopts a classical TIM (β/α)8-barrel tertiary-structure fold (Fig. 3 ▸ a). In addition to the eight helices that make up the TIM barrel, FnNanA has three additional α-helices (I, J and K) at the C-terminus (Fig. 3 ▸ a). The NanAs from H. influenzae (Barbosa et al., 2000 ▸) and P. multocida (Huynh et al., 2013 ▸) also have these three additional helices. We compared the structures of ligand-free FnNanA and ligand-bound FnNanA with a pyruvate Schiff base. The overall architecture of the ligand-bound FnNanA with a pyruvate Schiff base is similar to that of ligand-free FnNanA. There are no major structural changes in FnNanA upon the binding of pyruvate. All four subunits in ligand-free FnNanA are structurally similar, and superpose with a root-mean-square deviation (r.m.s.d.) of between 0.127 and 0.169 Å for 290 Cα atoms. The monomer of FnNanA bound to pyruvate superposes with an r.m.s.d. 0.836 Å onto monomers from the same asymmetric unit.
Figure 3.
The overall crystal structure of N-acetylneuraminate lyase from F. nucleatum. (a) Cartoon representation of FnNanA with individually coloured secondary structures. Secondary-structure elements are labelled sequentially (with lower case letters corresponding to β-strands and upper case letters corresponding to α-helices). The Lys161–pyruvate Schiff base intermediate in the active site is displayed as sticks with yellow C atoms. (b) The tertiary structure (tetramer) of FnNanA with each subunit shown in cartoon presentation in a different colour. The figures were generated using the PyMOL molecular-graphics system (DeLano, 2002 ▸).
The Cα atoms of chain A of the ligand-free N-acetylneuraminate lyases from F. nucleatum, H. influenzae (PDB entry 1f5z; Barbosa et al., 2000 ▸) and P. multocida (PDB entry 4imc; Huynh et al., 2013 ▸) superpose with r.m.s.d.s of 1.12 and 1.14 Å, respectively. The Cα atoms of chain A of the ligand-bound N-acetylneuraminate with a pyruvate Schiff base from F. nucleatum, P. multocida (PDB entry 4imd; Huynh et al., 2013 ▸) and S. aureus (PDB entry 4ah7; Timms et al., 2013 ▸) superposed with r.m.s.d.s of 0.54 and 0.78 Å, respectively. The quaternary structure of the N-acetylneuraminate lyase (PDB entry 5zjm) was analyzed and assembled into a tetramer (Fig. 3 ▸ b) using the PISA server (Krissinel & Henrick, 2007 ▸). All four crystallographically independent monomers are very similar. Small-angle X-ray scattering data of the soluble protein also concurred that the protein was a tetramer (data not shown).
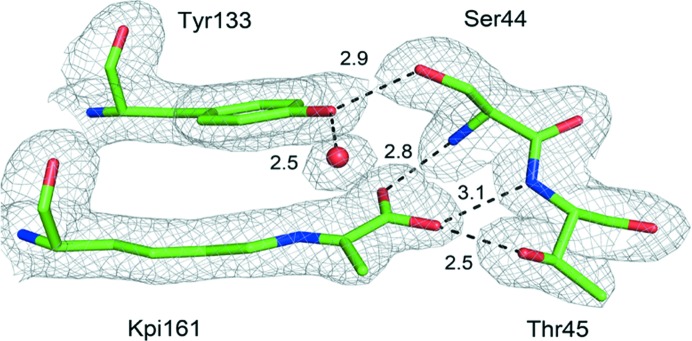
The observed electron-density map for ligand-bound FnNanA with a pyruvate Schiff base in chain A clearly showed that the N∊ atom of Lys161 is conjugated as a Schiff base to the C2 atom of pyruvate. The carboxylate group of the pyruvate makes hydrogen bonds to the backbone N atom of Ser44 and to the hydroxyl group of Thr45. In addition, the Tyr133 side chain makes a hydrogen bond to the hydroxyl group of Ser44 and a conserved water molecule (Fig. 4 ▸). Tyr133 lies almost parallel to the Schiff base intermediate and is engaged in hydrogen bonding for the Schiff base reaction. The atomic coordinates have been submitted to the Protein Data Bank (PDB) and the PDB codes for ligand-free FnNanA and ligand-bound FnNanA with a pyruvate Schiff base are 5zjm and 5zka, respectively.
Figure 4.
Electron-density map of the Schiff base formed by Lys161 with pyruvate. Complex showing the interaction of the pyruvate enamine with the enzyme. The pyruvate is covalently linked to Lys161 and makes hydrogen bonds to Ser44 and Thr45. The hydrogen-bonding network to Tyr133 and a conserved water molecule are also shown. The electron density represented by the grey mesh was contoured at the 1σ level, showing continuous density linking the C2 atom of pyruvate and the N∊ atom of Lys161. This figure was generated using the PyMOL molecular-graphics system (DeLano, 2002 ▸).
3.3. Sequence comparison based on structural alignment
Structure-based sequence alignment of FnNanA and four known N-acetylneuraminate lyase structures was carried out using the T-Coffee Expresso server (Fig. 5 ▸ a; Armougom et al., 2006 ▸). Sequence alignment showed that FnNanA contains conserved catalytic sites (Lys161 and Tyr133) and the conserved specific substrate (Neu5Ac) binding motif GXXGE (Barbosa et al., 2000 ▸; Ji et al., 2015 ▸; García García et al., 2012 ▸). The GXXGE motif, which is situated between positions 43 and 47 (FnNanA numbering), is involved in substrate recognition, and the XX residues are usually S and/or T. Apart from this, a group of amino acids (Asp187, Glu188 and Ser204) are also involved in binding the carbohydrate moiety.
Figure 5.
Structure-based alignment of F. nucleatum N-acetylneuraminate lyase (FnNAL) and selected N-acetylneuraminate lyases (NALs) of known structure. (a) The sequence identities to the N-acetylneuraminate lyases from H. influenzae (HiNAL; PDB entry 1f7b; Barbosa et al., 2000 ▸), P. multocida (PmNAL; PDB entry 4imd; Huynh et al., 2013 ▸), S. aureus (SaNAL; PDB entry 5a8g; Stockwell et al., 2016 ▸) and E. coli (EcNAL; PDB entry 1nal; Izard et al., 1994 ▸) are 74, 72, 57 and 36%, respectively. The structural alignment was computed using the T-Coffee Expresso server (Armougom et al., 2006 ▸) and the figure was produced with ESPript 3 (Robert & Gouet, 2014 ▸). Strictly conserved residues across NAL enzymes are shown on a red background. The secondary structure of FnNanA is shown at the top; coils represent helices and arrows represent β-strands. The residues involved in the active site are labelled with small black triangles. (b) Kinetic analysis of F. nucleatum N-acetylneuraminate lyase with Neu5Ac. The data were fitted to the Michaelis–Menten equation with an R 2 value of 0.99.
3.4. Enzyme assay
The data were fitted to the Michaelis–Menten kinetic model (Fig. 5 ▸ b). The Michaelis–Menten constant (K m) was found to be 6.0 ± 0.6 mM, V max was calculated to be 0.090 ± 0.003 nmol min−1 and k cat was determined to be 12.5 ± 0.5 s−1. A comparison with the Michaelis–Menten kinetic constants of related NanA enzymes reveals that the kinetic characteristics of this enzyme (both k cat and K m) are quite similar to those reported in the literature (Uchida et al., 1984 ▸; North et al., 2016 ▸; Table 5 ▸). We report the kinetic analysis of N-acetylneuraminate lyase from F. nucleatum.
Table 5. Kinetic parameters of FnNanA and previously characterized N-acetylneuraminate lyases from E. coli (EcNanA), P. multocida (PmNanA) and methicillin-resistant S. aureus (MRSA NanA).
4. Conclusion
In this paper, we present the crystal structures of ligand-free FnNanA and ligand-bound FnNanA with a pyruvate Schiff base. Analysis and comparison of the structures show a conserved TIM-barrel fold. Enzyme kinetics were determined and were observed to be similar to those of orthologs from other species. Atomic resolution structures of N-acetylneuraminate lyases from pathogenic bacteria such F. nucleatum will provide us with the structural information necessary for future antimicrobial development.
Supplementary Material
PDB reference: N-acetylneuraminate lyase from F. nucleatum, 5zjm
PDB reference: pyruvate Schiff-base intermediate, 5zka
Acknowledgments
We would like to acknowledge beamlines ID23 and ID30 at the European Synchrotron Radiation Source, Grenoble, France for X-ray data collection.
Funding Statement
This work was funded by Department of Biotechnology, Government of India grants BT/IN/Sweden/41/SR/2013, BT/PR12422/MED/31/287/214, BT/INF/22/SP22660/2017, BT/PR5081/INF/156/2012, and BBT/IN/Sweden/06/SR/2017-18.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Afonine, P. V., Grosse-Kunstleve, R. W., Echols, N., Headd, J. J., Moriarty, N. W., Mustyakimov, M., Terwilliger, T. C., Urzhumtsev, A., Zwart, P. H. & Adams, P. D. (2012). Acta Cryst. D68, 352–367. [DOI] [PMC free article] [PubMed]
- Aisaka, K., Igarashi, A., Yamaguchi, K. & Uwajima, T. (1991). Biochem. J. 276, 541–546. [DOI] [PMC free article] [PubMed]
- Armougom, F., Moretti, S., Poirot, O., Audic, S., Dumas, P., Schaeli, B., Keduas, V. & Notredame, C. (2006). Nucleic Acids Res. 34, W604–W608. [DOI] [PMC free article] [PubMed]
- Bairy, S. et al. (2018). Microb. Biotechnol. 11, 420–428. [DOI] [PMC free article] [PubMed]
- Barbosa, J. A. R. G., Smith, B. J., DeGori, R., Ooi, H. C., Marcuccio, S. M., Campi, E. M., Jackson, W. R., Brossmer, R., Sommer, M. & Lawrence, M. C. (2000). J. Mol. Biol. 303, 405–421. [DOI] [PubMed]
- Bouchet, V., Hood, D. W., Li, J., Brisson, J.-R., Randle, G. A., Martin, A., Li, Z., Goldstein, R., Schweda, E. K. H., Pelton, S. I., Richards, J. C. & Moxon, E. R. (2003). Proc. Natl Acad. Sci. USA, 100, 8898–8903. [DOI] [PMC free article] [PubMed]
- Campeotto, I., Carr, S. B., Trinh, C. H., Nelson, A. S., Berry, A., Phillips, S. E. V. & Pearson, A. R. (2009). Acta Cryst. F65, 1088–1090. [DOI] [PMC free article] [PubMed]
- DeLano, W. L. (2002). PyMOL. http://www.pymol.org.
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Evans, P. R. & Murshudov, G. N. (2013). Acta Cryst. D69, 1204–1214. [DOI] [PMC free article] [PubMed]
- García García, M. I., Sola Carvajal, A., García Carmona, F. & Sánchez Ferrer, Á. (2012). J. Agric. Food Chem. 60, 7450–7456. [DOI] [PubMed]
- Helmer, R. & Meyer, K. (1956). Proc. Natl Acad. Sci. USA, 42, 728–734.
- Huynh, N., Aye, A., Li, Y., Yu, H., Cao, H., Tiwari, V. K., Shin, D.-W., Chen, X. & Fisher, A. J. (2013). Biochemistry, 52, 8570–8579. [DOI] [PMC free article] [PubMed]
- Izard, T., Lawrence, M. C., Malby, R. L., Lilley, G. G. & Colman, P. M. (1994). Structure, 2, 361–369. [DOI] [PubMed]
- Ji, W., Sun, W., Feng, J., Song, T., Zhang, D., Ouyang, P., Gu, Z. & Xie, J. (2015). Sci. Rep. 23, 9341. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 133–144. [DOI] [PMC free article] [PubMed]
- Kantardjieff, K. A. & Rupp, B. (2003). Protein Sci. 12, 1865–1871. [DOI] [PMC free article] [PubMed]
- Kolenbrander, P. E., Palmer, R. J., Rickard, A. H., Jakubovics, N. S., Chalmers, N. I. & Diaz, P. I. (2006). Periodontol. 2000, 42, 47–79. [DOI] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Krüger, D., Schauer, R. & Traving, C. (2001). Eur. J. Biochem. 268, 3831–3839. [DOI] [PubMed]
- Martinez, J., Steenbergen, S. & Vimr, E. (1995). J. Bacteriol. 177, 6005–6010. [DOI] [PMC free article] [PubMed]
- Maru, I., Ohnishi, J., Ohta, Y. & Tsukada, Y. (2002). J. Biosci. Bioeng. 93, 258–265. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- North, R. A., Watson, A. J. A., Pearce, F. G., Muscroft-Taylor, A. C., Friemann, R., Fairbanks, A. J. & Dobson, R. C. J. (2016). FEBS Lett. 590, 4414–4428. [DOI] [PubMed]
- Pauluhn, A., Ahmed, H., Lorentzen, E., Buchinger, S., Schomburg, D., Siebers, B. & Pohl, E. (2008). Proteins, 72, 35–43. [DOI] [PubMed]
- Plater, A. R., Zgiby, S. M., Thomson, G. J., Qamar, S., Wharton, C. W. & Berry, A. (1999). J. Mol. Biol. 285, 843–855. [DOI] [PubMed]
- Plumbridge, J. & Vimr, E. (1999). J. Bacteriol. 181, 47–54. [DOI] [PMC free article] [PubMed]
- Ringenberg, M. A., Steenbergen, S. M. & Vimr, E. R. (2003). Mol. Microbiol. 50, 961–975. [DOI] [PubMed]
- Robert, X. & Gouet, P. (2014). Nucleic Acids Res. 42, W320–W324. [DOI] [PMC free article] [PubMed]
- Severi, E., Hood, D. W. & Thomas, G. H. (2007). Microbiology, 153, 2817–2822. [DOI] [PubMed]
- Stockwell, J., Daniels, A. D., Windle, C. L., Harman, T. A., Woodhall, T., Lebl, T., Trinh, C. H., Mulholland, K., Pearson, A. R., Berry, A. & Nelson, A. (2016). Org. Biomol. Chem. 14, 105–112. [DOI] [PMC free article] [PubMed]
- Sugahara, K., Sugimoto, K., Nomura, O. & Usui, T. (1980). Clin. Chim. Acta, 108, 493–498. [DOI] [PubMed]
- Terwilliger, T. C., Grosse-Kunstleve, R. W., Afonine, P. V., Moriarty, N. W., Zwart, P. H., Hung, L.-W., Read, R. J. & Adams, P. D. (2008). Acta Cryst. D64, 61–69. [DOI] [PMC free article] [PubMed]
- Theodossis, A., Walden, H., Westwick, E. J., Connaris, H., Lamble, H. J., Hough, D. W., Danson, M. J. & Taylor, G. L. (2004). J. Biol. Chem. 279, 43886–43892. [DOI] [PubMed]
- Timms, N., Windle, C. L., Polyakova, A., Ault, J. R., Trinh, C. H., Pearson, A. R., Nelson, A. & Berry, A. (2013). Chembiochem, 14, 474–481. [DOI] [PMC free article] [PubMed]
- Uchida, Y., Tsukada, Y. & Sugimori, T. (1984). J. Biochem. 96, 507–522. [DOI] [PubMed]
- Varki, A. (1992). Glycobiology, 2, 25–40. [DOI] [PMC free article] [PubMed]
- Varki, A. & Schauer, R. (2009). Essentials of Glycobiology, 2nd ed., edited by A. Varki, R. D. Cummings, J. D. Esko, H. H. Freeze, P. Stanley, C. R. Bertozzi, G. W. Hart & M. E. Etzler, pp. 199–218. New York: Cold Spring Harbor Laboratory Press. [PubMed]
- Vimr, E. R., Kalivoda, K. A., Deszo, E. L. & Steenbergen, S. M. (2004). Microbiol. Mol. Biol. Rev. 68, 132–153. [DOI] [PMC free article] [PubMed]
- Vimrt, E. R. & Troy, F. A. (1985). J. Bacteriol. 164, 845–853. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Wymer, N., Buchanan, L. V., Henderson, D., Mehta, N., Botting, C. H., Pocivavsek, L., Fierke, C. A., Toone, E. J. & Naismith, J. H. (2001). Structure, 9, 1–9. [DOI] [PubMed]
- Zhu, A., Romero, R. & Petty, H. R. (2010). Anal. Biochem. 396, 146–151. [DOI] [PMC free article] [PubMed]
- Zwart, P. H., Grosse-Kunstleve, R. W. & Adams, P. D. (2005). CCP4 Newsl. Protein Crystallogr. 43, 27–35.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: N-acetylneuraminate lyase from F. nucleatum, 5zjm
PDB reference: pyruvate Schiff-base intermediate, 5zka