Abstract
Lipid metabolism reprogramming emerges as a new hallmark of malignancies. Sterol regulatory element-binding proteins (SREBPs), which are central players in lipid metabolism, are endoplasmic reticulum (ER)-bound transcription factors that control the expression of genes important for lipid synthesis and uptake. Their transcriptional activation requires binding to SREBP cleavage-activating protein (SCAP) to translocate their inactive precursors from the ER to the Golgi to undergo cleavage and subsequent nucleus translocation of their NH2-terminal forms. Recent studies have revealed that SREBPs are markedly upregulated in human cancers, providing the mechanistic link between lipid metabolism alterations and malignancies. Pharmacological or genetic inhibition of SCAP or SREBPs significantly suppresses tumor growth in various cancer models, demonstrating that SCAP/SREBPs could serve as promising metabolic targets for cancer therapy. In this review, we will summarize recent progress in our understanding of the underlying molecular mechanisms regulating SCAP/SREBPs and lipid metabolism in malignancies, discuss new findings about SREBP trafficking, which requires SCAP N-glycosylation, and introduce a newly identified microRNA-29-mediated negative feedback regulation of the SCAP/SREBP pathway. Moreover, we will review recently developed inhibitors targeting the SCAP/SREBP pathway for cancer treatment.
Keywords: SCAP, SREBPs, Lipid metabolism, EGFR, miRNA-29, Metabolic targets
1. INTRODUCTION
Metabolic reprogramming is a new hallmark of cancer [1]. Increasing evidence has recently shown that alterations in lipid metabolism are often present in cancer cells and promote tumor growth. Lipids form the basic structures for the plasma membrane and for membranes of all cellular organelles. In addition, lipids serve as energy resources and function as important signaling molecules, regulating various cellular functions, such as migration, cell cycle, cell division and differentiation [2–7]. Markedly increased lipid requirement is associated with the rapid growth and proliferation of tumor cells [8–12]. Lipid synthesis and uptake are highly elevated in various cancers, representing a novel characteristic of human cancers [10–14]. SREBPs, the master transcription factors in lipid synthesis and uptake pathways, have been demonstrated to be highly upregulated in a variety of cancers and may be promising molecular targets for cancer therapy [9, 11–13, 15].
2. DISCOVERY AND PROPERTIES OF THE SREBP FAMILY
SREBPs are transcription factors of the basic helix-loop-helix-leucine zipper (bHLH-Zip) class [12]. They were discovered by Brown & Goldstein’s laboratory in the 1990s when they delineated the upstream regulators of low-density lipoprotein receptor (LDLR) and 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGCR) [16–20]. LDLR regulates cholesterol uptake by mediating the endocytosis of cholesterol-rich LDL [21], and HMGCR is the rate-limiting enzyme in the cholesterol biosynthesis pathway [22]. Both LDLR and HMGCR are transcriptionally regulated by a sterol-dependent mechanism through sterol regulatory-elements (SREs) located in their promoter regions [2, 23]. The NH2-terminal forms of SREBPs were purified from nuclear extracts of Hela cells and rat liver using double-stranded DNA fragments containing an SRE sequence (5ʹ-ATCACCCCAC-3ʹ), demonstrating that SREBPs bind to the SRE DNA fragments of LDLR and HMGCR to transcriptionally regulate their expression [16, 18]. Thereafter, Brown & Goldstein’s group further uncovered feedforward and feedback molecular mechanisms that regulate SREBP transcription, translation and activation processes [2, 23]. Moreover, they identified major downstream targets of SREBPs that are involved in lipid metabolism [24].
Three SREBP isoforms are expressed in mammalian cells, SREBP-1a, SREBP-1c and SREBP-2, which are transcribed by two genes, SREBF1 and SREBF2 [2, 17, 19]. SREBP-1a and −1c are encoded via alternative transcription start sites (TSS) by a single gene, SREBF1, resulting in two isoforms with a different exon 1, and different lengths as SREBP-1a has a 24 amino acids (aa)-longer NH2-teminus than SREBP-1c [25]. SREBP-1c mainly regulates the expression of genes required for fatty acid synthesis [26], while SREBP-1a is able to regulate fatty acid and cholesterol synthesis, and cholesterol uptake [27]. SREBP-1c is the predominant isoform expressed in most tissues, whereas SREBP-1a is highly expressed in specific tissues and cells, such as intestinal epithelial, heart, macrophage and bone marrow dendritic cells [28]. SREBP-2, encoded by the SREBF2 gene, is relatively specific to the regulation of cholesterol synthesis and uptake [17, 29]. SREBP-1c and SREBP-2 are the predominant isoforms in liver and most other tissues, and SREBP-1c is upregulated by insulin stimulation [24].
SREBPs contain three functional domains. Their NH2-terminal domain contains the bHLH-Zip motif, and an acidic transcriptional motif that binds co-activator specificity protein 1 (SP1) or nuclear transcription factor Y (NF-Y) to regulate gene expression [30]. The bHLH-Zip motif is involved in DNA binding and dimerization of the mature SREBP transcription factors. The acidic domain is essential for SREBP transcriptional activity, as its removal markedly reduces the transcriptional activity of SREBPs, although their bHLH-Zip motif can still bind to DNA [31]. The central portion of SREBPs, which is the membrane-binding region, consists of two hydrophobic, membrane-spanning segments separated by a hydrophilic loop that extends into the lumen of the ER. The COOH-terminal segments of SREBPs contain ~590 aa and function as regulatory domains for SREBP subcellular localization and translocation [32].
3. SREBP MATURATION AND REGULATION
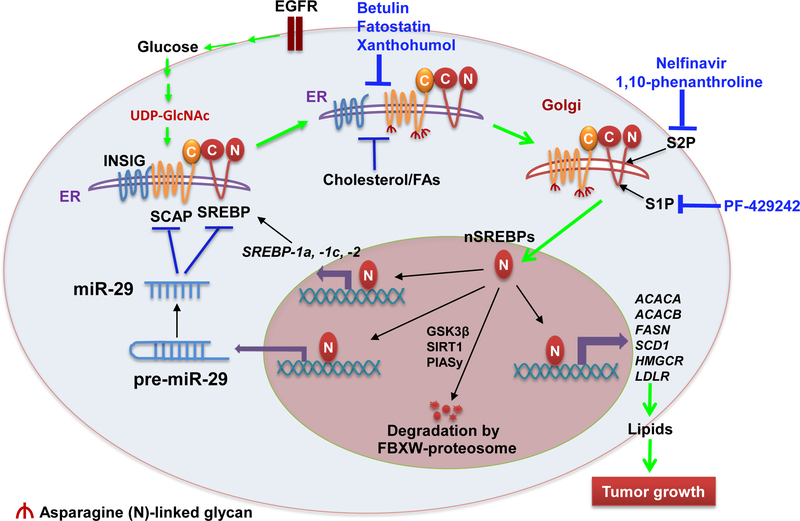
SREBPs are synthetized as 125 KDa inactive precursors, which bind to SREBP cleavage-activation protein (SCAP) after synthesis through their COOH-terminal domains and stay in the ER (Figure 1) [34]. The transcriptional activation of SREBPs requires that the SCAP/SREBP complex translocates from the ER to the Golgi for subsequent cleavage and release of the NH2-terminal transcription factor forms [33]. SCAP is a polytopic membrane-binding protein with 8 transmembrane helices. The long COOH-terminal extension of SCAP includes multiple copies of a WD-repeat sequence known to promote protein–protein interactions [32]. The NH2-terminal domain of SCAP binds to the ER-resident insulin-induced gene proteins (INSIGs), including INSIG1 and INSIG2, forming an INSIG/SCAP/SREBP complex that retains SREBPs in the ER (Figure 1) [35, 36]. INSIG1 is transcriptionally regulated by SREBPs and is abundant in cells [23, 24]. In contrast, INSIG2 is ubiquitously expressed at a low level in a variety of cells, suggesting that it serves as the regulator of SCAP/SREBP pathway at the basal level [35].
Figure 1.
Regulation of SCAP/SREBP activation in Cancer Cells.
In cancer cells, oncogenic EGFR signaling increases glucose uptake and enhances the synthesis of UDPGlcNAc, the end-product of the hexosamine synthesis pathway, promoting the N-glycosylation of SCAP, which enables SCAP dissociation from INSIG and leads to SCAP/SREBP trafficking from the ER to the Golgi. In the Golgi, SREBPs are sequentially cleaved by S1P and S2P proteases to release their NH2-terminal forms, which enter into the nucleus to activate the expression of key lipogenic genes, including themselves, forming a feedforward loop to activate lipid metabolism. Moreover, the newly synthesized INSIG1, cholesterol and unsaturated fatty acids mediated by SREBPs enhance the binding of INSIG and SCAP to retain SCAP/SREBP complex in the ER, forming a negative feedback loop to regulate SREBP activation. In addition, the nuclear SREBP forms are degraded by ubiquitin E3 ligase FBXW-mediated proteasome system, a process regulated by phosphorylation, acetylation and sumoylation by GSK3β, SIRT1 and PIAsy, respectively. Recently, miR-29 was found to be transcriptionally upregulated by SREBP-1, and in turn to inhibit SCAP and SREBP expression, mediating an additional negative feedback loop controlling this signaling pathway. Various inhibitors shown in blue, which inhibit SREBP translocation or maturation, have been tested in cancer cells and have shown promising anti-tumor effects. Abbreviation: ACACA, acetyl-coA carboxylase alpha; ACACB, acetyl-coA carboxylase beta; EGFR, epidermal growth factor receptor; ER, endoplasmic reticulum; FAs, fatty acids; FASN, fatty acid synthase; FBXW, F-box and WD repeat domain containing; GSK3β, glycogen synthase kinase-3 beta; HMGCR, hydroxymethylglutaryl-CoA reductase; INSIG, insulin-induced gene proteins; LDLR, low density lipoprotein receptor; PIASy, STAT Y; S1P, site-1 protease; S2P, site-2 protease; SCAP, SREBP cleavage-activating protein; SIRT1, sirtuin 1; SREBP, Sterol regulatory element-binding proteins; nSREBPs, nuclear forms of SREBPs.
Brown & Goldstein revealed that SCAP/SREBP trafficking and activation is regulated by a sterol-mediated negative feedback loop (Figure 1) [2, 23]. Cholesterol or oxysterol such as 25-hydroxycholesterol can bind to SCAP or INSIGs to strengthen their association, thereby preventing SREBPs to exit from the ER [37]. When sterol level decreases, SCAP will dissociate from INSIGs and facilitate the incorporation of SCAP/SREBP into coatomer II (COPII)-coated vesicles, which then transport the complex from the ER to the Golgi. In the Golgi, SREBPs will be sequentially cleaved by two membrane-bound proteases, site-1 protease (S1P) [38] and site-2 protease (S2P) [39], releasing the transcriptionally active NH2-terminal domains that can enter into the nucleus to activate the transcription of target genes [2, 23]. Meanwhile, INSIGs are recognized by ubiquitin ligase E3, TRC8 or GP78, and rapidly degraded after dissociation from SCAP [37, 40]. Interestingly, activation of SREBPs will promote the expression of INSIG1 and lipogenic genes, to restore INSIG1 protein and cholesterol levels, leading to the re-formation of the INSIG/SCAP/SREBP complex, limiting SREBP translocation [41]. A recent study showed that both SREBPs and SCAP associate with heat shock protein 90 (HSP90) after synthesis, which maintains their stability and interaction, whereas HSP90 inhibition results in the proteasome-dependent degradation of both SREBPs and SCAP [42]. In addition, Lee et al. have shown that ubiquitin regulatory X domain-containing protein 8 (UBXD8) binds to INSIG1 and promotes its degradation. In turn, the binding of UBXD8 to INSIG1 can be blocked by unsaturated fatty acids, thereby stabilizing INSIG1 and inhibiting SREBP-1 activation [43].
In addition to the tight regulation of the translocation process, SREBPs are also transcriptionally regulated by various transcription factors, including the mature NH2-terminal domains of SREBP-1 and SREBP-2, forming a feedforward loop to enhance their own expression (Figure 1) [44]. Multiple SRE motifs are present in the promoters of the SREBF1 and SREBF2 genes [45]. Moreover, NF-κB transcriptionally regulates SREBP-1a expression [46], and liver X receptor (LXR) transcriptionally activates SREBP-1c expression [47], which plays an important role in insulin-stimulated SREBP-1c expression [48]. SREBP-1c transcription and maturation could be inhibited by unsaturated fatty acids, particularly by polyunsaturated fatty acids [49]. SREBP-2 transcription could be regulated by thyroid hormone [50]. Nevertheless, the transcriptional regulation of SREBPs, particularly in cancer cells, is not fully understood, requiring further investigation.
In addition, the stability of the nuclear forms of SREBPs (nSREBPs) is regulated by various post-translational modifications, i.e., phosphorylation, acetylation and sumoylation (Figure 1). Phosphorylation of nSREBPs by glycogen synthase kinase-3 beta (GSK3β) results in their degradation mediated by the ubiquitin ligase E3 enzyme, FBXW7 [51]. Furthermore, activation of AMP-activated protein kinase (AMPK), which acts as an energy sensor, could enhance nSREBP degradation via phosphorylation [52]. In contrast, acetylation by CREB-binding protein (CBP)/p300 acetyltransferase stabilizes nSREBPs [53–55], while nSREBPs are destabilized by sirtuin 1 (SIRT1), which removes their acetylation modification [56]. In addition, sumoylation mediated by protein inhibitor of activated STAT Y (PIASy) enhances the degradation of nSREBP-1 [57]. In summary, the stabilization of nuclear SREBP forms is tightly regulated by multiple signals.
4. ACTIVATION OF SCAP/SREBPS IN CANCER
Rapidly proliferating tumor cells consume large amounts of energy and building blocks [58]. These high demands by cancer cells are met by the reprogramming of their metabolic processes by activated oncogenic signaling pathways [59]. Lipids, functioning as essential structural components of membranes and serving as important energy resources, are critical macromolecules for tumor growth. Recent studies have demonstrated that both lipid synthesis and uptake are significantly elevated in malignancies to support tumor growth [10–14]. SREBP-1 is highly expressed in glioblastoma (GBM) [60, 61], the most deadly brain tumor [62], and in prostate [63], endometrial [64], breast cancers [65, 66], hepatocellular carcinoma (HCC) [67], ovarian cancer [68], and pancreatic cancer (Table 1) [69]. The function of SREBP-1 has been investigated in multiple cancer cell lines including colon [70], lung [71] and pancreatic cancer cell lines [72]. SREBP-2 has been shown to be upregulated in prostate cancer patient tumor tissues (Table 1) [73], and elevated by ERBB4 signaling in breast cancer cells [74]. Moreover, SREBP-2 is also activated by Akt in Chinese hamster ovary-7 (CHO-7) and CHO cells [75].
Table 1:
Overexpression of SCAP/SREBPs in human cancers and testing their role in cancer animal models
Our previous studies demonstrated that SREBP-1 is highly upregulated by oncogenic EGFR signaling in GBM [13, 60, 61, 76–81]. We found that GBM tumors bearing amplified EGFR, or expressing EGFRvIII, the constitutively active form of the receptor that lacks a portion of the extracellular ligand-binding domain, were greatly dependent on SREBP-1-mediated lipid synthesis and uptake for rapid growth [61]. EGFR/EGFRvIII activates SREBP-1 through upregulation of PI3K/Akt signaling, promoting the expression of ATP citrate lyase (ACLY), acetyl-CoA carboxylase (ACC), fatty acid synthase (FASN), stearoyl-coenzyme A desaturase 1 (SCD1) and low density lipoprotein receptor (LDLR) to enhance fatty acid synthesis and cholesterol uptake [13, 60, 61, 80, 83].
We recently reported that glucose-mediated N-glycosylation of SCAP is essential for SCAP stability and dissociation from INSIG1, and mediates the activation of SREBP-1 by EGFR signaling (Figure 1) [77]. Genetically silencing SCAP expression or impairing its glycosylation via mutation significantly inhibited tumor growth and prolonged the survival of GBM-bearing mice. In contrast, overexpressing SCAP in GBM cells markedly enhanced tumor growth in mouse flank and brain (Table 1) [77]. This study demonstrated that glucose acts as an essential activator for SCAP/SREBP trafficking [77], while cholesterol functions as a key inhibitor of this process (Figure 1) [77, 82].
Oncogenic mutant of PI3K (H1047R) or K-Ras (G12V) has been shown to upregulate SREBP-1 and enhance lipogenesis through activation of mammalian target of rapamycin complex 1 (mTORC1) in breast epithelial cells [84]. Elevated mTORC1 signaling is correlated with increased mRNA and protein levels of SREBP-1 and of its downstream targets [85, 86]. In addition, loss of the tumor suppressor retinoblastoma protein (RB) promotes the transcription of SREBP-1 and SREBP-2 in an E2F-dependent manner [87]. In summary, increasing evidence demonstrates that SREBPs function as the central transition hubs that mediate signals from oncogenic pathways to the activation of lipid synthesis and uptake, promoting rapid tumor growth. Therefore, SREBPs have the potential to be efficient molecular targets for cancer therapy [13].
5. REGULATION OF SCAP/SREBPS BY MICRO-RNA
MicroRNAs (miRNAs) are small non-coding RNAs containing about 23 nucleotides. They regulate gene expression by binding to the 3’-untranslated regions (3’-UTR) of mRNAs, leading to the degradation or translation repression of the targeted mRNAs [88–90]. miRNAs have been identified and demonstrated to regulate nearly all cellular processes, i.e., cell cycle, apoptosis, autophagy, differentiation and metabolism [91–93]. The role of miRNAs in lipid metabolism has been extensively reviewed [92–96]. Here, we will focus on the role of several miRNAs, in the regulation of the SCAP/SREBP pathway.
We recently reported that miR-29 mediates a novel negative feedback loop that regulates the SCAP/SREBP-1 signaling pathway and lipid metabolism (Figure 1) [80–81]. The miRNA-29 family consists of three members, miR-29a, −29b and −29c, which share the same seed sequence [97, 98]. Analyzing a large cohort of tumor tissues from GBM patients with altered EGFR (amplification or mutation), we found that expression of all three mature miR-29s is positively correlated with SREBF1 gene expression [80, 81]. SREBP-1, via EGFR/PI3K/Akt signaling, transcriptionally upregulated the expression of all three miR-29s in GBM cells. In turn, they could all bind to the 3’-UTR of SREBF1 and SCAP mRNAs, inversely inhibiting their expression. Importantly, administration of miR-29 mimics inhibited SCAP/SREBP-1 and significantly reduced GBM tumor growth [80, 81], suggesting that miR-29s could be used to target GBM.
Both miR-185 and miR-342 have been shown to inhibit cell growth, migration and invasion in vitro and in vivo in prostate cancer cells by inhibiting SREBP-1 and −2 expression [99]. The expression of miR-185 and miR-342 is significantly downregulated in prostate cancer cells compared to non-cancerous epithelial cells [99]. Yang et al. reported that miR-185 controls cholesterol homeostasis through regulating SREBP-2 expression and activity [100].
Other miRNAs have been shown to affect tumor growth, such as miR-132, which suppresses cell growth, tumorigenesis, invasion and migration as well as promotes apoptosis of glioma cells by suppressing SREBP-1c that is related to SIRT1 [101]. Repression of INSIG1 by miR-24 promotes hepatic lipid accumulation and hyperlipidemia through activation of SREBPs [102]. Moreover, Osborne et al. recently identified a positive feedforward loop mediated by miR-96 and miR-182 to upregulate SREBPs in mouse liver. They demonstrated that SREBP-2 transcriptionally promotes the expression of miR-96 and miR-182, which induces the degradation of FBXW, an ubiquitin E3 ligase mediating the degradation of nuclear SREBP forms [103], thereby enhancing nuclear SREBPs and increasing lipid synthesis [104].
6. INHIBITING SCAP/SREBPS TO TREAT CANCER
Given the importance of SREBPs in the regulation of lipid metabolism and cancer growth, finding specific molecules that target SCAP/SREBPs to treat cancer has recently become a highly active field of research (Table 2) [13, 41].
Table 2:
The inhibitors targeting SCAP/SREBPs pathway in cancer cells
| Drug | Mechanism | Cancer type | In Vitro Cell line (IC50 μM) | In vivo xenograft mouse model (drug dose) | References |
|---|---|---|---|---|---|
| Fatostatin | Inhibition of ER-Golgi translocation of SCAP/SREBPs | ||||
| Pancreatic | MIA PaCa-2 (14.5) | None | [72] | ||
| Betulin | Inhibition of ER-Golgi translocation of SREBPs | ||||
| HCC | None | Diethylnitrosamine-induced mouse HCC (50 mg/kg) | [138] | ||
| Xanthohumol | Inhibition of ER-Golgi translocation of SCAP/SREBPs by binding to Sec23/24 | Normal liver | None | 75 or 150 mg/kg dietary Xanthohumol inhibits SREBP-1 target gene expression in the liver in diet-induced obese mice | [117] |
| PF-429242 | Inhibition of SREBP cleavage by inhibiting SIP | ||||
| Pancreatic | MIA PaCa-2 (24.5) | None | [72] | ||
| Nelfinavir | Inhibition of SREBP cleavage by inhibiting S2P | ||||
| Prostate | DU145 (−), PC3 (−) | None | [134] | ||
| 1,10-phenanthroline | Inhibition of SREBP cleavage by inhibiting S2P | Prostate | DU145 (−), PC3 (−) | None | [134] |
| BF175 | Inhibition of the transcription activity of SREBPs | Normal liver | None | BF175, 0.2% per weight of diet for 8 weeks, decreases the expression of SREBP target genes in mouse liver and reduces hepatic and blood levels of lipids | [137] |
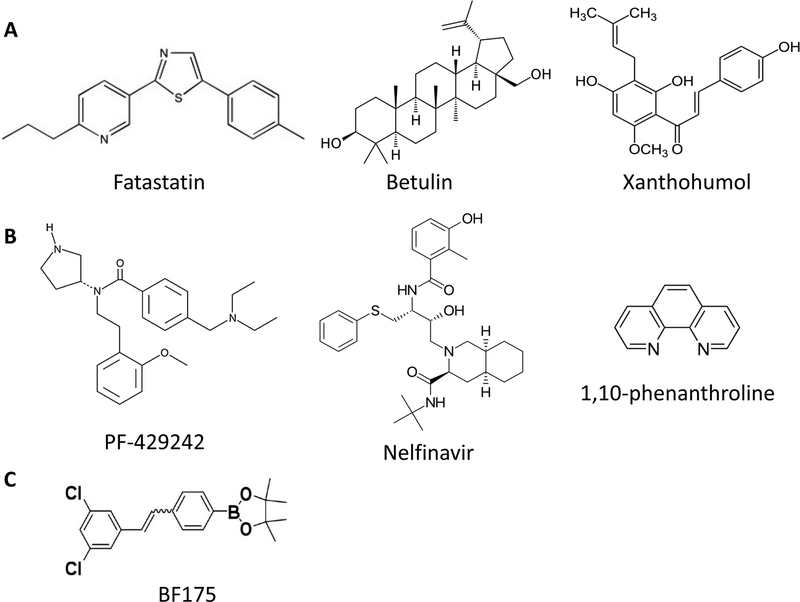
Two inhibitors, fatostatin and betulin, which suppress SCAP/SREBP translocation, have been extensively tested in cancer cells (Figure 1 and Figure 2) (Table 1) [105–111]. Fatostatin binds to SCAP, and inhibits its dissociation from INSIGs, thereby restricting the translocation of SREBPs to the ER and reducing lipogenesis [105, 112]. Fatostatin has been shown to inhibit cancer cell proliferation, invasion and migration, and to arrest cancer cells at the G2/M checkpoint in prostate cancer cells [109]. In addition, fatostatin alone or in combination with docetaxel suppresses the growth of androgen receptor-negative prostate cancers [111]. The combination of fatostatin and docetaxel leads to a greater proliferation inhibition and to apoptosis compared to treatment with single agents [111]. Moreover, fatostatin also inhibits the growth of pancreatic cancer MIA PaCa-2 cells by inhibiting SREBP-1 [72]. Similarly, betulin binds to SCAP and enhances its interaction with INSIGs, thereby suppressing SCAP/SREBP translocation [107]. Multiple studies have shown that betulin attenuates the growth of various cancers by inhibiting SREBP-1 [107, 110, 113–115].
Figure 2.
The structure of inhibitors in SCAP/SREBPs pathway.
A) Inhibitors suppressing SCAP/SREBP trafficking; B) Inhibitor suppressing S1P or S2P; C) Inhibitors suppressing the transcriptional activity of SREBPs.
Xanthohumol, a prenylated flavonoid found in hops, is a novel SREBP inhibitor (Figure 2) (Table 2) [116, 117]. Xanthohumol binds to Sec23/24 and blocks the incorporation of the SCAP/SREBP complex into COPII vesicles, thereby hindering ER-to-Golgi translocation of the complex [117]. Dietary xanthohumol reduced the maturation of hepatic SREBP-1 and transcription of its target genes [117]. Xanthohumol is also characterized as a broad-spectrum anti-tumor agent as it induces cancer cell apoptotic death and inhibits tumorigenesis through inhibition of STAT3 or NF-κB [118–125].
In addition to blocking SCAP/SREBP trafficking, suppressing the cleavage of SREBPs by inhibiting the S1P or S2P enzymes is another promising strategy to downregulate SREBP activity (Figure 1). PF-429242 is a reversible inhibitor of S1P, which significantly inhibits SREBP processing (Table 2) [126]. PF-429242 was selected through a high throughout screeningusing purified human S1P and a fluorescent assay that analyzes cleavage of the synthetic peptide Ac-VFRSLK-MCA (Figure 2) [127]. PF-429242 suppresses the proteolytic processing and nuclear translocation of SREBPs, thereby inhibiting cholesterol and fatty acid synthesis [127]. Beth et al. reported that PF-429242 suppressed GBM tumor growth by inducing apoptotic cell death by inhibiting SREBP activation [128]. PF-429242 also suppresses pancreatic cancer growth by inhibiting SREBP-1 and its downstream signaling cascade, FASN, HMGCR and SCD1 [72]. Nelfinavir is an inhibitor of S2P and suppresses the proteolysis of SREBPs (Table 2) [131]. Nelfinavir was firstly identified as a HIV-1 protease inhibitor (Figure 2) [130]. Studies show that nelfinavir treatment led to the accumulation of unprocessed SREBP-1 and increase of ER stress, leading to the apoptosis of liposarcoma and inhibition of castration-resistant prostate cancer cells [132–134]. Another S2P inhibitor, 1,10-phenanthroline, has the same effects in suppressing SREBP maturation and proliferation of prostate cancer cells as nelfinavir (Figure 2) (Table 2) [133].
Transcriptional activation of lipogenic gene expression requires nuclear SREBPs to bind to CREB-binding protein (CBP)/p300 acetyltransferase and activator-recruited co-factor 105 (ARC105, also named MED15) co-activators [135]. Yang’s group recently developed BF175, an inhibitor that can block the binding of MED15 to SREBP-1a, thereby inhibiting lipid synthesis and obesity in mouse models (Table 2) [136, 137]. BF175 reduces hepatic lipid content and decreases hepatic mRNA levels of SREBFs and their target genes in BF175 treated mice [137]. Thus, the effects of BF175 should be investigated in cancer cells.
CONCLUSION
In summary, lipid metabolism reprogramming has emerged as a novel hallmark of cancer [10–14]. Accumulating evidence has shown that SCAP/SREBPs play important roles in malignancies, connecting oncogenic signaling to lipid metabolism alterations, leading to rapid tumor growth (Figure 1) [13, 60, 61, 76–78, 80, 82]. Multiple studies have suggested that SCAP/SREBPs are very promising molecular targets in cancer treatment. A better understanding of the mechanisms underlying the trafficking and activation of SREBPs will provide optimal means to target SCAP/SREBPs. Developing effective inhibitors targeting lipid metabolism while avoiding toxicity in normal cells is the current challenge in cancer therapy. Moreover, identifying treatments that combine targeting SREBPs together with chemotherapy or immunotherapy may provide effective strategies for the treatment of cancer patients.
ACKNOWLEDGEMENTS:
This work was supported by the grants NIH NS079701 (DG), American Cancer Society Research Scholar Grant RSG-14-228-01-CSM (DG), OSUCCC Idea Grant (DG) and Translational Therapeutic Program seed grant (DG). The authors wish to thank Dr. Martine Torres for her editorial help.
Footnotes
CONFLICT OF INTEREST
The authors have declared that no competing interests exist.
REFERENCES
- [1].Hanahan D; Weinberg RA Hallmarks of cancer: the next generation. Cell, 2011, 144, (5), 646–674. [DOI] [PubMed] [Google Scholar]
- [2].Brown MS; Goldstein JL The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell, 1997, 89, (3), 331–340. [DOI] [PubMed] [Google Scholar]
- [3].Berfield AK; Andress DL; Abrass CK IGF-1–induced lipid accumulation impairs mesangial cell migration and contractile function. Kidney International, 2002, 62, (4), 1229–1237. [DOI] [PubMed] [Google Scholar]
- [4].Rysman E; Brusselmans K; Scheys K; Timmermans L; Derua R; Munck S; Van Veldhoven PP; Waltregny D; Daniels VW; Machiels J; Vanderhoydonc F; Smans K; Waelkens E; Verhoeven G; Swinnen JV De novo lipogenesis protects cancer cells from free radicals and chemotherapeutics by promoting membrane lipid saturation. Cancer Res, 2010, 70, (20), 8117–8126. [DOI] [PubMed] [Google Scholar]
- [5].Lochner M; Berod L; Sparwasser T Fatty acid metabolism in the regulation of T cell function. Trends in Immunology, 2014, 36, (2), 81–91. [DOI] [PubMed] [Google Scholar]
- [6].Muro E; Atilla-Gokcumen GE; Eggert US Lipids in cell biology: how can we understand them better? Molecular Biology of the Cell, 2014, 25, (12), 1819–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Furse S; Shearman GC Do lipids shape the eukaryotic cell cycle? Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids, 2018, 1863, (1), 9–19. [DOI] [PubMed] [Google Scholar]
- [8].DeBerardinis RJ; Lum JJ; Hatzivassiliou G; Thompson CB The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab, 2008, 7, (1), 11–20. [DOI] [PubMed] [Google Scholar]
- [9].Currie E; Schulze A; Zechner R; Walther TC; Farese RV Jr. Cellular fatty acid metabolism and cancer. Cell Metab, 2013, 18, (2), 153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Beloribi-Djefaflia S; Vasseur S; Guillaumond F Lipid metabolic reprogramming in cancer cells. Oncogenesis, 2016, 5, e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Rohrig F; Schulze A The multifaceted roles of fatty acid synthesis in cancer. Nat Rev Cancer, 2016, 16, (11), 732–749. [DOI] [PubMed] [Google Scholar]
- [12].Shimano H; Sato R SREBP-regulated lipid metabolism: convergent physiology - divergent pathophysiology. Nat Rev Endocrinol, 2017. [DOI] [PubMed] [Google Scholar]
- [13].Guo D; Bell EH; Mischel P; Chakravarti A Targeting SREBP-1-driven lipid metabolism to treat cancer. Curr Pharm Des, 2014, 20, (15), 2619–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Liu Q; Luo Q; Halim A; Song G Targeting lipid metabolism of cancer cells: A promising therapeutic strategy for cancer. Cancer Lett, 2017, 401, 39–45. [DOI] [PubMed] [Google Scholar]
- [15].Baenke F; Peck B; Miess H; Schulze A Hooked on fat: the role of lipid synthesis in cancer metabolism and tumour development. Dis Model Mech, 2013, 6, (6), 1353–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Briggs MR; Yokoyama C; Wang X; Brown MS; Goldstein JL Nuclear protein that binds sterol regulatory element of low density lipoprotein receptor promoter. I. Identification of the protein and delineation of its target nucleotide sequence. J Biol Chem, 1993, 268, (19), 14490–14496. [PubMed] [Google Scholar]
- [17].Hua X; Yokoyama C; Wu J; Briggs MR; Brown MS; Goldstein JL; Wang X SREBP-2, a second basic-helix-loop-helix-leucine zipper protein that stimulates transcription by binding to a sterol regulatory element. Proc Natl Acad Sci USA, 1993, 90, (24), 11603–11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wang X; Briggs MR; Hua X; Yokoyama C; Goldstein JL; Brown MS Nuclear protein that binds sterol regulatory element of low density lipoprotein receptor promoter. II. Purification and characterization. J Biol Chem, 1993, 268, (19), 14497–14504. [PubMed] [Google Scholar]
- [19].Yokoyama C; Wang X; Briggs MR; Admon A; Wu J; Hua X; Goldstein JL; Brown MS SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell, 1993, 75, (1), 187–197. [PubMed] [Google Scholar]
- [20].Wang X; Sato R; Brown MS; Hua X; Goldstein JL SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell, 1994, 77, (1), 53–62. [DOI] [PubMed] [Google Scholar]
- [21].Go G.-w.; Mani A Low-Density Lipoprotein Receptor (LDLR) Family Orchestrates Cholesterol Homeostasis. Yale J Biol Med, 2012, 85, (1), 19–28. [PMC free article] [PubMed] [Google Scholar]
- [22].Goldstein JL; Brown MS Regulation of the mevalonate pathway. Nature, 1990, 343, (6257), 425–430. [DOI] [PubMed] [Google Scholar]
- [23].Goldstein JL; DeBose-Boyd RA; Brown MS Protein sensors for membrane sterols. Cell, 2006, 124, (1), 35–46. [DOI] [PubMed] [Google Scholar]
- [24].Horton JD; Goldstein JL; Brown MS SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest, 2002, 109, (9), 1125–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Eberlé D; Hegarty B; Bossard P; Ferré P; Foufelle F SREBP transcription factors: master regulators of lipid homeostasis. Biochimie, 2004, 86, (11), 839–848. [DOI] [PubMed] [Google Scholar]
- [26].Hughes AL; Todd BL; Espenshade PJ SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell, 2005, 120, (6), 831–842. [DOI] [PubMed] [Google Scholar]
- [27].Shimano H; Shimomura I; Hammer RE; Herz J; Goldstein JL; Brown MS; Horton JD Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J Clin Invest, 1997, 100, (8), 2115–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Im S-S; Yousef L; Blaschitz C; Liu Janet Z.; Edwards Robert A.; Young Stephen G.; Raffatellu M; Osborne Timothy F. Linking Lipid Metabolism to the Innate Immune Response in Macrophages through Sterol Regulatory Element Binding Protein-1a. Cell Metabolism, 2011, 13, (5), 540–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Amemiya-Kudo M; Shimano H; Hasty AH; Yahagi N; Yoshikawa T; Matsuzaka T; Okazaki H; Tamura Y; Iizuka Y; Ohashi K; Osuga J; Harada K; Gotoda T; Sato R; Kimura S; Ishibashi S; Yamada N Transcriptional activities of nuclear SREBP-1a, −1c, and −2 to different target promoters of lipogenic and cholesterogenic genes. J Lipid Res, 2002, 43, (8), 1220–1235. [PubMed] [Google Scholar]
- [30].Shimano H; Horton JD; Shimomura I; Hammer RE; Brown MS; Goldstein JL Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J Clin Investig, 1997, 99, (5), 846–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sato R; Yang J; Wang X; Evans MJ; Ho YK; Goldstein JL; Brown MS Assignment of the membrane attachment, DNA binding, and transcriptional activation domains of sterol regulatory element-binding protein-1 (SREBP-1). J Biol Chem, 1994, 269, (25), 17267–17273. [PubMed] [Google Scholar]
- [32].Weber LW; Boll M; Stampfl A Maintaining cholesterol homeostasis: Sterol regulatory element-binding proteins. World J Gastroenterol, 2004, 10, (21), 3081–3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Brown MS; Radhakrishnan A; Goldstein JL Retrospective on Cholesterol Homeostasis: The Central Role of Scap. Annual Review of Biochemistry, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Sakai J; Nohturfft A; Cheng D; Ho YK; Brown MS; Goldstein JL Identification of Complexes between the COOH-terminal Domains of Sterol Regulatory Element-binding Proteins (SREBPs) and SREBP Cleavage-Activating Protein. J Biol Chem, 1997, 272, (32), 20213–20221. [DOI] [PubMed] [Google Scholar]
- [35].Yabe D; Brown MS; Goldstein JL Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins. Proc Natl Acad Sci USA, 2002, 99, (20), 12753–12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Yang T; Espenshade PJ; Wright ME; Yabe D; Gong Y; Aebersold R; Goldstein JL; Brown MS Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell, 2002, 110, (4), 489–500. [DOI] [PubMed] [Google Scholar]
- [37].Sun LP; Li L; Goldstein JL; Brown MS Insig required for sterol-mediated inhibition of Scap/SREBP binding to COPII proteins in vitro. J Biol Chem, 2005, 280, (28), 26483–26490. [DOI] [PubMed] [Google Scholar]
- [38].Sakai J; Rawson RB; Espenshade PJ; Cheng D; Seegmiller AC; Goldstein JL; Brown MS Molecular Identification of the Sterol-Regulated Luminal Protease that Cleaves SREBPs and Controls Lipid Composition of Animal Cells. Molecular Cell, 1998, 2, (4), 505–514. [DOI] [PubMed] [Google Scholar]
- [39].Rawson RB; Zelenski NG; Nijhawan D; Ye J; Sakai J; Hasan MT; Chang TY; Brown MS; Goldstein JL Complementation Cloning of S2P, a Gene Encoding a Putative Metalloprotease Required for Intramembrane Cleavage of SREBPs. Molecular Cell, 1997, 1, (1), 47–57. [DOI] [PubMed] [Google Scholar]
- [40].Jo Y; Lee PCW; Sguigna PV; DeBose-Boyd RA Sterol-induced degradation of HMG CoA reductase depends on interplay of two Insigs and two ubiquitin ligases, gp78 and Trc8. Proc Natl Acad Sci USA, 2011, 108, (51), 20503–20508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Soyal SM; Nofziger C; Dossena S; Paulmichl M; Patsch W Targeting SREBPs for treatment of the metabolic syndrome. Trends Pharmacol Sci, 2015, 36, (6), 406–416. [DOI] [PubMed] [Google Scholar]
- [42].Kuan YC; Hashidume T; Shibata T; Uchida K; Shimizu M; Inoue J; Sato R Heat Shock Protein 90 Modulates Lipid Homeostasis by Regulating the Stability and Function of Sterol Regulatory Element-binding Protein (SREBP) and SREBP Cleavage-activating Protein. J Biol Chem, 2017, 292, (7), 3016–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lee JN; Zhang X; Feramisco JD; Gong Y; Ye J Unsaturated Fatty Acids Inhibit Proteasomal Degradation of Insig-1 at a Postubiquitination Step. J Biol Chem, 2008, 283, (48), 33772–33783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Amemiya-Kudo M; Shimano H; Yoshikawa T; Yahagi N; Hasty AH; Okazaki H; Tamura Y; Shionoiri F; Iizuka Y; Ohashi K; Osuga J; Harada K; Gotoda T; Sato R; Kimura S; Ishibashi S; Yamada N Promoter analysis of the mouse sterol regulatory element-binding protein-1c gene. J Biol Chem, 2000, 275, (40), 31078–31085. [DOI] [PubMed] [Google Scholar]
- [45].Dif N; Euthine V; Gonnet E; Laville M; Vidal H; Lefai E Insulin activates human sterol-regulatory-element-binding protein-1c (SREBP-1c) promoter through SRE motifs. Biochem J, 2006, 400, (Pt 1), 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Im S-S; Yousef L; Blaschitz C; Liu JZ; Edwards RA; Young SG; Raffatellu M; Osborne TF Linking Lipid Metabolism to the Innate Immune Response in Macrophages through Sterol Regulatory Element Binding Protein −1a. Cell Metab, 2011, 13, (5), 540–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Repa JJ; Liang G; Ou J; Bashmakov Y; Lobaccaro JM; Shimomura I; Shan B; Brown MS; Goldstein JL; Mangelsdorf DJ Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev, 2000, 14, (22), 2819–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Chen W; Chen G; Head DL; Mangelsdorf DJ; Russell DW Enzymatic reduction of oxysterols impairs LXR signaling in cultured cells and the livers of mice. Cell Metab, 2007, 5, (1), 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Hannah VC; Ou J; Luong A; Goldstein JL; Brown MS Unsaturated fatty acids down-regulate srebp isoforms 1a and 1c by two mechanisms in HEK-293 cells. J Biol Chem, 2001, 276, (6), 4365–4372. [DOI] [PubMed] [Google Scholar]
- [50].Shin D-J; Osborne TF Thyroid Hormone Regulation and Cholesterol Metabolism Are Connected through Sterol Regulatory Element-binding Protein-2 (SREBP-2). J Biol Chem, 2003, 278, (36), 34114–34118. [DOI] [PubMed] [Google Scholar]
- [51].Bengoechea-Alonso MT; Ericsson J A phosphorylation cascade controls the degradation of active SREBP1. J Biol Chem, 2009, 284, (9), 5885–5895. [DOI] [PubMed] [Google Scholar]
- [52].Li Y; Xu S; Mihaylova MM; Zheng B; Hou X; Jiang B; Park O; Luo Z; Lefai E; Shyy JY; Gao B; Wierzbicki M; Verbeuren TJ; Shaw RJ; Cohen RA; Zang M AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metab, 2011, 13, (4), 376–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Shimano H; Horton JD; Hammer RE; Shimomura I; Brown MS; Goldstein JL Overproduction of cholesterol and fatty acids causes massive liver enlargement in transgenic mice expressing truncated SREBP-1a. J Clin Investig, 1996, 98, (7), 1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Toth JI; Datta S; Athanikar JN; Freedman LP; Osborne TF Selective coactivator interactions in gene activation by SREBP-1a and −1c. Mol Cell Biol, 2004, 24, (18), 8288–8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Yang F; Vought BW; Satterlee JS; Walker AK; Jim Sun ZY; Watts JL; DeBeaumont R; Saito RM; Hyberts SG; Yang S; Macol C; Iyer L; Tjian R; van den Heuvel S; Hart AC; Wagner G; Naar AM An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature, 2006, 442, (7103), 700–704. [DOI] [PubMed] [Google Scholar]
- [56].Walker AK; Yang F; Jiang K; Ji JY; Watts JL; Purushotham A; Boss O; Hirsch ML; Ribich S; Smith JJ; Israelian K; Westphal CH; Rodgers JT; Shioda T; Elson SL; Mulligan P; Najafi-Shoushtari H; Black JC; Thakur JK; Kadyk LC; Whetstine JR; Mostoslavsky R; Puigserver P; Li X; Dyson NJ; Hart AC; Naar AM Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev, 2010, 24, (13), 1403–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Sundqvist A; Bengoechea-Alonso MT; Ye X; Lukiyanchuk V; Jin J; Harper JW; Ericsson J Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCF(Fbw7). Cell Metab, 2005, 1, (6), 379–391. [DOI] [PubMed] [Google Scholar]
- [58].Vander Heiden MG; Cantley LC; Thompson CB Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science, 2009, 324, (5930), 1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Nagarajan A; Malvi P; Wajapeyee N Oncogene-Directed Alterations in Cancer Cell Metabolism. Trends in Cancer, 2016, 2, (7), 365–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Guo D; Prins RM; Dang J; Kuga D; Iwanami A; Soto H; Lin KY; Huang TT; Akhavan D; Hock MB; Zhu S; Kofman AA; Bensinger SJ; Yong WH; Vinters HV; Horvath S; Watson AD; Kuhn JG; Robins HI; Mehta MP; Wen PY; DeAngelis LM; Prados MD; Mellinghoff IK; Cloughesy TF; Mischel PS EGFR signaling through an Akt-SREBP-1-dependent, rapamycin-resistant pathway sensitizes glioblastomas to antilipogenic therapy. Sci Signal, 2009, 2, (101), ra82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Guo D; Reinitz F; Youssef M; Hong C; Nathanson D; Akhavan D; Kuga D; Amzajerdi AN; Soto H; Zhu S; Babic I; Tanaka K; Dang J; Iwanami A; Gini B; Dejesus J; Lisiero DD; Huang TT; Prins RM; Wen PY; Robins HI; Prados MD; Deangelis LM; Mellinghoff IK; Mehta MP; James CD; Chakravarti A; Cloughesy TF; Tontonoz P; Mischel PS An LXR agonist promotes glioblastoma cell death through inhibition of an EGFR/AKT/SREBP-1/LDLR-dependent pathway. Cancer Discov, 2011, 1, (5), 442–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Wen PY; Reardon DA Neuro-oncology in 2015: Progress in glioma diagnosis, classification and treatment. Nat Rev Neurol, 2016, 12, (2), 69–70. [DOI] [PubMed] [Google Scholar]
- [63].Ettinger SL; Sobel R; Whitmore TG; Akbari M; Bradley DR; Gleave ME; Nelson CC Dysregulation of sterol response element-binding proteins and downstream effectors in prostate cancer during progression to androgen independence. Cancer Res, 2004, 64, (6), 2212–2221. [DOI] [PubMed] [Google Scholar]
- [64].Li W; Tai Y; Zhou J; Gu W; Bai Z; Zhou T; Zhong Z; McCue PA; Sang N; Ji JY; Kong B; Jiang J; Wang C Repression of endometrial tumor growth by targeting SREBP1 and lipogenesis. Cell Cycle, 2012, 11, (12), 2348–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Bao J; Zhu L; Zhu Q; Su J; Liu M; Huang W SREBP-1 is an independent prognostic marker and promotes invasion and migration in breast cancer. Oncology Letters, 2016, 12, (4), 2409–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Zhu Z; Zhao X; Zhao L; Yang H; Liu L; Li J; Wu J; Yang F; Huang G; Liu J p54nrb/NONO regulates lipid metabolism and breast cancer growth through SREBP-1A. Oncogene, 2016, 35, (11), 1399–1410. [DOI] [PubMed] [Google Scholar]
- [67].Li C; Yang W; Zhang J; Zheng X; Yao Y; Tu K; Liu Q SREBP-1 Has a Prognostic Role and Contributes to Invasion and Metastasis in Human Hepatocellular Carcinoma. Int J Mol Sci 2014, 15, (5), 7124–7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Nie LY; Lu QT; Li WH; Yang N; Dongol S; Zhang X; Jiang J Sterol regulatory element-binding protein 1 is required for ovarian tumor growth. Oncol Rep, 2013, 30, (3), 1346–1354. [DOI] [PubMed] [Google Scholar]
- [69].Sun Y; He W; Luo M; Zhou Y; Chang G; Ren W; Wu K; Li X; Shen J; Zhao X; Hu Y SREBP1 regulates tumorigenesis and prognosis of pancreatic cancer through targeting lipid metabolism. Tumour Biol, 2015, 36, (6), 4133–4141. [DOI] [PubMed] [Google Scholar]
- [70].Schonberg SA; Lundemo AG; Fladvad T; Holmgren K; Bremseth H; Nilsen A; Gederaas O; Tvedt KE; Egeberg KW; Krokan HE Closely related colon cancer cell lines display different sensitivity to polyunsaturated fatty acids, accumulate different lipid classes and downregulate sterol regulatory element-binding protein 1. FEBS J, 2006, 273, (12), 2749–2765. [DOI] [PubMed] [Google Scholar]
- [71].Luo D; Xiao H; Dong J; Li Y; Feng G; Cui M; Fan S B7–H3 regulates lipid metabolism of lung cancer through SREBP1-mediated expression of FASN. Biochem Biophys Res Commun, 2017, 482, (4), 1246–1251. [DOI] [PubMed] [Google Scholar]
- [72].Siqingaowa; Sekar S; Gopalakrishnan V; Taghibiglou C Sterol regulatory element-binding protein 1 inhibitors decrease pancreatic cancer cell viability and proliferation. Biochem Biophys Res Commun, 2017, 488, (1), 136–140. [DOI] [PubMed] [Google Scholar]
- [73].Li X; Wu JB; Li Q; Shigemura K; Chung LWK; Huang W-C SREBP-2 promotes stem cell-like properties and metastasis by transcriptional activation of c-Myc in prostate cancer. Oncotarget, 2016, 7, (11), 12869–12884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Haskins JW; Zhang S; Means RE; Kelleher JK; Cline GW; Canfrán-Duque A; Suárez Y; Stern DF Neuregulin-activated ERBB4 induces the SREBP-2 cholesterol biosynthetic pathway and increases low-density lipoprotein uptake. Sci Signal, 2015, 8, (401), ra111–ra111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Luu W; Sharpe LJ; Stevenson J; Brown AJ Akt acutely activates the cholesterogenic transcription factor SREBP-2. BBA-MOL CELL RES, 2012, 1823, (2), 458–464. [DOI] [PubMed] [Google Scholar]
- [76].Guo D; Bell EH; Chakravarti A Lipid metabolism emerges as a promising target for malignant glioma therapy. CNS Oncol, 2013, 2, (3), 289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Cheng C; Ru P; Geng F; Liu J; Yoo JY; Wu X; Cheng X; Euthine V; Hu P; Guo JY; Lefai E; Kaur B; Nohturfft A; Ma J; Chakravarti A; Guo D Glucose-Mediated N-glycosylation of SCAP Is Essential for SREBP-1 Activation and Tumor Growth. Cancer Cell, 2015, 28, (5), 569–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Geng F; Cheng X; Wu X; Yoo JY; Cheng C; Guo JY; Mo X; Ru P; Hurwitz B; Kim SH; Otero J; Puduvalli V; Lefai E; Ma J; Nakano I; Horbinski C; Kaur B; Chakravarti A; Guo D Inhibition of SOAT1 Suppresses Glioblastoma Growth via Blocking SREBP-1-Mediated Lipogenesis. Clin Cancer Res, 2016, 22, (21), 5337–5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Guo D SCAP links glucose to lipid metabolism in cancer cells. Mol Cell Oncol, 2016, 3, (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Ru P; Hu P; Geng F; Mo X; Cheng C; Yoo JY; Cheng X; Wu X; Guo JY; Nakano I; Lefai E; Kaur B; Chakravarti A; Guo D Feedback Loop Regulation of SCAP/SREBP-1 by miR-29 Modulates EGFR Signaling-Driven Glioblastoma Growth. Cell Rep, 2016, 16, (6), 1527–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Ru P; Guo D microRNA-29 mediates a novel negative feedback loop to regulate SCAP/SREBP-1 and lipid metabolism. RNA Dis, 2017, 4, (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Guo D; Hildebrandt IJ; Prins RM; Soto H; Mazzotta MM; Dang J; Czernin J; Shyy JYJ; Watson AD; Phelps M; Radu CG; Cloughesy TF; Mischel PS The AMPK agonist AICAR inhibits the growth of EGFRvIII-expressing glioblastomas by inhibiting lipogenesis. Proc Natl Acad Sci USA, 2009, 106, (31), 12932–12937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Cheng C; Guo JY; Geng F; Wu X; Cheng X; Li Q; Guo D Analysis of SCAP N-glycosylation and Trafficking in Human Cells. J Vis Exp, 2016, (117). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Ricoult SJH; Yecies JL; Ben-Sahra I; Manning BD Oncogenic PI3K and K-Ras stimulate de novo lipid synthesis through mTORC1 and SREBP. Oncogene, 2016, 35, (10), 1250–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Porstmann T; Santos CR; Griffiths B; Cully M; Wu M; Leevers S; Griffiths JR; Chung Y-L; Schulze A SREBP Activity Is Regulated by mTORC1 and Contributes to Akt-Dependent Cell Growth. Cell Metabolism, 2008, 8, (3–3), 224–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Düvel K; Yecies JL; Menon S; Raman P; Lipovsky AI; Souza AL; Triantafellow E; Ma Q; Gorski R; Cleaver S; Vander Heiden MG; MacKeigan JP; Finan PM; Clish CB; Murphy LO; Manning BD Activation of a Metabolic Gene Regulatory Network Downstream of mTOR Complex 1. Molecular Cell, 2010, 39, (2), 171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Shamma A; Takegami Y; Miki T; Kitajima S; Noda M; Obara T; Okamoto T; Takahashi C Rb Regulates DNA Damage Response and Cellular Senescence through E2F-Dependent Suppression of N-Ras Isoprenylation. Cancer Cell, 2009, 15, (4), 255–269. [DOI] [PubMed] [Google Scholar]
- [88].Filipowicz W; Bhattacharyya SN; Sonenberg N Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet, 2008, 9, (2), 102–114. [DOI] [PubMed] [Google Scholar]
- [89].Bartel DP MicroRNAs: target recognition and regulatory functions. Cell, 2009, 136, (2), 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Ambros V The functions of animal microRNAs. Nature, 2004, 431, (7006), 350–355. [DOI] [PubMed] [Google Scholar]
- [91].Krutzfeldt J; Stoffel M MicroRNAs: a new class of regulatory genes affecting metabolism. Cell Metab, 2006, 4, (1), 9–12. [DOI] [PubMed] [Google Scholar]
- [92].Rottiers V; Näär AM MicroRNAs in Metabolism and Metabolic Disorders. Nat Rev Mol Cell Biol, 2012, 13, (4), 239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Desgagne V; Bouchard L; Guerin R microRNAs in lipoprotein and lipid metabolism: from biological function to clinical application. Clin Chem Lab Med, 2017, 55, (5), 667–686. [DOI] [PubMed] [Google Scholar]
- [94].Rotllan N; Fernandez-Hernando C MicroRNA Regulation of Cholesterol Metabolism. Cholesterol, 2012, 2012, 847849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Gomez de Cedron M; Ramirez de Molina A Microtargeting cancer metabolism: opening new therapeutic windows based on lipid metabolism. J Lipid Res, 2016, 57, (2), 193–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Aryal B; Singh AK; Rotllan N; Price N; Fernández-Hernando C MicroRNAs and lipid metabolism. Curr Opin Lipidol, 2017, 28, (3), 273–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Kriegel AJ; Liu Y; Fang Y; Ding X; Liang M The miR-29 family: genomics, cell biology, and relevance to renal and cardiovascular injury. Physiol Genomics, 2012, 44, (4), 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Jiang H; Zhang G; Wu JH; Jiang CP Diverse roles of miR-29 in cancer (review). Oncol Rep, 2014, 31, (4), 1509–1516. [DOI] [PubMed] [Google Scholar]
- [99].Li X; Chen Y-T; Josson S; Mukhopadhyay NK; Kim J; Freeman MR; Huang W-C MicroRNA-185 and 342 Inhibit Tumorigenicity and Induce Apoptosis through Blockade of the SREBP Metabolic Pathway in Prostate Cancer Cells. PLOS ONE, 2013, 8, (8), e70987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Yang M; Liu W; Pellicane C; Sahyoun C; Joseph BK; Gallo-Ebert C; Donigan M; Pandya D; Giordano C; Bata A; Nickels JT Identification of miR-185 as a regulator of de novo cholesterol biosynthesis and low density lipoprotein uptake. J Lipid Res, 2014, 55, (2), 226–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Li Y; Zhang J; He J; Zhou W; Xiang G; Xu R MicroRNA-132 cause apoptosis of glioma cells through blockade of the SREBP-1c metabolic pathway related to SIRT1. Biomedicine & Pharmacotherapy, 2016, 78, (Supplement C), 177–184. [DOI] [PubMed] [Google Scholar]
- [102].Ng R; Wu H; Xiao H; Chen X; Willenbring H; Steer CJ; Song G Inhibition of MicroRNA-24 Expression in Liver Prevents Hepatic Lipid Accumulation and Hyperlipidemia. Hepatology, 2014, 60, (2), 554–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Sundqvist A; Bengoechea-Alonso MT; Ye X; Lukiyanchuk V; Jin J; Harper JW; Ericsson J Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCFFbw7. Cell Metabolism, 2005, 1, (6), 379–391. [DOI] [PubMed] [Google Scholar]
- [104].Jeon TI; Esquejo RM; Roqueta-Rivera M; Phelan PE; Moon YA; Govindarajan SS; Esau CC; Osborne TF An SREBP-responsive microRNA operon contributes to a regulatory loop for intracellular lipid homeostasis. Cell Metab, 2013, 18, (1), 51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Choi Y; Kawazoe Y; Murakami K; Misawa H; Uesugi M Identification of Bioactive Molecules by Adipogenesis Profiling of Organic Compounds. J Biol Chem, 2003, 278, (9), 7320–7324. [DOI] [PubMed] [Google Scholar]
- [106].duct betulin. Eur J Pharm Sci, 2006, 29, (1), 1–13. [DOI] [PubMed] [Google Scholar]
- [107].Tang JJ; Li JG; Qi W; Qiu WW; Li PS; Li BL; Song BL Inhibition of SREBP by a small molecule, betulin, improves hyperlipidemia and insulin resistance and reduces atherosclerotic plaques. Cell Metab, 2011, 13, (1), 44–56. [DOI] [PubMed] [Google Scholar]
- [108].Williams KJ; Argus JP; Zhu Y; Wilks MQ; Marbois BN; York AG; Kidani Y; Pourzia AL; Akhavan D; Lisiero DN; Komisopoulou E; Henkin AH; Soto H; Chamberlain BT; Vergnes L; Jung ME; Torres JZ; Liau LM; Christofk HR; Prins RM; Mischel PS; Reue K; Graeber TG; Bensinger SJ An Essential Requirement for the SCAP/SREBP Signaling Axis to Protect Cancer Cells from Lipotoxicity. Cancer Research, 2013, 73, (9), 2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Li X; Chen YT; Hu P; Huang WC Fatostatin displays high antitumor activity in prostate cancer by blocking SREBP-regulated metabolic pathways and androgen receptor signaling. Mol Cancer Ther, 2014, 13, (4), 855–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Katarzyna Król S; Kiełbus M; Rivero-Müller A; Stepulak A Comprehensive Review on Betulin as a Potent Anticancer Agent. BioMed Research International, 2015, 2015, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Li X; Wu JB; Chung LWK; Huang W-C Anti-cancer efficacy of SREBP inhibitor, alone or in combination with docetaxel, in prostate cancer harboring p53 mutations. Oncotarget, 2015, 6, (38), 41018–41032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Kamisuki S; Mao Q; Abu-Elheiga L; Gu Z; Kugimiya A; Kwon Y; Shinohara T; Kawazoe Y; Sato S.-i.; Asakura K; Choo H-YP; Sakai J; Wakil SJ; Uesugi M A Small Molecule That Blocks Fat Synthesis By Inhibiting the Activation of SREBP. Chemistry & Biology, 2009, 16, (8), 882–892. [DOI] [PubMed] [Google Scholar]
- [113].Chintharlapalli S; Papineni S; Ramaiah SK; Safe S Betulinic Acid Inhibits Prostate Cancer Growth through Inhibition of Specificity Protein Transcription Factors. Cancer Research, 2007, 67, (6), 2816. [DOI] [PubMed] [Google Scholar]
- [114].Dehelean CA; Feflea S; Gheorgheosu D; Ganta S; Cimpean AM; Muntean D; Amiji MM Anti-angiogenic and anti-cancer evaluation of betulin nanoemulsion in chicken chorioallantoic membrane and skin carcinoma in Balb/c mice. J Biomed Nanotechnol, 2013, 9, (4), 577–589. [DOI] [PubMed] [Google Scholar]
- [115].Lee JH; Jeon YG; Lee KH; Lee HW; Park J; Jang H; Kang M; Lee HS; Cho HJ; Nam DH; Kwak C; Kim JB RNF20 Suppresses Tumorigenesis by Inhibiting SREBP1c-PTTG1 Axis in Kidney Cancer. Mol Cell Biol, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Stevens JF; Page JE Xanthohumol and related prenylflavonoids from hops and beer: to your good health! Phytochemistry, 2004, 65, (10), 1317–1330. [DOI] [PubMed] [Google Scholar]
- [117].Miyata S; Inoue J; Shimizu M; Sato R Xanthohumol Improves Diet-induced Obesity and Fatty Liver by Suppressing Sterol Regulatory Element-binding Protein (SREBP) Activation. J Biol Chem, 2015, 290, (33), 20565–20579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Monteiro R; Calhau C; Silva AO; Pinheiro-Silva S; Guerreiro S; Gartner F; Azevedo I; Soares R Xanthohumol inhibits inflammatory factor production and angiogenesis in breast cancer xenografts. J Cell Biochem, 2008, 104, (5), 1699–1707. [DOI] [PubMed] [Google Scholar]
- [119].Doddapattar P; Radović B; Patankar JV; Obrowsky S; Jandl K; Nusshold C; Kolb D; Vujić N; Doshi L; Chandak PG; Goeritzer M; Ahammer H; Hoefler G; Sattler W; Kratky D Xanthohumol ameliorates atherosclerotic plaque formation, hypercholesterolemia, and hepatic steatosis in ApoE-deficient mice. Mol Nutr Food Res, 2013, 57, (10), 1718–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Kim SY; Lee IS; Moon A 2-Hydroxychalcone and xanthohumol inhibit invasion of triple negative breast cancer cells. Chem Biol Interact, 2013, 203, (3), 565–572. [DOI] [PubMed] [Google Scholar]
- [121].Jiang W; Zhao S; Xu L; Lu Y; Lu Z; Chen C; Ni J; Wan R; Yang L The inhibitory effects of xanthohumol, a prenylated chalcone derived from hops, on cell growth and tumorigenesis in human pancreatic cancer. Biomed Pharmacother, 2015, 73, 40–47. [DOI] [PubMed] [Google Scholar]
- [122].Chen P-H; Chang C-K; Shih C-M; Cheng C-H; Lin C-W; Lee C-C; Liu A-J; Ho K-H; Chen K-C The miR-204–3p-targeted IGFBP2 pathway is involved in xanthohumol-induced glioma cell apoptotic death. Neuropharmacology, 2016, 110, (Part A), 362–375. [DOI] [PubMed] [Google Scholar]
- [123].Dokduang H; Yongvanit P; Namwat N; Pairojkul C; Sangkhamanon S; Yageta MS; Murakami Y; Loilome W Xanthohumol inhibits STAT3 activation pathway leading to growth suppression and apoptosis induction in human cholangiocarcinoma cells. Oncol Rep, 2016, 35, (4), 2065–2072. [DOI] [PubMed] [Google Scholar]
- [124].Saito K; Matsuo Y; Imafuji H; Okubo T; Maeda Y; Sato T; Shamoto T; Tsuboi K; Morimoto M; Takahashi H; Ishiguro H; Takiguchi S Xanthohumol inhibits angiogenesis by suppressing nuclear factor-kappaB activation in pancreatic cancer. Cancer Sci, 2018, 109, (1), 132–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Viola K; Kopf S; Rarova L; Jarukamjorn K; Kretschy N; Teichmann M; Vonach C; Atanasov AG; Giessrigl B; Huttary N; Raab I; Krieger S; Strnad M; de Martin R; Saiko P; Szekeres T; Knasmuller S; Dirsch VM; Jager W; Grusch M; Dolznig H; Mikulits W; Krupitza G Xanthohumol attenuates tumour cell-mediated breaching of the lymphendothelial barrier and prevents intravasation and metastasis. Arch Toxicol, 2013, 87, (7), 1301–1312. [DOI] [PubMed] [Google Scholar]
- [126].Hawkins JL; Robbins MD; Warren LC; Xia D; Petras SF; Valentine JJ; Varghese AH; Wang IK; Subashi TA; Shelly LD; Hay BA; Landschulz KT; Geoghegan KF; Harwood HJ Pharmacologic inhibition of site 1 protease activity inhibits sterol regulatory element-binding protein processing and reduces lipogenic enzyme gene expression and lipid synthesis in cultured cells and experimental animals. J Pharmacol Exp Ther, 2008, 326, (3), 801–808. [DOI] [PubMed] [Google Scholar]
- [127].Hay BA; Abrams B; Zumbrunn AY; Valentine JJ; Warren LC; Petras SF; Shelly LD; Xia A; Varghese AH; Hawkins JL; Van Camp JA; Robbins MD; Landschulz K; Harwood HJ Jr. Aminopyrrolidineamide inhibitors of site-1 protease. Bioorg Med Chem Lett, 2007, 17, (16), 4411–4414. [DOI] [PubMed] [Google Scholar]
- [128].Caruana BT; Skoric A; Brown AJ; Lutze-Mann LH Site-1 protease, a novel metabolic target for glioblastoma. Biochem Biophys Res Commun, 2017, 490, (3), 760–766. [DOI] [PubMed] [Google Scholar]
- [129].Nakakuki M; Kawano H; Notsu T; Imada K; Mizuguchi K; Shimano H A novel processing system of sterol regulatory element-binding protein-1c regulated by polyunsaturated fatty acid. J Biochem, 2014, 155, (5), 301–313. [DOI] [PubMed] [Google Scholar]
- [130].Fatima R-B; Federico G HIV Protease Inhibition: Limited Recent Progress and Advances in Understanding Current Pitfalls. Curr Top Med Chem, 2004, 4, (9), 991–1007. [DOI] [PubMed] [Google Scholar]
- [131].Ye J; Rawson RB; Komuro R; Chen X; Davé UP; Prywes R; Brown MS; Goldstein JL ER Stress Induces Cleavage of Membrane-Bound ATF6 by the Same Proteases that Process SREBPs. Molecular Cell, 2000, 6, (6), 1355–1364. [DOI] [PubMed] [Google Scholar]
- [132].Guan M; Fousek K; Jiang C; Guo S; Synold T; Xi B; Shih C-C; Chow WA Nelfinavir Induces Liposarcoma Apoptosis through Inhibition of Regulated Intramembrane Proteolysis of SREBP-1 and ATF6. Clin Cancer Res, 2011, 17, (7), 1796. [DOI] [PubMed] [Google Scholar]
- [133].Guan M; Fousek K; Chow WA Nelfinavir inhibits regulated intramembrane proteolysis of sterol regulatory element binding protein-1 and activating transcription factor 6 in castration-resistant prostate cancer. FEBS J, 2012, 279, (13), 2399–2411. [DOI] [PubMed] [Google Scholar]
- [134].Guan M; Su L; Yuan YC; Li H; Chow WA Nelfinavir and nelfinavir analogs block site-2 protease cleavage to inhibit castration-resistant prostate cancer. Sci Rep, 2015, 5, 9698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Yang F; Vought BW; Satterlee JS; Walker AK; Jim Sun ZY; Watts JL; DeBeaumont R; Mako Saito R; Hyberts SG; Yang S; Macol C; Iyer L; Tjian R; van den Heuvel S; Hart AC; Wagner G; Näär AM An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature, 2006, 442, (7103), 700–704. [DOI] [PubMed] [Google Scholar]
- [136].Das BBC; Zhao X; Tang X-Y; Yang F Design, Synthesis and Biological Study of Pinacolylboronate-Substituted Stilbenes as Novel Lipogenic Inhibitors. Bioorganic Med Chem Lett 2011, 21, (18), 5638–5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Zhao X; Xiaoli; Zong H; Abdulla A; Yang EST; Wang Q; Ji J-Y; Pessin JE; Das BC; Yang F Inhibition of SREBP Transcriptional Activity by a Boron-Containing Compound Improves Lipid Homeostasis in Diet-Induced Obesity. Diabetes, 2014, 63, (7), 2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Li N; Zhou ZS; Shen Y; Xu J; Miao HH; Xiong Y; Xu F; Li BL; Luo J; Song BL Inhibition of the sterol regulatory element-binding protein pathway suppresses hepatocellular carcinoma by repressing inflammation in mice. Hepatology, 2017, 65, (6), 1936–1947. [DOI] [PubMed] [Google Scholar]
- [139].Griffiths B; Lewis CA; Bensaad K; Ros S; Zhang Q; Ferber EC; Konisti S; Peck B; Miess H; East P; Wakelam M; Harris AL; Schulze A Sterol regulatory element binding protein-dependent regulation of lipid synthesis supports cell survival and tumor growth. Cancer Metab, 2013, 1, (1), 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Wen YA; Xiong X; Zaytseva YY; Napier DL; Vallee E; Li AT; Wang C; Weiss HL; Evers BM; Gao T Downregulation of SREBP inhibits tumor growth and initiation by altering cellular metabolism in colon cancer. Cell Death Dis, 2018, 9, (3), 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Li J; Yan H; Zhao L; Jia W; Yang H; Liu L; Zhou X; Miao P; Sun X; Song S; Zhao X; Liu J; Huang G Inhibition of SREBP increases gefitinib sensitivity in non-small cell lung cancer cells. Oncotarget, 2016, 7, (32), 52392–52403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Krycer JR; Phan L; Brown AJ A key regulator of cholesterol homoeostasis, SREBP-2, can be targeted in prostate cancer cells with natural products. Biochem J, 2012, 446, (2), 191–201. [DOI] [PubMed] [Google Scholar]
- [143].Damle AA; Pawar YP; Narkar AA Anticancer activity of betulinic acid on MCF-7 tumors in nude mice. Indian J Exp Biol, 2013, 51, (7), 485–491. [PubMed] [Google Scholar]
- [144].Blanchet M; Seidah NG; Labonte P SKI-1/S1P inhibition: a promising surrogate to statins to block hepatitis C virus replication. Antiviral Res, 2012, 95, (2), 159–166. [DOI] [PubMed] [Google Scholar]