Abstract
Heparan sulfate (HS) is a complex linear polysaccharide that modulates a wide range of biological functions. Elucidating the structure-function relationship of HS has been challenging. Here we report the generation of a HS mutant mouse lung endothelial cell library by systematic deletion of HS genes expressing in the cell. We applied this library to answer several fundamental questions about HS biology including: 1) determining that strictly defined fine structure of HS, not its overall sulfation degree, is more important for FGF2-FGFR1 signaling; 2) defining the epitope features of commonly used anti-HS phage display antibodies; and 3) delineating the fine inter-regulation networks of HS modification and chain length by HS genes in mammalian cells and at a cell type specific level. Our mutant cell library will enable robust and systematic interrogation of the roles and related structures of HS in a cellular context.
Introduction
Heparan sulfate (HS) is a linear polysaccharide with complex structures. HS varies considerably in size, position and degree of sulfation, and epimerization of uronic acid in different cells, tissues and developmental stages. Such structural complexity and spatial and temporal expression patterns form the basis of the multifaceted functions of HS.
Biochemical studies using small-sized HS oligosaccharides and chemically modified heparins, a highly sulfated form of HS, have shown that HS interacts with protein ligands through its unique binding sites, which consist of relatively small tracts of variable modifications, including N−, 6-O and 3-O−sulfation of glucosamine residues (NS, 6S and 3S, correspondingly), 2-O−sulfaton of uronic acid (2S) and epimerization of glucuronic acid (GlcA) to iduronic acid (IdoA), in specific arrangements (Fig.1a)1–3. Extension of the biochemical findings to a cellular level is essential for a better understanding of HS structure-function relationships and its unique biological and pathological roles. However this has been challenging due to the length of native HS and its propensity to interact simultaneously with a ligand and the ligand`s receptor or harboring multiple copies of binding sites of a ligand1,4,5. In addition, the relative importance of fine structures versus overall sulfation of HS in interaction with protein ligand also remains a fundamental question in HS biology6–9. Furthermore, anti-HS phage display antibodies have been widely used to probe specific HS structures in situ, but their native HS epitope structures remain unclear. Better characterization of the HS epitope features may yield valuable new information for future antibody-based studies.
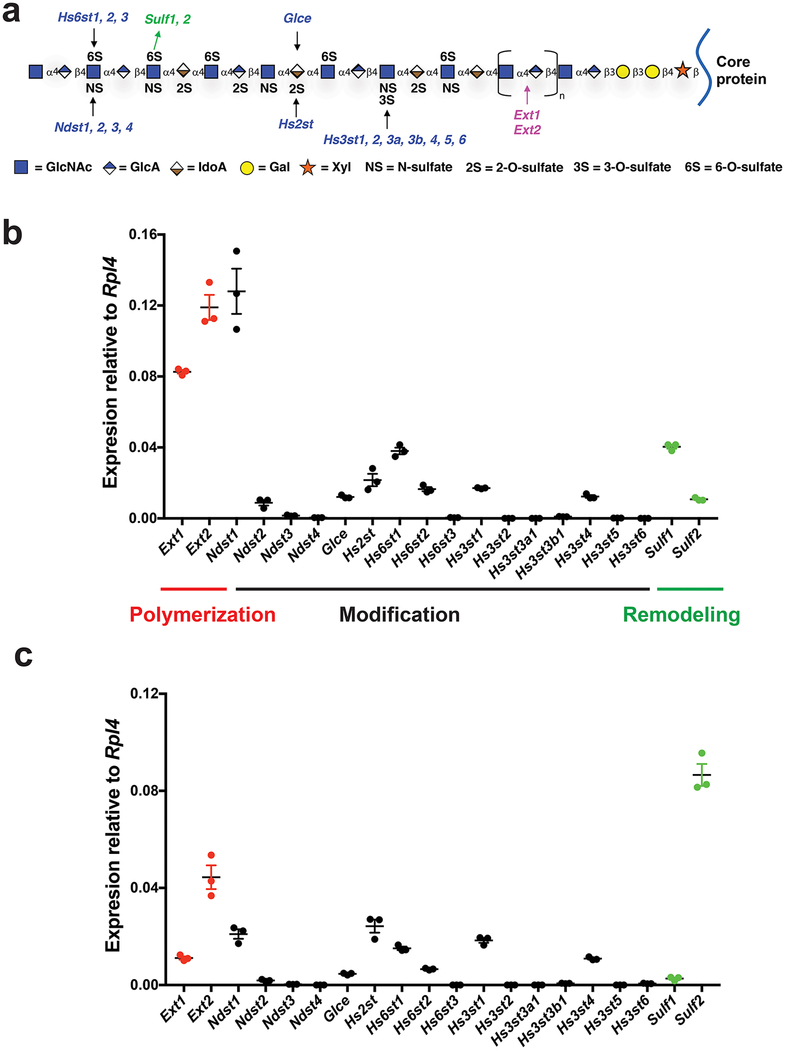
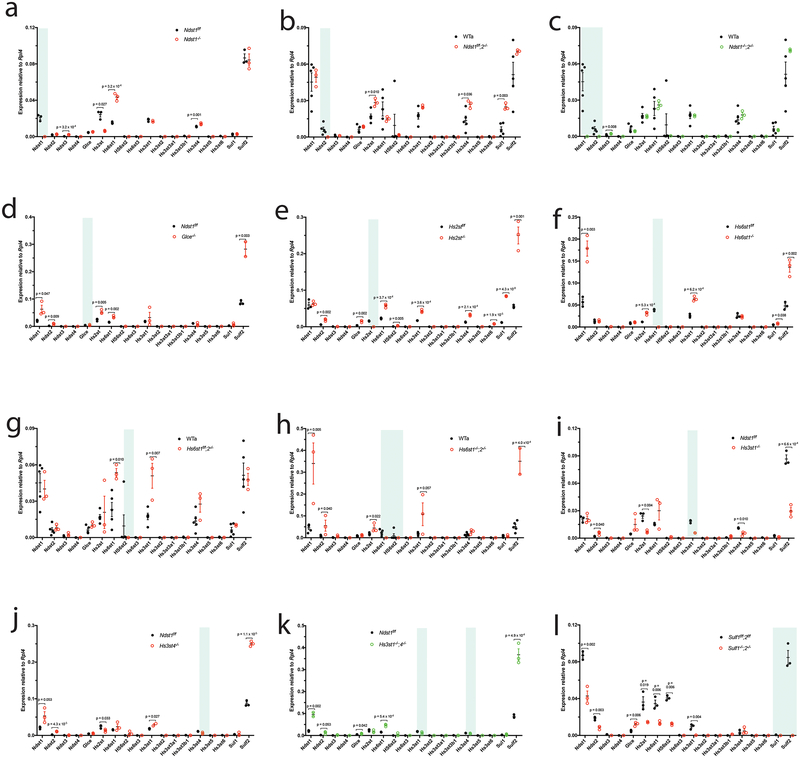
Figure 1. HS biosynthetic and remodeling gene expression in MLECs.
a. HS structure and biosynthetic/remodeling genes. Each sugar residue is depicted by a geometric symbol.. b, c. Expression patterns of HS polymerization, modification, and remodeling genes in primary (b) and immortalized (c) MLECs were determined by qRT-PCR analysis. The representative data from 3 independent experiments are presented as mean ± SD. GlcNAc, N-acetylglucosamine; GlcA, glucuronic acid; IdoA, iduronic acid; GalNAc, N-acetylgalactosamine; Gal, galactose; Xyl, xylose.
Here we report the development of a HS mutant mouse lung endothelial cell (MLEC) library that enabled to address some of these aforementioned fundamental HS biology issues, and represents a novel test platform to systematically interrogate the roles and related structures of HS in a cellular context.
Results
Generation of a HS mutant MLEC library
To generate a comprehensive HS mutant MLEC library that targets all the expressing HS genes in the cell, we first carried out qRT-PCR analysis to profile HS gene expression in primary MLECs. In mammals, there are as many as 20 HS-specific genes functioning to biosynthesize HS and two genes involving in postbiosynthetic remodeling. Among these 24 HS genes, eleven are expressed including the chain polymerization genes Exotosin-1 (Ext1) and Ext2, the modification genes N-deacetylase/N-sulfotransferase-1 (Ndst1) and Ndst2, C5-epimerase (Glce), HS 2-O-sulfotransferease (Hs2st), HS 6-O-sulfotransferease-1 (Hs6st1), Hs6st2, HS 3-O-sulfotransferease-1 (Hs3st1) and Hs3st4, and the remodeling genes HS 6-O-endosulfatase-1 (Sulf1) and Sulf2 (Fig. 1b).
We targeted all the expressing HS genes. By cell immortalization and cloning (Supplementary Fig. 1), we derived MLEC lines directly from 8–10 weeks-old Ext1f/f, Ndst1f/f, Ndst1f/f;2−/−, Hs2stf/f, Hs6st1f/f, Hs6st1f/f;2−/− and Sulf1f/f;2f/f mice. These cell lines were further subjected to transient Cre recombinase expression to derive their corresponding mutant Ext1−/−, Ndst1−/−, Ndst1−/−;2−/−, Hs2st−/−, Hs6st1−/−, Hs6st1−/−;2−/− and Sulf1−/−;2−/− cell lines. All 7 directly derived MLEC lines express endothelial cell markers CD31 and VEGFR2, confirming their endothelial cell identity (Supplementary Fig. 2). The HS gene expression patterns in the 5 wildtype MLEC lines Ext1f/f, Ndst1f/f, Hs2stf/f, Hs6st1f/f and Sulf1f/f;2f/f are similar to primary MLECs (Fig. 1b, c and Supplementary Fig. 3), indicating that the derived HS mutant cell lines may closely reflect HS structure alteration in vivo upon targeted gene deletion. We did not collect the mice deficient for Glce, Hs3st1 or Hs3st4 because of their embryonic lethality or unavailability. Instead, we transiently co-transfected the wildtype (WT) Ndst1f/f MLEC line with Cas9 and gRNA specific for Glce, Hs3st1 or Hs3st4 (Supplementary Fig. 1,4). After screening for induced genomic insertion/deletion (indel) mutations and confirming the indel by sequencing the gRNA targeted regions, we obtained the Glec−/−, Hs3st1−/−, Hs3st4−/− and Hs3st1−/−;4−/− MLEC lines (Supplementary Fig. 4). In total, by deleting expressing HS genes individually or in combination, we developed a HS mutant MLEC library containing 18 cell lines and harboring alterations of all HS modification types (Table 1).
Table 1. HS disaccharide composition of derived MLEC lines and their Mw.
Eighteen MLEC lines were generated, including five wildtype (with the HS-specific genes are conditionally targeted) and 13 HS mutant cell lines. The HS mutant cell lines were derived from conditionally targeted HS gene alleles by Cre recombinase treatment (Cre-LoxP gene targeting approach) or by directly targeting using gRNA (CRISPR-Cas9 approach). WTa, average of the 5 immortalized wildtype MLEC lines (Ext1f/f, Ndst1f/f, Hs2stf/f, Hs6st1f/f and Sulf1f/f;2f/f). *, the data were pooled from the 5 WT MLEC lines as control for the mutant cell lines that were directly derived from the corresponding conventional HS mutant mice. #, the parental Ndst1f/f MLEC line was used as the WT control for CRISPR-Cas9-derived HS mutant cell lines. NAc, N-acetyl group; NS, N-sulfate; 2S, 2-O-sulfate; 6S, 6-O-sulfate; t-S; total sulfate. The data were summarized from 3 independent experiments and are presented as mean ± SD except for the Mw analysis which was measured only one time per each cell line.
| Cell lines | Gene editing | NAc | NS | 2S | 6S | t-S | Mw (KDa) |
|---|---|---|---|---|---|---|---|
| Ext1f/f | Cre-LoxP | 33.47 ± 0.64 | 56.28 ± 0.35 | 21.64 ± 0.71 | 14.63 ± 0.38 | 92.55 | 14 |
| Ext1−/− | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | |
| Ndst1f/f | Cre-LoxP | 32.61 ± 3.38 | 58.69 ± 3.28 | 17.29 ± 1.95 | 16.35 ± 1.67 | 94.07 | 15 |
| Ndst1−/− | 70.71 ± 2.51 | 22.75 ± 2.59 | 5.18 ± 1.04 | 10.08 ± 1.02 | 38.02 | 15 | |
| WTa* | Direct derivation And Cre-LoxP | 33.54 ± 2.54 | 56.77 ± 2.04 | 17.72 ± 2.91 | 17.02 ± 2.47 | 92.66 | 14.4 |
| Ndst1f/f;2−/− | 48.85 ± 8.48 | 39.14 ± 7.38 | 10.30 ± 6.91 | 16.36 ± 1.12 | 65.80 | 13 | |
| Ndst1−/−;2−/− | 91.50 ± 0.76 | 2.93 ± 0.56 | 2.53 ± 0.90 | 3.47 ± 1.13 | 10.58 | 17 | |
| WT (Ndst1f/f)# | CRISPR-Cas9 | 32.61 ± 3.38 | 58.69 ± 3.28 | 17.29 ± 1.95 | 16.35 ± 1.67 | 94.07 | 15 |
| Glce−/− | 23.12 ± 0.27 | 72.50 ± 0.25 | 6.84 ± 0.26 | 33.53 ± 0.58 | 107.73 | 14 | |
| Hs2stf/f | Cre-LoxP | 31.99 ± 2.59 | 59.06 ± 3.10 | 18.67 ± 2.82 | 17.55 ± 2.82 | 95.28 | 14 |
| Hs2st−/− | 21.92 ± 3.77 | 72.23 ± 2.71 | 0 | 29.96 ± 5.23 | 102.20 | 17 | |
| Hs6st1f/f | Cre-LoxP | 37.92 ± 5.36 | 54.41 ± 5.26 | 17.58 ± 2.10 | 15.03 ± 1.36 | 87.02 | 12 |
| Hs6st1−/− | 36.06 ± 5.20 | 58.05 ± 5.76 | 19.16 ± 2.17 | 8.40 ± 0.30 | 85.61 | 16 | |
| WTa* | Direct derivation and Cre-LoxP | 33.54 ± 2.54 | 56.77 ± 2.04 | 17.72 ± 2.91 | 17.02 ± 2.47 | 92.66 | 14.4 |
| Hs6st1f/f;2−/− | 37.03 ± 0.48 | 54.45 ± 0.32 | 13.29 ± 0.65 | 17.78 ± 0.25 | 85.52 | 12 | |
| Hs6st1−/−;2−/− | 31.80 ± 1.54 | 66.30 ± 1.12 | 26.41 ± 1.82 | 0.08 ± 0.14 | 92.79 | 16 | |
| WT (Ndst1f/f)# | CRISPR-Cas9 | 32.61 ± 3.38 | 58.69 ± 3.28 | 17.29 ± 1.95 | 16.35 ± 1.67 | 94.07 | 15 |
| Hs3st1−/− | 34.52 ± 3.86 | 54.98 ± 3.41 | 20.10 ± 1.57 | 16.95 ± 0.58 | 94.24 | 11 | |
| Hs3st4−/− | 37.30 ± 3.11 | 55.65 ± 2.51 | 18.03 ± 1.47 | 10.73 ± 1.28 | 84.15 | 16 | |
| Hs3st1−/−;4−/− | 39.62 ± 3.16 | 51.25 ± 4.06 | 18.88 ± 2.90 | 11.26 ± 2.45 | 92.79 | 15 | |
| Sulf1f/f;2f/f | Cre-LoxP | 31.71 ± 0.55 | 55.38 ± 0.51 | 18.59 ± 0.34 | 20.42 ± 0.36 | 94.39 | 15 |
| Sulf1−/−;2−/− | 34.96 ± 0.18 | 50.51 ± 1.32 | 14.18 ± 1.50 | 27.21 ± 0.96 | 91.91 | 18 |
Characterization of HS structure expression in mutant MLECs
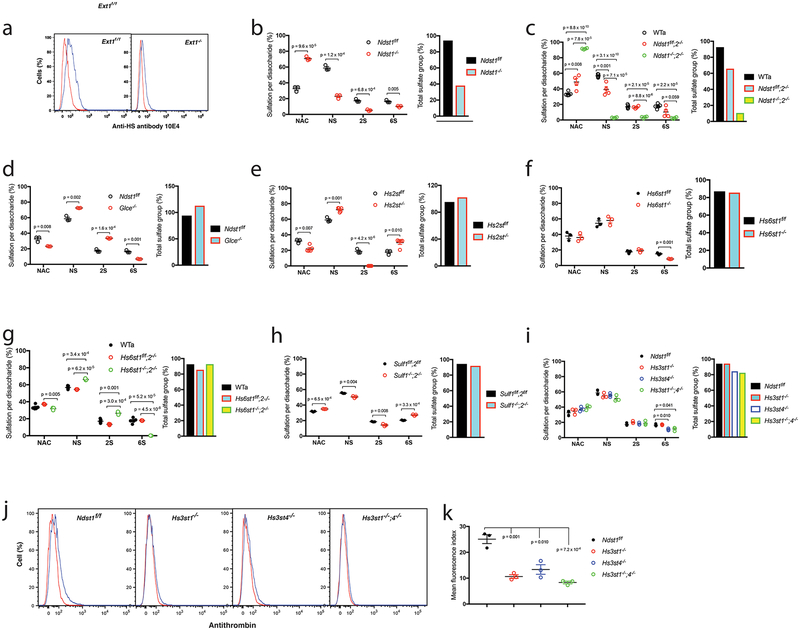
We next characterized the generated mutant cells to understand HS alteration upon the HS gene deletion. As we reported previously10–12, Ext1 deletion diminishes HS expression as reflected by diminished anti-HS antibody 10E4 staining (Fig. 2a). The other HS mutant cell lines all express HS and were analyzed for HS disaccharide composition ny digesting isolated HS with heparinases I-III and separation of the resulting disaccharide with an anion exchange column in HPLC system (Supplementary data). For the Cre-loxP-derived HS mutants we used their corresponding floxed cell lines as WT control. For the Ndst1f/f;2−/−, Ndst1−/−;2−/−, Hs6st1f/f;2−/− and Hs6st1−/−;2−/− lines that were directly derived from conventional gene knockout mice, the data pooled from the 5 WT MLEC lines (Ext1f/f, Ndst1f/f, Hs2stf/f, Hs6st1f/f and Sulf1f/f;2f/f) (WTa) were applied as their WT control. The Glce−/−, Hs3st1−/−, Hs3st4−/− and Hs3st1−/−;4−/− lines were generated using CRISPR-cas9 technology, their parent Ndst1f/f line was applied as WT control.
Figure 2. HS expression in the generated mutant MLEC lines.
a. Cell surface anti-HS antibody 10E4 binding was analyzed by flow cytometry. b-i. HS disaccharide composition analysis. HS isolated from mutant MLECs and their controls were digested with heparinases I-III, and the resulting disaccharides were separated by HPLC and quantified. The same type sulfate groups, including NS, 2S and 6S, of the separated disaccharides were combined to assess the levels of each sulfation modification type. The data were summarized from 3 independent experiments and are presented as mean ± SEM. WTa, the wildtype control data were summarized from similarly generated 5 wildtype MLEC lines (Ext1f/f, Ndst1f/f, Hs2stf/f, Hs6st1f/f and Sulf1f/f;2f/f); The total sulfate groups were calculated by adding up NS, 2S and 6S. j, k. Cell surface antithrombin binding. The wildtype control (Ndst1f/f) and Hs3st mutant MLECs were stained with biotinylated antithrombin and cell surface bound antithrombin was quantified by flow cytometry after further staining the cells with fluorescein-tagged streptavidin. The representative histograms from 3 independent experiments are shown (j). The quantitation of mean fluorescence index data were summarized from 3 independent experiments and are presented as mean ± SD (k). Statistical analyses were performed using two-sided, Student`s t-test.
In the Ndst family, the Ndst1−/− HS shows a 40–60% reduction in NS, 2S and 6S (Ndst1f/f: NS 58.7%, 2S 17.3%, 6S 16.4% vs Ndst1−/−: NS 22.7%, 2S 5.2%, 6S 10.0%) (Fig. 2b, Table 1), Ndst2−/− HS shows less reduction in the modifications, and Ndst1−/−;2−/− HS shows diminished sulfation modifications (WTa: NS 56.8%, 2S 17.7%, 6S 17.0% vs Ndst1f/f;2−/−: NS 39.1%, 2S 10.3%, 6S 16.4% vs Ndst1−/−;2−/−: 2.9% NS, 2S 2.5%, 6S 3.5%) (Fig. 2c, Table 1). The overall sulfation is reduced by 59.6% for Ndst1−/− HS, 29.0% for Ndst2−/− HS and 88.6% for Ndst1−/−;2−/− HS. The Ndst mutants can be used to determine the contribution of both NS and overall sulfation to ligand binding and downstream signaling activation in the cells. However, caution needs to be taken since Ndst−deficient HS has reductions in 2S and 6S.
Glce and Hs2st are the only members in their gene family in mouse. Glce−/− HS possesses reduction in 2S and increases in NS and 6S (Ndst1f/f: NS 58.7%, 2S 17.3%, 6S 16.4% vs Glce−/−: NS 72.5%, 2S 6.8%, 6S 33.5%) (Fig. 2d, Table 1). Hs2st−/− HS shows a disaccharide composition alteration similar to Glce−/− HS except for complete lack of 2S (Hs2stf/f: NS 59.1%, 2S 18.7%, 6S 17.6% vs Hs2st1−/−: NS 72.2%, 2S 0%, 6S 30.0%) (Fig. 2e, Table 1). Glce−/− and Hs2st−/− HS show increased overall sulfation, 14.5% and 7.3%, respectively. The Glce−/− and Hs2st−/− cells can be applied to specifically determine the necessity of IdoA and 2S versus the overall sulfation for ligand binding and downstream signaling, respectively.
The 6S is co-determined by Hs6sts and Sulfs. Hs6st1−/− HS shows reduced 6S (Hs6st1f/f: NS 54.4%, 2S 17.6%, 6S 15.0% vs Hs6st1−/−: NS 58.1%, 2S 19.2%, 6S 8.4%) (Fig. 2f, Table 1). Hs6st2−/− HS has normal disaccharide composition, whereas Hs6st1−/−;2−/− HS completely lacks 6S accompanied with increases in NS and 2S (WTa: NS 56.8%, 2S 17.7%, 6S 17.0% vs Hs6st1f/f;2−/−: NS 54.5%, 2S 13.3%, 6S 17.8% vs Hs6st1−/−;2−/−: NS 66.3%, 2S 26.4%, 6S 0.08%) (Fig. 2g, Table 1). Sulf1−/−;2−/− HS shows increase in 6S with reductions in NS and 2S, showing an effect opposite to Hs6st1 and Hs6st2 double deletion (Sul1f/f;2f/f: NS 55.4%, 2S 18.6%, 6S 20.4% vs Sulf1−/−2−/−: NS 50.5%, 2S 14.2%, 6S 27.2%) (Fig. 2h, Table 1). The Hs6st1−/−, Hs6st2−/−, Hs6st1−/−;2−/− and Sulf1−/−;2−/− HS have normal overall sulfation level. The Hs6st−/− and Sulf−/− cells can be applied to specifically determine the necessity of the 6S, higher 6S vs overall sulfation in ligand binding and downstream signaling in the cells.
In the Hs3st mutants, Hs3st1−/− HS has normal HS disaccharide composition, Hs3st4−/− and Hs3st1−/−;4−/− HS show slight reduction in 6S (Ndst1f/f: NS 58.7%, 2S 17.3%, 6S 16.4% vs Hs3st1−/−: NS 55.0%, 2S 20.1%, 6S 17.0% vs Hs3st4−/−: NS 55.7%, 2S 18.0%, 6S 10.7% vs Hs3st1−/−;4−/−: NS 51.3%, 2S 18.9%, 6S 11.3%) (Fig. 2i, Table 1). 3S cannot be detected in the disaccharide composition assay and was assessed by binding of antithrombin, a ligand that strictly requires HS-3S for binding. Hs3st1−/− and Hs3st4−/− MLECs both show reduced antithrombin binding, and the binding is further reduced on Hs3st1−/−;4−/− cells (Fig. 2j, k), reflecting that Hs3st1 and Hs3st4 deletion each reduces 3S. The Hs3st4−/−, not Hs3st1−/−, and Hs3st1−/−;4−/− HS show slightly reduced overall sulfation (Hs3st4−/−, −10.6%; Hs3st1−/−;4−/−, −12.3%) (Fig. 2i). The Hs6st1−/−, Hs6st2−/−, Hs6st1−/−;2−/− and Sulf1−/−;2−/− HS have normal overall sulfation, the combined examination of the Hs6st, Sulf and Hs3st mutant cells will allow determining and differentiating the importance of 3S, 6S and overall sulfation in ligand binding and downstream signaling activation in the cells.
Together, the generated MLEC library harbors alterations of all HS modification types and may allow determining the necessity of specific modification and overall HS sulfation level required for ligand binding and its downstream signaling activation.
The importance of fine structure versus sulfation level of HS in FGF2-FGFR1 signaling
HS functions as a co-receptor for FGF signaling by interacting with FGF and FGFR to form functional FGF/HS/FGFR ternary complexes on cell surface. FGF2 is one of the most studied HS-binding ligands, yet there is still debate on the relative importance of HS specific modification versus overall sulfation level required for FGF2 binding. Affinity binding and crystallography studies have demonstrated the necessity of NS and 2S for FGF2 binding, and 6S is only required for bridging FGF2 and FGFR for effective signaling activation but not for FGF2 binding13–16. Biochemical studies also demonstrated that distinct HS structures differentially regulate FGF2 signaling via different FGFRs in a manner not related to sulfation level, supporting that sulfation patterns are crucial17. On the other hand, some biochemical studies6,7,18, especially, a genetic study of HS mutant Drosophila8, suggested that the overall sulfation level appears more important for FGF signaling than strictly defined HS fine structures. Considering that HS deficiency in vivo potentially affects multiple signaling pathways simultaneously, direct stimulation of HS mutant MLECs, which express only FGFR1 at comparable levels among the cell lines (Supplementary Fig. 5, 6, 7), with FGF2 may allow to better address this fundamental issue.
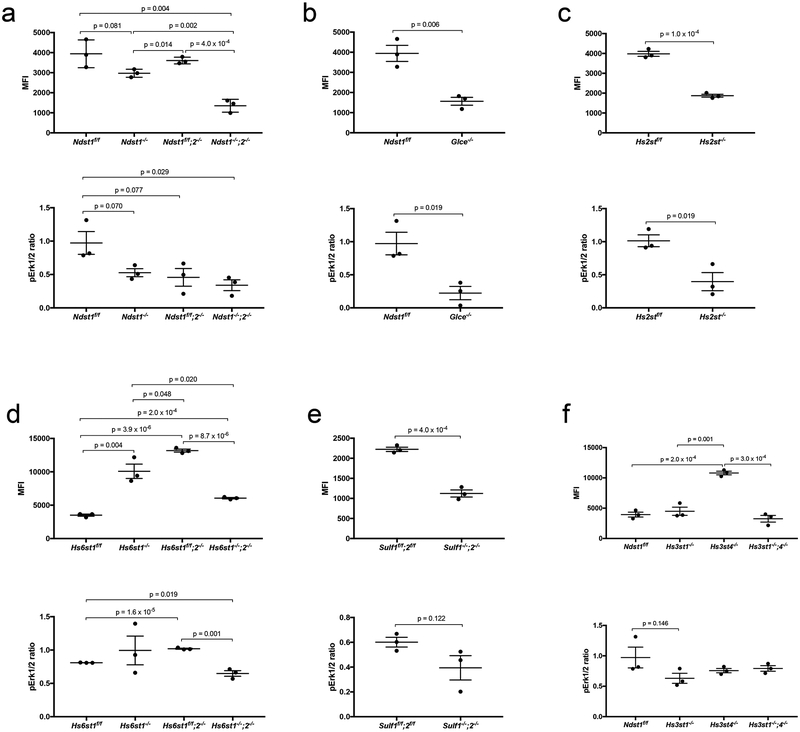
We assessed cell surface FGF2 binding by flow cytometry after staining the cells with biotinylated FGF2. FGFR1 activation was assessed by measuring Erk1/2 phosphorylation. Deletion of Ndst1, Ndst2 or both Ndst1 and Ndst2 reduces NS and the overall sulfation levels. Upon FGF2 staining, the reductions of cell surface FGF2 binding on the Ndst1−/−, Ndst1f/f;2−/− and Ndst1−/−;2−/− MLECs correlate with their NS and overall sulfation reduction levels, and with attenuated Erk1/2 phosphorylation (Fig. 3a; Table 2; Supplementary Fig. 8, 9), implying that NS, overall sulfation level or both is required for HS to facilitate FGF2-FGFR1 signaling. Since the Ndst1 deletion also reduces 2S and 6S, examinations of Hs2st−/− and Hs6st−/− cells is needed to better determine the importance of HS-NS in FGF2-FGFR1 signaling.
Figure 3. Effects of HS structure alteration on FGF2 binding and downstream Erk1/2 activation.
The HS mutant MLECs and their wildtype controls were incubated with biotinylated FGF2, and the cell surface bound FGF2 was quantified by flow cytometry after further staining the cells with fluorescein-tagged streptavidin. In parallel, the cells were stimulated with FGF2, and the resulting FGFR1 signaling activation was assessed by measuring increased pErk1/2 level. The data were summarized from 3 independent experiments and are presented as mean ± SD. Statistical analyses were performed using two-sided, Student`s t-test.
Table 2. Effects of HS biosynthetic/remodeling gene deletion on FGF2 binding and FGF2-FGFR1 signaling activation.
FGF2-FGFR1 signaling activation was assessed by measuring downstream Erk1/2 phosphorylation. ↑, increases cell surface FGF2 binding or FGF2-FGFR1 signaling activation; ↓, attenuates cell surface FGF2 binding or FGF2-FGFR1 signaling activation; ~, slight alteration which does not reach a statistical significance.
| Mutant | FGF2-FGFR1 signaling | |
|---|---|---|
| FGF2 binding | Erk1/2 phosphorylation | |
| Ndst1−/− | ~↓ | ~↓ |
| Ndst2−/− | ~↓ | ~↓ |
| Ndst1−/−;2−/− | ↓ | ↓ |
| Glce−/− | ↓ | ↓ |
| Hs2st−/− | ↓ | ↓ |
| Hs6st1−/− | ↑ | − |
| Hs6st1f/f;2−/− | ↑ | ↑ |
| Hs6st1−/−;2−/− | ↑ | ↓ |
| Hs3st1−/− | − | − |
| Hs3st4−/− | ↑ | − |
| Hs3st1−/−;4−/− | − | − |
| Sulf1−/−;2−/− | ↓ | − |
Deletion of Glce and Hs2st diminishes the epimerization and 2S, respectively, but slightly increases the overall sulfation of the mutant HS. The Glce−/− and Hs2st−/− MLECs both show significantly reduced FGF2 binding and attenuated downstream signaling (Fig. 3b, c; Table 2; Supplementary Fig. 8, 9), indicating that IdoA and 2S modifications are more important than overall sulfation level for FGF2-FGFR1 signaling.
Deletion of Hs6st1 only reduces 6S, deletion of Hs6st2 does not affect HS disaccharide composition, whereas Hs6st1 and Hs6st2 double deletion diminishes 6S accompanied with increases of NS and 2S. The deletion of Hs6st1, Hs6st2 or both does not alter overall sulfation. The Hs6st1−/−, Hs6st1f/f;2−/− and Hs6st1−/−;2−/− mutants show increased cell surface FGF2 binding, which correlates with unchanged, increased and attenuated Erk1/2 phosphorylation, respectively (Fig. 3d, Table 2; Supplementary Fig. 8, 9). Double deletion of Sulf1 and Sulf2 increases 6S, but does not alter the overall sulfation. The Sulf1−/−;2−/− MLECs show reduced FGF2 binding and slightly attenuated Erk1/2 phosphorylation (Fig. 3e; Table 2; Supplementary Fig. 8, 9). Therefore, examination of the Hs6st and Sulf mutants demonstrated that 6S is more important than the overall sulfation for FGF2 signaling and highlighted proper 6S is required for HS to effectively facilitate FGF2 signaling. These observations also exemplified that cell surface FGF2 binding capacity does not necessarily to positively correlate with downstream signaling activation. Furthermore, Hs6st2−/− cells have normal HS disaccharide composition but show altered FGF2 binding and downstream signaling activation, suggesting that Hs6st2 deletion alters the sulfated domains in HS.
Reduced cell surface antithrombin binding indicates that deletion of Hs3st1, Hs3st4, or Hs3st1 and Hs3st4 each reduces 3S. Deletion of Hs3st4, not Hs3st1, and double deletion of Hs3st1 and Hs3st4 also significantly reduces 6S and slightly reduces the overall sulfation. The cell surface FGF2 binding is increased on Hs3st4−/− MLECs, and remains unchanged on Hs3st1−/− and Hs3st1−/−;4−/− MLECs, whereas all the Hs3st mutant MLECs show normal Erk1/2 phosphorylation upon FGF2 stimulation (Fig. 3f; Table 2; Supplementary Fig. 8, 9). These results reveal that 3S is dispensable for HS to facilitate FGF2 signaling and demonstrate again that increased cell surface FGF2 binding does not positively correlate with enhanced FGF2 signaling activation.
Our results demonstrate that NS, IdoA and 2S are essential for HS to bind FGF2, but only in the presence of proper 6S modification can this binding effectively facilitate FGF2-FGFR1 signaling activation. Our data supports that HS fine structure is more important than overall sulfation for FGF2-FGFR1 signaling.
Characterization of anti-HS phage display antibody epitopes
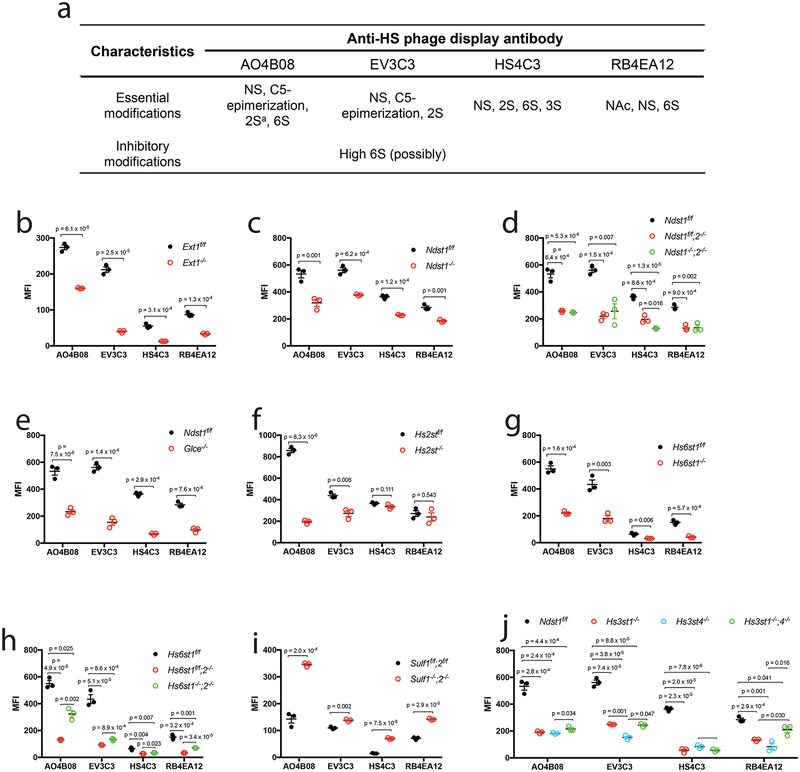
Anti-HS phage display antibodies are commonly used to probe HS structure expression in tissue in situ19. However, their putative epitopes have only been determined based on their binding capacity to chemically modified heparins and a limited number of synthetic HS oligosaccharides20,21. To address this unmet need, we examined our mutant library using the common anti-HS phage display antibodies AO4B08, EV3C3V, RB4EA12, and HS4C3 (Fig. 4a)19–24. The mutant MLECs were analyzed for antibody binding to cell surface HS by flow cytometry. All the four antibodies show positive staining of WT Ext1f/f cells (Fig. 4b). Epitopes recognized by AO4B08 and EV3C3 are more abundant than the ones recognized by HS4C3 and RB4EA12. The antibody binding to Ext1−/− cells was largely lost for EV3C3, Hs4C3 and RB4EA12, and partially lost for AO4B08 (Fig. 4b), showing that the antibodies indeed bind native HS.
Figure 4. Binding of anti-HS phage display antibody to mutant HS on endothelial cell surface.
a. Summarized characteristics of the HS modifications involved in binding or inhibiting the binding of the anti-HS phage display antibodies in reported biochemical studies. a, antibody AO4B08 requires an internal IdoA2S residue for binding. b-j. Cell surface binding of the antibody. The HS mutant MLECs and their wildtype controls were incubated with the VSV-G-tagged anti-HS phage display antibody, and the cell surface bound antibody was quantified by flow cytometry after further staining the cells with biotinylated anti-VSV-G antibody and fluorescein-tagged streptavidin. The data were summarized from 3 independent experiments and are presented as mean ± SD. Statistical analyses were performed using two-sided, Student`s t-test.
Previous biochemical studies established that the AO4B08-binding epitope contains NS, 6S, IdoA and an internal 2-O−sulfated IdoA residue (Fig. 4a)20,21,24. In agreement with these reports, we observed that binding of AO4B08 is reduced on Ndst1−/−, Ndst1f/f;2−/−, Ndst1−/−;2−/−, Glce−/−, Hs2st−/−, Hs6st1−/−, Hs6st1f/f;2−/− and Hs6st1−/−;2−/− MLECs, whereas increased on Sulf1−/−;2−/− MLECs (Fig. 4c–i). The binding of AO4B08 to Hs3st1−/−, Hs3st4−/− and Hs3st1−/−;4−/− on MLECs are also reduced (Fig. 4j), indicating that the AO4B08 epitope contains 3S.
Biochemical studies have also shown that the EV3C3 epitope contains NS, IdoA and 2S, and prefers to low level of 6S for better binding (Fig. 4a)20. We observed that EV3C3 have reduced binding on Ndst1−/−, Ndst1f/f;2−/−, Ndst1−/−;2−/−, Glce−/−, Hs2st−/−, Hs6st1−/−, Hs6st1f/f;2−/− and Hs6st1−/−;2−/− MLECs (Fig. 4c–h), showing that EV3C3 epitope does contain NS, IdoA, 2S and 6S. Interestingly, EV3C3 binding is increased to Sulf1−/−;2−/− MLECs, which express HS with increased 6S (Fig. 4i), showing that higher 6S does not interfere with EV3C3 binding. These observations indicate that 6S is an essential component of EV3C3 epitope. The bindings of EV3C3 to Hs3st1−/−, Hs3st4−/− and Hs3st1−/−;4−/− MLECs are also reduced (Fig. 4j), revealing that the EV3C3 epitope contains 3S.
Biochemical studies have shown that the HS4C3 epitope contains NS, 2S, 6S and 3S (Fig. 4a)19,22. In agreement with these observations, we found that HS4C3 exhibits reduced binding on Ndst1−/−, Ndst1f/f;2−/−, Ndst1−/−;2−/−, Hs6st1−/−, Hs6st1f/f;2−/−, Hs6st1−/−;2−/−, Hs3st1−/−, Hs3st4−/− and Hs3st1−/−;4−/− MLECs, but increased binding on Sulf1−/−;2−/− MLECs (Fig. 4c, d, g-j). However, the binding of HS4C3 to Hs2st−/− MLECs is unchanged (Fig. 4f), indicating that the HS4C3 epitope does not necessarily to contain 2S modification. In addition, the binding of HS4C3 to Glce−/− MLECs is reduced (Fig. 4E), indicating that IdoA is an essential component of the HS4C3 epitope.
Previous biochemical studies determined that RB4EA12 epitope contains N−acetylated and N−sulfated glucosamine residues with 6S modification (Fig. 4a)20. This is supported by reduced RB4EA12 binding on Ndst1−/−, Ndst1f/f;2−/−, Ndst1−/−;2−/−, Hs6st1−/−, Hs6st1f/f;2−/−, Hs6st1−/−;2−/− MLECs and increased binding on Sulf1−/−;2−/− MLECs (Fig. c, d, g-i). Hs2st−/− MLECs show normal RB4EA12 binding (Fig. 4f), indicating that the native HS epitope recognized by RB4EA12 does not necessarily to contain 2S, in agreement with the biochemical analysis report. RB4EA12 show reduced binding to Glce−/−, Hs3st1−/−, Hs3st4−/− and Hs3st1−/−;4−/− MLECs (Fig. 4e, j), revealing that the RB4EA12 epitope also contains IdoA and 3S.
Our HS mutant cell staining study determined the structural features of the native HS epitopes recognized by the four anti-HS phage antibodies, which, in most cases, are in good agreement with the reported biochemical studies (Supplementary Table 1). However, several discrepancies were observed including the requirement of 6S for EV3C3 and the lack of a required 2S for HS4C3. In addition, 3S was observed to be a component of native HS epitopes for AO4B08, EV3C3 and RE4EA12. The 3S might be additive since the Hs3st1−/−, Hs3st4−/− or Hs3st1−/−;4−/− MLECs only show reduced the antibody binding. Furthermore, the epitopes for HS4C3 and RB4EA12 also contain IdoA.
Characterization of the HS modification network in HS fine structure expression
HS biosynthesis is generally termed as a sequential process that is initiated by copolymerases Ext1/Ext2, followed by modification reactions that is initiated by Ndsts and proceeded by Glce, Hs2st, Hs6sts and Hs3sts. The biosynthesized HS is further subjected to extracellular remodeling by Sulf5,9. In HS composition analysis, we noticed that inactivation of an individual HS enzyme also affects upstream and/or downstream modifications that are not catalyzed by the mutated enzyme, indicating an internal HS modification network regulation exists in MLECs. We therefore correlated the disaccharide composition of the mutant HS with HS gene expression in the mutant cells to better understand the inter-modification network regulation in MLECs.
Deletion of Ndst1 upregulates Ndst3, Hs6st1 and Hs3st4 but downregulates Hs2st (Fig. 5a). Ndst2 deletion does not affect other Ndst expression, and upregulates Hs2st, Hs3st4 and Sulf1 (Fig. 5b), showing that Ndst1 and Ndst2 have distinct regulation on other modification gene expressions. The Ndst1 deletion reduces NS and 6S but upregulates Ndst3 and Hs6st1. Similarly, Ndst2 deletion has no effect on 2S and 6S even upregulates Hs2st and Sulf1. These observations indicate that, at the NS step, the modification network regulation occurs at transcriptional level but contributes minimally to HS fine structure expression. Furthermore, simultaneous deletion of Ndst1 and Ndst2 diminishes the network regulation except for slight Ndst3 upregulation (Fig. 5c; Supplementary Fig. 10a), indicating the modification network regulation essentially depends on Ndst expression.
Figure 5. Inter-regulation among HS modification and remodeling genes in HS fine structure expression.
The expressions of HS modification and remodeling genes in the HS mutant MLECs and their wildtype controls were quantified by qRT-PCR analysis and compared. The data were summarized from 3 independent experiments and are present as mean ± SD. Statistical analyses were performed using two-sided, Student`s t-test. The targeted genes in the mutant MLECs are highlighted by light blue.
Glce deletion upregulates Ndst1, Ndst2, Hs2st, Hs6st1 and Sulf2 which correlate with increased NS and 6S in HS (Fig. 5d), indicating an inhibitory regulation of Glce on NS and 6S by downregulating Ndst1, Ndst2 and Hs6st1. Increased 6S in Glce−/− HS also indicates that the Sulf2 upregulation contributes minimally to 6S modification.
Hs2st deletion upregulates Ndst2, Glce, Hs6st1, Hs6st2, Hs3st1, Hs3st4, Hs3st6, Sulf1 and Sulf2 (Fig. 5e), showing a profound inhibition of Hs2st on other modification genes. Similar to Glce, Hs2st deletion increases NS and 6S, indicating that Hs2st inhibits NS and 6S by downregulating Ndst2, Hs6st1 and Hs6st2 and the contributions of the upregulated Sulf1 and Sulf2 remain minimal.
Hs6st1 deletion upregulates Ndst1, Hs2st, Hs3st1, Sulf1 and Sulf2 but does not affect NS and 2S (Fig. 5f). The Hs6st2 deletion upregulates Hs6st1 and Hs3st1, and does not alter 6S, NS and 2S (Fig. 5g; Supplementary Fig. 10b). However, simultaneous deletion of Hs6st1 and Hs6st2 upregulates Ndst1, Ndst2, Hs2st, Hs3st1 and Sulf2 (Fig. 5h), which correlate with increased NS and 2S, revealing inhibition of Hs6st on NS and 2S modification by suppression of Ndst1, Ndst2 and Hs2st expression.
Hs3st1 deletion slightly upregulates Ndst2 but downregulates Hs2st and Hs3st4 (Fig. 5i), and does not alter HS composition. Hs3st4 deletion slightly upregulates Ndst1, Ndst2 and Hs3st1, slightly downregulates Hs2st, and highly upregulates Sulf2 (Fig. 5j), which correlates with reduced 6S. Simultaneous deletion of Hs3st1 and Hs3st4 shows regulatory effects similar to Hs3st4 deletion (Fig. 5k, Supplementary Fig. 10c–e) except for slight Hs6st1 upregulation, indicating that, within the Hs3st family expressed in MLECs, Hs3st4 plays the major role in negative regulation of the modification network and shows an overall role in increasing 6S.
Simultaneous deletion of Sulf1 and Sulf2 downregulates Ndst1, Ndst2 and Hs2st, and upregulates Glce and Hs3st1 (Fig. 5l), which correlate with reduced NS and 2S, indicating a positive regulation of Sulf for NS and 2S and a negative regulation for epimerization and 3S. Furthermore, the 6S increase in Sulf1−/−;2−/− HS indicates that the overall 6S level is critically regulated by Sulfs in MLECs.
Together, our systematic study reveals that: 1). In MLECs Ndsts are essentially required for the inter-modification network regulation to occur, in a good agreement with the modification initiation function of Ndsts in HS biosynthesis; 2). Glce, Hs2st, Hs6sts and Hs3sts reciprocally inhibit non-self modifications at gene expression and HS composition levels; and 3). Sulfs regulate positively NS and 2S, but negatively epimerization and 3S.
Regulation of HS chain length by HS gene expression
Ndst2, but not Ndst1, was reported to regulate HS chain length25. Currently we lack a systematic view of the HS chain length regulation by HS genes. In PAGE analysis of intact HS, we determined that Ndst1−/− and Hs3st1−/−;4−/− HS have normal chain length, whereas Ndst2−/−, Glce−/−, Hs6st2−/− and Hs3st1−/− HS become shorter, and Ndst1−/−;2−/−, Hs2st−/−, Hs6st1−/−, Hs6st1−/−;2−/− and Hs3st4−/− HS become longer (Supplementary Fig. 11). Comparing single and double gene deficient HS reveals that, in the same family genes, including Ndst1 and Ndst2, Hs6st1 and Hs6sts, and Hs3st1 and Hs3st4, the two expressing genes have opposite regulation of HS chain length and reciprocally inhibit each other. All examined HS genes regulate HS chain length and these can be positive or negative as well as direct or indirect.
Discussion
Examination of HS mutant cells is an effective and straightforward approach for determining the function and structure-function relationships of native HS in a biological setting. This type of study was originally explored using chemical mutagenesis-generated HS mutant CHO cell lines26–28. Currently only four CHO HS mutant cell lines are available. CHO cells do not express Hs3sts and cannot be applied to examine 3S-related studies29,30. CHO cells also lack endogenous expression of many HS-dependent signaling receptors, such as FGFR, receptors for vascular endothelial growth factor (VEGF) and Slit; very limited cellular function information has been obtained using these CHO mutants. Our examination of Ndst1−/− and Ndst1f/f MLEC lines determined that NS is required for HS to function as a co-receptor for VEGF and Slit3 signaling31–34. The success of these previous studies and the FGF signaling co-receptor study described here demonstrate that our HS mutant MLEC library represents a powerful platform to examine the roles and the structure-function relationship of HS in a cellular context, which could be correlated with in vivo findings in corresponding mutant mice. In addition, our study demonstrates CRISPR-Cas9 technology as a highly efficient approach to generate HS mutant MLECs and is expected to be applicable to other cell types to generate HS mutants.
HS functions as a co-receptor for FGF signaling by interacting with both FGF and FGFR. A number of biochemical studies showed that interactions of HS with FGFs require unique structures in which NS, 2S and 6S contribute to generate specific sulfation patterns15–17,35–37. Crystallographic studies observed that 2S and 6S form hydrogen bonds with heparin binding residues of FGFs and/or FGFRs to induce dimerization of FGFRs14. Meanwhile, other studies reported that FGF binding to HS is dictated primarily by overall sulfation level rather than by the precise positioning of various sulfate groups7,8,18. We observed that Glce−/−, Hs2st−/−, Hs6st−/− and Sulf−/− MLECs, which only express FGFR1, show altered FGF2 binding and downstream signaling activation although the mutants have normal or slightly increased HS overall sulfation. Our results support that HS fine structure is more important than overall sulfation for FGF2-FGFR1 signaling activation.
HS phage display antibodies have been widely applied to probe HS structure in various tissues in situ19,21,38. However, their native HS epitope structures remain largely unclear21,39. Using our HS mutant cell library we found that: 1) EV3C3 epitope involves 6S and natural increase of 6S does not inhibit antibody binding; 2) HS4C3 epitope does not necessarily contain 2S but requires IdoA; and 3) except for the HS4C3, the epitopes for AO4B08, EV3C3 and RB4EA12 also require 3S. The involvement of rare 3S suggests that the antibodies may function in the “all-or-nothing” high-specific binding mode40, supporting the application of the antibodies as high-affinity probes for specific HS structures in situ. Further studies with a comprehensive, synthesized, 3S-containing HS library may help further define the epitope structures.
Studies of Hs2st−deficient CHO cells and mouse fibroblasts biosynthesis41,42 and HS-deficient Drosophila8 and Caenorhabditis elegans43 revealed an inter-regulatory modification network in HS biosynthesis. By systematic HS modification gene perturbation in C. elegans, an inter-regulation model of modification enzyme in HS biosynthesis in metazoans was proposed – Glce, Hs2st and Hs6sts inhibit NS, Glce stimulates both 2S and 6S, and Hs2st and Hs6st inhibits 6S and 2S, respectively, and 6S is also inhibited by Sulf143. We observed similar inter-regulation in MLECs except for Glce, which stimulates 2S but inhibits 6S, and Sulfs also stimulate 2S. The discrepancy may be due to the fact that we only examined MLECs whereas the C. elegans study examined the whole model organism. We also examined Ndsts and Hs3sts, and uncovered that the inter-regulation network essentially depends on Ndst expression and Hs3st4 stimulates 6S. In addition, we also observed that HS gene generally regulates, either elongates or shortens, HS chain, and furthermore, HS genes within same family normally act reciprocally to regulate HS chain length. These studies provide a comprehensive and systematic view of the inter-regulation of HS genes in mammalian cells at specific cell type, transcription and HS structure levels. HS expression is cell type-specific; similar studies with other cell types may determine if our observed inter-regulation in HS expression is common in mammalian cells.
In current study, HS structure analyses determined disaccharide composition and chain length, not the sequence which plays the central role underpinning structure-function relationship of HS. This should be examined once capable technology becomes available in future.
Online Methods
Cell derivation, immortalization, and gene knockout
The Ext1f/f11,44, Ndst1f/f32,45, Ndst1f/f;2−/−31,46, Hs2stf/f47, Hs6st1f/f48, Hs6st1f/f;2−/−49, and Sulf1f/f;2f/f50 mice in C57BL6 background were bred in accordance with regulation in University of Georgia (UGA) and approved by UGA IACUC. The primary MLECs were isolated from adult mice and were immortalized as we reported previously11. Briefly, lung was dissected from mice after perfusion, cut into small pieces and digested with collagenase and dispase. The resultant cell suspension was subject to phase separation using Histopaque 1088 (Sigma-Aldrich). The endothelial cells were enriched at the interface of PBS and histopaque. The cells were further purified using anti-CD31 antibody-conjugated magnetic beads (Miltenyi Biotech). The isolated cells were cultured in gelatin-coated plates in high glucose DMEM supplemented with 20% FBS (Atlanta Biologicals), 30 μg/ml endothelial cell growth supplement (Sigma-Aldrich) and 50 μg/ml heparin (Sigma-Aldrich). The primary cells were immortalized by transfecting a plasmid encoding SV40 large T-antigen and subject to single cell cloning culture. After 7 days, single cell clones were picked out and expand for future use. A plasmid encoding Cre recombinase was introduced into the cells by transiently transfection to knockout out the floxed genes in the obtained cell clones. The knockout cell lines were obtained by single cell cloning culture and targeted gene deletion confirmed by genomic DNA genotyping as reported previously45–49,51. The genotype condition and primers are listed in Supplementary Table 2.
CRISPR-Cas9 procedure to generate knockout cell line
Guide RNAs (gRNA) for mouse Glce, Hs3st1 and Hs3st4 were designed using the online CRISPR Design Tool (http://tools.genome-engineering.org) (Supplementary Fig. 4) and inserted into the pCRISPR-LvSG03 plasmid (Genecopeia, USA). The gRNA and Cas9 plasmids were co-transfected into the Ndst1f/f MLEC line. The cells were selected by puromycin (10 μg/ml) for 3–7 days in culture, and then were single cell sorted into 96-well culture plates. The cell clones were screened for the targeted gene indel mutation by PCR amplification of the edited regions, followed by enzyme mismatch assay after nuclease T7E1 digestion52. The selected clones were further validated by sequencing the targeted regions. The sequence chromatograms of the pooled PCR products of the targeted regions were decoded by either DSDecodeM53 or CRISP-ID54 software online.
FGF2 biotinylation
A heparin-Sepharose CL-6B column was pre-equilibrated with binding buffer PB150 (17.9 mM Na2HPO4, 2.1 mM NaH2PO4, 150 mM NaCl, pH7.8) and then loaded with FGF2 in the binding buffer. The flow-through was reloaded onto the column twice to ensure complete binding. After washing 4 times with PB150, sulfo-NHS-biotin was added according to the manufacturer`s instruction and incubated for 1 h at RT. Reaction was terminated by adding glycine (100 mM glycine in PB150). After washing the column with gradient NaCl solutions (150 mM, 600 mM, and 1 M) in 20 mM PB buffer (44.75 mM Na2HPO4, 5.25 mM NaH2PO4, pH7.8), biotinylated-FGF2 was eluted with 4M NaCl PB buffer, collected and stored at −80°C before use.
Flow cytometry analysis
MLECs were collected after digestion with PBS supplemented with 2 mM EDTA and 1% BSA (PBS-EB), and adjusted at 106 cells/ml. For FGF2 binding, the dissociated cells were incubated with biotinylated FGF2 (20 μg/ml) on ice for 30 min. Following, the cells were incubated with eFluor®450-conjugated streptavidin and were then subjected to flow cytometry analysis (LSRII, BD Biosciences). For endothelial cell marker analysis, the cells were incubated with FITC-conjugated anti-mouse CD31 (eBioscience) or PE-conjugated anti-mouse VEGFR2 antibody (BD Pharmingen) on ice for 30 min and were then subjected to flow cytometry analysis. For HS phage display antibody staining, 106 MLECs in 100 μl PBS-EB were firstly incubated with 10 μl antibody (AO4B08, HS4C3, RB4EA12, and EV3C3) on ice for 1 h and then incubated with biotinylated anti-VSV-G antibody (Abcam) on ice for 1 h after washing. After further incubating with streptavidin-conjugated eFluor®450 (eBioscience) and propidium iodide (25 μg/ml) for 30 min on ice, the cells were subjected to flow cytometry analysis. The flow cytometry data was processed and analyzed using FlowJo software.
Real time polymerization chain reaction (RT-PCR) analysis
Cells in confluence were washed with PBS 3 times and then flash frozen in liquid nitrogen and stored at −80°C until use. Total RNA isolation, cDNA synthesis, qRT-PCR reactions, and data analysis were performed as previously reported55. Ribosomal Protein L4 (RPL4) was included as internal control for normalization of individual gene expression.
HS Disaccharide analysis56,57
Cells were cultured in a 10-cm dish in high glucose DMEM supplemented with 10% FBS and penicillin (100 U/ml) and streptomycin (100 μg/ml). After washing with PBS-EB, the cells were lysed with 1 ml 0.1 M NaOH. Following, 8 μl acetic acid was added. The cell lysate was added with Pronase (2 mg/ml, Sigma-Aldrich) and digested overnight at 37°C. After inactivation at 95°C for 10 min, the released glycosaminoglycan was enriched by passing through a Q-column, desalting and then digested with heparin lyases I, II, and III (Sigma-Aldrich) overnight at 37 °C. The resultant HS disaccharides were collected by filtering the digested glycosaminoglycan though a 3000 MW cut-off filter (Thermo-Fisher), labeled with 2-aminobenzide and then subjected to separation by an Agilent 1260 HPLC system with a Propac PA1 (4 × 250 mm, DIONEX) column connected with a Propac PA1 Guard column (4 × 50 mm, DIONEX). The separated disaccharides were detected using a fluorescence detector with excitation at 348 nm and emission at 440 nm.
PAGE analysis of intact HS extracted from cell samples58,59
1) HS extraction: Cell samples were first treated with 100 μl protein extraction reagent under sonication for 20 min. In order to remove chondroitin sulfate and hyaluronic acid, recombinant chondroitin lyase and hyaluronidase (100 mU each) and 500 μl digestion buffer (50 mM ammonium acetate, 2 mM calcium chloride) were added to the reaction buffer and incubated under 37 °C for 2 h. Next, 0.1 ml actinase E solution (20 mg/mL) was mixed with each sample to perform proteolysis at 55 °C for 24 h. HS was then recovered and purified by Vivapure MINI Q spin column. The obtained product solution was desalted by passing through 3 KDa MWCO spin columns, lyophilized and finally re-dispersed in 10 μl DI water for further use.
2) PAGE: A analytical PAGE gel with 5 ml of 15% total acrylamide monomer resolving solution was allowed to polymerize for 30 min with 5 μl of TEMED and 30 μl of 10% (w/v) ammonium persulfate. Above the polymerized resolving gel, 2 mL of 5% total acrylamide monomer stacking gel was cast. An aliquot of 5 μl of purified HS was loaded in a solution of 10 μg/mL (w/v) Phenol Red and 25% (w/v) sucrose. Electrophoresis was conducted for 30 min at a constant voltage of 200 V.
Western blotting
Cells (2 × 105 cells/well) were seeded in 6-well plate and cultured overnight in high DMEM supplemented with 10% FBS. Prior to stimulation, the cells were starved in DMEM serum-free culture condition for 1 h. The cells were incubated with DMEM supplemented with FGF2 (5 ng/ml) for 15 min and then lysed with RIPA buffer containing protease and phosphatase inhibitors (Sigma-Aldrich). The resultant cell lysate was resolved on 7.5% SDS-PAGE, transferred onto nitrocellulose membrane, and blotted with anti-phospho-Erk1/2 (Cell Signaling Technology) and anti-Erk1/2 (Cell Signaling Technology) antibodies at 1:1000. The HS-dependent FGF2 induced intracellular signaling activation was calculated as Erk1/2 activation ratio = [(p-Erk1/2 - Erk1/2)FGF2 - (p-Erk1/2 - Erk1/2) control]mutant / [(p-Erk1/2 -Erk1/2) FGF2 -(p-Erk1/2 - Erk1/2) control]wildtype. For FGFR expression analysis, the untreated cell lysate was subject to 8% SDS-PAGE separation, transferred to nitrocellulose membrane, and blotted with anti-FGFR1 (D8E4, Cell Signaling Technology) or anti-FGFR2 (D4L2V, Cell Signaling Technology) antibodies in 1:1000 dilution to determine the protein expression levels of FGFR1 and FGFR2 in the cell lines. The β-actin in the cell lysate was also determined to serve as internal control by blotting the membrane with a monoclonal anti-β-actin antibody (A2228). The results were imaged using KwikQuant™ imager (Kindle Bioscience, LLC, USA). The quantitation was based on the densitometry of the western blot bands.
Statistical analysis
The statistical analysis was carried out using two-sided student`s t-test with p<0.05 considering statistical significance.
Supplementary Material
Acknowledgement
The research was supported by NIH (R21HL131553, P41GM103390, 5R01HL093339 and U01CA225784 to L.W.) and by AHA (15POST21260001,17SDG33660550 to H.Q). S.S. and S.W. were supported by the Oversea Visiting Scholar Program for Middle-aged and Young Teachers in Shanghai Municipal Universities. We thank Karen Howard for her English revision of the manuscript. We also thank Donald Bernsteel and Dr. Huabei Guo in Dr. Michael Pierce lab at the University of Georgia for providing the HT-29 cells, and Jamie Barber and Julie Nelson in the flow cytometry core at the University of Georgia for their technical assistance.
Footnotes
Resource availability. All cell lines are available on request under a standard material transfer agreement with the University of Georgia for academic research purposes.
Reporting Summary. Further information on experimental design is available in the Life Sciences Reporting Summary linked to this article.
Data availability. All data generated or analyzed during this study are included in this article and/or associated Supplementary Information files. The raw data files are available upon request.
Competing Financial Interests Statement. No competing financial interests.
Reference
- 1.Bernfield M et al. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem 68, 729–777 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Esko JD & Lindahl U Molecular diversity of heparan sulfate. J Clin Invest 108, 169–173, doi: 10.1172/JCI13530 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang L, Brown JR, Varki A & Esko JD Heparin’s anti-inflammatory effects require glucosamine 6-O-sulfation and are mediated by blockade of L- and P-selectins. J Clin Invest 110, 127–136 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bishop JR, Schuksz M & Esko JD Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature 446, 1030–1037 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Xu D & Esko JD Demystifying heparan sulfate-protein interactions. Annu Rev Biochem 83, 129–157, doi: 10.1146/annurev-biochem-060713-035314 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jemth P et al. Biosynthetic oligosaccharide libraries for identification of protein-binding heparan sulfate motifs. Exploring the structural diversity by screening for fibroblast growth factor (FGF)1 and FGF2 binding. J Biol Chem 277, 30567–30573, doi: 10.1074/jbc.M203404200 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Kreuger J et al. Fibroblast growth factors share binding sites in heparan sulphate. Biochem J 389, 145–150, doi: 10.1042/BJ20042129 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamimura K et al. Specific and flexible roles of heparan sulfate modifications in Drosophila FGF signaling. J Cell Biol 174, 773–778, doi: 10.1083/jcb.200603129 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindahl U & Li JP Interactions between heparan sulfate and proteins-design and functional implications. Int Rev Cell Mol Biol 276, 105–159, doi: 10.1016/S1937-6448(09)76003-4 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Kraushaar DC, Yamaguchi Y & Wang L Heparan sulfate is required for embryonic stem cells to exit from self-renewal. J Biol Chem 285, 5907–5916, doi: 10.1074/jbc.M109.066837 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiu H et al. Quantitative phosphoproteomics analysis reveals broad regulatory role of heparan sulfate on endothelial signaling. Mol Cell Proteomics 12, 2160–2173, doi: 10.1074/mcp.M112.026609 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kraushaar DC et al. Heparan sulfate facilitates FGF and BMP signaling to drive mesoderm differentiation of mouse embryonic stem cells. J Biol Chem 287, 22691–22700, doi: 10.1074/jbc.M112.368241 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faham S, Hileman RE, Fromm JR, Linhardt RJ & Rees DC Heparin structure and interactions with basic fibroblast growth factor. Science 271, 1116–1120 (1996). [DOI] [PubMed] [Google Scholar]
- 14.Schlessinger J et al. Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol Cell 6, 743–750 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Turnbull JE, Fernig DG, Ke Y, Wilkinson MC & Gallagher JT Identification of the basic fibroblast growth factor binding sequence in fibroblast heparan sulfate. J Biol Chem 267, 10337–10341 (1992). [PubMed] [Google Scholar]
- 16.Ashikari-Hada S et al. Characterization of growth factor-binding structures in heparin/heparan sulfate using an octasaccharide library. J Biol Chem 279, 12346–12354, doi: 10.1074/jbc.M313523200 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Guimond SE & Turnbull JE Fibroblast growth factor receptor signalling is dictated by specific heparan sulphate saccharides. Curr Biol 9, 1343–1346 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Jastrebova N et al. Heparan sulfate-related oligosaccharides in ternary complex formation with fibroblast growth factors 1 and 2 and their receptors. J Biol Chem 281, 26884–26892, doi: 10.1074/jbc.M600806200 (2006). [DOI] [PubMed] [Google Scholar]
- 19.van Kuppevelt TH, Dennissen MA, van Venrooij WJ, Hoet RM & Veerkamp JH Generation and application of type-specific anti-heparan sulfate antibodies using phage display technology. Further evidence for heparan sulfate heterogeneity in the kidney. J Biol Chem 273, 12960–12966 (1998). [DOI] [PubMed] [Google Scholar]
- 20.Dennissen MA et al. Large, tissue-regulated domain diversity of heparan sulfates demonstrated by phage display antibodies. J Biol Chem 277, 10982–10986, doi: 10.1074/jbc.M104852200 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Thompson SM et al. Heparan sulfate phage display antibodies identify distinct epitopes with complex binding characteristics: insights into protein binding specificities. J Biol Chem 284, 35621–35631, doi: 10.1074/jbc.M109.009712 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ten Dam GB et al. 3-O-sulfated oligosaccharide structures are recognized by anti-heparan sulfate antibody HS4C3. J Biol Chem 281, 4654–4662, doi: 10.1074/jbc.M506357200 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Jenniskens GJ, Oosterhof A, Brandwijk R, Veerkamp JH & van Kuppevelt TH Heparan sulfate heterogeneity in skeletal muscle basal lamina: demonstration by phage display-derived antibodies. J Neurosci 20, 4099–4111 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurup S et al. Characterization of anti-heparan sulfate phage display antibodies AO4B08 and HS4E4. J Biol Chem 282, 21032–21042, doi: 10.1074/jbc.M702073200 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Deligny A et al. NDST2 (N-Deacetylase/N-Sulfotransferase-2) Enzyme Regulates Heparan Sulfate Chain Length. J Biol Chem 291, 18600–18607, doi: 10.1074/jbc.M116.744433 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bai X, Wei G, Sinha A & Esko JD Chinese hamster ovary cell mutants defective in glycosaminoglycan assembly and glucuronosyltransferase I. The Journal of biological chemistry 274, 13017–13024 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Esko JD, Rostand KS & Weinke JL Tumor formation dependent on proteoglycan biosynthesis. Science 241, 1092–1096 (1988). [DOI] [PubMed] [Google Scholar]
- 28.Esko JD, Stewart TE & Taylor WH Animal cell mutants defective in glycosaminoglycan biosynthesis. Proc Natl Acad Sci U S A 82, 3197–3201 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, Lawrence R, Frazier BA & Esko JD in Methods in Enzymology Vol. Volume 416 205–221 (Academic Press, 2006). [DOI] [PubMed] [Google Scholar]
- 30.Xu X et al. The genomic sequence of the Chinese hamster ovary (CHO)-K1 cell line. Nat Biotech 29, 735–741, doi:http://www.nature.com/nbt/journal/v29/n8/abs/nbt.1932.html#supplementary-information (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wijelath E et al. Multiple mechanisms for exogenous heparin modulation of vascular endothelial growth factor activity. J Cell Biochem 111, 461–468, doi: 10.1002/jcb.22727 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang B et al. Heparan sulfate deficiency disrupts developmental angiogenesis and causes congenital diaphragmatic hernia. J Clin Invest 124, 209–221, doi: 10.1172/JCI71090 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiu H, Xiao W, Yue J & Wang L Heparan sulfate modulates Slit3-induced endothelial cell migration. Methods Mol Biol 1229, 549–555, doi: 10.1007/978-1-4939-1714-3_43 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Zhang B et al. Repulsive axon guidance molecule Slit3 is a novel angiogenic factor. Blood 114, 4300–4309, doi: 10.1182/blood-2008-12-193326 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakato H & Kimata K Heparan sulfate fine structure and specificity of proteoglycan functions. Biochim Biophys Acta 1573, 312–318 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Habuchi H, Habuchi O & Kimata K Sulfation pattern in glycosaminoglycan: does it have a code? Glycoconj J 21, 47–52, doi: 10.1023/B:GLYC.0000043747.87325.5e (2004). [DOI] [PubMed] [Google Scholar]
- 37.Maccarana M, Casu B & Lindahl U Minimal sequence in heparin/heparan sulfate required for binding of basic fibroblast growth factor. J Biol Chem 268, 23898–23905 (1993). [PubMed] [Google Scholar]
- 38.Lensen JF et al. Localization and functional characterization of glycosaminoglycan domains in the normal human kidney as revealed by phage display-derived single chain antibodies. J Am Soc Nephrol 16, 1279–1288, doi: 10.1681/ASN.2004050413 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Powell AK, Yates EA, Fernig DG & Turnbull JE Interactions of heparin/heparan sulfate with proteins: appraisal of structural factors and experimental approaches. Glycobiology 14, 17R–30R, doi: 10.1093/glycob/cwh051 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Li JP & Kusche-Gullberg M Heparan Sulfate: Biosynthesis, Structure, and Function. Int Rev Cell Mol Biol 325, 215–273, doi: 10.1016/bs.ircmb.2016.02.009 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Bai X & Esko JD An animal cell mutant defective in heparan sulfate hexuronic acid 2-O-sulfation. J Biol Chem 271, 17711–17717 (1996). [DOI] [PubMed] [Google Scholar]
- 42.Merry CL et al. The molecular phenotype of heparan sulfate in the Hs2st−/− mutant mouse. J Biol Chem 276, 35429–35434 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Townley RA & Bulow HE Genetic analysis of the heparan modification network in Caenorhabditis elegans. J Biol Chem 286, 16824–16831, doi: 10.1074/jbc.M111.227926 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Method reference
- 44.Inatani M, Irie F, Plump AS, Tessier-Lavigne M & Yamaguchi Y Mammalian brain morphogenesis and midline axon guidance require heparan sulfate. Science 302, 1044–1046 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Wang L, Fuster M, Sriramarao P & Esko JD Endothelial heparan sulfate deficiency impairs L-selectin- and chemokine-mediated neutrophil trafficking during inflammatory responses. Nat Immunol 6, 902–910, doi:ni1233 [pii] 10.1038/ni1233 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Forsberg E et al. Abnormal mast cells in mice deficient in a heparin-synthesizing enzyme. Nature 400, 773–776, doi: 10.1038/23488 (1999). [DOI] [PubMed] [Google Scholar]
- 47.Stanford KI et al. Heparan sulfate 2-O-sulfotransferase is required for triglyceride-rich lipoprotein clearance. J Biol Chem 285, 286–294, doi: 10.1074/jbc.M109.063701 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Izvolsky KI, Lu J, Martin G, Albrecht KH & Cardoso WV Systemic inactivation of Hs6st1 in mice is associated with late postnatal mortality without major defects in organogenesis. Genesis 46, 8–18, doi: 10.1002/dvg.20355 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Sugaya N, Habuchi H, Nagai N, Ashikari-Hada S & Kimata K 6-O-sulfation of heparan sulfate differentially regulates various fibroblast growth factor-dependent signalings in culture. J Biol Chem 283, 10366–10376, doi: 10.1074/jbc.M705948200 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Tran TH, Shi X, Zaia J & Ai X Heparan sulfate 6-O-endosulfatases (Sulfs) coordinate the Wnt signaling pathways to regulate myoblast fusion during skeletal muscle regeneration. J Biol Chem 287, 32651–32664, doi: 10.1074/jbc.M112.353243 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nagai N et al. Involvement of heparan sulfate 6-O-sulfation in the regulation of energy metabolism and the alteration of thyroid hormone levels in male mice. Glycobiology 23, 980–992, doi: 10.1093/glycob/cwt037 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Vouillot L, Thélie A & Pollet N Comparison of T7E1 and Surveyor Mismatch Cleavage Assays to Detect Mutations Triggered by Engineered Nucleases. G3: Genes|Genomes|Genetics 5, 407–415, doi: 10.1534/g3.114.015834 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu W et al. DSDecode: A Web-Based Tool for Decoding of Sequencing Chromatograms for Genotyping of Targeted Mutations. Molecular Plant 8, 1431–1433, doi: 10.1016/j.molp.2015.05.009 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Dehairs J, Talebi A, Cherifi Y & Swinnen JV CRISP-ID: decoding CRISPR mediated indels by Sanger sequencing. Scientific Reports 6, 28973, doi: 10.1038/srep28973 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nairn AV et al. Glycomics of Proteoglycan Biosynthesis in Murine Embryonic Stem Cell Differentiation. Journal of Proteome Research 6, 4374–4387, doi: 10.1021/pr070446f (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Volpi N & Linhardt RJ High-performance liquid chromatography-mass spectrometry for mapping and sequencing glycosaminoglycan-derived oligosaccharides. Nat. Protocols 5, 993–1004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skidmore MA, Guimond SE, Dumax-Vorzet AF, Yates EA & Turnbull JE Disaccharide compositional analysis of heparan sulfate and heparin polysaccharides using UV or high-sensitivity fluorescence (BODIPY) detection. Nat Protoc 5, 1983–1992, doi: 10.1038/nprot.2010.145 (2010). [DOI] [PubMed] [Google Scholar]
- 58.Park Y, Yu G, Gunay NS & Linhardt RJ Purification and characterization of heparan sulphate proteoglycan from bovine brain. Biochem J 344 Pt 3, 723–730 (1999). [PMC free article] [PubMed] [Google Scholar]
- 59.Chen Y, Reddy M, Yu Y, Zhang F & Linhardt RJ Glycosaminoglycans from chicken muscular stomach or gizzard. Glycoconj J 34, 119–126, doi: 10.1007/s10719-016-9737-4 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.