Abstract
Background
Circulating free DNA sequencing (cfDNA-Seq) can portray cancer genome landscapes but highly sensitive and specific technologies are necessary to accurately detect mutations with often low variant frequencies.
Methods
We developed a customizable hybrid-capture cfDNA-Seq technology using off-the-shelf molecular barcodes and a novel duplex DNA-molecule identification tool for enhanced error correction.
Results
Modelling based on cfDNA-yields from 58 patients showed this technology, requiring 25 ng cfDNA, could be applied to >95% of patients with metastatic colorectal cancer (mCRC). cfDNA-Seq of a 32-gene/163.3kbp target region detected 100% of single nucleotide variants with 0.15% variant frequency in spike-in experiments. Molecular barcode error correction reduced false positive mutation calls by 97.5%. In 28 consecutively analyzed patients with mCRC, 80 out of 91 mutations previously detected by tumor tissue sequencing were called in the cfDNA. Call rates were similar for point mutations and indels. cfDNA-Seq identified typical mCRC driver mutations in patients where biopsy sequencing had failed or did not include key mCRC driver genes. Mutations only called in cfDNA but undetectable in matched biopsies included a subclonal resistance driver mutation to anti-EGFR antibodies in KRAS, parallel evolution of multiple PIK3CA mutations in two cases, and TP53 mutations originating from clonal hematopoiesis. Furthermore, cfDNA-Seq off-target read analysis allowed simultaneous genome-wide copy number profile reconstruction in 20 of 28 cases. Copy number profiles were validated by low-coverage whole genome sequencing.
Conclusions
This error-corrected ultra-deep cfDNA-Seq technology with a customizable target region and publicly available bioinformatics tools enables broad insights into cancer genomes and evolution.
Keywords: cancer genomics, circulating tumor DNA, liquid biopsy, molecular barcodes, sequencing error correction
Introduction
Many tumors release cell free DNA (cfDNA) into the circulation, allowing the analysis of cancer genetic aberrations from blood samples [1–6]. Such ‘liquid biopsies’ can inform tailored therapies [7] or predict recurrences after surgery [8, 9]. cfDNA analysis also permits subclonal mutation detection that is often missed by biopsies due to spatial intratumor heterogeneity [10, 11]. Genetic techniques with high analytical sensitivity and low false positive error rates are crucial for accurate cfDNA-Seq due to low tumor-derived cfDNA fractions and low abundances of subclonal mutations. Digital droplet PCR (ddPCR) and BEAMing assays can accurately detect point mutations present at frequencies ≤0.1% but are restricted to the analysis of a small number of genomic loci [8, 12]. Targeted next generation sequencing (NGS) can interrogate larger regions such as gene panels but the error rate of NGS complicates the calling of mutations with variant allele frequencies (VAFs) <5% [13]. Error correction through random molecular barcodes (MBC) has been incorporated into NGS cfDNA assays to reduce this error rate [14, 15] and has enabled mutation calling with VAFs ≤0.1%. However, these methods have often used amplicon sequencing, which can hamper coverage of entire genes due to primer design restrictions. Some methods have employed solution hybrid-capture, which is ideal to target entire genes, but used bespoke or proprietary rather than off-the-shelf reagents and publicly available bioinformatics tools, limiting their broad application for clinical or research purposes.
Here we assessed how novel, commercially available off-the-shelf MBC reagents combined with customized capture-based target enrichment technology could be optimized for ultra-deep error-corrected cfDNA-Seq. We developed a duplex-DNA molecule calling tool to improve the calling accuracy and assessed concordance of mutation calls from cfDNA with clinical grade tumor tissue sequencing in patients with metastatic colorectal cancer (mCRC).
Methods
Patients and samples
Plasma samples and clinical data were available from the FOrMAT trial (Feasibility Of Molecular characterization Approach to Treatment [16], Chief Investigator: N Starling ClinicalTrials.gov NCT02112357). Healthy donor (HD) cfDNA was obtained through the Tissue Collection Framework to Improve Outcomes in Solid Tumours (Chief Investigator: T Powles). Both trials were approved by UK ethics committees and all patients provided written informed consent. Details of clinical trials, patients, samples, sample processing and experimental techniques are provided in the online Supplemental Methods file.
cfDNA sequencing
SureSelectXT-HS (Agilent) was used to prepare sequencing libraries using our optimized protocol (online Supplemental Methods file) and a custom designed SureSelect bait-library (online Supplemental Table 1). Sequencing libraries were clustered using the cBot and sequenced with paired-end 75 reads on an Illumina HiSeq2500 in rapid mode.
SureCall software (version 4.0.1.45, Agilent) was used to trim and align fastq reads to the hg19 reference genome with default parameters and for MBC de-duplication, permitting one base mismatch within each MBC. Consensus families comprising of single reads were removed, on-target depths were assessed and variants were called with SureCall SNPPET.
To identify variants supported by duplexes we developed the freely available duplexCaller bioinformatics tool [17].
All variant positions identified in patient cfDNA were assessed in six HD samples using bam-readcount [18]. Most called variants were absent in HD samples (online Supplemental Table 2) but mutations with VAF less than double that of an identical variant in HD were removed as false positives.
BAM files resulting from MBC de-duplication before removal of single-read consensus families were used to generate genome-wide DNA copy number profiles with CNVkit [19], with Antitarget average size set to 30 kb. HD samples were used as the normal reference pooled dataset.
Low coverage whole genome sequencing (lcWGS)
Genomic libraries were constructed from 10 ng cfDNA with the NEBNext Ultra II kit and sequenced with 100bp single-end reads on HiSeq2500 in rapid mode (0.42x median coverage). Data was aligned (hg19 reference genome) with Bowtie (v0.12.9), and processed as described [20]. logRatios were normalised against a gender-matched pooled dataset from HD cfDNA (9 male, 8 female) before segmentation and median centering.
ddPCR
ddPCR was performed in case 8 (BRAF V600E) and to validate discordant variants between cfDNA and tumor tissue. 4 of 11 such cases had sufficient remaining cfDNA to validate subclonal variants (online Supplemental Methods file).
cfDNA sequencing with a commercial kit
25 ng, 17 ng and 25 ng cfDNA (cases 3, 15, and 23, respectively) were processed using the Roche AVENIO Expanded kit as per the manufacturer’s protocol. Libraries were sequenced with 151 bp paired-end reads on Illumina NextSeq500 to 2,689-6,420x depth after de-duplication. Data was analyzed using the Roche AVENIO ctDNA Analysis Software v1.0.0 with default parameters.
Results
cfDNA sequencing optimization
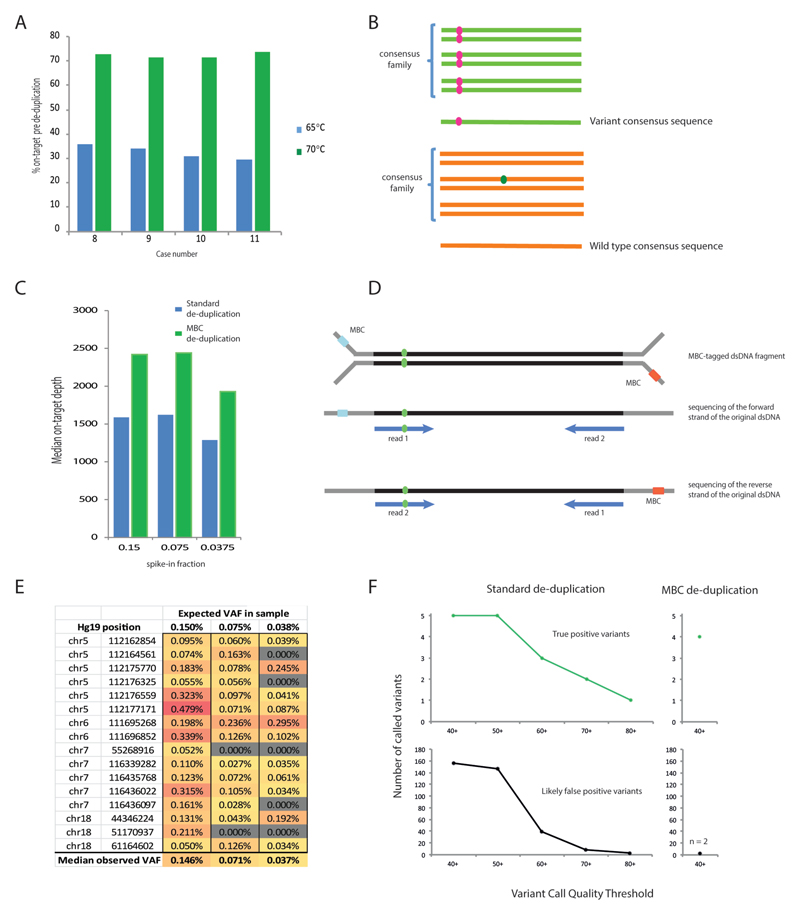
Modelling based on cfDNA yields from 58 patients with mCRC showed that 25 ng of cfDNA could be extracted from 20-30 ml blood from >95% of cases (online Supplemental Figure 2C). 25 ng was therefore chosen as our standard cfDNA input quantity. We designed a solution hybrid-capture panel targeting 32 genes including all major CRC driver genes, (163.3 kb, online Supplemental Table 1) and used Agilent SureSelectXT-HS kit, which tags each DNA strand with a random 10-base MBC, for sequencing library preparation. The SureSelectXT-HS protocol was optimized to perform reliably with 25 ng cfDNA input (online Supplemental Methods file). The fraction of on-target reads is usually low when using small targeted sequencing panels and low input DNA, so we first assessed how the on-target fraction could be optimized by varying the stringency of the post-capture wash. Two library preparations were started in parallel from each of four cfDNA samples, using the 1.5 h fast-hybridization protocol. Then, post-capture washes were performed at 65°C in one library and at 70°C in the other. Sequencing generated similar read numbers (65°C: 92,820,887; 70°C: 102,582,694 median reads/sample) and the on-target fraction significantly (p=0.0011) increased from 30-35% to 71-74% with the 70°C protocol (Figure 1A). Hence, the more stringent conditions were chosen for our standard protocol. Target exon coverage was even with this solution hybrid-capture technique and was not subject to the gaps commonly seen with commercial amplicon sequencing designs (online Supplemental Figure 3). This would be particularly advantageous for the analysis of tumor suppressor genes where driver mutations often spread across large parts of the gene.
Figure 1.
(A) Percentage of reads on-target before de-duplication in samples prepared with 65°C vs 70°C post-capture washes. (B) Graphic depicting the principles of MBC error correction. Reads with the same MBC that map to the identical genomic location are grouped into a consensus family. If a variant (pink) occurs in all reads then the consensus read sequence will be variant for that base (top). However if a variant (green) is only detected in a small fraction of the reads in the family, it will be disregarded and the consensus read sequence will be wild-type (bottom). (C) cfDNA mixing experiment: 25 ng mixes of donor A spiked into donor B at 0.15%, 0.075% and 0.0375%. (D) Illustration of duplex read pair detection. A double stranded cfDNA fragment (black) containing a variant (green) is depicted, ligated to Y-shaped MBC-tagged adapters (grey). (E) Expected and observed variant allele frequencies (VAF) and genomic positions for the 16 SNPs in the cfDNA mixing experiment. (F) Impact of MBC error correction on true positive and false positive calls. The top panels show the number of true positive variants (expected SNPs) that were bioinformatically called in the mixing experiment with standard de-duplication (left) and MBC de-duplication (right) using different variant call quality thresholds. The lower panel shows the number of likely false positive variant calls (not observed in the deep sequencing of either cfDNA sample used in the mix) for standard de-duplication (left) and MBC de-duplication (right).
We next used MBCs to de-duplicate sequencing data and perform error correction. SureCall creates families of reads with matched MBC that also align to the same genomic position and then identifies the most likely consensus sequence for each family (Figure 1B). This reduces random errors arising during PCR and sequencing, as these are not common to all reads of a family. Consensus families contained a median of 8 to 15 supporting reads in samples sequenced with the optimized protocol (online Supplemental Figure 4), which was within the optimal range for barcode error correction [21]. After MBC de-duplication, the median on-target depth with the 70°C protocol was 1,782x. This was theoretically sufficient to achieve a detection limit as low as 1 mutated DNA fragment in 1,782 molecules (0.056%). However, the analytical sensitivity for de novo mutation detection is lower in practice since more than one read is required to support robust bioinformatics calling. Thus, we designed a mixing experiment to test the ability to detect and bioinformatically call mutations with low VAFs.
Assay sensitivity and specificity
cfDNA from two donors that differed in 16 homozygous single nucleotide polymorphisms (SNPs) within the targeted region were used to prepare a dilution series with 0.15%, 0.075% and 0.0375% cfDNA from donor A spiked into cfDNA from donor B. Sequencing a median of 74,030,118 reads/sample generated a median on-target depth of 21,651x before de-duplication. Data from each sample was then processed in two ways: first, we used MBCs for de-duplication and calling of consensus sequences; second, we performed standard de-duplication using only the genomic position of each read pair. The median on-target depth was higher after MBC de-duplication (MBC 2,420x versus 1,587x with standard de-duplication; Figure 1C). This was anticipated as different MBCs tag distinct DNA fragments that would otherwise be counted as duplicates. For example, the forward and reverse strands of each original ‘duplex’ dsDNA molecule were separately tagged by MBC and so were retained as independent consensus families (Figure 1D). Standard de-duplication cannot distinguish these reads from PCR duplicates.
We first investigated whether the spiked-in SNPs could be re-identified in the MBC de-duplicated BAM files using the Integrative Genomics Viewer (IGV) [22] and tried to understand patterns associated with true positive variants. All 16 SNPs were detected in the 0.15% mix, 14/16 at 0.075% and 11/16 at 0.0375% mixing ratios (Figure 1E). Thus, our ultra-deep cfDNA-Seq assay allowed robust detection of variants at 0.15% and retained a high detection capability at 0.075%. We then assessed if MBC error correction improved the bioinformatics calling accuracy of ultra-low frequency variants, which is more challenging than re-identification of known variants. While interrogating sequencing data manually in IGV, we had observed that all true variants were at least supported by two consensus families mapping to the same genomic position but differing in whether the variant was seen in read 1 or read 2 in paired-end sequencing (Figure 1D). These were highly likely to represent the forward and reverse strand of the double-stranded input cfDNA molecule as observed previously [15]. Based on this observation, we developed the duplexCaller bioinformatics tool that identified variants supported by duplex reads (online Supplemental Methods file) and added the requirement for such a ‘duplex-configuration’ to be present to accept a mutation as genuine. The presence of a variant in at least one additional family with a different alignment position was also added to the post-call filters to assure high specificity. Thus, a variant had to be present in ≥3 consensus DNA families in order to be accepted as a mutation call in the MBC de-duplicated data. For a meaningful comparison, mutations in the standard de-duplicated data were also required to be present in ≥3 reads.
We then compared SureCall calls for the mixing experiment on standard-versus MBC-deduplicated data and quantified how many of the homozygous SNPs from sample A that were present at 0.15% in the cfDNA mixture were called. Although samples A and B differed at 16 homozygous SNP positions, only the 9 variant SNPs in spiked-in sample A could be assessed for capability to call at low frequency against the reference genome. The other 7 SNPs were reference wild-type in spiked-in sample A and so could not be called. Mutation calling after standard de-duplication with low stringency caller settings (variant call quality threshold [VCQT]=40) detected 5/9 homozygous SNPs (Figure 1F) but also generated 156 additional calls. These additional variants were likely false positives, since they had not been identified by deep sequencing of the individual cfDNA samples used in the mixing experiment. Stepwise increase of the VCQT reduced false positives but this was accompanied by a loss of analytical sensitivity. When the same data were called using MBCs and a low stringency VCQT=40 (Figure 1F), 4 of the spiked-in SNPs were called with only 2 likely false positive variants. We assessed why calling with MBC error correction failed to identify the 5 other SNPs. Each of these had VAFs <0.1% when visualized in IGV [23], which was below the minimum VAF of 0.1% that can be called by SureCall. We also assessed the number of false positive calls in standard de-duplicated data at the maximum VCQT that identified the same four true positive variants detected with MBC: 81 likely false positives were called compared to just 2 using MBC. Hence at the same analytical sensitivity, de-duplication using the MBCs dramatically decreased false positives by 97.5%. Mutation calling in 6 HD samples subjected to cfDNA-Seq only identified heterozygous and homozygous SNPs but no mutations with lower frequency (online Supplemental Table 3), further supporting the high analytical specificity of this MBC technology.
Concordance of cfDNA- and tumor-sequencing in mCRC patients
cfDNA from 28 patients with mCRC were consecutively analyzed. Seven were sequenced with the 65°C protocol and 21 with the 70°C protocol. The median sequencing depth was higher with 70°C (2,087x) than 65°C (1,205x) (Figure 2A).
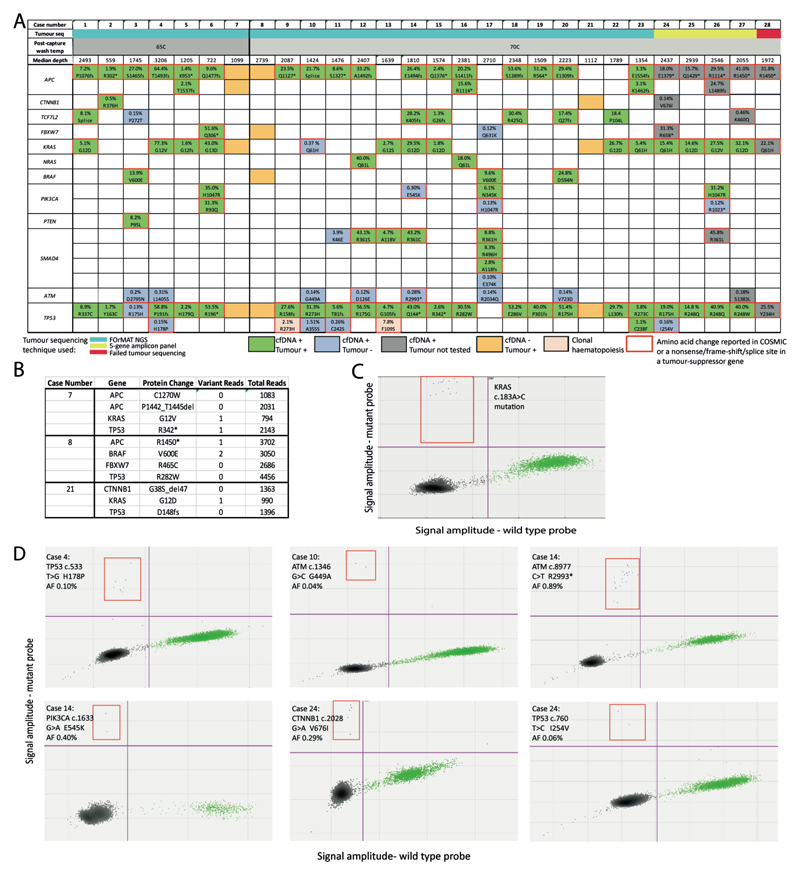
Figure 2.
(A) Concordance of mutations identified by cfDNA-Seq and by sequencing of tumor material. Mutations identified in both cfDNA-Seq and tumor sequencing are colored green. Novel variants called by cfDNA-Seq and not by tumor sequencing are colored blue. Variants not detected by cfDNA-Seq that were detected in tumor sequencing are colored orange. Pink indicates clonal hematopoiesis. Red outlines indicate mutations reported as tumorigenic in COSMIC. Variants in grey have been identified in the cfDNA of patients that either had been sequenced using the limited 5-gene amplicon panel or failed FOrMAT sequencing. Percentages indicate VAF in cfDNA. (B) Read depth and number of consensus family reads supporting each of the 11 variants in cases 7, 8, and 21 that had not been called in cfDNA but had previously been detected in tumor tissue. Median VAF 0.066%. (C) ddPCR validation of the KRAS c.183A>C mutation that results in the amino acid change Q61H in case 10. Green dots: droplets with wild-type DNA, blue dots (outlined by the red quadrant): droplets with mutant DNA, black dots: droplets that have no incorporated DNA. (D) ddPCR validation of 6 subclonal mutations called in cfDNA but not in tumor tissue.
We then analyzed the concordance and discordance of mutation calls within the target regions common to the tumor biopsy sequencing assay and our cfDNA-Seq panel. Biopsies of 23 cases had been sequenced with the FOrMAT NGS panel (online Supplemental Table 4) and four biopsies had been subjected to routine clinical amplicon sequencing of 5 genes (BRAF, KRAS, NRAS, PIK3CA and TP53). One case had failed tissue sequencing.
88% (80/91) of all mutations that had been found by tumor sequencing were called in the cfDNA (Figure 2A). All 11 mutations not called in cfDNA were from only 3 cases. Inspection of the sequencing data on IGV revealed that 5/11 mutations were present in cfDNA at VAFs below the SureCall detection limit (Figure 2B). Sufficient cfDNA remained from case 8 for orthogonal analysis by ddPCR. Using manufacturer-validated ddPCR-probes for the BRAF V600E mutation we identified 2,830 wild type DNA fragments but no mutated fragments (data not shown). This confirmed that the absence of sufficiently abundant tumor-derived cfDNA molecules, rather than technical failure, explained the inability to detect mutations.
We next assessed mutations called by cfDNA-Seq in genes that had not been sequenced in corresponding tumor tissue. APC mutations were detected in each of 4 cases whose tumors had only been analyzed with the 5-gene amplicon panel (Figure 2A). Furthermore, one mutation was found in each of FBXW7, CTNNB1, TCF7L2, ATM and SMAD4. We also detected mutations in APC, TP53 and KRAS in case 28 that had failed prior tumor tissue sequencing attempts. In total, 11 of these 13 mutations (85%) encoded protein changes previously reported in the COSMIC cancer mutation database [24] and all variants in the tumour suppressor genes APC and FBXW7 were truncating and hence likely driver mutations. This demonstrated that our assay could detect biologically and clinically important cancer mutations directly from cfDNA.
We then investigated mutations that had been called in cfDNA but were absent when the same gene had been analyzed in tumor tissue: 7 in TP53, 7 in ATM, 3 in PIK3CA, 2 in SMAD4 and one each in KRAS, FBXW7 and TCF7L2. All four mutations called in the oncogenes KRAS and PIK3CA were canonical cancer driver mutations. 8/18 mutations (44%) located in tumor suppressor genes were nonsense mutations or encoded for amino acid changes found recurrently in cancer [24], suggesting that these were also driver mutations. Together, 54.5% (12/22) of variants detected only in cfDNA were likely cancer driver mutations. The VAFs of mutations that were only detected in cfDNA but not in tumor tissue were a mean 105-fold lower than the VAF of the most abundant mutation detected in the same cfDNA sample (online Supplemental Figure 1); these variants likely originated from small cancer subclones. However, two TP53 mutations present in cfDNA but not in matched tumor tissue (Cases 9, 13) were also detected with similar VAF in DNA from blood cells (online Supplemental Table 5). These TP53 mutations hence originated from a clonal expansion of blood cells [9], termed clonal hematopoiesis [25, 26].
An activating mutation in KRAS (Q61H) was detected with a VAF of 0.37% in cfDNA but not in the matched tumor (case 10). This was the only patient that had received treatment with the anti-EGFR antibody cetuximab prior to blood collection and the KRAS mutation was likely a driver of acquired resistance that evolved during therapy [27]. ddPCR testing of cfDNA provided orthogonal validation (Figure 2C), showing that our technology is suitable for the detection of subclonal resistance driver mutations. Suspected driver mutations in PIK3CA were frequently discordant with 3/7 mutations only detectable in cfDNA (E545K, H1046R, R1023*). Two cases (17,26) harbored parallel evolution events, as further activating PIK3CA mutations were present in the tumors and the cfDNA. These results are consistent with studies showing that intratumor heterogeneity of PIK3CA mutations is common in mCRCs whereas heterogeneity is rare for mutations in APC and, in tumors not previously treated with anti-EGFR antibodies, for KRAS, NRAS and BRAF mutations [28].
Mutations in ATM tumor suppressor gene were called in 8/28 cfDNA samples. Sequencing of matched tumor showed wild-type sequence in seven of these and one tumor had only been sequenced with the 5-gene panel. All ATM mutations had low VAFs (median: 0.17%) and only 2/8 encode protein changes previously catalogued in cancer [24], making it difficult to interpret their functional relevance. No ATM mutations were called in 6 healthy donors, indicating that the mutation calls in cfDNA from mCRC patients are unlikely the result of a high false positive call rate in this gene.
Next, we used ddPCR to validate further subclonal mutations called in cfDNA but not in tumor tissue. All subclonal variants with VAF <2% from samples where sufficient cfDNA material was available and where a custom ddPCR-assay could be designed were assessed (online Supplemental Methods file). ddPCR validated all 6 tested mutations and VAFs were similar to those found by our error-corrected cfDNA technology (Figure 2D, online Supplemental Table 6).
Additionally, we re-sequenced three cfDNA samples containing low VAF (<2%) mutations (cases 3, 15, 23) with the commercially available AVENIO ctDNA kit. 9/10 point mutations in genes targeted by both panels were concordant (online Supplemental Table 7). The low frequency TP53 R175H variant in case 3 was not called by AVENIO software but was seen to be present upon manual review of the BAM file. Three indels in APC (cases 3,23) were not called by AVENIO analysis. This comparison further confirmed the reliable performance of our customizable cfDNA assay.
Genome wide DNA copy number aberration analysis
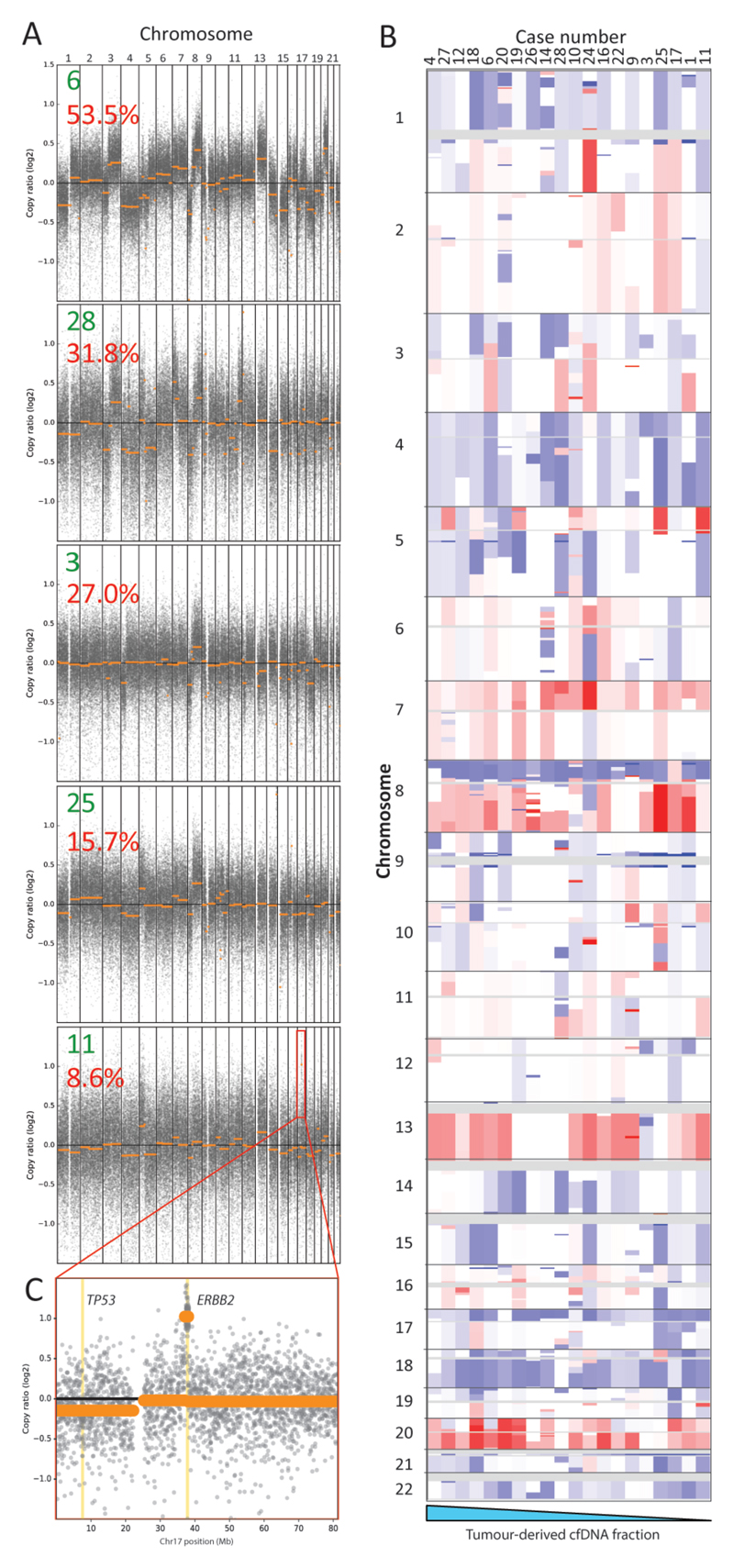
We finally assessed if we could maximise the information gain from a targeted cfDNA assay through simultaneous reconstruction of genome-wide copy number aberration (CNA) profiles. Applying the CNVkit-package [19] that uses off-target reads to infer copy number changes, we generated genome-wide CNA profiles for 20/28 cases (71%) (Figure 3A-B). Chromosome arm losses (Chr17p and 18q) and gains (Chr1q, 7, 8q, 13 and 20), which are typical for mCRC, were observed [29]. All 8 samples with a flat CNA profile had very low maximum VAFs ≤5.6%. A high-level targetable amplification involving the ERBB2 oncogene was detected despite a low tumor-derived cfDNA fraction (8.6% VAF) in case 11 (Figure 3C). This amplification had also been detected in the matched tumor, validating the ability to profile CNAs with our cfDNA-Seq technology. No other amplifications had been detected in tumor biopsies with the FOrMAT NGS panel. Low-coverage whole genome sequencing is an established approach for genome wide copy number profiling and we applied this to 18 samples with sufficient cfDNA. This independent validation showed a median weighted Spearman correlation of 0.886 with the profiles generated from cfDNA-Seq using CNVkit (online Supplemental Figure 5).
Figure 3.
(A) Genome wide copy number aberrations can be detected from targeted cfDNA-Seq, even where tumor content is low. Representative log copy ratio plots for five cases (green number) in our cohort with tumor content ranging from 53.5% to 8.6% (red number indicates max VAF) are shown. (B) Genome wide heat map of segmented copy number raw log ratio data after amplitude normalization. Gains are red and losses are blue. Profiles are ordered (left to right) from highest to lowest tumor content (based on maximum VAF) for all 20 cases that had a visible CNA profile. (C) Focused log copy ratio plot of chromosome 17 for case 11 which had a high level amplification of ERBB2.
Discussion
Our ultra-deep and error-corrected cfDNA-Seq protocol that uses off-the-shelf MBCs in combination with a custom-designed solution hybrid capture panel detected 100% of the known variants with VAFs of 0.15% in a mixing experiment. The use of MBC error correction and the requirement for variants to be supported by a duplex-pair of consensus families reduced false positive mutation calls by 97.5% while maintaining true positives. We developed the DuplexCaller bioinformatics tool, which can be run directly after MBC de-duplication to facilitate mutation calling; all bioinformatics tools for the analysis of data generated with this technology are hence freely available. Our approach did not rely on background error correction models that are constructed from large numbers of healthy donor samples and are therefore impractical for applications requiring frequently changing custom gene panels, including clinical assay development.
Importantly, the 1.5 h fast-hybridization step (standard protocol: 16h) used in our assay dramatically reduces library preparation time which is advantageous when fast turnaround is critical. Increasing the wash temperature after capture dramatically reduced off target reads. The higher temperature likely relaxes the target/bait-bond in hybridised molecules with a higher number of mismatches, reducing the non-specific carry over of DNA fragments into the library.
cfDNA-Seq of 28 mCRC patients demonstrated that 88% of mutations detected by clinical grade tumor tissue sequencing were also called in cfDNA. This detection capability is similar to that reported for MBC-error corrected cfDNA-Seq with a 5-gene assay using amplicons (87.2%) [1] and a 54-gene assay using target-capture (85%) [14, 30]. Furthermore, indels are more difficult to call than point mutations. Yet, our cfDNA assay called 23/26 indels (88.5%) that were known based on tumor sequencing, showing a similar performance to point mutation detection (87.7% called).
cfDNA-Seq detected several additional driver mutations not reported by tumor sequencing. Seven were in TP53. Two were also observed in the matched blood cells, indicating that they originated from clonal hematopoiesis. The discovery of clonal hematopoiesis in 7% of our cohort demonstrates the importance of sequencing DNA extracted from blood cells to avoid misinterpreting such variants as cancer-associated mutations. In one patient who received cetuximab therapy, we detected a KRAS Q61H variant that was absent from the matched tumor and likely represents the evolution of a drug resistant subclone. Multiple PIK3CA activating mutations detected in two anti-EGFR therapy naive patients represent parallel evolution events. These examples show that our cfDNA assay can provide insights into cancer evolution. Because the minimally invasive nature of cfDNA-Seq allows application at multiple time-points, this could be used to monitor the evolution of subclonal drug resistance driver mutations without prior knowledge of specific loci where resistance mutations will occur. We finally demonstrate that cfDNA-Seq allows genome-wide CNA reconstruction and validate this against low-coverage genome sequencing. As the number of targeted therapies increases, custom target enrichment panels that can be readily adapted and scaled for the tumor type and therapeutic agent in question could be used to investigate the full tumor genomic landscape of point mutations, indels and CNAs. This would facilitate the identification of novel resistance mechanisms. Importantly, this ultra-sensitive cfDNA-Seq technology can also address the subset of 20% of patients with mCRC who cannot be molecularly profiled due to unobtainable or inadequate biopsy tissues [16, 31].
In conclusion, this cfDNA-Seq approach with customizable and off-the-shelf reagents showed a similar performance to published techniques that use bespoke reagents and more complex analyses.
Supplementary Material
Acknowledgements
We would like to thank all patients participating in the FOrMAT clinical trial and the clinical research team members at the Royal Marsden Hospital who supported the sample collection. The study was supported by charitable donations from Tim Morgan to the Institute of Cancer Research, from Philip Moodie to The Royal Marsden Cancer Charity and by a Clive and Ann Smith Fellowship. The study received funding by Cancer Research UK, a Wellcome Trust Strategic Grant (105104/Z/14/Z), the Royal Marsden Hospital/Institute of Cancer Research National Institute for Health Research Biomedical Research Centre for Cancer and by a Cancer Research UK Clinical PhD Studentship.
Footnotes
Disclosure Declaration
The authors had pre-marketing access to Agilent SureSelectXT-HS reagents. BH is an employee of Agilent. The other authors received no financial support or compensation from Agilent.
Statement of Author Contributions
SM, LJB and MG conceived the study and wrote the manuscript; SM, LJB and BG processed samples; SM, LJB and BH developed the cfDNA-Seq assay; DK and SL developed the DuplexCaller tool; SM, LJB, DK, AW, MND and MG analyzed the data; SYM, MD, AP, AO, IR, RB, DW, AWoth, KvL, IC, DC, NS and TP provided clinical data and samples. KF and NM sequenced the cfDNA libraries, PZP, DGDC, SH and MH provided tumor biopsy sequencing data from the FORMAT panel and ran the Avenio analysis, NT and BOL provided support for ddPCR.
Copyright
“This is an un-copyedited authored manuscript copyrighted by the American Association for Clinical Chemistry (AACC). This may not be duplicated or reproduced, other than for personal use or within the rule of 'Fair Use of Copyrighted Materials' (section 107, Title 17, U.S. Code) without permission of the copyright owner, AACC. The AACC disclaims any responsibility or liability for errors or omissions in this version of the manuscript or in any version derived from it by the National Institutes of Health or other parties. The final publisher-authenticated version of the article is available at http://www.clinchem.org.”
Data access
Sequencing fastq files have been deposited into the NCBI Sequence Read Archive (SRA submission code SUB3510375).
References
- 1.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra224. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14(9):531–548. doi: 10.1038/nrclinonc.2017.14. [DOI] [PubMed] [Google Scholar]
- 3.Heitzer E, Ulz P, Belic J, Gutschi S, Quehenberger F, Fischereder K, et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med. 2013;5(4):30. doi: 10.1186/gm434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haber DA, Velculescu VE. Blood-Based Analyses of Cancer: Circulating Tumor Cells and Circulating Tumor DNA. Cancer Discov. 2014;4(6):650–661. doi: 10.1158/2159-8290.CD-13-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leary RJ, Sausen M, Kinde I, Papadopoulos N, Carpten JD, Craig D, et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med. 2012;4(162):162ra154. doi: 10.1126/scitranslmed.3004742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368(13):1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 7.Mok T, Wu YL, Lee JS, Yu CJ, Sriuranpong V, Sandoval-Tan J, et al. Detection and Dynamic Changes of EGFR Mutations from Circulating Tumor DNA as a Predictor of Survival Outcomes in NSCLC Patients Treated with First-line Intercalated Erlotinib and Chemotherapy. Clin Cancer Res. 2015;21(14):3196–3203. doi: 10.1158/1078-0432.CCR-14-2594. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Murillas I, Schiavon G, Weigelt B, Ng C, Hrebien S, Cutts RJ, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med. 2015; 7(302):302ra133. doi: 10.1126/scitranslmed.aab0021. [DOI] [PubMed] [Google Scholar]
- 9.Phallen J, Sausen M, Adleff V, Leal A, Hruban C, White J, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med. 2017;9(403) doi: 10.1126/scitranslmed.aan2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature. 2017;545(7655):446–451. doi: 10.1038/nature22364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet. 2014;46(3):225–233. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diehl F, Li M, He Y, Kinzler KW, Vogelstein B, Dressman D. BEAMing: single-molecule PCR on microparticles in water-in-oil emulsions. Nat Methods. 2006;3(7):551–559. doi: 10.1038/nmeth898. [DOI] [PubMed] [Google Scholar]
- 13.Perakis S, Speicher MR. Emerging concepts in liquid biopsies. BMC Med. 2017;15(1):75. doi: 10.1186/s12916-017-0840-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lanman RB, Mortimer SA, Zill OA, Sebisanovic D, Lopez R, Blau S, et al. Analytical and Clinical Validation of a Digital Sequencing Panel for Quantitative, Highly Accurate Evaluation of Cell-Free Circulating Tumor DNA. PLoS One. 2015;10(10):e0140712. doi: 10.1371/journal.pone.0140712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newman AM, Lovejoy AF, Klass DM, Kurtz DM, Chabon JJ, Scherer F, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol. 2016;34(5):547–555. doi: 10.1038/nbt.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moorcraft SY, Gonzalez de Castro D, Cunningham D, Jones T, Walker BA, Peckitt C, et al. Investigating the feasibility of tumour molecular profiling in gastrointestinal malignancies in routine clinical practice. Ann Oncol. 2018;29(1):230–236. doi: 10.1093/annonc/mdx631. [DOI] [PubMed] [Google Scholar]
- 17.GitHub. duplexCaller. [Accessed January 2018]; https://github.com/dkleftogi/duplexFiltering/blob/master/duplexCaller.py.
- 18.GitHub. bam-readcount. [Accessed October 2017]; https://github.com/genome/bam-readcount.
- 19.Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS Comput Biol. 2016;12(4):e1004873. doi: 10.1371/journal.pcbi.1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baslan T, Kendall J, Rodgers L, Cox H, Riggs M, Stepansky A, et al. Genome-wide copy number analysis of single cells. Nat Protoc. 2012;7(6):1024–1041. doi: 10.1038/nprot.2012.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kennedy SR, Schmitt MW, Fox EJ, Kohrn BF, Salk JJ, Ahn EH, et al. Detecting ultralowfrequency mutations by Duplex Sequencing. Nat Protoc. 2014;9(11):2586–2606. doi: 10.1038/nprot.2014.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson James T, T H, Winckler Wendy, Guttman Mitchell, Lander Eric S, Getz Gad, Mesirov Jill P. Integrative Genomics Viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forbes SA, Beare D, Bindal N, Bamford S, Ward S, Cole CG, et al. COSMIC: High-Resolution Cancer Genetics Using the Catalogue of Somatic Mutations in Cancer. Curr Protoc Hum Genet. 2016;91:10 11 11–10 11 37. doi: 10.1002/cphg.21. [DOI] [PubMed] [Google Scholar]
- 25.Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20(12):1472–1478. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coombs CC, Zehir A, Devlin SM, Kishtagari A, Syed A, Jonsson P, et al. Therapy-Related Clonal Hematopoiesis in Patients with Non-hematologic Cancers Is Common and Associated with Adverse Clinical Outcomes. Cell Stem Cell. 2017;21(3):374–382.e374. doi: 10.1016/j.stem.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, et al. Emergence of KRAS mutations and acquired resistance to anti EGFR therapy in colorectal cancer. Nature. 2012;486(7404):532–536. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brannon AR, Vakiani E, Sylvester BE, Scott SN, McDermott G, Shah RH, et al. Comparative sequencing analysis reveals high genomic concordance between matched primary and metastatic colorectal cancer lesions. Genome Biol. 2014;15(8):454. doi: 10.1186/s13059-014-0454-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.TCGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim ST, Lee WS, Lanman RB, Mortimer S, Zill OA, Kim K-M, et al. Prospective blinded study of somatic mutation detection in cell-free DNA utilizing a targeted 54-gene next generation sequencing panel in metastatic solid tumor patients. Oncotarget. 2015;6(37):40360–40369. doi: 10.18632/oncotarget.5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khakoo S, Georgiou A, Gerlinger M, Cunningham D, Starling N. Circulating tumour DNA, a promising biomarker for the management of colorectal cancer. Crit Rev Oncol Hematol. 2018;122:72–82. doi: 10.1016/j.critrevonc.2017.12.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.