Abstract
Isocitrate dehydrogenase (IDH) mutations are a prognostic factor in diffuse glioma. However, the mechanism by which these mutations improve prognosis are not clear. In a subset of IDH-mutant glioma, remodeling of the methylome results in the glioma-CpG island methylator phenotype (G-CIMP) and transcriptional reorganization. In this study, we focus on G0/G1 switch 2 (G0S2), which is highly downregulated in G-CIMP glioma. We found that G0S2 expression tended to increase as the WHO grade increased, and G0S2 knockdown inhibited glioma invasion. Additionally, we revealed that the overexpression of the DNA demethylase Ten-eleven translocation 2 (TET2) in IDH1-plasmid transfected glioblastoma multiforme (GBM) cells restored G0S2 expression. These results indicate that G0S2 is epigenetically silenced in IDH1-mutant glioma. In addition, the stereotactic delivery of glioma cells with decreased G0S2 expression in the mouse brain resulted in prolonged survival. The Cancer Genome Atlas (TCGA) analysis also indicated that survival is longer in the lower G0S2 expression group than in the higher G0S2 expression group. Moreover, G0S2 expression was higher in recurrent tumor specimens than at the initial diagnosis in the same patient. These results provide one explanation for the improved survival in IDH1-mutant glioma as well as a new epigenetic target for glioma treatment.
Introduction
Glioma, the most common primary tumor in the central nervous system, is classified into four grades according to its malignancy based on histopathological and molecular features established by the World Health Organization (WHO). WHO grade IV glioblastoma multiforme (GBM) is the most malignant and has the worst prognosis among all glioma. Although the current standard treatment includes the maximal removal of the tumor, followed by adjuvant chemotherapy and radiation therapy, the median overall survival is only 16.8 months [1]. Treatment approaches and survival have not improved for decades.
Moreover, as glioma cells invade eloquent areas and migrate undetectably toward normal outer brain tissues, it is difficult to resect the entire glioma by surgery. Accordingly, most gliomas recur and progress to more malignant WHO grades than that at primary diagnosis. Unfortunately, there is no standard therapy for recurrent glioma. Thus, new treatment targets are urgently needed.
Most GBM (~90%) occur in elderly people and develop rapidly (primary GBM or de novo GBM). However, a subset of GBM that progresses from a lower grade (WHO grade II/III) glioma (termed secondary GBM) occurs in younger people and has a significantly better prognosis than primary GBM [2, 3, 4].
Although primary and secondary GBM are indistinguishable in terms of pathological findings, they have different genomic backgrounds. Genome-wide analyses have shown that novel somatic mutations in isocitrate dehydrogenase 1 (IDH1) occur in 12% of all patients with GBM [5]. An IDH1 mutation at the arginine residue in codon 132, most commonly the R132H mutation [5, 6], promotes the direct catalysis of α-ketoglutarate (α-KG) to 2-hydroxyglutarate (2-HG). 2-HG is a competitive inhibitor of α-KG-dependent dioxygenases, including histone demethylases and the Ten-eleven translocation (TET) family of 5-methlycytosine (5mC) hydroxylases and hence results in genome-wide DNA methylation [7, 8]. IDH mutations are enriched in WHO grade II and III astrocytoma and oligodendroglioma and in secondary GBM (>75%), but are not common in primary GBM (5%) [6, 9, 10, 11].
In addition, the updated fourth edition of the WHO Classification of CNS Tumors published in 2016 includes well-established molecular genetic parameters for subclassification [12]. In this classification, IDH1 mutations, the codeletion status of chromosome arms 1p and 19q, and the histone 3 mutational status can be used to distinguish between biologically distinct glioma [13, 14]. Moreover, it has been reported that the genomic status better predicts outcome than histologic grade [15].
Patients with WHO grade III anaplastic astrocytoma and GBM carrying IDH1 mutations have a significantly longer median overall survival than patients with wild-type IDH1 [6]. However, the mechanism underlying the improved prognosis in patients with glioma carrying IDH1 mutations is not fully understood.
Concurrent genome-wide DNA methylation and DNA promoter CpG island hypermethylation leads to transcriptional silencing in cancer [16]. The frequency of promoter methylation is higher in secondary GBM than in primary GBM [17]. A distinct subset of glioma displaying DNA hypermethylation, i.e., glioma-CpG island methylator phenotype (G-CIMP), has a characteristic profile; it is more prevalent among lower grade glioma, strongly associated with IDH1 mutation, diagnosed at a younger age, and has a significantly better prognosis [18]. However, the functional importance of this altered epigenetic state remains unclear. In comparisons between G-CIMP-positive tumors and G-CIMP-negative tumors, G0/G1 switch 2 (G0S2) exhibits the strongest differential expression [18], yet the mechanism and function of G0S2 in glioma are unclear. These observations prompted us to hypothesize that the epigenetic silencing of G0S2 in G-CIMP may improve prognosis in patients with IDH1 mutations. In this study, we revealed the functional role of G0S2 in glioma and elucidated one explanation for the increased survival in glioma with IDH1 mutation.
Materials and methods
Cell culture
U87 and U251 cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured at 37°C in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum. Normal human astrocytes were obtained from Gibco (N7805-100, Gaithersburg, MD, USA) and cultured in complete astrocyte medium according to the manufacturer’s instructions.
RNA preparation and qRT-PCR analysis
Total RNA was prepared using the RNeasy Mini Kit (74104; Qiagen, Hilden, Germany) or TRIzol Reagent (15596026; Invitrogen, Carlsbad, CA, USA) according to the manufacturers’ instructions. Total RNA was converted into cDNA using the High Capacity cDNA Reverse Transcription Kit (4368814; Thermo Fisher, Waltham, MA, USA) and amplified according to the manufacturer’s protocol. RT-PCR was performed using Fast SYBR Green Master Mix (4385612; Thermo Fisher), with β-actin as an internal control, using the following primers: G0S2 (#1; forward 5′-CCGCTGACATCTAGAACTGACCTA-3′ and reverse 5′-CAGCAAAACTCAATCCCAAACTC-3′, #2; forward 5′-GCTGACATCTAGAACTGACCTA-3′ and reverse 5′- CAGCAAAACTCAATCCCAAACTC-3′) and β-actin (forward 5′- GCGGGAAATCGTGCGTGACA-3′ and reverse 5′-AAGGAAGGCTGGAAGAGTGC-3′).
Analysis of G0S2 expression in tumor samples
Either signed consent forms or exemptions were obtained for all human samples. The study of human samples was approved by the Osaka University Institutional Review Board (17022–3). Tumor specimens were obtained during surgery and classified according to the histological grade of nervous system tumors published by the WHO in 2007. All tissue samples used in this study were acquired with informed consent, and all experiments were performed in accordance with relevant guidelines and regulations of the Graduate School of Medicine of Osaka University. Human astrocytes were used as normal controls. Gene expression levels were analyzed by qRT-PCR, as described above.
siRNA/plasmid transfection
siRNA/plasmid was transfected into U251 using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Gene expression levels were analyzed by qRT-PCR, as described above. G0S2 siRNAs used were (Invitrogen #147175, #147176, and #181705, referred to as #1, #2, and #3 in this study). G0S2 siRNA #1 used, unless otherwise specified.
On day 0, U251 cells were plated in a 60-mm dish and cultured at 37°C. On day 1, cells were 70–90% confluent, and siRNA/plasmid transfection was performed. On day 3, cells were extracted.
Construct of G0S2 plasmid
The G0S2 sequence was amplified from U251 cDNA as a template using the following primers: (forward 5′- GGGAAGATGGTGAAGCTGTA -3′ and reverse 5′- CTGGTCTCCCACAGTTCCTA -3′). After ligation with pGEM-T Easy Vector (A1360; Promega, Wisconsin Madison, USA), G0S2 cds was amplified at the BglII site and EcoRI site with the following primers: (forward 5′- CGCAGATCTATGGAAACGGTCCAGGAGCTG -3′ and reverse 5′- AATGAATTCGGGAGGCGTGCTGCCGGTTGG -3′). The resulting double-stranded oligo-DNA was subcloned to the pAcGFP1-N1 vector (632469; Takara Bio Inc, Japan) at the BglII and EcoRI sites. The insert was confirmed by both restriction enzyme digestion and DNA sequencing.
Rescue experiment for G0S2 siRNA
Using the above G0S2 overexpression plasmid as a template, a siRNA-resistant form of G0S2 was generated by changing the targeted sequence of the siRNA to 5′-AAATGGTCAAACTGTATGT-3′ (4 mutant nucleotides are underlined) using the KOD-Plus-Mutagenesis Kit (Toyobo, Osaka, Japan). The plasmid was confirmed by DNA sequencing.
Production of a lentiviral vector expressing G0S2 shRNA
The shRNA sequence was made using G0S2 siRNA sense and antisense sequences with one nucleotide exchanged at the BglII site (GATCT -> GATCC) and XbaI site (G0S2 shRNA: 5′-GATCCCCAGATGGTGAAGCTGTACGTACGTGTGCTGTCCGTACGTACAGCTTCACCATCTTTTTTGGAAAT, control shRNA: 5′-GATCCCCACTACCGTTGTTAAGGTGACGTGTGCTGTCCGTCACCTTAACAACGGTAGTTTTTTGGAAAT) and their complementary strands were synthesized (Fasmac) and annealed in TE buffer by heating to 100°C for 2–3 min followed by cooling to room temperature. The resulting double-stranded oligo-DNA was subcloned to the pENTR4-H1 entry vector at the BglII and XbaI sites. The insert was confirmed by both restriction enzyme digestion and DNA sequencing. Next, pENTR4-H1-shRNA was cloned into the lentiviral vector (CS-RfA-CG) using Gateway LR Clonase II Enzyme Mix (Invitrogen) according to the manufacturer’s instructions. The insert was confirmed by both restriction enzyme digestion and DNA sequencing. These control or G0S2 shRNA-expressing lentiviral vector stocks were generated by the transfection of 293T cells. The 293T cells were cultured in DMEM supplemented with 10% fetal bovine serum. 293T cells were plated in 100-cm2 dishes in 10 ml of the medium and transfected the following day with 5 μg of the pCAG-HIV plasmid, 5 μg of the pCMV-VSV-G-RSV-Rev plasmid, and 10 μg of each lentiviral vector plasmid. Lentiviral vectors were collected on day 5 post-transfection and filtered through a 0.45-μm pore size filter and concentrated by ultracentrifugation at 4°C for 2 h at 48,820 × g.
U87 cells were plated on 100-cm2 dishes at a density of 1 × 106 cells in 9 ml of medium per dish. Transductions were carried out in the presence of 4 μg/ml Polybrene for Lenti-control or G0S2 shRNA. After incubation for 12 h, the transduction medium was replaced with fresh DMEM.
Transwell invasion assay
Transwell inserts (6.5 mm, 8-μm pore size membrane) were coated with vitronectin (10 μg/mL in PBS). The assay was largely performed as previously reported [19]. At 2 days after siRNA transfection, U251 cells were harvested with Trypsin-EDTA (0.25%), washed with PBS, and resuspended in invasion assay buffer at 4 × 105 cells/ml. Cell aliquots of 100 μl were plated on the upper filter surface and incubated at 37°C for 3 h. Filters were then washed, and cells on the upper surface were removed using cotton swabs. Cells on the lower filter surface were fixed in 4% paraformaldehyde for 30 min, blocked in PBS with 0.1% Triton-X and 5% BSA for 1 h, and stained with DAPI. Five random selected fields were evaluated with a 100× oil immersion lens; the number of invaded cells was compared with cells counts in the control group. An average of five fields at 100× total magnification was used for the quantitative analysis.
In vivo tumorigenicity assay
All experiments were performed with the approval of the Animal Studies Ethics Committee of the Osaka University Hospital (J4822), and all experiments were performed in accordance with the Guide for the Care and Use of Laboratory Animals of the Graduate School of Medicine of Osaka University. Five-week-old NOD-SCID mice (Charles River) were randomly divided into two groups (three mice per group). Under anesthesia, a 1-mm burr hole was made (2 mm to the midline and 2 mm posterior to the bregma) using a microskull drill. A total of 2 × 105 cells (U87 transfected with Lenti-control or G0S2 shRNA) suspended in 1 μl of PBS were slowly and smoothly injected to a depth of 3 mm via the skull hole using a 10-μl Hamilton syringe with a 30-gauge needle at a rate of 0.5 μl/min into the subcortex of the mouse brain. The needle was retained in place for 2 min. The scalp was sutured. Survival was recorded every 2 days over the course of the study.
At 4 weeks post-injection, anesthetized mice were perfused. The brains were removed and fixed in 4% paraformaldehyde for 24 h and then cryoprotected in 30% sucrose. Frozen coronal sections (30 μm) were made. For immunofluorescence, sections were incubated overnight at 4°C with the rabbit monoclonal anti-GFP antibody (1:1000 dilution, A-11122; Invitrogen). Following washing, the sections were treated with Alexa Fluor 488 goat anti-rabbit IgG (H+L) secondary antibody (1:1000 dilution, A-11008; Invitrogen).
Immunohistochemistry for G0S2
Immunohistochemical analyses of G0S2 were performed using paraffin blocks of specimens. The sections were deparaffinized in xylene and rehydrated in a descending ethanol-to-water gradient series. Endogenous peroxidase was blocked by exposure to 1% H2O2 at room temperature for 1 h. Sections were soaked for antigen retrieval in pH 6.0 citrate buffer at 125°C for 5 min. After cooling to room temperature, sections were blocked with 10% goat serum in PBS for 30 min at room temperature. Sections were incubated for 2 h at room temperature with a primary antibody against G0S2 (1:200; Novus, St. Louis, MO, USA) followed by incubation for 30 min at room temperature with Anti-rabbit IgG labeled with biotin (1:1000). The sections were prepared for the chromogen reaction with 0.05% diaminobenzidine and 0.01% H2O2, and were counterstained with hematoxylin.
Analysis of the cancer genome Atlas dataset
For the WHO grade II–IV glioma analysis, G0S2 gene expression based on RNASeq data (TOIL RSEM log2(normalized_count+1)), DNA methylation β values (the ratio of methylation-specific and demethylation-specific fluorophores), average Methylation450K data, IDH1 binary non-silent mutation (broad), primary tumor (sample_type) for brain lower grade glioma (LrGG, WHO grade II/III), and glioblastoma multiforme (GBM, WHO grade IV) samples were extracted from the TCGA Pan-Cancer (PANCAN) dataset using the University of California Santa Cruz Xena Browser (http://xena.ucsc.edu/). Excluding null data, 554 matching samples were available (509 lower grade glioma and 45 primary GBM, 157 wt IDH1 and 397 mut IDH1). For the lower grade (WHO grade II/III) glioma analysis, G0S2 gene expression based on RNASeq (polyA+ IlluminaHiSeq) data, DNA methylation β values, average Methylation450K data, IDH1 mutation found (0 = No, 1 = Yes), WHO grade II/III (neoplasm histologic grade), and primary tumor (sample_type) samples were extracted. Excluding null data, 125 matching samples were obtained (57 WHO grade II and 68 WHO grade III), dividing into two groups according to the G0S2 expression level (“G0S2 low” and “G0S2 high”). The survival ratio was compared with respect to G0S2 expression within the same WHO grade. For the GBM analysis, G0S2 gene expression based on RNASeq (polyA+ IlluminaHiSeq) data and primary tumor (sample_type) samples were extracted from the TCGA Glioblastoma (GBM) dataset. Excluding null data, 154 matching samples were obtained, the survival ratio was compared with respect to G0S2 expression among the GBM samples.
Additionally, the G0S2 mutation status was extracted from the glioma dataset {Merged Cohort of LGG and GBM (TCGA, Cell 2016)} using cBioPortal for Cancer Genomics (www.cbioportal.org), and 1102 samples were available.
Statistical analysis
Data are presented as means ± SEM (standard error of the mean). Comparisons between groups were performed using Student’s t-tests and one-way ANOVA followed by Tukey–Kramer tests. Statistical significance was set at p < 0.05. JMP statistical package was used for the statistical analyses.
The statistical analyses of TCGA datasets using Welch's t-test, Pearson's and Spearman's rank correlation were collected from the University of California Santa Cruz Xena Browser (http://xena.ucsc.edu/)
Results
Glioma with low G0S2 expression is associated with a longer overall survival in the TCGA dataset
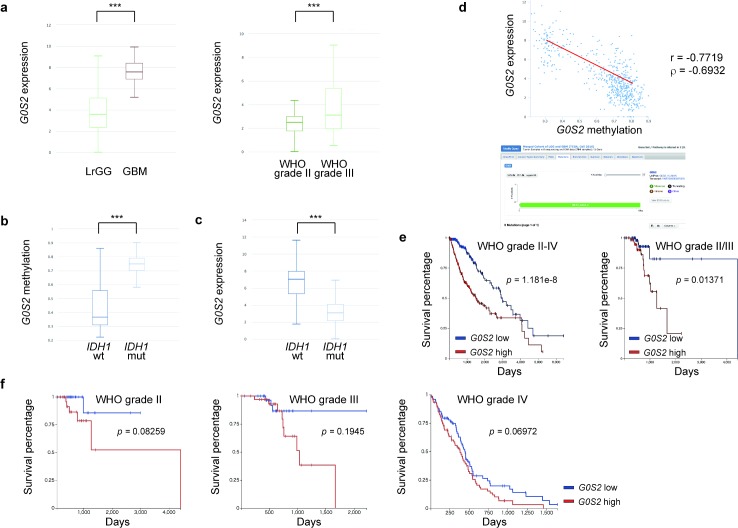
Using the Cancer Genome Atlas (TCGA) database, we found that G0S2 expression elevated as the WHO grade increased among WHO grade II–IV gliomas. Relative G0S2 expression was higher in GBM than in lower grade gliomas (7.57 vs. 3.59, p = 1269e-16), and was higher in WHO grade III glioma than in WHO grade II glioma (3.12 vs. 2.49, p = 0.006330) among the lower grade gliomas (Fig 1A). Mutant IDH1 was statistically significantly related to both higher G0S2 methylation (0.749 vs. 0.366, p = 9.439e-53) and lower G0S2 expression (3.1 vs. 7.03, p = 0.000) compared to wild-type IDH1 among WHO grade II–IV gliomas (Fig 1B and 1C). G0S2 gene expression was negatively associated with G0S2 methylation (Pearson’s r = -0.7719, Spearman’s rho = -0.6932) within WHO grade II–IV gliomas (Fig 1D). Interestingly, G0S2 mutations were not reported in 1102 gliomas among TCGA WHO grade II–IV gliomas (Fig 1D), supporting the notion that G0S2 is epigenetically silenced in gliomas harboring IDH1 mutations. In addition, the lower G0S2 expression group clearly showed better over survival than the higher G0S2 expression group for all WHO grade II–IV gliomas (p = 1.181e-8) and for WHO grade II/III lower grade gliomas (p = 0.01371) (Fig 1E). Furthermore, the lower G0S2 expression group tended to exhibit a better prognosis than the higher G0S2 expression group among the same WHO grade (Fig 1F). These observations suggest that G0S2 is a new potential diagnostic marker for glioma.
Fig 1. Analysis of the TCGA dataset.
(a) As the WHO grade increased, G0S2 expression significantly increased. (left) G0S2 expression was higher in primary GBM (7.57, n = 45) than in lower grade gliomas (LrGG, WHO grade II/III, 3.59, n = 509). (right) G0S2 expression was higher in WHO grade III (3.12, n = 68) than WHO grade II (2.49, n = 57). ***p < 0.001 (p = 1269e-16, left; p = 0.00633, right) using Welch’s t-test. (b) The G0S2 methylation β values were higher with IDH1 mutations (mut) (0.749, n = 397) than with wild-type (wt) IDH1 (wt) (0.366, n = 157) among WHO grade II–IV gliomas. ***p < 0.001 (p = 9.439e-53) using Welch’s t-test. (c) G0S2 expression was lower in samples with IDH1 mutations (3.1, n = 397) than wild-type IDH1 (7.03, n = 157) among WHO grade II–IV gliomas. ***p < 0.001 (p = 0.000). Welch’s t-test. (d) (upper) G0S2 gene expression was negatively associated with G0S2 methylation (Pearson’s r = -0.7719, Spearman’s rho = -0.6932). (lower) There were no G0S2 mutations in 1102 gliomas among the TCGA WHO grade II–IV glioma cohort. (e) (left) The lower G0S2 expression group (n = 273) clearly showed better overall survival than the higher G0S2 expression group (n = 275) in WHO grade II–IV glioma. Log-rank test, p = 1.181e-8. (right) The lower G0S2 expression group (n = 63) clearly showed better overall survival than the higher G0S2 expression group (n = 62) in WHO grade II/III glioma. Log-rank test, p = 0.01371. (f) The lower G0S2 expression group tended to have a better prognosis than the higher G0S2 expression group within the same WHO grade glioma (left, WHO grade II, n = 28, 29; middle, WHO grade III, n = 34, 34; right, WHO grade IV, n = 76, 76, where n = lower expression, higher expression).
Decreased G0S2 expression in glioma with IDH1 mutation
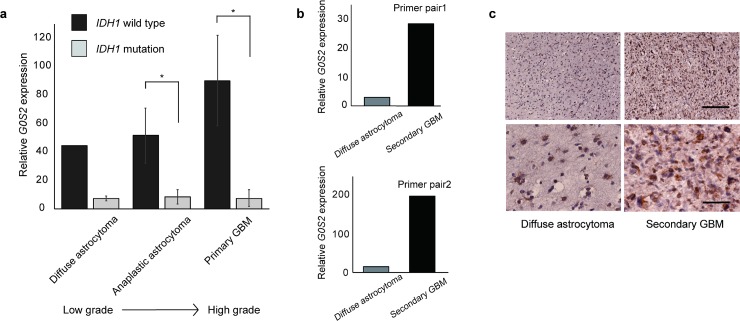
To verify the role of G0S2 in glioma malignancy, we measured G0S2 gene expression in WHO grade II diffuse astrocytoma, grade III anaplastic astrocytoma, and grade IV primary GBM tissue samples (Fig 2A). G0S2 levels were significantly higher for wild-type IDH1 than for mutant IDH1 among WHO grade III/IV gliomas. Although the correlation was not analyzed owing to the limited number of samples (n = 2, wild-type IDH1 WHO grade II specimens), G0S2 mRNA levels tended to be elevated as the WHO grade increased. These findings suggest that reduced expression of G0S2 contributes to better pathological features in glioma with IDH1 mutation.
Fig 2. Relative G0S2 expression was downregulated in gliomas with mutant IDH1.
(a) Relative G0S2 expression by RT-PCR was quantitatively analyzed in primary glioma samples (n = 26). The data were normalized by β-actin, used as the internal control, followed by normalization by cultured human astrocyte. G0S2 mRNA was significantly higher in gliomas with wild-type IDH1 than with mutant IDH1 for WHO grade III/IV glioma. Although there was no significant difference owing to the limited specimen number, G0S2 mRNA tended to increase as the WHO grade increased. Wild-type (wt) IDH1 diffuse astrocytoma, n = 2; mutant (mut) IDH1 diffuse astrocytoma, n = 3; wt IDH1 anaplastic astrocytoma, n = 3; mut IDH1 anaplastic astrocytoma, n = 5; wt IDH1 primary GBM, n = 10; mut IDH1 primary GBM, n = 3. *p < 0.05, Student’s t-test. (b) G0S2 expression was higher at the recurrent diagnosis than at the initial diagnosis in the same patient. Relative G0S2 expression was quantified by RT-PCR using normal human astrocytes as a control. G0S2 expression was elevated in recurrent glioma, diagnosed as WHO grade IV secondary GBM, compared to initial glioma, diagnosed as WHO grade II diffuse astrocytoma, in the same patient. β-Actin was used as an internal control. n = 1. (c) Representative images of the sections of diffuse astrocytoma (DA) and secondary GBM (sGBM) immunostained with an anti-G0S2 antibody. The specimens were prepared from the same case. G0S2 expression was higher in sGBM than in DA. Scale bar: 200 μm (low-magnification, upper panel), 50 μm (high-magnification, lower panel).
Further, mRNA expression of G0S2 was higher in recurrent malignant glioma, diagnosed as WHO grade IV secondary GBM, than in initial glioma, diagnosed as WHO grade II diffuse astrocytoma, within the same patient (Fig 2B). G0S2 mRNA expression was higher in the same human high-grade glioma tissues (secondary GBM) than in low-grade glioma tissues (diffuse astrocytoma; DA) (Fig 2C). These results suggest that increased G0S2 gene expression is associated with the malignancy of glioma.
Knockdown of G0S2 reduces the invasion of glioma cells
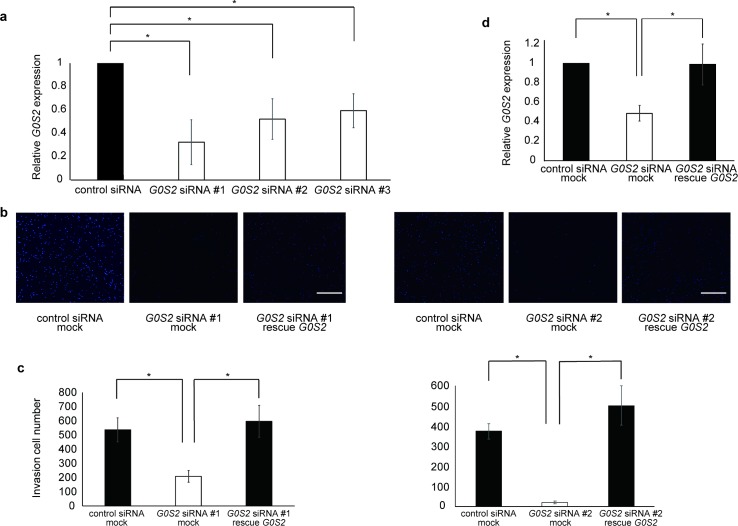
Next, we analyzed the role of G0S2 in glioma invasion by a transwell assay. To perform loss-of-function experiments using small interfering RNA (siRNA), we examined the knockdown efficacy of G0S2 siRNA. Efficient G0S2 mRNA downregulation was observed in G0S2 siRNA #1-, 2-, and 3-transfected cells, but not in the control, non-target siRNA-transfected cells (Fig 3A). siRNA-mediated knockdown of G0S2 in the U251 malignant glioma cell line significantly suppressed cellular invasion (Fig 3B and 3C). To determine the specificity, the cells were transfected with both G0S2 siRNA and the RNA interference (RNAi)-resistant G0S2 rescue plasmid, and G0S2 expression was recovered to the control level (Fig 3D). Reconstituting G0S2 expression with the G0S2 rescue plasmid restored the cellular invasion ability (Fig 3B and 3C). These results indicate that the inhibition of G0S2 expression suppresses the invasion ability of glioma cells.
Fig 3. Repression of G0S2 expression inhibits glioma cellular invasion.
(a) Each G0S2 siRNA knocked down gene expression. n = 3. *p < 0.05 using Student’s t-test. (b) The cells were transfected with the indicated siRNA #1, #2 and plasmid and invasion ability was measured using transwell assays. Representative images of the cells stained with DAPI are shown. (c) The number of invaded cells was determined. (d) Invasion ability was blocked by G0S2 siRNA, and re-exacerbated by G0S2 rescue. n = 5. *p < 0.05 by one-way ANOVA followed by a Tukey–Kramer test. Scale bar: 200 μm.
Antitumor effect of G0S2 knockdown in U87 glioma xenograft-bearing mice
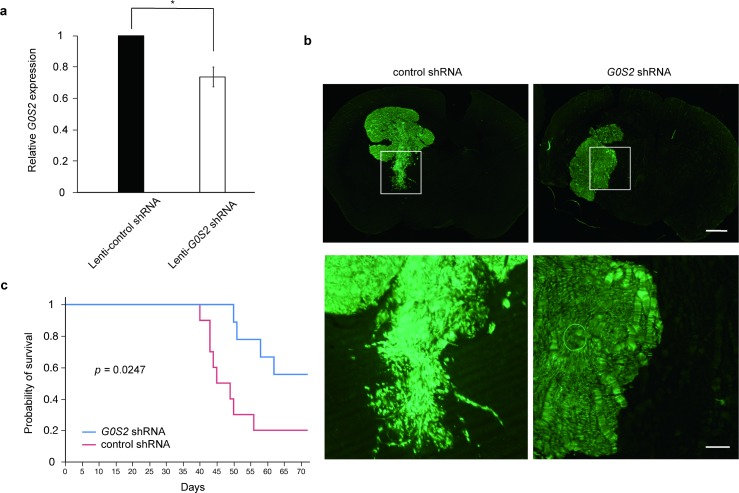
Our in vitro experiments indicated that the downregulation of G0S2 expression can efficiently attenuate the invasion of glioma cells. Therefore, we further examined the antitumor effect of G0S2 in vivo using a xenograft model. We first transplanted both U251 and U87 cells into mice brains, and found that U87 cells, but not U251 cells, tended to survive in the brain. We examined the knockdown efficacy of Lenti-G0S2 shRNA in U87 cells (Fig 4A). U87 GBM cells infected with the lentivirus encoding G0S2 shRNA in mouse brains exhibited reduced cell invasion toward surrounding normal brain tissues (Fig 4B). Importantly, mice in which G0S2-silenced GBM cells were injected into the brain exhibited prolonged overall survival (Fig 4C).
Fig 4. Suppression of G0S2 improved the survival rate in a glioma xenograft model.
(a) Lenti-G0S2 shRNA-infected U87 with knocked down gene expression. n = 3. *p < 0.05 by Student’s t-test. (b) Although Lenti-G0S2 shRNA-GFP U87 cells infiltrated normal brain tissues, the outer rim of Lenti–control shRNA–GFP U87 cells became smooth. Regions of interest are enlarged in the lower panel. Scale bar: 1 mm (low-magnification, upper panel), 200 μm (high-magnification, lower panel). (c) Mice injected with U87 cells with lentiviral expression of G0S2 shRNA-GFP exhibited prolonged overall survival compared with those with control shRNA-GFP. Median OS; G0S2 shRNA (n = 9) 64.6 days, control shRNA (n = 10) 51.4 days. P = 0.0247 by log rank tests.
IDH1 mutation epigenetically represses the transcription of G0S2
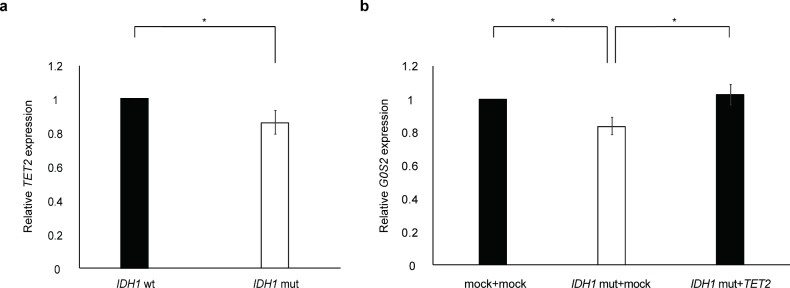
Since 2-HG, which is increased in IDH-mutant glioma, directly inhibits the activity of TET DNA demethylase, many genes in cancer are epigenetically silenced by promoter hypermethylation [8,20]. Of note, the G0S2 promoter contains a CpG island [21], suggesting that G0S2 down-regulation in the mutant IDH1 background results from epigenetic silencing. To test this hypothesis, we examined whether GBM cells transfected with a mutant IDH1 plasmid (pcDNA3-Flag-IDH1-R132H, a gift from Yue Xiong, #62907; Addgene, Cambridge, MA, USA) decreased the G0S2 mRNA level, and DNA hypomethylation by TET2 overexpression could restore the G0S2 mRNA level. It was reported that expression of IDH1 mutation lead to decrease in 5hmC [22], and that 5hmC decrease was associated with TET2 gene expression [23]. Actually, TET2 expression was decreased in GBM cells transfected with the mutant IDH1 plasmid (Fig 5A). Co-transfection of the mutant IDH1 plasmid with the TET2 plasmid (FH-TET2-pEF, a gift from Anjana Rao, Addgene #41710) recovered G0S2 expression (Fig 5B). These results suggest that IDH1 mutation represses G0S2 by DNA methylation.
Fig 5. G0S2 is epigenetically silenced in IDH1 mutant glioma cells.
(a) TET2 expression was decreased in U251MG glioma cells transfected with the IDH1 R132H mutant (mut) compared with wild-type (wt) IDH1. n = 5. *p < 0.05 by Student’s t-test. (b) After IDH R132H plasmid transfection, on day 2, cells were extracted. Quantification of relative G0S2 expression by RT-PCR. Although the forced induction of the IDH1 mutation plasmid to the U251 cells decreased G0S2 expression, transfecting both the IDH1 mutation plasmid and the TET2 overexpression plasmid restored G0S2 expression. The pcDNA-Flag plasmid was used as a mock plasmid.
Discussion
Mutations and the overexpression of several oncogenes have been identified in glioma [24, 25, 26]. The IDH R132H mutation, in particular, is thought to play a key role in gliomagenesis. This point mutation results not only in a loss of function, i.e., an inability to catalyze the conversion of isocitrate to α-ketoglutarate, but also in a gain of function, i.e., the ability to catalyze the NADPH-dependent reduction of α -ketoglutarate to 2-hydroxyglutarate (2-HG). Excess 2-HG contributes to the progression of epigenetic alterations, including DNA methylation at promoter CpG islands, and finally increases the risk of developing glioma [7]. Chao et al. reported that 2-HG accumulation can inhibit H3K9 demethylase KDM4C (also known as JMJD2C) and that the inhibition of histone demethylation impairs the differentiation of non-transformed cells into terminally differentiated cells [27]. In addition, Flavahan et al. found that IDH mutations promote gliomagenesis by disrupting the chromosomal topology and allowing aberrant regulatory interactions that induce PDGFRA expression [28]. Hypermethylation of the O6-methylguanine-DNA methyltransferase (MGMT) promoter-associated CpG island has been reported as a favorable prognostic indicator. MGMT encodes an O6-methylguanine methyltransferase that removes alkyl groups from the O-6 position of guanine. Patients with MGMT hypermethylation show sensitivity to alkylating agents, such as temozolomide, with improved outcomes [29]. Rivera et al. reported that the methylation status of the MGMT promoter also predicts the response to radiotherapy in the absence of adjuvant alkylating agents in patients with newly diagnosed GBM [30].
IDH mutations are associated with improved survival in patients with glioma [6], and are a useful prognostic marker in clinical settings. However, the mechanism underlying the improved prognosis in patients with IDH mutations has not been determined. In this study, we elucidated the role of G0S2 in glioma cell invasion and identified a mechanism by which patients with glioma carrying IDH mutations and G-CIMP show better prognosis.
At present, only histopathological findings can be used to distinguish between WHO grade II and WHO grade III gliomas. According to our analysis, survival was longer for the lower G0S2 expression group than for the higher G0S2 expression group among the patients with WHO grade II–IV gliomas and WHO grade II/III gliomas in the TCGA dataset. Moreover, G0S2 expression increased as the WHO grade increased. We also demonstrated that the expression of G0S2 was higher in post-operative recurrence diagnosed as WHO grade IV secondary GBM compared with initial glioma diagnosed as WHO grade II diffuse astrocytoma in the same patient. It should be noted that there may be a contamination of normal brain parenchyma when using surgical material from low grade gliomas, resulting in lower relative G0S2 expression levels in the samples we examined. These data suggest that G0S2 is correlated with malignant transformation and can be a new marker when glioma progresses to a higher WHO grade or recurrence. It is necessary to evaluate a larger cohort to validate the correlation between G0S2 and WHO grade diagnosis. It is difficult to evaluate the mechanism underlying glioma malignant transformation, such as progression from WHO grade II/III to WHO grade IV, in vitro or in vivo; accordingly, an effective therapy for glioma recurrence is lacking. If a patient is diagnosed WHO grade II/III low-grade glioma, specifically suppressing G0S2 methylation has the potential to prevent malignant transformation.
The inhibition of 2-HG has been proposed as a possible treatment strategy for IDH-mutant glioma [31]. Although DNA methyltransferase inhibitors and histone deacetylase inhibitors are in clinical trials and show promise for the treatment of hematopoietic malignancies [32, 33, 34], treatment with vorinostat, a histone deacetylase (HDAC) inhibitor, combined with standard chemoradiation in newly diagnosed GBM in a phase II cohort did not meet efficacy objectives [35, 36]. With respect to the molecular landscape, it is thought that focusing on specific alterations is more advantageous than conventional treatment strategies [37]. Our results further suggest that G0S2 methylation may be a potential therapeutic target.
In other cancers, G0S2 is correlated with cancer invasion [38], apotosis [39], and metabolism [40]. In a previous study of the genome-wide DNA methylation status of invasive cancer cell lines derived from breast, liver, and prostate cancers, G0S2 mRNA was significantly upregulated compared to expression in non-invasive cell lines, and its depletion decreased cell invasion in these three invasive cell lines [38]. Previous study reported that U87 cells were used for analyzing invasion ability [41]. We demonstrated that glioma harboring mutant IDH1 epigenetically represses G0S2 expression, thereby suppressing surrounding cell invasion and resulting in a better prognosis. Consistent with our results, G0S2 expression was down-regulated epigenetically by DNA hypermethylation in non-small cell lung cancer cell lines [38, 39]. G0S2 is induced by tumor necrosis factor alpha (TNFA), whose activation also requires NFkB. G0S2 promotes apoptosis by interacting with BCL2 and by preventing the formation of protective BCL2 and BAX heterodimers [39]. Kim et al. have demonstrated that the G0S2 gene is significantly upregulated in BRAF V600E cells compared to wild-type BRAF human thyroid cells, and this expression change caused by the BRAF V600E mutation may have an important role in thyroid cancer development [42]. These observations are consistent with our results, demonstrating that G0S2 downregulation is related to the suppression of glioma cell invasion (Fig 3).
Clearly, as the characteristics of glioma with IDH mutations are related to both the inactivation of several tumor suppressor genes and the overexpression of other genes, although it should be noted that G0S2 is not an only marker for the better prognosis of IDH mutation glioma, the epigenetic down-regulation of the G0S2 is not the only factor determining the biological characteristics of glioma, however, our results showed that epigenetic G0S2 downregulation following IDH1 mutation effectively inhibits the invasion of GBM cells both in vitro and in vivo. Unlike genetic mutations, epigenetic changes, such as promoter methylation, are reversible and can be targeted by drugs. Future studies should elucidate the downstream pathways involved in the suppression of invasion by G0S2.
In conclusion, we identified one mechanism by which IDH mutations improve prognosis in glioma. Continued investigations are necessary to shed light on the roles of IDH mutations.
Acknowledgments
We thank the members of Yamashita Laboratory (Laboratory of Molecular Neuroscience, Graduate School of Medicine, Osaka University) for their helpful suggestions and discussions, Dr. Hiroyuki Miyoshi (RIKEN) for providing the CS-RfA-CG lentiviral vector, Dr. Yue Xiong for providing pcDNA3-Flag-IDH1-R132H, and Dr. Anjana Rao for providing FH-TET2-pEF.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by a Grant-in-Aid for Scientific Research (S) from the Japan Society for the Promotion of Science (17H06178) to T.Y., and Grant Number JP17K16644 to T.F., http://www.jsps.go.jp/english/index.html. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Chinot OL, Wick W, Mason W, Henriksson R, Saran F, Nishikawa R, et al. Bevacizumab plus radiotherapy–temozolomide for newly diagnosed glioblastoma. New England Journal of Medicine, 2014, 3708: 709–722. 10.1056/NEJMoa1308345 [DOI] [PubMed] [Google Scholar]
- 2.Ohgaki H, Kleihues P. The definition of primary and secondary glioblastoma. Clinical cancer research, 2013, 194: 764–772. 10.1158/1078-0432.CCR-12-3002 [DOI] [PubMed] [Google Scholar]
- 3.Adamson C, Kanu OO, Mehta AI, Di C, Lin N, Mattox AK, et al. Glioblastoma multiforme: a review of where we have been and where we are going. Expert opinion on investigational drugs, 2009, 188: 1061–1083. 10.1517/13543780903052764 [DOI] [PubMed] [Google Scholar]
- 4.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes & development, 2007, 2121: 2683–2710. [DOI] [PubMed] [Google Scholar]
- 5.Parsons DW, Jones S, Zhang X, Lin JCH, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science, 2008, 3215897: 1807–1812. 10.1126/science.1164382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. New England Journal of Medicine, 2009, 3608: 765–773. 10.1056/NEJMoa0808710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature, 2009, 4627274: 739 10.1038/nature08617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer cell, 2011, 191: 17–30. 10.1016/j.ccr.2010.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, Von Deimling A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta neuropathologica, 2008, 1166: 597–602. 10.1007/s00401-008-0455-2 [DOI] [PubMed] [Google Scholar]
- 10.Bleeker FE, Lamba S, Leenstra S, Troost D, Hulsebos T, Vandertop WP, et al. IDH1 mutations at residue p. R132 (IDH1R132) occur frequently in high‐grade gliomas but not in other solid tumors. Human mutation, 2009, 301: 7–11. 10.1002/humu.20937 [DOI] [PubMed] [Google Scholar]
- 11.Hartmann C, Meyer J, Balss J, Capper D, Mueller W, Christians A, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta neuropathologica, 2009, 1184: 469–474. 10.1007/s00401-009-0561-9 [DOI] [PubMed] [Google Scholar]
- 12.Louis DN, Perry A, Reifenberger G, Von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta neuropathologica, 2016, 1316: 803–820. 10.1007/s00401-016-1545-1 [DOI] [PubMed] [Google Scholar]
- 13.Reuss DE, Kratz A, Sahm F, Capper D, Schrimpf D, Koelsche C, et al. Adult IDH wild type astrocytomas biologically and clinically resolve into other tumor entities. Acta neuropathologica, 2015, 1303: 407–417. 10.1007/s00401-015-1454-8 [DOI] [PubMed] [Google Scholar]
- 14.Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell, 2016, 1643: 550–563. 10.1016/j.cell.2015.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eckel-Passow JE, Lachance DH, Molinaro AM, Walsh KM, Decker PA, Sicotte H, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. New England Journal of Medicine, 2015, 37226: 2499–2508. 10.1056/NEJMoa1407279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones PA, Baylin SB. The epigenomics of cancer. Cell, 2007, 1284: 683–692. 10.1016/j.cell.2007.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohgaki H, Kleihues P. Genetic pathways to primary and secondary glioblastoma. The American journal of pathology, 2007, 1705: 1445–1453. 10.2353/ajpath.2007.070011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer cell, 2010, 175: 510–522. 10.1016/j.ccr.2010.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soroceanu L, Manning TJ, Sontheimer H. Modulation of glioma cell migration and invasion using Cl− and K+ ion channel blockers. Journal of Neuroscience, 1999, 1914: 5942–5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robertson KD. DNA methylation and human disease. Nature Reviews Genetics, 2005, 68: 597 10.1038/nrg1655 [DOI] [PubMed] [Google Scholar]
- 21.Russell L, Forsdyke DR. A human putative lymphocyte G0/G1 switch gene containing a CpG-rich island encodes a small basic protein with the potential to be phosphorylated. DNA and cell biology, 1991, 108: 581–591. 10.1089/dna.1991.10.581 [DOI] [PubMed] [Google Scholar]
- 22.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature, 2012, 4837390: 479–483. 10.1038/nature10866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang H, Liu Y, Bai F, Zhang JY, Ma SH, Liu J, et al. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene, 2012, 325: 663–669 10.1038/onc.2012.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorovets D, Kannan K, Shen R, Kastenhuber ER, Islamdoust N, Campos C, et al. IDH mutation and neuroglial developmental features define clinically distinct subclasses of lower grade diffuse astrocytic glioma. Clinical Cancer Research, 2012, 189: 2490–2501. 10.1158/1078-0432.CCR-11-2977 [DOI] [PubMed] [Google Scholar]
- 25.Brennan CW, Verhaak RGW, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell, 2013, 1552: 462–477. 10.1016/j.cell.2013.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guan X, Vengoechea J, Zheng S, Sloan AE, Chen Y, Brat DJ, et al. Molecular subtypes of glioblastoma are relevant to lower grade glioma. PLoS One, 2014, 93: e91216 10.1371/journal.pone.0091216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature, 2012, 4837390: 474 10.1038/nature10860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature, 2016, 5297584: 110 10.1038/nature16490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hegi ME, Diserens AC, Gorlia T, Hamou MF, De Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. New England Journal of Medicine, 2005, 35210: 997–1003. 10.1056/NEJMoa043331 [DOI] [PubMed] [Google Scholar]
- 30.Rivera AL, Pelloski CE, Gilbert MR, Colman H, De La Cruz C, Sulman EP, et al. MGMT promoter methylation is predictive of response to radiotherapy and prognostic in the absence of adjuvant alkylating chemotherapy for glioblastoma. Neuro-oncology, 2009, 122: 116–121. 10.1093/neuonc/nop020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frezza C, Tennant DA, Gottlieb E. IDH1 mutations in gliomas: when an enzyme loses its grip. Cancer cell, 2010, 171: 7–9. 10.1016/j.ccr.2009.12.031 [DOI] [PubMed] [Google Scholar]
- 32.Borthakur G, Huang X, Kantarjian H, Faderl S, Ravandi F, Ferrajoli A, et al. Report of a phase 1/2 study of a combination of azacitidine and cytarabine in acute myelogenous leukemia and high-risk myelodysplastic syndromes. Leukemia & lymphoma, 2010, 511: 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santos FPS, Kantarjian H, Garcia-Manero G, Issa JP, Ravandi F. Decitabine in the treatment of myelodysplastic syndromes. Expert review of anticancer therapy, 2010, 101: 9–22. 10.1586/era.09.164 [DOI] [PubMed] [Google Scholar]
- 34.Mercurio C, Minucci S, Pelicci PG. Histone deacetylases and epigenetic therapies of hematological malignancies. Pharmacological research, 2010, 621: 18–34. 10.1016/j.phrs.2010.02.010 [DOI] [PubMed] [Google Scholar]
- 35.Bezecny P. Histone deacetylase inhibitors in glioblastoma: pre-clinical and clinical experience. Medical oncology, 2014, 316: 985 10.1007/s12032-014-0985-5 [DOI] [PubMed] [Google Scholar]
- 36.Galanis E, Anderson SK, Miller CR, Sarkaria JN, Jaeckle K, Buckner JC, et al. Phase I/II Trial of Vorinostat Combined with Temozolomide and Radiation Therapy for Newly Diagnosed Glioblastoma: Final Results of Alliance N0874/ABTC 02. Neuro-oncology, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prados MD, Byron SA, Tran NL, Phillips JJ, Molinaro AM, Ligon KL, et al. Toward precision medicine in glioblastoma: the promise and the challenges. Neuro-oncology, 2015, 178: 1051–1063. 10.1093/neuonc/nov031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheishvili D, Stefanska B, Yi C, Li CC, Yu P, Arakelian A, et al. A common promoter hypomethylation signature in invasive breast, liver and prostate cancer cell lines reveals novel targets involved in cancer invasiveness. Oncotarget, 2015, 632: 33253 doi: 10.18632/oncotarget.5291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Welch C, Santra MK, El-Assaad W, Zhu X, Huber WE, Keys RA, et al. Identification of a protein, G0S2, that lacks Bcl-2 homology domains and interacts with and antagonizes Bcl-2. Cancer research, 2009, 6917: 6782–6789. 10.1158/0008-5472.CAN-09-0128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heckmann BL, Zhang X, Xie X, Liu J. The G0/G1 switch gene 2 (G0S2): regulating metabolism and beyond. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids, 2013, 18312: 276–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang JB, Dong DF, Wang MD, Gao K. IDH1 overexpression induced chemotherapy resistance and IDH1 mutation enhanced chemotherapy sensitivity in Glioma cells in vitro and in vivo. Asian Pacific journal of cancer prevention, 2014, 151: 427–432. [DOI] [PubMed] [Google Scholar]
- 42.Kim BA, Jee HG, Yi JW, Kim SJ, Chai YJ, Choi JY, et al. Expression Profiling of a Human Thyroid Cell Line Stably Expressing the BRAFV600E Mutation. Cancer Genomics-Proteomics, 2017, 141: 53–67. doi: 10.21873/cgp.20018 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.