X-ray ptychography and fluorescence imaging reveal 3D elemental composition and ultrastructure in frozen-hydrated green algae.
Abstract
Accurate knowledge of elemental distributions within biological organisms is critical for understanding their cellular roles. The ability to couple this knowledge with overall cellular architecture in three dimensions (3D) deepens our understanding of cellular chemistry. Using a whole, frozen-hydrated Chlamydomonas reinhardtii cell as an example, we report the development of 3D correlative microscopy through a combination of simultaneous cryogenic x-ray ptychography and x-ray fluorescence microscopy. By taking advantage of a recently developed tomographic reconstruction algorithm, termed GENeralized Fourier Iterative REconstruction (GENFIRE), we produce high-quality 3D maps of the unlabeled alga’s cellular ultrastructure and elemental distributions within the cell. We demonstrate GENFIRE’s ability to outperform conventional tomography algorithms and to further improve the reconstruction quality by refining the experimentally intended tomographic angles. As this method continues to advance with brighter coherent light sources and more efficient data handling, we expect correlative 3D x-ray fluorescence and ptychographic tomography to be a powerful tool for probing a wide range of frozen-hydrated biological specimens, ranging from small prokaryotes such as bacteria, algae, and parasites to large eukaryotes such as mammalian cells, with applications that include understanding cellular responses to environmental stimuli and cell-to-cell interactions.
INTRODUCTION
Imaging biological material in a native or near-native state is central to modern high-resolution microscopy. Recent developments in optical fluorescence microscopy allow for real-time imaging of cellular processes at subwavelength resolution (1). On the other hand, cryo-electron tomography in combination with focused ion beam milling now offers exquisite views inside cells, resolving organelles, and even individual protein complexes, without the need for additional staining (2, 3). However, these techniques are not without their limitations: The need for fluorescent labels allows for only a preselected subset of cell’s components to be imaged in light microscopy, and the low penetration depth of electrons through soft matter limits cryo-electron tomography to either small, non-eukaryotic cells or thin sections. Therefore, there is a need for additional complementary techniques that can image thick specimens under a near-native state, without the need for sectioning, and can also provide imaging contrast by means of variable x-ray absorption, fluorescence, and phase shift from endogenous composition to avoid the need for dehydrating or chemically staining specimens.
X-ray microscopy is one such technique, as it provides sufficient penetrating power to image whole cells with high spatial resolution, high contrast, and in three dimensions (3D) (4, 5). Similar to other methods, it is also compatible with cryogenic imaging of frozen-hydrated samples for minimal chemical and structural modification of the specimen. Moreover, x-ray fluorescence microscopy (XFM) allows one to measure intrinsic trace element distributions with parts-per-million sensitivity in cells without the need to add or genetically encode specific fluorescence labels (6–9). However, the resolution of existing cryo x-ray microscopes is limited by the optics used, to about 30-nm resolution in 2D (10) and about 60 nm in 3D (5).
Coherent diffractive imaging (CDI) offers a means to surpass the aforementioned limitations. CDI collects scattered x-rays from noncrystalline samples or nanocrystals using bright coherent x-rays (11, 12). When these diffraction patterns are collected over a large scattering angle and are sufficiently oversampled (13), they can be computationally inverted to produce high-resolution images using iterative phase retrieval algorithms (14, 15). In CDI, the effective numerical aperture of the imaging system can be increased, resulting in higher resolution images than conventional x-ray microscopy. Moreover, by properly selecting the x-ray wavelength, CDI permits imaging of thick biological samples without chemical labeling or sectioning due to x-ray’s large penetration depth and natural contrast arising from internal electron density (16–25). However, phase retrieval in CDI requires isolated objects or finite illumination due to the algorithmic constraint. Ptychography, a scanning CDI method (26), has proven to be a powerful method to overcome this constraint for imaging extended objects such as whole cells (27–29) and biological materials (30, 31) in 2D and 3D. Furthermore, ptychography can be combined with XFM to simultaneously image the internal structures and trace elements distributions within cells.
Previous x-ray tomography studies have imaged Chlamydomonas reinhardtii using soft x-ray microscopy (4, 32) and x-ray ptychography (33), with organelle segmentations performed via thresholding based on estimated mass density, but without clear elemental identification. Moreover, while simultaneous ptychography and XFM have been applied to simultaneously image a dehydrated diatom (34) and frozen-hydrated algae (29, 35) in 2D, its extension to 3D has not been realized because of the limited number of tomographic projections obtained and the challenges of projection alignment.
Here, we report the implementation of a correlative tomography technique, combining simultaneously acquired ptychography and XFM images with a new tomography algorithm, GENeralized Fourier Iterative REconstruction (GENFIRE), to obtain tomograms of an intact, cryogenically preserved C. reinhardtii cell. GENFIRE iterates between real and reciprocal space to search for the best-possible 3D reconstruction that is concurrently consistent with the measured images and general physical constraints such as the sample boundary and positivity (36, 37). From the GENFIRE reconstructions, we map out the 3D cellular locations of key algal organelles and confirm their identities with 3D XFM elemental maps.
METHODS
C. reinhardtii sample preparation and cryogenic preservation
C. reinhardtii cells, approximately 10 μm in diameter, were grown to the early exponential phase in tris-acetate-phosphate medium at 296 K on a rotary shaker (100 rpm). Five microliters of cell suspensions was dropped onto a plasma-treated Si3N4 window (200 nm thick, 1.5 mm by 1.5 mm membrane area). The window was then mounted onto a Vitrobot Mark IV plunge freezer (FEI), where the temperature and humidity were set and equilibrated at 22°C and 100%, respectively. The window was blotted for 2 s with a blot force of 0 and then immediately plunged into a liquid ethane bath cooled by liquid nitrogen (LN2). Cells were embedded in an ice layer with a thickness of 1.2 ± 0.2 μm. The cryogenically prepared samples were observed using a cryogenic light microscope and then stored in LN2 until they were retrieved for use in the x-ray microscope.
Ptychography and XFM data acquisition
Our experiment was carried out at the Bionanoprobe beamline, a hard x-ray nanoprobe with cryogenic sample environment and transfer capabilities, located at the Advanced Photon Source (APS) in Argonne National Laboratory (38). Before data collection, a Si3N4 membrane supporting frozen-hydrated cells was mounted on a sample cartridge and then loaded into a LN2-cooled transfer chamber in a cryogenic workstation. The transfer chamber was connected to the Bionanoprobe and delivered the shuttle to the main high-vacuum (10−7 torr) chamber. A sample exchange robot was used to grip the sample cartridge from the shuttle and deliver it to the sample stage that was cooled conductively from a LN2 reservoir. During data collection, a cryogenic sample environment was maintained by a Cu cold finger and the LN2 reservoir inside the vacuum chamber. The sample was maintained at ~110 K by conductive cooling. Furthermore, a cold shield made of Al was used around the sample to minimize radiation heat loss. More details about the setup and thermal condition characterization can be found elsewhere (38). For x-ray measurements, a monochromatic x-ray beam at 5.5 keV of energy was spectrally filtered using a Si <111> double-crystal monochromator (DCM). Details of the experimental setup are shown in Fig. 1. A Fresnel zone plate with an outermost zone width of 70 nm was used to focus the coherent x-ray beam down to a spot size of approximately 90 nm, with a depth of focus of approximately 102 μm. The cryogenically preserved sample was placed at the focus position and was scanned in “fly-scan” mode for data collection (35, 39–41). In this fly-scan mode, the fast scan was set along the horizontal (x) direction, where a piezo-scanning stage was set to move across a scan line at a constant velocity by a Delta Tau Turbo PMAC2 Ultralite VME motion control system. The control system used feedback provided by a laser interferometer system reading piezo-scanning stage positions to match the desired position and send triggers to detectors.
Fig. 1. Experimental schematics for simultaneous x-ray fluorescence and ptychography measurements.
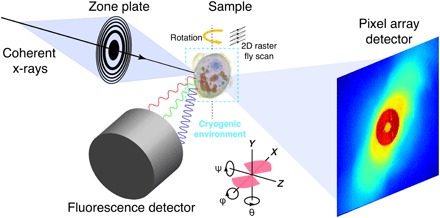
A coherent monochromatic x-ray beam was focused by a Fresnel zone plate into a spot of ~90 nm on a sample. The sample, preserved in the cryogenic environment, was raster fly-scanned in the x-y plane. During the scan, fluorescent signals and diffraction patterns were simultaneously recorded by a fluorescence detector and a pixel array detector, respectively. After finishing a 2D scan, the sample was rotated to a new angle until completing the whole 3D scan. Bottom schematic shows the orientation of 3D reconstructions with respect to experimental setup, where pink regions contain measured projection data between −68° and 56°.
As the sample was scanned, a collimated four-element (four independent detection areas) silicon drift detector (Hitachi Vortex-ME4, mounted at 90° to the incident x-ray beam) and a Dectris Pilatus 100K hybrid pixel array detector (2 m downstream of the sample) were simultaneously triggered for every 50 nm sample motion to record both the fluorescence spectra (each element of the detector records a fluorescence spectrum that consists of 2048 energy channels) and ptychographic diffraction patterns, respectively. The corresponding pixel time for each acquisition was 65 ms, including 4 ms data readout time between triggers in the Pilatus detector. A single projection scan covered a 10 μm by 10 μm field of view and took about 48 min to collect ~40,000 data points. For tomography, the sample was rotated in 2° angular spacings, so that a total of 63 projections at different angles were acquired in an angular range of −68° to 56° over a period of 3 days, including experimental interruptions.
The x-ray flux of the focused coherent beam was of the order of 3 × 108 photons/s. For each projection, the estimated radiation dose deposited to the sample with 65-ms exposure time per 50 nm2 pixel was about 2.1 × 107 gray (Gy). In the tomography scan consisting of 63 projections, there was 1.3 × 109 Gy in total imparted dose to the sample, except for one specific region which will be discussed below. A 1.3 × 109 Gy is above the 4.3 × 107 Gy dose at which the diffraction quality for atom-to-atom spatial correlation starts to degrade, as observed in x-ray macromolecular crystallography (42), but well below the dose ~3 × 1013 Gy, which was estimated for single metal atom detection in electron microscopy (43). Although chemical bonds are broken, no notable mass loss or redistribution has been observed at 30- to 100-nm length scales when studying frozen-hydrated specimens at cryogenic temperatures with a dose of ~109 Gy (10, 44).
3D GENFIRE reconstructions of the XFM data
2D fluorescence maps (P, S, Cl, K, and Ca) were compiled from the recorded fluorescence spectra using MAPS software (45), where the spectrum from each scan pixel was fitted with modified Gaussians. For XFM tomography, the best 53 of the 63 XFM projections (10 projections were excluded due to their obvious image distortion) were manually selected from each of the five fluorescence channels (figs. S1 to S5). Hot pixels in the fluorescence images caused by readout errors were removed. Background subtraction was performed on each projection as follows. First, the image was smoothed with a Gaussian kernel, and a threshold was applied to define a region outside of the cell. This region was used to calculate a mean background, which was subsequently subtracted from each image. Since the isolated spherical features in the P channel had the highest signal-to-noise ratio, we used the center of mass method to align the P images to a common tilt axis (46). Masks created by thresholding smoothed projection images were applied to further isolate XFM signals from the surrounding. After alignment and masking of the P channel data, we used the same parameters to align the K, Ca, S, and Cl projections to the common axis, as all XFM data were acquired simultaneously and thus shared the same set of orientation parameters.
For the P channel XFM dataset, GENFIRE reconstruction was performed using positivity and support constraints and with an oversampling ratio of 2 over 100 iterations, which constituted one GENFIRE macroiteration. After each macroiteration, one angular refinement macroiteration was performed to optimize tilt angles. In each angular refinement macroiteration, an angular search range between −2° and 2° with 0.5° increments was used to optimize all three Euler angles as the reconstructed volume was back projected at different angles and compared with measured projections by cross-correlation. A total of five GENFIRE macroiterations followed by angular refinements were performed to obtain the final reconstructed 3D volume. The refined angles obtained from P XFM were then used to reconstruct the K, Ca, Cl, and S XFM and ptychography datasets using the same reconstruction parameters. The convergence of the reconstruction process was monitored by an error function, defined as the difference between the back-projected images from the reconstruction and the measured XFM projections. For more details on the implementation of GENFIRE and angular refinement, see (37).
3D GENFIRE reconstruction of the ptychography data
2D ptychographic projections were reconstructed using 200 iterations of the extended ptychographic iterative engine algorithm (47) implemented in a custom software package developed for fly-scan ptychography (39). This algorithm used graphical processing units to speed up data processing (48). For reconstruction, the central 256 pixels × 256 pixels of each diffraction pattern were used, resulting in an image pixel of 10.2 nm. Because the x-ray monochromator was not stable at the beginning of the experiment, the reconstruction quality of the first few scans was poor. After excluding these scans and other low-quality projections, the best 47 ptychographic phase projections were used for tomographic reconstruction (fig. S6). The spatial resolution of 2D phase images was about 30 nm, estimated by Fourier shell correlation (49) on two identical scans at 0° (fig. S7). Note that since the magnitude image of each ptychographic projection was very weak due to the use of hard x-rays, we applied the phase images for the tomographic reconstruction.
The 47 phase projections were background-subtracted, aligned by the center of mass method (46), and masked and binned by two before GENFIRE reconstruction. GENFIRE was run using positivity and support constraints, with an oversampling ratio of 2 over 50 iterations, and using initial input angles obtained via angular refinement of P XFM data. Because the specimen was thinner than the depth of focus associated with both the ptychography and fluorescence images, we were able to use ptychography to produce linear, 2D projection maps of the specimen’s net phase shift [proportional to projected mass, as used in other demonstrations of ptychographic tomography (30)] and fluorescence to produce linear, 2D projection maps of elemental content. Thus, both imaging methods provide linear 2D projections at each rotation angle, as required for standard tomography. The GENFIRE technique then provides an improved approach to assemble and align these projection images and modalities in the Fourier domain to yield a 3D image by inverse Fourier transform. Five macroiterations of GENFIRE and angular refinement were performed to obtain the final reconstructed 3D volume. The convergence of GENFIRE was monitored by the same metric as the XFM reconstructions.
RESULTS
3D XFM and ptychography reconstructions
Figure 2 (A to D) shows GENFIRE 3D XFM and ptychography phase-contrast reconstructions, respectively. The tomographic reconstructions of the elemental XFM densities indicate that P and Ca are particularly enriched within circular structures that are mainly distributed within the cytoplasm and in close proximity to the cell wall. These structures are determined to be acidocalcisomes, also known as polyphosphate bodies (50–52). They are electron-dense organelles, with sizes ranging from 0.2 to 1 μm, that are rich in Ca and P (Fig. 2B). They have proton and calcium pumps and exhibit enzymatic activities in their limiting membranes (50). North of the acidocalcisomes, S XFM volume shows the location of the pyrenoid, which is situated at the inner side of the cup-shaped chloroplast visible in the ptychography reconstruction. The pyrenoid is electron-dense and contains relatively higher S concentration than its surrounding, presumably due to the predominant composition of S-rich RuBisCO enzymes required for photosynthesis. The low S density halo surrounding the pyrenoid is indicative of the starch sheath.
Fig. 2. GENFIRE x-ray fluorescence and ptychographic tomography reconstructions.
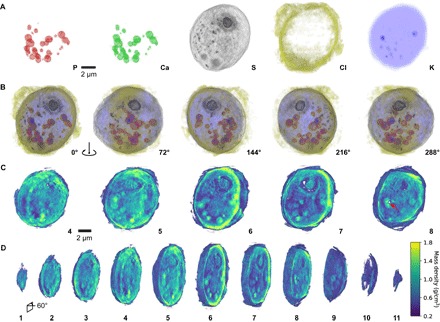
(A) 0° projection view of reconstructed 3D volumes of P (red), Ca (green), S (black), Cl (yellow), and K (blue) channels. (B) Projected views of the composite reconstructed XFM volume, rotated in 72° increments, showing clear localization of pyrenoid (S channel) near the top and acidocalcisomes near the bottom (P and Ca channels). (C) Select ~1-μm slabs of the reconstructed ptychography volume, cut into the volume at 60° experimental projection angle, showing the pyrenoid, chloroplast, various electron-dense spherical organelles, and a damaged spot in slab 8 due to x-ray overexposure (red arrow). (D) Complete set of cutouts from the same ptychography volume, viewed at 60° angle. Numbers correspond to those in (C). The color bar is for (C) and (D).
The 3D Cl XFM distribution shows most Cl localized in the periphery of the alga. This extracellular Cl distribution most likely reflects that the cell was in good hydrated condition at the time of the experiment, as the cell was still surrounded by a layer of tris-acetate-phosphate cell medium immediately before plunge freezing. The 3D K distribution is homogeneous throughout the entire cell, further suggesting that the cell is reasonably preserved with most K ions present in native sites. The ~1-μm cutouts of the ptychography phase-contrast reconstruction show the pyrenoid and numerous electron-dense bodies inside the cell (Fig. 2, C and D). Mass density calculation follows the procedures outlined in (33) and is obtained using calculated electron density derived from the phase tomography result.
During data acquisition, the focused x-ray beam dwelled on one spot of the cell for an extended amount of time at the 0° position as the shutter was unintentionally left open. This resulted in a region with abnormally high radiation exposure and consequent mass loss. This region can be seen in the eighth cutout in Fig. 2C (red arrow) as a ~0.6 μm wide and ~2.2 μm long void near the polyphosphate bodies on the left side of the cell.
Figure 3A shows 2-μm-thick slabs of the superimposed XFM and ptychography volumes and highlights colocalization of various organelles inside the cell. The addition of ultrastructure information from ptychography to XFM provides structural context for the fluorescent signals and helps confirm the identities and locations of acidocalcisomes and pyrenoid inside the algae. The correlated volumes also provide clues into the possible location of the nucleus in the low-density region near the center of the cell, as there is little to no fluorescent signal in that area. The high density and the absence of strong fluorescent signal near the cell edge also suggest the location of cup-shaped chloroplast.
Fig. 3. Superimposed x-ray fluorescence and ptychography volumes.
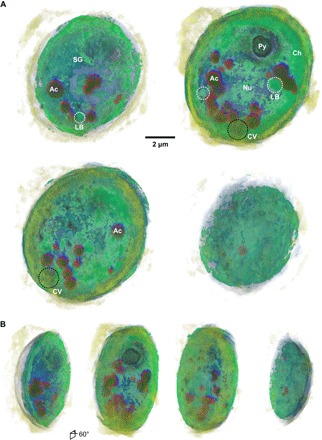
(A) Two-micrometer-thick cutouts through the superimposed volumes showing spatial correlation between x-ray fluorescent signals and electron-dense organelles. P-rich (dark green) and Ca-rich (red) acidocalcisomes (Ac) can be seen, as well as starch granules (SG), pyrenoid (Py), and chloroplast (Ch). The void in the center of the cell suggests the location of low contrast nucleus (Nu). Two dark spots in the S volume (black) near the bottom of the cell (black dashed circles) suggest the locations of contractile vacuoles (CVs). Some ~1-μm-diameter electron-dense organelles suggest the presence of lipid bodies (white dashed circles). Scale bar, 2 μm. (B) Same cutouts as (A) displayed at 60°.
Correlative S XFM and ptychography information helps define the cell boundary, and the two dense, dark S clusters near the bottom of the cell suggest the location of contractile vacuoles, which are known to play a role in osmoregulation and contain S-rich ion channels. Moreover, there are several small bodies, about 250 to 400 nm in diameter, with relatively high concentrations of S (see also S channel in Fig. 2A) that are localized to one edge of the cell. Together, they suggest the location of mitochondria, which plays a role in Fe–S biogenesis (53). Figure 3B displays the same 2-μm-thick slabs at 60° to provide another view of the correlated volume.
Comparison results between GENFIRE and filtered back projection
When the number of projections is limited, due to experimental and radiation dose-related constraints, this gives rise to incomplete sampling; the missing wedge problem. Under these circumstances, GENFIRE (36, 37) produces better 3D reconstructions than other tomographic methods such as filtered back projection (FBP), equal slope tomography (54), and simultaneous iterative reconstruction technique. Figure 4 (A to E) shows the projections of FBP reconstruction along the missing wedge direction for ptychography, P, S, Ca, and K distributions, respectively. The corresponding GENFIRE reconstructions are shown in Fig. 4 (F to J). GENFIRE’s Fourier-based iterative approach provides a better recovery of missing data and more detailed information on cellular ultrastructure in all cases.
Fig. 4. GENFIRE and FBP reconstruction comparison and angular refinement.
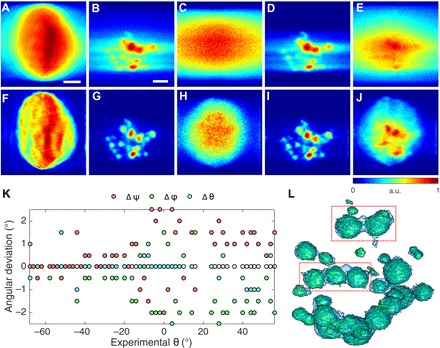
(A to E) FBP reconstructions for ptychography phase contrast, P, S, Ca, and K channels, respectively, projected from the missing wedge direction (i.e., x direction). (F to J) GENFIRE reconstructions of the corresponding volumes shown in (A) to (E), projected from the same missing wedge direction, showing a much better recovery of missing information. Scale bars, 2 μm. a.u., arbitrary units. (K) P channel angular refinement results revealing angular deviations from the recorded tilt axes [ϕ deviation (green), ψ deviation (red), and θ deviation (cyan)]. To see angular orientation with respect to experimental setup, see Fig. 1. (L) Improvements in the P channel 3D reconstruction as a result of angular refinement. Light blue and light green volumes are before and after angular refinement, respectively. Red boxes highlight volumes where angular refinement helped resolve individual acidocalcisomes better.
3D reconstruction improvement with angular refinement
Before alignment, unprocessed projection images showed noticeable out-of-plane rotation and drift from tilt axis due to the imperfect motor stage movement. Inaccuracies in projection alignment in any direction will result in lower reconstruction resolution. At worst, this may lead to misinterpretation of reconstruction artifacts in the cell as true cellular features. To algorithmically correct this experimental error in projection tilt angles, an angular refinement procedure was incorporated into the GENFIRE tomographic reconstruction workflow (37). Because x-ray fluorescence and ptychography data were acquired simultaneously in this experiment, we used the P channel projections for initial angular refinement because they had the best signal-to-noise ratio and used the resulting updated angles for all other reconstructions. We found angular deviations to be between ±2° in φ and ψ (i.e., out-of-plane rotation) and ±1.5° along the rotation axis (Fig. 4K). Figure 4L shows noticeable improvement in reconstruction quality after angular refinement. The blue and green solid volumes represent the reconstructions before and after the angular refinement, respectively. Individual polyphosphate bodies in close proximity are better differentiated and isolated after the refinement.
3D resolution estimation
We quantify the 3D resolution of the reconstructions by plotting the density variations across cellular features in three axial directions (Fig. 5, A and C) and with 3D power spectrum analysis (Fig. 5, B and D). Both analyses indicate a resolution of ~125 nm along the x and y axes and ~140 nm along the z direction for the 3D fluorescence reconstruction. The spatial resolution of fluorescence microscopy cannot exceed the Rayleigh resolution corresponding to the beam spot size (~90 nm). The final 3D reconstruction resolution would degrade due to a low signal-to-noise ratio of the images, a limited number of projections, the missing wedge, and the tilt angle misalignment. The angular refinement feature and iterative reconstruction in GENFIRE allow us to alleviate these issues and achieve a final 3D resolution that approaches (but does not exceed) the Rayleigh resolution. The 3D ptychography reconstruction achieves a resolution of ~45 nm along the x and y axes and ~55 nm along the z direction, thus taking advantage of the well-known gain in resolution beyond the illumination spot size that ptychography provides. The decrease in resolution along the z direction is expected due to the limited number and angular range of projections.
Fig. 5. Quantification of 3D reconstruction resolution.
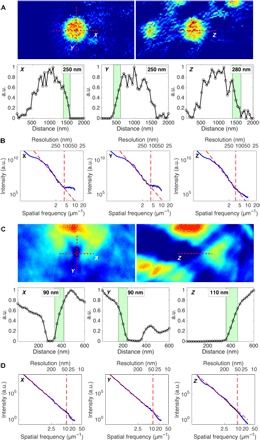
(A) One-pixel-thick layer of a reconstructed acidocalcisome in the P channel XFM volume along x and y (top left) and z directions (top right). Line scan profiles along the dashed lines shown in the top images gives a resolution of ~125, ~125, and ~140 nm along x, y, and z directions, respectively (bottom). (B) 3D power spectrum analysis of XFM along three axial directions, with cutoff spatial frequency at azimuthally averaged signal deviating from the power-law relationship, showing good agreement in the resolution estimation. (C) One-pixel layer through the burnt hole in ptychography reconstruction in x and y (top left) and z (top right). Line scan indicates a resolution of ~45 nm along x and y directions and ~55 nm along the z direction. (D) 3D power spectrum analysis of ptychography reconstruction along three axial directions. To see angular orientation with respect to the experimental setup, see Fig. 1.
DISCUSSION
Revealing 3D cellular ultrastructure via correlative imaging without chemical labeling
Simultaneous tomography using XFM and ptychography provides complimentary information about C. reinhardtii. GENFIRE 3D XFM reconstructions localize and confirm the identity of various organelles based on the distribution of trace elements (Fig. 2, A and B). In a complementary fashion, GENFIRE reconstruction of x-ray ptychography phase images produces a high-resolution 3D map of alga ultrastructure with contrast dictated by components’ electron densities. The 3D location of the pyrenoid, chloroplast, and various isolated dense bodies can be identified (Fig. 2, C and D). In addition, the 3D reconstruction even localizes a small cellular volume that is affected by beam-induced radiation damage (Fig. 2C, red arrow). All this mapping and identification is realized without the requirement of sectioning or labeling that may compromise cell integrity and introduce unwanted artifacts into cellular structures.
When both ptychography and XFM 3D volumes are overlaid, many interesting observations can be made. First, some of the electron-dense bodies found in the ptychography volume correlate well with P and Ca XFM signals and can be confidently identified as acidocalcisomes (Fig. 3A); in the past, this identification usually involved different experiments using tools such as electron microscopy and x-ray microanalysis to get the structural and elemental information (50, 55). In this work, however, both types of correlative information are acquired simultaneously.
Second, in addition to occupying the cell interior, K XFM volume also exhibits higher concentrations in some distinct regions that correlate well with acidocalcisomes in the cytosol (Fig. 3A). This suggests that each acidocalcisome may be in a different biochemical state and that surface ionic pumps could be undergoing K exchange with the cytosolic environment. As the resolution of this correlative 3D imaging technique continues to improve in the future, it will be able to provide even finer detailed elemental and chemical analysis into the complex biochemical activities of individual organelles in their native cellular states.
Third, S XFM volume not only delineates the cell boundary but also labels the location of the pyrenoid. In addition, some small S clusters suggest potential locations of mitochondria, and two faint S clusters near the bottom of the cell point to the possible locations of the contractile vacuoles, which contain S-rich H+-pyrophosphatase and vacuolar adenosine triphosphatase enzymes (56) and are involved in osmoregulation. Ultrastructure information from contractile vacuoles may not show up in ptychography due to their relatively homogeneous density compare to their surroundings; however, through S x-ray fluorescence, the locations of those organelles with respect to the cell can be visualized. This important complementary advantage between ptychography and XFM will provide researchers with even more insights into complex structural biology than either method can do alone.
Last, the white dashed circles in Fig. 3A highlights some regions of the ptychography volume that appears to be electron-dense bodies but do not contain Ca or P, suggesting that they are different organelles. This unexpected finding, along with the organelle’s size, location, and visibility in ptychography volume, led us to believe that at least some of those bodies are algal lipid bodies. The size of those dense bodies ranges from 0.5 to 1 μm in diameter and are localized near the chloroplast (57), which agree well with the properties of lipid bodies in literature. Also, since lipid bodies are known to have good contrast in x-ray (4, 58–60), they are expected to be visible in ptychography volume. This ability to confirm the identities of individual organelles in 3D ptychography using XFM rather than relying on mass density thresholding, as demonstrated in previous x-ray 3D imaging works on C. reinhardtii (32, 33), highlights the power of this correlative microscopy tool for studying complex biological structures.
Optimizing 3D reconstruction with GENFIRE angular refinement
The GENFIRE algorithm was used to produce high-quality 3D XFM and ptychography reconstructions from a limited number of 2D projections. GENFIRE’s angular refinement feature allows us to overcome errors in the recorded rotation angles due to imperfections in the rotation stage, resulting in higher quality reconstructions. Angular refinement can be thought of as a way to provide information on the experimental setup’s overall stability. Our angular refinement result shows small θ deviations from recorded angles in the negative rotational angles (Fig. 4K), which increased noticeably as the stage tilted into the positive angular range. This suggests that the stage stability differed between the positive and negative angles during the 3-day data acquisition period. This type of diagnostic information could help devise better tomographic data collection schemes in the future to ensure that the highest quality data can be obtained.
Simultaneous data collection streamlines data processing
In addition to considerably shortening the data collection time, the correlative microscopy method demonstrated here can also shorten postprocessing workflow and prevent additional sources of reconstruction errors. Since all data are acquired simultaneously, the computational projection image alignment procedure performed on one dataset can be applied to the rest. This is helpful for aligning XFM datasets, where object boundary in some channels is not always defined. Likewise, image segmentation parameters obtained from separating the object from background in one dataset can be shared with all others. Efficient postprocessing workflow, such as those mentioned above, is especially important for correlative tomography reconstructions, where multiple tomograms with different information content need to be processed efficiently and consistently to obtain results in a timely manner.
CONCLUSION
In this work, we demonstrate an application of 3D correlative microscopy by combining GENFIRE with x-ray ptychography and XFM tomography to image a whole frozen-hydrated C. reinhardtii in 3D. XFM provides trace element distributions of various organelles within the alga, and they are coupled with ptychography to reveal complex ultrastructure information. This simultaneous data acquisition process streamlines the experiment and saves researchers considerable amount of time in sample transfer between different instruments and in searching for the exact specimen on the sample substrate to do correlative imaging. It reduces the total radiation dose accumulated on the sample from the sequential imaging approach and alleviates damage from exposing the sample to different imaging conditions. Furthermore, we also demonstrate GENFIRE’s superior 3D reconstruction capability when dealing with limited number of projections. This presents yet another avenue to reduce the amount of tomographic data necessary to achieve high-quality 3D reconstructions and can further minimize radiation damage imparted on the sample.
One of the limitations of this initial demonstration is the very long time period used to collect the data. There are a number of speedups that we plan on using in the future. First, scan trajectories with better photon utilization will be used. The fly-scan scheme used in this experiment only implemented continuous motion along one scan axis, while the other axis was still in step-scan mode. By applying continuous motion throughout the whole projection scan (61) or even the whole 3D scan [e.g., spiral tomography (62)], the scan time can be reduced by avoiding the overhead between scan lines. For instance, in this experiment, the overhead was about 5 min for one 2D projection scan. Second, a double-multilayer monochromator with a spectral bandwidth of 10−2 delivers ~20 times more flux than the DCM source used in the present measurement, which can speed up this correlative 3D measurement. This improvement will be significant as the APS is upgraded, which will provide at least 100 times more coherent flux. Last, using the dose fractionation theorem (63), the total photons for a 3D measurement can be divided into many tilt angles, so each 2D projection will be acquired rapidly with high-throughput detectors. This means that more angular projections can be obtained to achieve higher quality 3D reconstruction while reducing the missing wedge effect.
Another limitation of this method concerns imaging large specimens such as thick whole mammalian cells, which may introduce depth of focus problems when acquiring tomography projection data. To scale up and generalize the range of applications achievable by ptychography, additional techniques have been developed, such as multislice ptychography (64, 65), which allows ptychography to go beyond the depth of focus limits and image thick biological specimens with high resolution in 3D (66, 67). A detailed discussion of the thickness limits of x-ray microscopy and its relationship to electron microscopy of biological specimens is available (68). Moreover, ongoing efforts in synchrotron source upgrades and faster and larger data handling will decrease data acquisition time and allow scientists to scale up the type and size of biological specimen they can image using ptychography.
With the improvements described above, we believe that the combination of x-ray ptychography for structural information, x-ray fluorescence for trace element information, and GENFIRE for synthesizing 2D data into 3D will become an important microscopy tool in structural biology. By combining with new developments in dose-reduced imaging, this technique will allow researchers to image whole, unsectioned eukaryotic cells and elucidate each component’s elemental compositions, all the while preserving structural integrity. As this tool continues to develop, we envision that it can find a nice niche between fluorescence light microscopy, where selective intracellular components and macroscopic intercellular phenomena are imaged in lower resolution, and cryo-electron microscopy, where fine internal ultrastructure of thinned subregions is imaged in higher resolution. This will open the door to study a myriad of biological problems where high-resolution elemental identification and complex internal structure information are required in 3D.
Supplementary Material
Acknowledgments
We thank J. A. Rodriguez for stimulating discussions. Molecular graphics and analyses were performed with the UCSF Chimera package (69). Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311). Funding: This work was supported by the NIH NIGMS under grant no. 1R01GM104530 and STROBE: A National Science Foundation Science and Technology Center under grant no. DMR 1548924. J.M. thanks the partial support of the U.S. Department of Energy (DOE; DE-SC0010378) for the development of GENFIRE. The Bionanoprobe is funded by NIH/NCRR High End Instrumentation (HEI) grant (1S10RR029272-01) as part of the American Recovery and Reinvestment Act (ARRA). Use of the APS, an Office of Science User Facility operated for the DOE, Office of Science by Argonne National Laboratory, was supported by the DOE under contract no. DE-AC02-06CH11357. Author contributions: J.M. and C.J. directed the project. Q.J. and J.D. prepared the samples. J.D., S.C., and Y.P.H. collected the ptychography and XFM data. J.D., S.C., Y.S.G.N., and S.V. performed the reconstructions of ptychographic and XFM 2D projections. M.G.-J. and A.P. performed the tomography reconstructions. Y.H.L., M.G.-J., A.P., and J.M. conducted the data analyses. J.D., M.G.-J., Y.H.L., C.J., and J.M. contributed to the discussion and writing of the manuscript with comments from other authors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/11/eaau4548/DC1
Fig. S1. Experimental phosphorus channel x-ray fluorescence projections.
Fig. S2. Experimental calcium channel x-ray fluorescence projections.
Fig. S3. Experimental sulfur channel x-ray fluorescence projections.
Fig. S4. Experimental chlorine channel x-ray fluorescence projections.
Fig. S5. Experimental potassium channel x-ray fluorescence projections.
Fig. S6. Experimental ptychographic projections.
Fig. S7. Resolution of 2D ptychographic projections.
Movie S1. Reconstructed x-ray fluorescence volume of C. reinhardtii.
REFERENCES AND NOTES
- 1.Sahl S. J., Hell S. W., Jakobs S., Fluorescence nanoscopy in cell biology. Nat. Rev. Mol. Cell Biol. 18, 685–701 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Lučić V., Rigort A., Baumeister W., Cryo-electron tomography: The challenge of doing structural biology in situ. J. Cell Biol. 202, 407–419 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engel B. D., Schaffer M., Kuhn Cuellar L., Villa E., Plitzko J. M., Baumeister W., Native architecture of the Chlamydomonas chloroplast revealed by in situ cryo-electron tomography. eLife 4, e04889 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weiß D., Schneider G., Niemann B., Guttmann P., Rudolph D., Schmahl G., Computed tomography of cryogenic biological specimens based on X-ray microscopic images. Ultramicroscopy 84, 185–197 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Larabell C. A., Le Gros M. A., X-ray tomography generates 3-D reconstructions of the yeast, saccharomyces cerevisiae, at 60-nm resolution. Mol. Biol. Cell 15, 957–962 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paunesku T., Vogt S., Maser J., Lai B., Woloschak G., X-ray fluorescence microprobe imaging in biology and medicine. J. Cell. Biochem. 99, 1489–1502 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Fahrni C. J., Biological applications of X-ray fluorescence microscopy: Exploring the subcellular topography and speciation of transition metals. Curr. Opin. Chem. Biol. 11, 121–127 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Majumdar S., Peralta-Videa J. R., Castillo-Michel H., Hong J., Rico C. M., Gardea-Torresdey J. L., Applications of synchrotron μ-XRF to study the distribution of biologically important elements in different environmental matrices: A review. Anal. Chim. Acta 755, 1–16 (2012). [DOI] [PubMed] [Google Scholar]
- 9.de Jonge M. D., Holzner C., Baines S. B., Twining B. S., Ignatyev K., Diaz J., Howard D. L., Legnini D., Miceli A., McNulty I., Jacobsen C. J., Vogt S., Quantitative 3D elemental microtomography of Cyclotella meneghiniana at 400-nm resolution. Proc. Natl. Acad. Sci. U.S.A. 107, 15676–15680 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schneider G., Cryo X-ray microscopy with high spatial resolution in amplitude and phase contrast. Ultramicroscopy 75, 85–104 (1998). [DOI] [PubMed] [Google Scholar]
- 11.Miao J., Ishikawa T., Robinson I. K., Murnane M., Beyond crystallography: Diffractive imaging using coherent x-ray light sources. Science 348, 530–535 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Miao J., Charalambous P., Kirz J., Sayre D., Extending the methodology of X-ray crystallography to allow imaging of micrometre-sized non-crystalline specimens. Nature 400, 342–344 (1999). [Google Scholar]
- 13.Miao J., Sayre D., Chapman H. N., Phase retrieval from the magnitude of the Fourier transforms of nonperiodic objects. J. Opt. Soc. Am. A 15, 1662–1669 (1998). [Google Scholar]
- 14.Shechtman Y., Eldar Y. C., Cohen E., Chapman H. N., Phase retrieval with application to optical imaging: A contemporary overview. IEEE Signal Process. Mag. 32, 87–109 (2015). [Google Scholar]
- 15.Marchesini S., Invited article: A unified evaluation of iterative projection algorithms for phase retrieval. Rev. Sci. Instrum. 78, 011301 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Miao J., Hodgson K. O., Ishikawa T., Larabell C. A., LeGros M. A., Nishino Y., Imaging whole Escherichia coli bacteria by using single-particle x-ray diffraction. Proc. Natl. Acad. Sci. U.S.A. 100, 110–112 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shapiro D., Thibault P., Beetz T., Elser V., Howells M., Jacobsen C., Kirz J., Lima E., Miao H., Neiman A. M., Sayre D., Biological imaging by soft x-ray diffraction microscopy. Proc. Natl. Acad. Sci. U.S.A. 102, 15343–15346 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishino Y., Takahashi Y., Imamoto N., Ishikawa T., Maeshima K., Three-dimensional visualization of a human chromosome using coherent X-ray diffraction. Phys. Rev. Lett. 102, 018101 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Jiang H., Song C., Chen C.-C., Xu R., Raines K. S., Fahimian B. P., Lu C.-H., Lee T.-K., Nakashima A., Urano J., Ishikawa T., Tamanoi F., Miao J., Quantitative 3D imaging of whole, unstained cells by using X-ray diffraction microscopy. Proc. Natl. Acad. Sci. U.S.A. 107, 11234–11239 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song C., Jiang H., Mancuso A., Amirbekian B., Peng L., Sun R., Shah S. S., Hong Zhou Z., Ishikawa T., Miao J., Quantitative Imaging of Single, Unstained Viruses with Coherent X Rays. Phys. Rev. Lett. 101, 158101 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Kimura T., Joti Y., Shibuya A., Song C., Kim S., Tono K., Yabashi M., Tamakoshi M., Moriya T., Oshima T., Ishikawa T., Bessho Y., Nishino Y., Imaging live cell in micro-liquid enclosure by X-ray laser diffraction. Nat. Commun. 5, 3052 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallagher-Jones M., Bessho Y., Kim S., Park J., Kim S., Nam D., Kim C., Kim Y., Young Noh D., Miyashita O., Tama F., Joti Y., Kameshima T., Hatsui T., Tono K., Kohmura Y., Yabashi M., Samar Hasnain S., Ishikawa T., Song C., Macromolecular structures probed by combining single-shot free-electron laser diffraction with synchrotron coherent X-ray imaging. Nat. Commun. 5, 3798 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Bergh M., Huldt G., Tîmneanu N., Maia F. R. N. C., Hajdu J., Feasibility of imaging living cells at subnanometer resolutions by ultrafast X-ray diffraction. Q. Rev. Biophys. 41, 181–204 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Seibert M. M., Ekeberg T., Maia F. R. N. C., Svenda M., Andreasson J., Jönsson O., Odić D., Iwan B., Rocker A., Westphal D., Hantke M., DePonte D. P., Barty A., Schulz J., Gumprecht L., Coppola N., Aquila A., Liang M., White T. A., Martin A., Caleman C., Stern S., Abergel C., Seltzer V., Claverie J.-M., Bostedt C., Bozek J. D., Boutet S., Alan Miahnahri A., Messerschmidt M., Krzywinski J., Williams G., Hodgson K. O., Bogan M. J., Hampton C. Y., Sierra R. G., Starodub D., Andersson I., Bajt S., Barthelmess M., Spence J. C. H., Fromme P., Weierstall U., Kirian R., Hunter M., Bruce Doak R., Marchesini S., Hau-Riege S. P., Frank M., Shoeman R. L., Lomb L., Epp S. W., Hartmann R., Rolles D., Rudenko A., Schmidt C., Foucar L., Kimmel N., Holl P., Rudek B., Erk B., Hömke A., Reich C., Pietschner D., Weidenspointner G., Strüder L., Hauser G., Gorke H., Ullrich J., Schlichting I., Herrmann S., Schaller G., Schopper F., Soltau H., Kühnel K.-U., Andritschke R., Schröter C.-D., Krasniqi F., Bott M., Schorb S., Rupp D., Adolph M., Gorkhover T., Hirsemann H., Potdevin G., Graafsma H., Nilsson B., Chapman H. N., Hajdu J., Single mimivirus particles intercepted and imaged with an X-ray laser. Nature 470, 78–81 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlichting I., Miao J., Emerging opportunities in structural biology with X-ray free-electron lasers. Curr. Opin. Struct. Biol. 22, 613–626 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfeiffer F., X-ray ptychography. Nat. Photonics 12, 9–17 (2018). [Google Scholar]
- 27.Giewekemeyer K., Thibault P., Kalbfleisch S., Beerlink A., Kewish C. M., Dierolf M., Pfeiffer F., Salditt T., Quantitative biological imaging by ptychographic x-ray diffraction microscopy. Proc. Natl. Acad. Sci. U.S.A. 107, 529–534 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gallagher-Jones M., Sato Baraldi Dias C., Pryor A. Jr, Bouchmella K., Zhao L., Hung Lo Y., Borba Cardoso M., Shapiro D., Rodriguez J., Miao J., Correlative cellular ptychography with functionalized nanoparticles at the Fe L-edge. Sci. Rep. 7, 4757 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng J., Vine D. J., Chen S., Nashed Y. S. G., Jin Q., Phillips N. W., Peterka T., Ross R., Vogt S., Jacobsen C. J., Simultaneous cryo X-ray ptychographic and fluorescence microscopy of green algae. Proc. Natl. Acad. Sci. U.S.A. 112, 2314–2319 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dierolf M., Menzel A., Thibault P., Schneider P., Kewish C. M., Wepf R., Bunk O., Pfeiffer F., Ptychographic X-ray computed tomography at the nanoscale. Nature 467, 436–439 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Zanette I., Enders B., Dierolf M., Thibault P., Gradl R., Diaz A., Guizar-Sicairos M., Menzel A., Pfeiffer F., Zaslansky P., Ptychographic X-ray nanotomography quantifies mineral distributions in human dentine. Sci. Rep. 5, 9210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hummel E., Guttmann P., Werner S., Tarek B., Schneider G., Kunz M., Frangakis A. S., Westermann B., 3D ultrastructural organization of whole chlamydomonas reinhardtii cells studied by nanoscale soft x-ray tomography. PLOS ONE 7, e53293 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diaz A., Malkova B., Holler M., Guizar-Sicairos M., Lima E., Panneels V., Pigino G., Bittermann A. G., Wettstein L., Tomizaki T., Bunk O., Schertler G., Ishikawa T., Wepf R., Menzel A., Three-dimensional mass density mapping of cellular ultrastructure by ptychographic X-ray nanotomography. J. Struct. Biol. 192, 461–469 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Vine D. J., Pelliccia D., Holzner C., Baines S. B., Berry A., McNulty I., Vogt S., Peele A. G., Nugent K. A., Simultaneous X-ray fluorescence and ptychographic microscopy of Cyclotella meneghiniana. Opt. Express 20, 18287–18296 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Deng J., Vine D. J., Chen S., Jin Q., Nashed Y. S. G., Peterka T., Vogt S., Jacobsen C., X-ray ptychographic and fluorescence microscopy of frozen-hydrated cells using continuous scanning. Sci. Rep. 7, 445 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Y., Chen C.-C., Scott M. C., Ophus C., Xu R., Pryor A., Wu L., Sun F., Theis W., Zhou J., Eisenbach M., Kent P. R. C., Sabirianov R. F., Zeng H., Ercius P., Miao J., Deciphering chemical order/disorder and material properties at the single-atom level. Nature 542, 75–79 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Pryor A., Yang Y., Rana A., Gallagher-Jones M., Zhou J., Hung Lo Y., Melinte G., Chiu W., Rodriguez J. A., Miao J., GENFIRE: A generalized Fourier iterative reconstruction algorithm for high-resolution 3D imaging. Sci. Rep. 7, 10409 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen S., Deng J., Yuan Y., Flachenecker C., Mak R., Hornberger B., Jin Q., Shu D., Lai B., Maser J., Roehrig C., Paunesku T., Gleber S. C., Vine D. J., Finney L., VonOsinski J., Bolbat M., Spink I., Chen Z., Steele J., Trapp D., Irwin J., Feser M., Snyder E., Brister K., Jacobsen C., Woloschak G., Vogt S., The Bionanoprobe: Hard X-ray fluorescence nanoprobe with cryogenic capabilities. J. Synchrotron Radiat. 21, 66–75 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng J., Deng J., Yuan Y., Flachenecker C., Mak R., Hornberger B., Jin Q., Shu D., Lai B., Maser J., Roehrig C., Paunesku T., Gleber S. C., Vine D. J., Finney L., VonOsinski J., Bolbat M., Spink I., Chen Z., Steele J., Trapp D., Irwin J., Feser M., Snyder E., Brister K., Jacobsen C., Woloschak G., Vogt S., Continuous motion scan ptychography: Characterization for increased speed in coherent x-ray imaging. Opt. Express 23, 5438–5451 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pelz P. M., Guizar-Sicairos M., Thibault P., Johnson I., Holler M., Menzel A., On-the-fly scans for X-ray ptychography. Appl. Phys. Lett. 105, 251101 (2014). [Google Scholar]
- 41.Huang X., Lauer K., Clark J. N., Xu W., Nazaretski E., Harder R., Robinson I. K., Chu Y. S., Fly-scan ptychography. Sci. Rep. 5, 9074 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Owen R. L., Rudiño-Piñera E., Garman E. F., Experimental determination of the radiation dose limit for cryocooled protein crystals. Proc. Natl. Acad. Sci. U.S.A. 103, 4912–4917 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leapman R. D., Detecting single atoms of calcium and iron in biological structures by electron energy-loss spectrum-imaging. J. Microsc. 210, 5–15 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Beetz T., Jacobsen C., Soft X-ray radiation-damage studies in PMMA using a cryo-STXM. J. Synchrotron Radiat. 10, 280–283 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Vogt S., MAPS : A set of software tools for analysis and visualization of 3D X-ray fluorescence data sets. J. Phys. IV 104, 635–638 (2003). [Google Scholar]
- 46.Scott M. C., Chen C.-C., Mecklenburg M., Zhu C., Xu R., Ercius P., Dahmen U., Regan B. C., Miao J., Electron tomography at 2.4-ångström resolution. Nature 483, 444 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Maiden A. M., Rodenburg J. M., An improved ptychographical phase retrieval algorithm for diffractive imaging. Ultramicroscopy 109, 1256–1262 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Nashed Y. S. G., Vine D. J., Peterka T., Deng J., Ross R., Jacobsen C., Parallel ptychographic reconstruction. Opt. Express 22, 32082–32097 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Heel M., Schatz M., Fourier shell correlation threshold criteria. J. Struct. Biol. 151, 250–262 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Ruiz F. A., Marchesini N., Seufferheld M., Govindjee, Docampo R., The Polyphosphate Bodies of Chlamydomonas reinhardtii Possess a Proton-pumping Pyrophosphatase and Are Similar to Acidocalcisomes. J. Biol. Chem. 276, 46196–46203 (2001). [DOI] [PubMed] [Google Scholar]
- 51.Docampo R., Moreno S. N. J., Acidocalcisomes. Cell Calcium 50, 113–119 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lander N., Cordeiro C., Huang G., Docampo R., Polyphosphate and acidocalcisomes. Biochem. Soc. Trans. 44, 1–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stehling O., Lill R., The role of mitochondria in cellular iron–sulfur protein biogenesis: Mechanisms, connected processes, and diseases. Cold Spring Harb. Perspect. Biol. 5, a011312 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miao J., Förster F., Levi O., Equally sloped tomography with oversampling reconstruction. Phys. Rev. B 72, 052103 (2005). [Google Scholar]
- 55.Komine Y., Eggink L. L., Park H., Hoober J. K., Vacuolar granules in Chlamydomonas reinhardtii: Polyphosphate and a 70-kDa polypeptide as major components. Planta 210, 897–905 (2000). [DOI] [PubMed] [Google Scholar]
- 56.Robinson D. G., Hoppenrath M., Oberbeck K., Luykx P., Ratajczak R., Localization of Pyrophosphatase and V-ATPase in Chlamydomonas reinhardtii. Bot. Acta 111, 108–122 (1998). [Google Scholar]
- 57.Wang Z. T., Ullrich N., Joo S., Waffenschmidt S., Goodenough U., Algal lipid bodies: Stress induction, purification, and biochemical characterization in wild-type and starchless Chlamydomonas reinhardtii. Eukaryot. Cell 8, 1856–1868 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Uchida M., Sun Y., McDermott G., Knoechel C., Le Gros M. A., Parkinson D., Drubin D. G., Larabell C. A., Quantitative analysis of yeast internal architecture using soft X-ray tomography. Yeast 28, 227–236 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uchida M., McDermott G., Wetzler M., Le Gros M. A., Myllys M., Knoechel C., Barron A. E., Larabell C. A., Soft X-ray tomography of phenotypic switching and the cellular response to antifungal peptoids in Candida albicans. Proc. Natl. Acad. Sci. U.S.A. 106, 19375–19380 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McDermott G., Le Gros M. A., Knoechel C. G., Uchida M., Larabell C. A., Soft X-ray tomography and cryogenic light microscopy: The cool combination in cellular imaging. Trends Cell Biol. 19, 587–595 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Odstrčil M., Holler M., Guizar-Sicairos M., Arbitrary-path fly-scan ptychography. Opt. Express 26, 12585–12593 (2018). [DOI] [PubMed] [Google Scholar]
- 62.de Jonge M. D., Kingston A. M., Afshar N., Garrevoet J., Kirkham R., Ruben G., Myers G. R., Latham S. J., Howard D. L., Paterson D. J., Ryan C. G., McColl G., Spiral scanning X-ray fluorescence computed tomography. Opt. Express 25, 23424–23436 (2017). [DOI] [PubMed] [Google Scholar]
- 63.Hegerl R., Hoppe W., Influence of electron noise on three-dimensional image reconstruction. Z. Für Naturforschung A. 31, 1717–1721 (1976). [Google Scholar]
- 64.Öztürk H., Yan H., He Y., Ge M., Dong Z., Lin M., Nazaretski E., Robinson I. K., Chu Y. S., Huang X., Multi-slice ptychography with large numerical aperture multilayer Laue lenses. Optica 5, 601–607 (2018). [Google Scholar]
- 65.Tsai E. H. R., Usov I., Diaz A., Menzel A., Guizar-Sicairos M., X-ray ptychography with extended depth of field. Opt. Express 24, 29089–29108 (2016). [DOI] [PubMed] [Google Scholar]
- 66.Li P., Maiden A., Multi-slice ptychographic tomography. Sci. Rep. 8, 2049 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maiden A. M., Humphry M. J., Rodenburg J. M., Ptychographic transmission microscopy in three dimensions using a multi-slice approach. J. Opt. Soc. Am. A 29, 1606–1614 (2012). [DOI] [PubMed] [Google Scholar]
- 68.Du M., Jacobsen C., Relative merits and limiting factors for x-ray and electron microscopy of thick, hydrated organic materials. Ultramicroscopy 184, 293–309 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/11/eaau4548/DC1
Fig. S1. Experimental phosphorus channel x-ray fluorescence projections.
Fig. S2. Experimental calcium channel x-ray fluorescence projections.
Fig. S3. Experimental sulfur channel x-ray fluorescence projections.
Fig. S4. Experimental chlorine channel x-ray fluorescence projections.
Fig. S5. Experimental potassium channel x-ray fluorescence projections.
Fig. S6. Experimental ptychographic projections.
Fig. S7. Resolution of 2D ptychographic projections.
Movie S1. Reconstructed x-ray fluorescence volume of C. reinhardtii.


