SUMMARY
Activation of Class I phosphoinositide 3-kinase (PI3K) leads to formation of phosphatidylinositol-3,4,5-trisphophate (PIP3) and phosphatidylinositol-3,4-bisphophate (PI34P2), which spatiotemporally coordinate and regulate myriad of cellular processes. By simultaneous quantitative imaging of PIP3 and PI34P2 in live cells, we here show that they have distinctively different spatiotemporal distribution and history in response to growth factor stimulation, which allows them to selectively induce the membrane recruitment and activation of Akt isoforms. PI34P2 selectively activates Akt2 at both the plasma membrane and early endosomes whereas PIP3 selectively stimulates Akt1 and Akt3 exclusively at the plasma membrane. These spatiotemporally distinct activation patterns of Akt isoforms provide a mechanism for their differential regulation of downstream signaling molecules. Collectively, our studies show that different spatiotemporal dynamics of PIP3 and PI34P2 and their ability to selectively activate key signaling proteins allow them to mediate Class I PI3K signaling pathways in a spatiotemporally specific manner.
Graphical Abstract
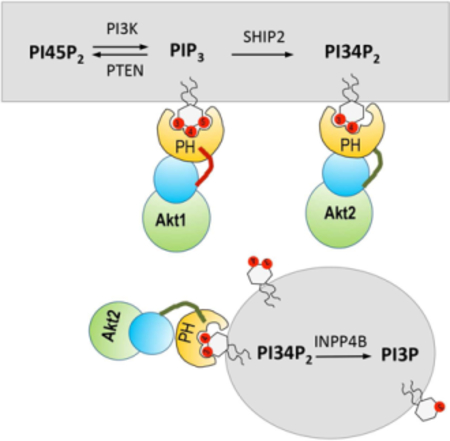
eTOC Blurb
The PI3K signaling is mediated by two lipids, PIP3 and PI34P2. By means of quantitative lipid imaging, we determined the spatiotemporal dynamics of PIP3 and PI34P2 in live cells and elucidated how they specifically control the cellular activities of different Akt isoforms and their downstream signaling proteins.
INTRODUCTION
Activation of Class I phosphoinositide 3-kinases (PI3K) leads to 3’-phosphorylation of phosphatidylinositol-4,5-bisphophate (PI45P2) in the plasma membrane (PM), producing phosphatidylinositol-3,4,5-trisphophate (PIP3). PIP3 is subsequently converted to phosphatidylinositol-3,4-bisphophate (PI34P2) by lipid phosphatases, including SHIP and INPP5 (Vanhaesebroeck et al., 2012). Dysregulated Class I PI3K signaling has been linked to various human diseases, including cancer (Fruman and Rommel, 2014; Thorpe et al., 2015) and inflammatory diseases (Hawkins and Stephens, 2015), and thus Class I PI3K signaling pathways are major targets for drug development. PIP3 is a potent signaling lipid that activates myriad of cellular processes (Cantley, 2002; Vanhaesebroeck et al., 2012). PIP3 carries out its signaling functions primarily by facilitating PM recruitment of cellular proteins with PIP3-binding domains and motifs, most notably the pleckstrin homology (PH) domain (DiNitto and Lambright, 2006). Interestingly, many PIP3-binding PH domains (e.g., Akt, phosphoinositide-dependent kinase 1 (PDK1), ARNO, and DAPP1/Bam32 PH domains) also effectively bind PI34P2 (DiNitto and Lambright, 2006; Franke et al., 1997; Manna et al., 2007), suggesting that PI34P2 may either supplement or substitute for the signaling activity of PIP3. The presence of cellar proteins with PI34P2-specific PH domains, including TAPP1, TAPP2, and lamellipodin/RAPH1, also points to the potential importance of PI34P2-mediated cell signaling (Hawkins and Stephens, 2016; Li and Marshall, 2015). Despite numerous studies on Class I PI3K signaling pathways, the amplitude, the duration, and the interplay of PIP3 and PI34P2 signals after Class I PI3K activation have not been quantified in a spatiotemporally resolved manner in living cells. In particular, the exact signaling function of PI34P2 remains elusive despite recent spike in investigation of its role in membrane trafficking (Boucrot et al., 2015; Posor et al., 2013; Renard et al., 2015) and cell signaling (Braccini et al., 2015; Marat et al., 2017). The aim of this study is to elucidate the differential roles of PIP3 and PI34P2 in Class I PI3K signaling pathways with a focus on Akt activation.
Akt is a primary mediator of Class I PI3K cell signaling pathways (Manning and Cantley, 2007; Manning and Toker, 2017). The Akt kinase family is composed of three homologous isoforms, Akt1, Akt2 and Akt3, encoded by separate genes. Akt1 is ubiquitously expressed whereas Akt2 and Akt3 are the major isoforms expressed in insulin response tissues and the brain, respectively. Cellular Akt activities are tightly regulated by their membrane recruitment (Manning and Cantley, 2007; Manning and Toker, 2017) and post-translational modification (Risso et al., 2015). For Akt1 whose PH domain binds both PIP3 and PI34P2 (Franke et al., 1997), Class I PI3K-mediated production of PIP3 and PI34P2 leads to its PM recruitment and colocalization with other kinases, resulting in phosphorylation of Ser308 in the activation loop by PDK1 and at Ser473 in the hydrophobic motif by the mammalian target of rapamycin complex 2 (mTORC2). However, questions as to if and how PIP3 and PI34P2 differentially regulate three Akt isoforms remain largely unanswered. To address these questions, we simultaneously quantified PIP3 and PI34P2 after growth factor stimulation of Class I PI3K by in situ quantitative lipid imaging technology (Liu et al., 2017; Liu et al., 2014; Yoon et al., 2011). Our study demonstrates that cellular PIP3 and PI34P2 have distinctively different spatiotemporal distribution and history, which allows them to regulate Akt isoforms and their downstream signaling proteins in a spatiotemporally specific manner.
RESULTS
Preparation and characterization of orthogonal PIP3 and PI34P2 sensors
Many analytical methods have been developed to quantify cellular PIP3 and PI34P2 levels (Clark et al., 2011; Kielkowska et al., 2014; Malek et al., 2017); however, none allows spatiotemporally resolved quantification of PIP3 and PI34P2 in live cells. To simultaneously determine the PIP3 and PI34P2 concentrations in mammalian cells by our lipid imaging technology (Liu et al., 2017; Liu et al., 2014; Yoon et al., 2011), we developed orthogonal ratiometric lipid sensors for PIP3 and PI34P2 (Fig. 1a and Supplemental Fig. 1a). Our PIP3 sensor was prepared from the engineered tandem PH domains of myosin X (eMyoX-tPH) (Lu et al., 2011) containing a single cysteine that was chemically labeled with a solvatochromic fluorophore, acrylodan (DAN-eMyoX-tPH). The PI34P2 sensor was prepared by labeling the engineered C-terminal PH domain of Tapp1 (eTapp1-cPH) with another solvatochromic fluorophore, NR3 (NR3-eTapp1-cPH) (Liu et al., 2014). Due to their high lipid head-group specificity and minimal spectral overlap, these sensors allow robust simultaneous quantification of PI34P2 and PIP3 (Supplemental Fig. 1b). Prior to cellular quantification, the sensors were calibrated using PIP3 or PI34P2-containing giant unilamellar vesicles whose lipid composition stipulates that of the cytofacial PM or early endosomes (EEs) (Supplemental Fig. 1c).
Figure 1.
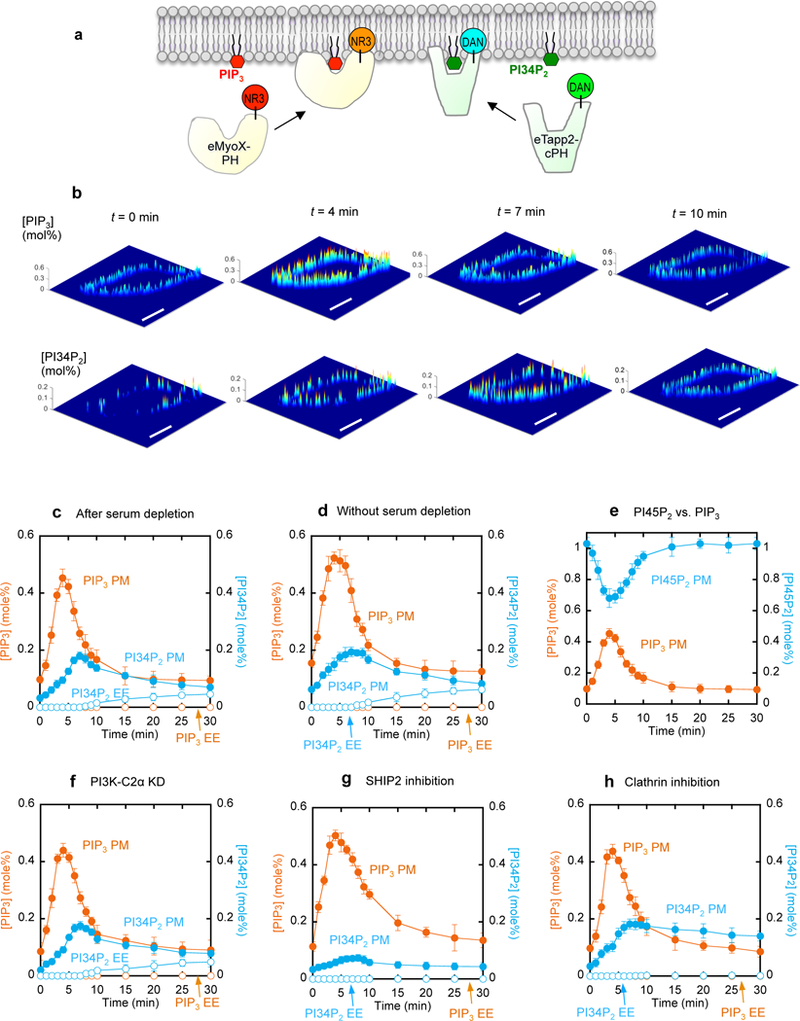
Simultaneous in situ quantification of PIP3 and PI34P2 at the PM and EEs of NIH 3T3 cells after PDGF-BB (50 ng/ml) stimulation. a. Basic strategy for simultaneous quantification using our orthogonal PIP3 and PI34P2 sensors (DAN-eMyoX-PH and NR3-eTapp1-cPH). b. Spatial distribution of PIP3 and PI34P2 at the PM. Each image shows spatially resolved [PIP3] or [PI34P2] on the cross-section of a representative cell at a given time after PDGF stimulation. A pseudo-coloring scheme with red representing the highest and blue the lowest concentration is used to illustrate the spatial concentration heterogeneity. Scale bars represent 20 µm. c.-d. Time-dependent changes of average [PIP3] (orange) and [PI34P2] (blue) at the PM (closed symbols) and EE (open symbols) in serum-depleted (c) and serum-sufficient (d) media. Spatial averaging of [PIP3] and [PI34P2] was performed over the PM and EEs of multiple cells at a given time. e. Time-dependent changes of average [PIP3] (orange) and [PI45P2] (blue) at the PM in response to PDGF. f-h. Effects of PI3K-C2α knockdown (f), SHIP2 inhibition (g), and clathrin inhibition (h) on [PIP3] (orange) and [PI34P2] (blue) at the PM (closed symbols) and EE (open symbols) after PDGF stimulation. SHIP2 inhibition was performed with 20 µM AS1949490 for 1 h whereas clathrin inhibition with 30 µM Pitstop® 2 for 15 min. See Supplemental Fig. 4c for the degree of PI3K-C2α expression after siRNA knockdown. Each data represents the mean ± S.D. from three to five independent experiments (n = 50 cells for Fig. 1c and 1d, 30 for Fig. 1e, 26 for Fig. 1f, 25 for Fig. 1g, and 30 for Fig. 1h). All experiments except Fig. 1d were performed with the serum-free medium.
In situ simultaneous quantitative imaging of PIP3 and PI34P2
Class I PI3K is stimulated by diverse growth factors and G protein-coupled receptor agonists in various mammalian cells (Vanhaesebroeck et al., 2012). In this study we focused on platelet-derived growth factor (PDGF)-induced stimulation of Class I PI3K in NIH 3T3 cells as a model system. We microinjected DAN-eMyoX-tPH and NR3-eTapp1-cPH into the cells and simultaneously monitored fluorescence intensity signals by the sensors in four detector channels after PDGF stimulation (Supplemental Fig. 2a and 2b), from which PIP3 and PI34P2 concentrations were calculated (Fig. 1b). Even after serum depletion for 24 h, NIH 3T3 cells had basal levels of heterogeneously distributed PIP3 and PI34P2 at the PM (Fig. 1b). When spatially averaged, the PIP3 and PI34P2 concentrations at the PM were 0.10 mole% and 0.033 mole%, respectively (Fig. 1c). These PM signals represent genuine basal levels of PIP3 and PI34P2 in NIH 3T3 cells because they were significantly higher without serum depletion and suppressed upon Class I PI3K inhibition (Fig. 1d and Supplemental Fig. 2c and 2d). Also, it is unlikely that non-lipid-related interactions contribute to the signals because mutations of key lipid binding residues in the sensors abrogated their PM localization (Supplemental Fig. 2e and 2f).
Addition of PDGF to the cells caused a rapid increase in PIP3 concentration at the PM, which again showed lateral heterogeneity (Fig. 1b). When spatially averaged, the PIP3 concentration at the PM reached a maximum (0.45 mole%) at 4 min after stimulation, then declined to a nearbasal level after 15 min (Fig. 1c). The spatially averaged PI34P2 concentration at the PM rose more slowly than the PIP3 concentration, reaching a maximum (0.18 mole%) at 7 min, and also decreased more slowly (Fig. 1c). As a result, PI34P2 and PIP3 levels remained comparable at the PM after 10 min. A similar trend was observed under serum-sufficient conditions, except that higher levels of PIP3 and PI34P2 were detected throughout the course of stimulation (Fig. 1d). Furthermore, the increase in PIP3 at the PM was spatiotemporally counterbalanced by the decrease in PI45P2 (Fig. 1e), demonstrating that the observed PIP3 increase truly reflects the local Class I PI3K-mediated conversion of PI45P2 into PIP3. Lastly, similar patterns of PIP3 and PI34P2 distribution at the PM were observed when other mammalian cell lines were stimulated with PDGF (Supplemental Fig. 2g and 2h) and when NIH 3T3 cells were stimulated with other growth factors (Supplemental Fig. 2i and 2j).
It has been reported that PIP3 (Jethwa et al., 2015) and PI34P2 (Braccini et al., 2015; Li Chew et al., 2015; Marat et al., 2017; Watt et al., 2004) exist at endomembranes. In particular, two recent reports indicate that separate pools of PI34P2 are produced from phosphatidylinositol-4-phosphate (PI4P) by Class II PI3Ks, i.e., PI3K-C2α and its liver-specific isoform PI3K-C2γ, respectively, at EEs of thyroid cancer cells (Li Chew et al., 2015) and hepatocytes (Braccini et al., 2015), respectively. To check this possibility, we carefully examined the presence of PIP3 and PI34P2 at intracellular organelles of NIH 3T3 cells. First of all, we could not detect any PIP3 signal at endomembranes for up to 30 min after stimulation (Fig. 1c and Supplemental Fig 3a). We were able to detect some PI34P2 signals after 10 min in cytosolic punctae (Supplemental Fig. 3b), which were confirmed as EEs by an EE marker, EEA1 (Supplemental Fig. 4a). Ratiometric quantification of these signals showed that the spatially averaged PI34P2 concentration at EEs started to rise slowly after 10 min, reaching 0.045 mole% at 30 min, which corresponds to 25% of the maximal PI34P2 concentration at the PM (Fig. 1c). Knockdown of Class II PI3K-C2α in NIH 3T3 cells had little effect on PDGF-mediated PI34P2 production at either the PM or EEs (Fig. 1f and Supplemental Fig. 3b), precluding the possibility that PI34P2 is produced from PI4P by PI3K-C2α after PDGF stimulation. We then treated NIH 3T3 cells with a specific inhibitor (AS1949490) of SHIP2 that is a ubiquitous isoform of the inositol polyphosphate 5-phosphatase SHIP family (Eramo and Mitchell, 2016; Rodgers et al., 2017). SHIP2 inhibition enhanced the amplitude and the duration of the PIP3 signal (Fig. 1g) and drastically suppressed the PI34P2 level at the PM (Fig. 1g and Supplemental Fig 3b), establishing SHIP2 as a main phosphatase converting PIP3 to PI34P2 at the PM of NIH 3T3 cells. Furthermore, SHIP2 inhibition eliminated the PI34P2 signals at EEs (Fig. 1g and Supplemental Fig 3b). In conjunction with the lack of PIP3 molecule at EEs (Fig. 1c and Supplemental Fig 3a), this result suggests that PI34P2 molecules at EEs might be delivered from the PM via endocytosis. Indeed, when NIH 3T3 cells were pretreated with 30 µM of Pitstop® 2, a cell-permeable inhibitor of clathrin (von Kleist et al., 2011), the endosomal PI34P2 signal was eliminated while the PI34P2 signal at the PM remained largely undiminished after its peak at 7 min (Fig. 1h and Supplemental Fig. 3b). Pitstop® 2 had little effect on the cellular PIP3 levels (Fig. 1h). These results strongly support the notion that clathrin-mediated endocytosis serves as a primary mechanism to selectively move PI34P2 from the PM to EEs.
We also investigated the role of another lipid phosphatase, INPP4B, which was reported to degrade PI34P2 to phosphatidylinositol-3-phophate (PI3P) (Li Chew et al., 2015), which may be synergistically augmented by PTEN at the PM (Malek et al., 2017). First, RNAi knockdown of INPP4B in NIH 3T3 cells had a much smaller effect on PI34P2 (and PIP3) at the PM than SHIP2 inhibition (Supplemental Fig. 3b and 5a). Also, INPP4B overexpression in NIH 3T3 cells only modestly reduced the amplitudes of PIP3 and PI34P2 signals at the PM (Supplemental Fig. 3b and 5d). Interestingly, INPP4B knockdown increased the PI34P2 levels at EEs by 40% (Supplemental Fig. 3b and 5a) while its overexpression depleted PI34P2 at EEs (Supplemental Fig. 3b and 5d). To check the potential synergistic action of PTEN and INPP4B, we performed RNAi knockdown of INPP4B in Pten−/− mouse embryonic fibroblast (MEF) cells (Nogueira et al., 2008), which produced a 50% higher PIP3 level at the PM than NIH 3T3 cells upon PDGF stimulation but similar PI34P2 levels to NIH 3T2 cells (Supplemental Fig. 5g). As was the case with NIH 4T3 cells, INPP4B knockdown in Pten−/− MEF cells increased the PI34P2 level at EEs by ≈40% but had little effect on its PM level (Supplemental Fig 5h). Collectively, these results are consistent with the notion that INPP4B is involved in degrading PI34P2 primarily at EEs, independent of PTEN, under our experimental conditions.
Differential responses of Akt isoforms to PIP3 and PI34P2 signals
To elucidate how differently PIP3 and PI34P2 induce cellular responses, we measured the PM recruitment of Akt isoforms via their PH domains, which is the hallmark of Akt activation (Manning and Cantley, 2007). Upon PDGF stimulation, all enhanced green fluorescence protein (EGFP)-tagged Akt PH domains moved to the PM (Fig. 2a) with similar kinetic patterns. Their kinetic curves for PM localization have a maximum at 7 min and a shoulder peak at 4 min (Fig. 2b), which coincide with the maximal PI34P2 and PIP3 signals, respectively (see Fig. 1c). Given that the maximal PI34P2 level is only 40% of that of PIP3, these results show that PI34P2 plays a more dominant role than PIP3 in PM recruitment of the Akt PH domains.
Figure 2.
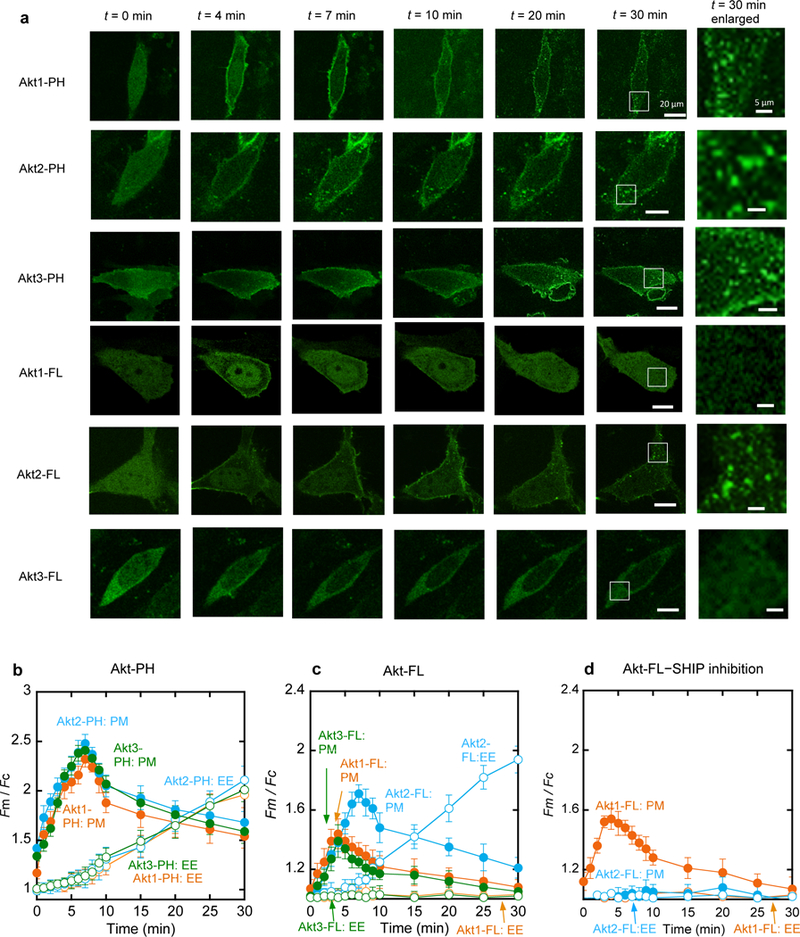
Kinetics of membrane translocation of EGFP-tagged Akt isoforms and their PH domains in NIH 3T3 cells in response to PDGF-BB (50 ng/ml) stimulation. a. Representative fluorescence images of cells expressing Akt isoforms and their PH domains at a given time after PDGF stimulation. Images at 30 min are enlarged (white boxes indicate the enlarged regions) to demonstrate the localization of proteins at EE. Scale bars represent 20 µm for regular images and 5 µm for enlarged images. b. Quantification of the PM (filled symbols) and EE (open symbols) translocation of Akt1-PH (orange), Akt2-PH (blue), and Akt3-PH (green). c. Quantification of the PM (filled symbols) and EE (open symbols) translocation of Akt1-FL (orange), Akt2-FL (blue), and Akt3-FL (green) after PDGF stimulation. d. Effects of SHIP2 inhibition by AS1949490 (20 µM for 1 h) on the kinetics of the PM (filled symbols) and EE (open symbols) translocation of Akt1-FL (orange) and Akt2-FL (blue). Fm/Fc indicates the ratio of the average EGFP intensity at the PM to that in the cytosol. Each data represents the mean ± S.D. from three to five independent experiments (n = 42 cells for Akt1-PH, 35 for Akt2-PH, 35 for Akt3-PH, 45 for Akt1-FL, 38 for Akt2-FL, 31 for Akt3-FL, 28 for Akt1-FL with SHIP2 inhibition, and 29 for Akt2-FL with SHIP2 inhibition). Cells expressing similar levels of Akt proteins were selected for quantification. Scale bars indicate 20 µm.
We then monitored the PM recruitment of EGFP-tagged full-length (FL) Akt isoforms after PDGF stimulation. Interestingly, Akt isoforms showed distinctively different kinetic patterns (Fig. 2a and Fig. 2c). Akt2 basically followed the same kinetics as Akt2-PH and it showed only a modestly lower degree of PM translocation than Akt2-PH, indicating that PI34P2 primarily drives its PM recruitment via its PH domain. In stark contrast, Akt1 and Akt3 showed greatly reduced PM translocation than their PH domains and their maximal PM recruitment occurred at 4 min, followed by a gradual decrease, suggesting that Akt1 and Akt3 respond only to the change in the PIP3 concentration and that their PH domains are not fully exposed.
We also monitored the translocation of Akt isoforms and their PH domains to EEs (Fig. 2a-2c). All Akt PH domains showed similar kinetics of EE localization (Fig. 2b), which is reminiscent of the kinetics of PI34P2 formation at EEs (Fig. 1c). It is thus evident that all Akt PH domains spontaneously respond to the PI34P2 formation both at the PM and at EEs. As was the case with PM translocation, Akt isoforms exhibited distinctively different EE localization patterns. Akt1 and Akt3 showed no endomembrane localization for up to 30 min after PDGF stimulation in NIH 3T3 cells whereas Akt2 displayed a definite degree of localization to the cytosolic punctae (Fig. 2a, Fig. 2c, and Supplemental Fig. 4b), which were colocalized with EEA1, an EE marker (Supplemental Fig. 4a).
To further test the notion that Akt1 and Akt3 are exclusively recruited to the PM by PIP3 whereas Akt2 is targeted to both the PM and EEs primarily by PI34P2, we first measured the effects of modulating the lipid phosphatase activities on subcellular localization of Akt isoforms. SHIP2 inhibition facilitated PM recruitment of Akt1 to the degree that is comparable to the increase in PIP3 while virtually blocking that of Akt2 (Fig. 2d). SHIP2 inhibition also abrogated translocation of Akt2 to EEs (Fig. 2d and Supplemental Fig. 4b). Consistent with relatively modest effects of INPP4B on the PIP3 and PI34P2 levels at the PM (Supplemental Fig. 5a and 5d), the PM recruitment of Akt1 and Akt2 was not significantly altered by INPP4B modulation (Supplemental Fig. 5b and 5e). However, localization of Akt2 to EEs was significantly enhanced by INPP4B knockdown (Supplemental Fig. 4b and 5c) while being largely suppressed by INPP4B overexpression (Supplemental Fig. 4b and 5f).
We then determined the dynamic co-localization of Akt isoforms with PIP3 and PI34P2 in the PM by dual-color single molecule tracking analysis (Koyama-Honda et al., 2005; Park et al., 2016; Sheng et al., 2012). Since endogenous PIP3 and PI34P2 cannot be directly tracked, we employed tetramethylrhodamine (TMR)-labeled SNAP-eMyox-tPH and SNAP-eTapp1-cPH to track PIP3 and PI34P2 molecules, respectively. EGFP-Akt1 and EGFP-Akt2 molecules showed a low degree of dynamic co-localization with either TMR-SNAP-eMyox-tPH or TMR-SNAP-eTapp1-cPH prior to PDGF stimulation. Upon PDGF stimulation, however, EGFP-Akt1 and EGFP-Akt2 displayed enhanced dynamic co-localization specifically with TMR-SNAP-eMyox-tPH and TMR-SNAP-eTapp1-cPH, respectively (Fig. 3a). Detailed kinetic analysis of colocalization showed that EGFP-Akt1 and EGFP-Akt3 had the highest degree of localization with the PIP3 sensor at 4 min after PDGF stimulation (Fig. 3b, 3d and 3h) when the PIP3 concentration was the highest (see Fig. 1b and Fig. 1c). EGFP-Akt2 had the maximal localization with the PI34P2 sensor at 7 min (Fig. 3c and 3h) when the maximal level of PI34P2 is present at the PM (see Fig. 1b and Fig. 1c). Under the same conditions, EGFP-Akt1 (and Akt3) and EGFP-Akt2, respectively, showed consistently low colocalization with the PI34P2 and the PIP3 sensors, respectively (Fig. 3d, 3e, and 3f).
Figure 3.
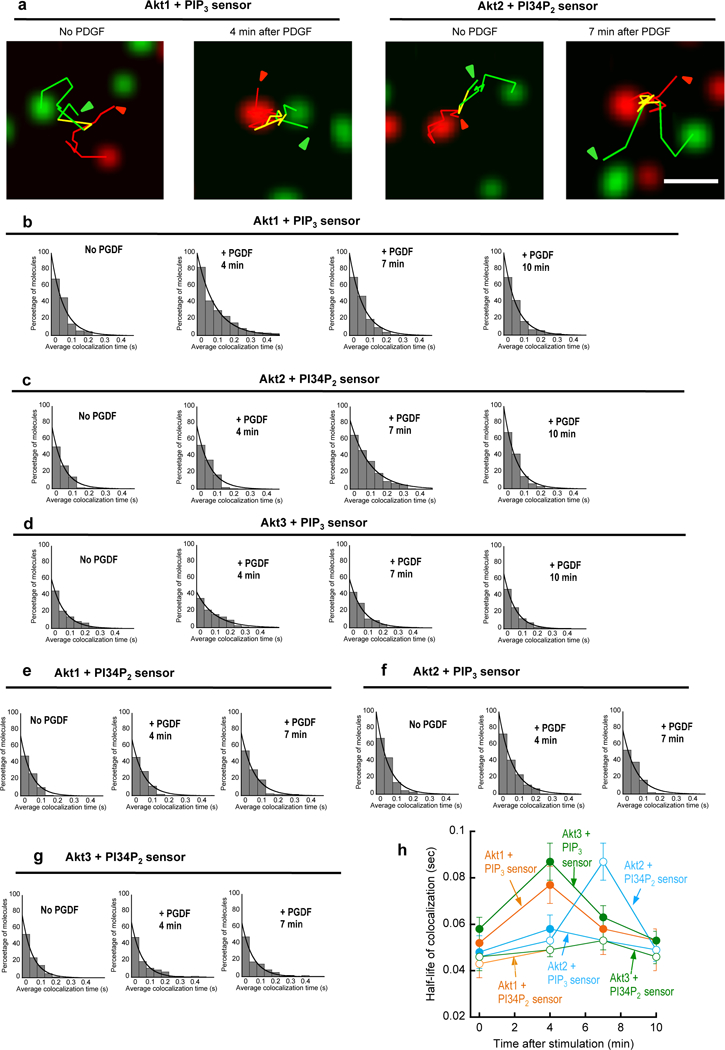
Single-molecule tracking of EGFP-Akt FL and SNAP-TMR-labeled eMyox-PH/eTapp1-PH at the PM of NIH 3T3 cells. a. Representative images of Akt1 and eMyox-PH (two left panels), and Akt2 and eTapp1-PH (two right panels) in a NIH 3T3 cell before and after PDGF-BB (50 ng/ml) stimulation. Green, red, and yellow lines are trajectories of Akt1/2, eMyox-PH/eTapp1-PH, and colocalized molecules, respectively. Arrows indicate the starting points of tracking. The scale bar represents 1 µm. b.-g. Time courses of dynamic colocalization of Akt1 and eMyox-PH (b), Akt2 and eTapp1-cPH (c), Akt3 and eMyox-PH (d), Akt1 and eTapp1-cPH (e), Akt2 and eMyox-PH (f), and Akt3 and eTapp1-cPH (g). The histograms display percentages of Akt molecules spending a given colocalzation time (>0.2 sec) with eMyox-PH (or eTapp1-PH) on the PM. h. Time courses of the half-life of colocalization for Akt isoforms and PIP3/PI34P2 sensors. Colocalization of the PIP3 sensor with Akt1 (red), Akt2 (cyan), and Akt3 (green) is shown in closed symbols whereas that of the PI34P2 sensor with Akt is in open symbols. Error bars indicate S.D. (n = 80–100 cells).
Differential membrane binding properties of Akt isoforms.
To understand the origin of differential responses of Akt isoforms to PIP3 and PI34P2 signals, we quantitatively measured the affinity of Akt isoforms and their PH domains for PIP3- and PI34P2-containing vesicles by surface plasmon resonance (SPR) analysis (Table 1). In agreement with a more prominent effect of PI34P2 in PM recruitment of Akt PH domains, all isolated PH domains have ca. two-fold higher affinity for PI34P2-containing vesicles than for PIP3-containing vesicles. In general, Akt proteins have lower membrane affinity than their PH domains, which agrees with their relatively lower degree of PM translocation upon PDGF stimulation (see Fig. 2b and 2c). It should be noted, however, that Akt proteins still have high enough membrane affinity (i.e., submicromolar Kd), indicating that the PH domains in the FL Akt are not as fully occluded as previously reported (Calleja et al., 2007; Parikh et al., 2012). As for lipid selectivity, Akt2 has the same PI34P2 selectivity as its PH domain whereas Akt1 and Akt3 have definite PIP3 selectivity. This disparate intrinsic PIP3 versus PI34P2 selectivity of Akt isoforms should account for their differential responses to cellular PIP3 and PI34P2 levels (see Fig. 2c). Lastly, Akt proteins expressed in E. coli have essentially the same membrane binding properties as those expressed in insect cells, suggesting that potential post-translational modifications of Akt do not significantly affect their intrinsic membrane binding properties.
Table 1.
Membrane binding properties of Akt isoforms and their PH domains determined by SPR analysis.
| Kd (nM) for vesiclesa | Relative affinityb (PI34P2/PIP3) |
||
|---|---|---|---|
| PC/PS/PI34P2 (77:20:3) | PC/PS/PIP3 (77:20:3) | ||
| Akt1 PHc | 310b ± 45 | 580 ± 80 | 1.9 |
| Akt2 PHc | 220 ± 30 | 440 ± 50 | 2.0 |
| Akt3 PHc | 340 ± 60 | 610 ± 65 | 1.8 |
| Akt1 FLd | 960 ± 150 | 470 ± 70 | 0.5 |
| Akt2 FLd | 410 ± 80 | 610 ± 100 | 1.5 |
| Akt3 FLd | 900 ± 100 | 460 ± 90 | 0.5 |
| Akt1 FLe | 880 ± 160 | 510 ± 210 | 0.6 |
| Akt2 FLe | 360 ± 150 | 600 ± 200 | 1.7 |
| Akt3 FLe | 940 ± 260 | 390 ± 120 | 0.4 |
Data represent mean ± S.D. from triplicate equilibrium SPR measurements.
Kd for PIP3 vesicles /Kd for PI34P2 vesicles.
Akt PH domains were expressed as C-terminal EGFP-tagged proteins for enhanced stability. EGFP-tagged Akt PH domains and non-tagged Akt PH domains have essentially the same membrane binding properties.
Akt full-length proteins expressed in E. coli.
Akt full-length proteins expressed in insect cells.
Molecular basis for differential membrane binding properties of Akt isoforms
All Akt isoforms have the same domain arrangement, the N-terminal PH domain and the C-terminal catalytic domain, which are connected by a short linker region (Fig. 4a) (Manning and Cantley, 2007; Manning and Toker, 2017). The catalytic domain also contains a regulatory region with the turn and the hydrophobic motifs at the C-terminus. The PH and catalytic domains of Akt isoforms show high sequence similarity but their linker sequences are variable. For example, the PH and catalytic domains of Akt1 and Akt2 show >80% sequence similarity but their linker sequences have ≈50% similarity. Multiple reports have indicated that the variable linker region might confer isoform-specific function on each isoform (Chin and Toker, 2010; Kim et al., 2008). To test the possibility that the linker region is involved in lipid specificity of Akt isoforms, we prepared a number of chimeric constructs for Akt isoforms (Fig. 4a). For instance, Akt1 FL-L2 and Akt2 FL-L1 are the chimeric Akt1 and Akt2, respectively, containing the linker for Akt2 (L2) and Akt1 (L1), respectively. We also prepared extended Akt1-PH, Akt2-PH, and Akt3-PH with either the L1 or the L2 linker (e.g., Akt1-PH-L2).
Figure 4.
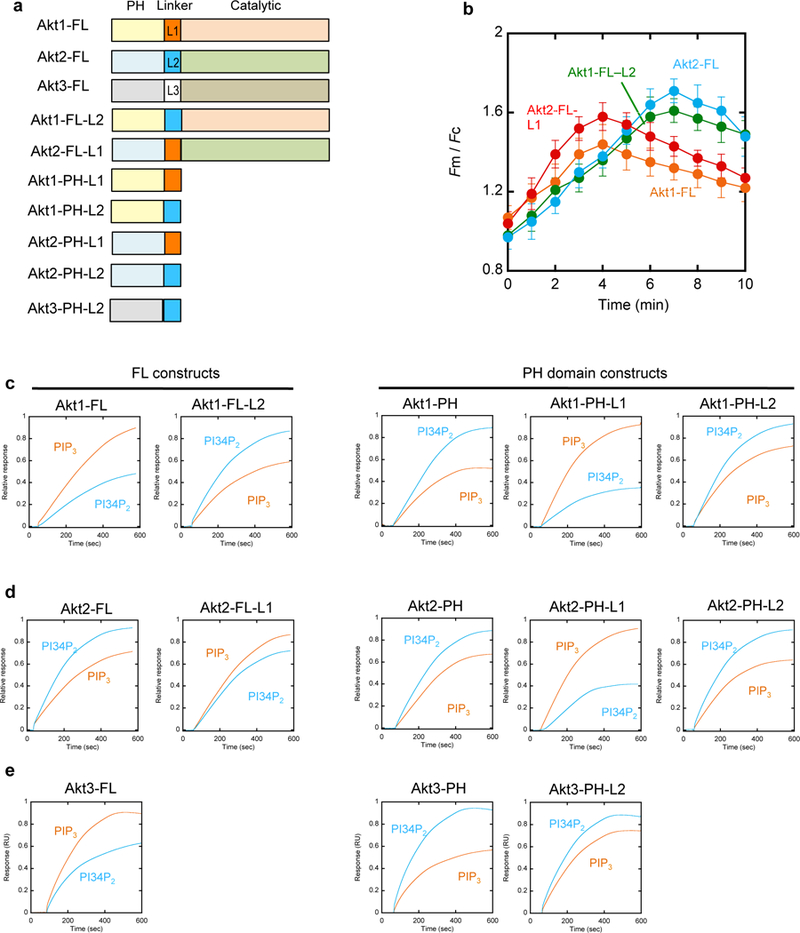
Different membrane binding properties of various Akt constructs. a. Schematic illustration of Akt constructs. b. Quantification of the PM translocation of Akt1-FL (orange), Akt2-FL (blue), Akt1-FL-L2 (green), and Akt2-FL-L1 (red) after PDGF stimulation. Fm/Fc indicates the ratio of the average EGFP intensity at the PM to that in the cytosol. Each data represents the mean ± S.D. from three to five independent experiments (n = 45, 38, and 35 cells for Akt1, Akt2, and Akt2-FL-L1, respectively). c.-e. SPR sensorgrams for various Akt1 (c), Akt2 (d), and Akt3 (e) constructs. All proteins were bacterially expressed and their representative sensorgrams (from triplicate measurements) for POPC/POPS/PIP3 (77:20:3) and POPC/POPS/PI34P2 (77:20:3) large unilamellar vesicles are shown in orange and blue, respectively.
Akt1 FL-L2 exhibited a strikingly similar kinetic pattern of PM translocation to Akt2-FL whereas Akt2 FL-L1 behaved similarly to Akt1-FL (Fig. 4b). This suggests that the linker region is primarily responsible for the differential membrane binding properties of Akt isoforms. More specifically, the L1 linker seems to confer PIP3 selectivity on the Akt constructs. To further test this notion, we measured the vesicle binding of all chimera constructs by the SPR analysis. As summarized in Table 1, Akt1-PH, Akt2-PH, Akt3-PH, and Akt2-FL prefer PI34P2-containing vesicles to PIP3-containing vesicles whereas Akt1-FL and Akt3-FL show the opposite lipid selectivity (see also Fig. 4c-4e). Under the same conditions, the chimeric Akt1 FL-L2 shows PI34P2 selectivity (Fig. 4c) while chimeric Akt2 FL-L1 displays PIP3 selectivity (Fig. 4d). Furthermore, all chimeric Akt PH domains with a linker extension showed the lipid selectivity of their linkers (4c-4e). Collectively, these results clearly show that the variable linker region dictates the differential membrane binding properties of Akt isoforms. In particular, the L1 (and L3) linker imparts the unique PIP3 selectivity to Akt1-PH (and Akt3-PH) that intrinsically prefers PI34P2 to PIP3. These results also suggest that the specific lipid binding site of Akt1 (and Akt3) comprises the PH domain and the linker region.
Effects of PIP3 and PI34P2 on cellular activities of Akt isoforms.
We then measured how different membrane binding properties of Akt1 and Akt2 affect their cellular activities. We first monitored the activation of Akt1 and Akt2 in living cells after PDGF stimulation using fluorescence resonance energy transfer (FRET)-based biosensors for Akt1 and Akt2, ReAktion1 (Ananthanarayanan et al., 2007) and ReAktion2 (Zhou et al, unpublished). These isoform-specific biosensors allow for real-time monitoring of Akt activation in terms of the change in the cyan fluorescence proteins (CFP)-to-yellow fluorescence proteins (YFP) FRET accompanying conformational changes of Akt caused by phosphorylation and activation (Supplemental Fig. 6a). We transfected ReAktion1 and ReAktion2 into NIH 3T3 cells, respectively, and after PDGF stimulation, quantified Akt1 and Akt2 activation in terms of the corrected and normalized FRET changes (nFRET) (Wang et al., 2014). Time-dependent plots of cellular nFRET show that Akt1 is activated primarily at the PM whereas Akt2 is activated at both the PM and EEs (Fig. 5a). Most importantly, the kinetics of Akt1 activation at PM is almost identical to that of the PIP3 formation (see Fig. 1c) at PM and that of PM translocation of Akt1 (see Fig. 2a and Fig. 2c). Likewise, Akt2 activation coincides with the PI34P2 formation (see Fig. 1c) at and Akt2 translocation (see Fig. 2a and Fig. 2c) to the PM and EEs (Fig. 5a). We also measured the effect of the SHIP2 inhibition on Akt activation. Akt1 was more active after SHIP2 inhibition at the PM (in terms of both magnitude and duration) but Akt2 showed drastically reduced activity at the PM and EEs (Fig. 5b and Supplemental Fig. 6b).
Figure 5.
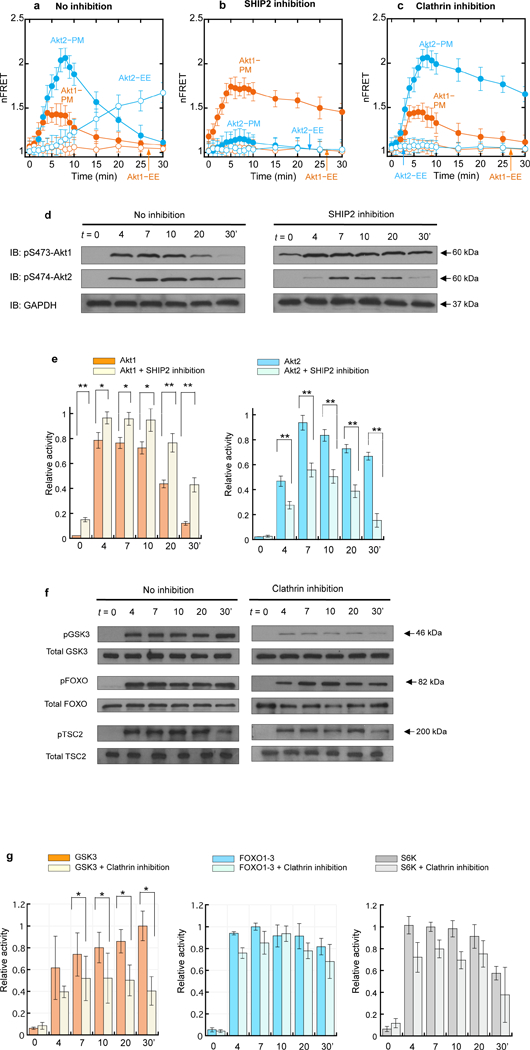
Kinetics of activation for Akt isoforms and downstream signaling proteins in NIH 3T3 cells. a. Kinetics of activation of Akt1 (orange) and Akt2 (blue) biosensors after PDGF stimulation. b. Effects of SHIP2 inhibition (20 µM AS1949490 for 2 h) on subcellular Akt1 (orange) and Akt2 (blue) activation. c. Effects of a Pitstop® (30 µM for 15 min) on subcellular Akt1 (orange) and Akt2 (blue) activation. nFRET at PM and EE indicate the CFP-to-YFP FRET values corrected against the filter bleed-through and normalized against the CFP signals at the two different cell membranes. Each data represents the mean ± S.D. from three independent experiments (n = 22, 20, and 24 cells for Fig. 5a, 5b, and 5c, respectively). d. The time course of phosphorylation of S473 and S474 of endogenous Akt1 and Akt2, respectively, in NIH 3T3 cells in response to PDGF stimulation before and after SHIP2 inhibition (20 µM AS1949490 for 2 h). Representative western blots (n = 3) show Akt phosphorylation detected by immunoblotting (IB) with isoform-specific phosphor-S473/4 (pS473/4) antibodies. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a loading control. e. Quantitative densitometry analysis of the pS473 and pS474 bands shown in Fig. 5d. f. The time course of phosphorylation of GSK3, FOXO1–3, and TSC2, respectively, in NIH 3T3 cells in response to PDGF stimulation before and after treatment with 30 µM Pitstop® 2 for 15 min. Representative western blots (n = 3) show phosphorylation of the three proteins detected by immunoblotting (IB) with indicated antibodies. g. Quantitative densitometry analysis of the pGSK3, pFOXO1–3, and pS6K bands shown in Fig. 5f. Relative activity was calculated by normalizing density values against the maximal density for pAkt1, pAkt2, pGSK3, pFOXO1–3, and pS6K, respectively, after GAPDH correction. Error bars indicate S.E. Statistical significance was calculated by the Student’s t-test. Significance levels are *p < 0.05 and **p < 0.01.
To demonstrate the physiological relevance of these activation data, we monitored the time-dependent mTORC2-mediated phosphorylation of the hydrophobic motif serine (S473 for Akt1 and S474 for Akt2) of endogenous Akt1 and Akt2 in NIH 3T3 cells using isoform-specific antibodies against pS473 and pS474, respectively. First, we performed immunoblotting to quantitatively monitor the degree of Akt phosphorylation. Akt1 reached the maximal degree of phosphorylation at 4 min after PDGF stimulation (Fig. 5d and 5e) that started to decrease after 10 min, which agrees with the kinetics of its activation at PM monitored by the Akt1 biosensor (Fig. 5a). S474 phosphorylation of Akt2 increased more slowly and reached a plateau at 7–10 min that persisted up to 30 min after stimulation (Fig. 5d and 5e). This time course seems to reflect the combined activity of Akt2 at the PM and EEs (Fig. 5a). Furthermore, SHIP2 inhibition increased the phosphorylation of endogenous Akt1 while greatly reducing the endogenous Akt2 phosphorylation, especially at a later stage (Fig. 5d and 5e).
We also performed time-lapse immunofluorescence staining of endogenous pS473-Akt1 and pS474-Akt2 in response to PDGF stimulation (Supplemental Fig. 7a). pS473-Akt1 was primarily localized to the PM at 4 min after stimulation whereas pS474-Akt2 showed dual localization, first at the PM (at 7 min) and then at EEs (after 20 min). Overall, these results demonstrate that PIP3 and PI34P2 not only selectively trigger membrane translocation of Akt1 (and Akt3) and Akt2, respectively, but also induce their selective and immediate phosphorylation and activation under physiological conditions.
Lastly, we investigated how the unique endosomal activity of Akt2 regulates downstream signaling pathways by taking advantage of our finding that Pitstop® 2 selectively blocks delivery of PI34P2 from the PM to EEs (see Fig. 1h and Supplemental Fig. 3b). Pre-treatment of NIH 3T3 cells with Pitstop® 2 specifically suppressed the Akt2 activity at EEs while greatly elongating the high Akt2 activity at the PM (Fig. 5c and Supplemental Fig. 6c). It had no effect on Akt1 activity. Having established that Pitstop® 2 selectively inhibits the Akt2 activity at EEs, we then measured the effects of Pitstop® 2 treatment on phosphorylation of three major downstream effectors of Akt, glycogen synthase kinase 3 (GSK3), forkhead box O (FOXO) transcription factors, and tuberous sclerosis complex2 (TSC2) that regulates the mTORC1 activity (Manning and Cantley, 2007; Manning and Toker, 2017). As shown in Fig. 5f and 5g, Pitstop® 2 significantly reduced (≈60%) GSK3 phosphorylation while exerting minimal effects on phosphorylation of FOXO1–3 and TSC2. This selective inhibition of GSK3 phosphorylation despite unaltered Ak1 activity and sustained Akt2 activity at the PM (see Fig. 5c) suggests that the Akt2 activity at EEs might specifically modulate the GSK3 branch of the PI3K-Akt signaling pathways in response to PDGF stimulation. Time-lapse immunofluorescence staining of endogenous pGSK3 in NIH 3T3 cells showed primary punctate endosomal localization mainly at 20 min after PDGF stimulation and afterward, which is synchronized with endosomal localization of the PI34P2 sensor (Supplemental Fig. 7b), supporting the notion that endosomal Akt2 is primarily involved in GSK3 activation.
DISCUSSION
Since the discovery of Class I PI3K, PIP3 has been generally thought to be a major player in downstream signaling pathways. PI34P2 was initially regarded as a byproduct of PIP3 that might supplement the signaling functions of PIP3 (Hawkins and Stephens, 2016; Li and Marshall, 2015). Recently, much attention has been given to the cellular functions of PI34P2 produced from PI4P by Class II PI3K (Marat et al., 2017; Posor et al., 2013). Although Class I PI3K-derived PI34P2 has been recently reported to control fast endophilin-mediated endocytosis (Boucrot et al., 2015; Renard et al., 2015), the question as to how PI34P2 specifically regulates Class I PI3K signaling still remains largely unanswered. We have addressed this critical question by simultaneously quantifying PIP3 and PI34P2 in response to growth factor stimulation and correlating their spatiotemporal dynamics with subcellular localization and activation of Akt isoforms that are central players in PI3K signaling pathways.
Our study shows that PDGF stimulation triggers rapid, laterally heterogeneous enrichment of PIP3 at the PM, followed by a delayed localized increase in PI34P2. Based on the average PI45P2 concentration at the PM (i.e., 1.0 mole) of NIH 3T3 cells (Fig. 1e) (Liu et al., 2014; Yoon et al., 2011), our results show that up to 45% of PI45P2 is converted to PIP3 by Class I PI3K, from which 40% is degraded to PI34P2 at the PM (Fig. 6), which suggests that 60% of PIP3 is converted back to PI45P2 by PTEN. Genetic ablation of PTEN significantly increases the PM PIP3 level with a minor effect on the PM PI34P2 level, confirming the primary role of PTEN as the 3’-phosphatase of PIP3. Also, pharmacological inhibition of SHIP2 virtually eliminates PI34P2 in the PM while enhancing both the amplitude and the duration of PIP3 signaling, verifying that it mainly drives the dephosphorylation of PIP3 to PI34P2 at the PM under our experimental conditions. In contrast, INPP4B does not appear to play a significant role in the turnover of PIP3 and PI34P2 at the PM in PDGF-mediated PI3K signaling. These results thus suggest that the relatively rapid decline in the PIP3 level at the PM is caused by the combined action of SHIP2 and PTEN (Fig. 6).
Figure 6.
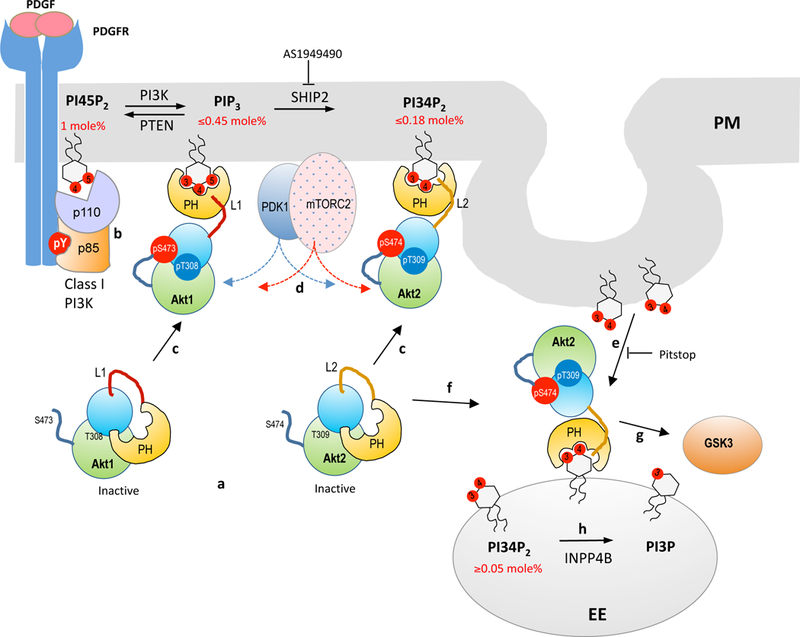
A proposed model for PIP3 and PI34P2 formation and isoform-specific activation of Akt during PDGF-mediated Class I PI3K signaling. a. Akt isoforms remain auto-inhibited in the resting state, with the active site in the C-terminal catalytic domain (shown in blue and green lobes) blocked by the N-terminal PH domain (yellow). The lipid binding site of the PH domain is partially open to allow spontaneous response to PIP3 and PI34P2 formation at the PM. b. Class I PI3K activation leads to rapid (maximal at ≈4 min) formation of PIP3 (up to 45% of PI45P2) which is then (maximal at ≈7 min) converted to PI34P2 (up to 18% of PI45P2) by SHIP2. c. Akt1 (and Akt3) is selectively recruited to PM by PIP3 whereas Akt2 by PI34P2. In both cases, Akt translocation is spatiotemporally coordinated with lipid formation. The variable linker region (L1 and L2) dictates the lipid specificity of Akt isoforms. d. Selective PM recruitment of Akt isoforms by PIP3 and PI34P2 leads to immediate activation through phosphorylation of T308 (T309 for Akt2) in the activation T loop by PDK1 and S473 (S474 for Akt2) by mTORC2. PDK1 and mTORC2 are also reported to be recruited to PM by PIP3. e. PI34P2 is selectively enriched in vesicles and transferred to EEs by endocytosis. f. Akt2 is selectively recruited to EEs by this endosomal pool of PI34P2 that reaches about 30% of the maximal PI34P2 at the PM at 30 min after PDGF stimulation. g. Endosomally activated Akt2 may play a key role in phosphorylating and regulating GSK3. h. Endosomal PI34P2 is converted to PI3P by INPP4B, terminating Akt2 activation.
Our study also provides new mechanistic and quantitative insight into the origin and the fate of PI34P2 at EEs. Our analysis shows that in response to PDGF stimulation in NIH 3T3 cells PI34P2 is formed much more slowly at EEs than at the PM: however, at 30 min after stimulation the PI34P2 level at EEs reaches about 30% of the maximal PI34P2 at PM, implying an important role of endosomal PI34P2 at a later stage of Class I PI3K activation. Our results suggest that PI34P2 is mainly formed from PIP3 at the PM by SHIP2, subsequently transported to EEs via clathrin-mediated endocytosis, and eventually converted to PI3P by INPP4B at EEs during PDGF-mediated PI3K signaling (Fig. 6). The kinetics of PI34P2 appearance at EEs is consistent with the reported kinetics of clathrin-mediated endocytosis as monitored with transferrin (Ciechanover et al., 1983). Although it is not fully understood why PI34P2 is preferentially transported to EEs over PIP3 by endocytosis, one can speculate that PI34P2 is enriched in budding clathrin-coated vesicles (Boucrot et al., 2015; Renard et al., 2015) as a mechanism to eliminate PI34P2 signals from the PM (Fig. 6). Collectively, our study quantitatively defines the spatiotemporal history of PIP3 and PI34P2 and the roles of various lipid phosphatases in the Class I PI3K signaling pathways. Furthermore, it shows that PIP3 is a relatively short-lived signal localized at the PM whereas PI34P2 is a more sustained signal that operates at both the PM and EE.
Our study shows that differential subcellular membrane targeting behaviors of Akt isoforms can be explained unambiguously in terms of their PIP3 versus PI34P2 selectivity and the asynchronous kinetics of PIP3 and PI34P2 formation and degradation at the PM and EEs (Fig. 6). The SPR analysis of Akt isoforms shows that distinct membrane localization behaviors of Akt isoforms largely reflect their intrinsic membrane binding properties and may not significantly depend on other factors, such as different post-translational modification or protein-protein interactions. It has been reported that the PH domain and catalytic domain of Akt tightly interact with each other through hydrophobic interactions, which keeps the protein in an inactive “PH-in” conformation (Calleja et al., 2007; Parikh et al., 2012) in which both the lipid binding pocket of the PH domain and the activation loop in the catalytic domain are occluded. Although it has been postulated that PM binding of Akt would expose the activation loop for PDK1 phosphorylation (Calleja et al., 2007; Calleja et al., 2009), it remains unclear as to how an intact Akt molecule with the occluded PH domain initially binds PIP3/PI34P2-containing cell membranes. Our SPR data show that the intact Akt isoforms, while less active than their isolated PH domains, have high enough membrane affinity that they can directly bind to the PM in response to a rise in PIP3 and PI34P2 (Fig. 6). Most importantly, our results show that the variable linker region plays a key role in defining the lipid specificity of Akt isoforms. The linker for Akt2 allows the intact protein to retain the membrane binding properties of its PH domain whereas those for Akt1 and Akt3 alter the lipid binding properties of their respective PH domains. Our SPR study on various PH-linker constructs shows that the PH domain and the linker together constitute a structurally and functionally independent lipid-binding module and as such they serve as an isoform-specific membrane-targeting regulatory unit.
Our studies also provide critical new insight into the spatiotemporal dynamics of Akt activation. It has been long thought that PM-recruited Akt isoforms are phosphorylated by PDK1 and mTORC2 and that activated Akt can then regulate various downstream proteins either at PM or in other subcellular locations (Manning and Cantley, 2007; Manning and Toker, 2017). This traditional view has been recently challenged by reports supporting compartmentalized and site-specific Akt activation and signaling. For instance, it has been reported that the phosphorylated Akt1 is active only at the PM (Ebner et al., 2017) whereas Akt2 is specifically activated at EEs by PI34P2 locally generated from PI4P (Braccini et al., 2015; Li Chew et al., 2015). Our ability to accurately modulate and quantify cellular PIP3 and PI34P2 levels in a spatiotemporally resolved manner and to quantitatively correlate these data with the membrane translocation and activation of Akt isoforms allowed us to investigate how Akt isoforms are activated and subsequently regulate the activities of downstream effector proteins in a spatiotemporally specific manner. An excellent spatiotemporal correlation between the cellular PIP3 and PI34P2 levels and the Akt activation monitored by Akt biosensors and by endogenous Akt phosphorylation shows that the PIP3- and PI34P2-dependent PM translocation of Akt1 and Akt2, respectively, leads to their immediate and direct phosphorylation by PDK1 and mTORC2 (Fig. 5 and Fig. 6). However, Akt1 and Akt2 follow distinctively different paths thereafter. Spatiotemporal dynamics and the magnitude of S473 phosphorylation of endogenous Akt1 suggests that Akt1 is active primarily at the PM while PIP3 is present at PM. This would confine Akt1 activation and action both spatially and temporally. In contrast, Akt2 is recruited first to the PM and then to EEs with an increase in the endosomal PI34P2 level and remains active at EEs while PI34P2 is present at EEs. This should allow Akt2 to act on its diverse downstream proteins with more spatiotemporal flexibility. Our evidence for the selective endosomal activation of GSK3 by Akt2 supports this notion. A similar role for endosomal Akt2 in regulation of glucose transport was reported during insulin-mediated PI3K signaling (Braccini et al., 2015).
In summary, our work demonstrates a new independent signaling function of PI34P2, i.e., isoform- and site-specific membrane targeting and activation of Akt2. It should be noted that although the PDGF signaling pathway serves as an excellent model system for Class I PI3K signaling, growth factor signaling pathways are highly complex and diverse. Thus, spatiotemporal dynamics of PIP3 and PI34P2 metabolism and Akt activation in other growth factor signaling pathways might be different from our results. Furthermore, there are other cellular proteins whose PH domains can bind both PIP3 and PI34P2 and thus respond to spatiotemporal dynamics of PIP3 and PI34P2. The present study provides a conceptual framework and experimental approaches with which to further investigate the membrane targeting and activation of various proteins in diverse Class I PI3K signaling pathways. This study also suggests that one can specifically modulate individual downstream signaling pathways of Class I PI3K by controlling the cellular levels of PI34P2 and PIP3 or the cellular activities of Akt isoforms in a spatiotemporally specific manner.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Wonhwa Cho (wcho@uic.edu).
METHOD DETAILS
PI45P2, PI34P2 and PIP3 sensors preparation
The Tapp1-cPH domain (amino acids 182–303) was subcloned into the pGEX 4T-1 vector. Two surface-exposed cysteine residues, Cys-198 and Cys-246, were mutated to Ala and Val-204 was mutated to Cys to introduce a single cysteine for fluorophore labeling in the membrane-binding region. To improve membrane affinity of the protein, Met-205 was mutated to Trp and Ala was inserted between Val-204 and Met-205, yielding the final PI34P2 sensor, eTapp1-cPH. For the PIP3 sensor, tandem PH domains of human myosin X (MyoXPH: a.a.1170–1385) was also cloned into pGEX 4T-1 vector. Cys-1174 and Cys-1344 were mutated to Ala and Leu-1231 was mutated to cysteine to introduce a single cysteine for fluorophore labeling in the membrane-binding region. To improve membrane affinity of the protein, Thr-1230 was mutated to Ala, yielding MyoXPH-C1174A/T1230A/L1231C/C1344A (eMyoX-PH). All eTapp1-PH and eMyox-PH proteins were expressed as GST-tagged proteins in E. coli BL21 RIL codon plus (Stratagene) cells and purified using the glutathione resin (GenScript). These proteins were labeled at the single cysteine site by acrylodan or NR3 on the column as described previously (Cho et al., 2017). The PI45P2 sensor was prepared from the engineered epsin 1 ENTH domain as described previously (Yoon et al., 2011).
Bacterial expression and purification of Akt proteins.
All constructs were produced using the pRSET B vector (Thermo Fisher). All Akt proteins have a N-terminal His6-tag and Akt PH domains and (PH domain + linker) chimeras also contain a C-terminal EGFP. All proteins were expressed in E. coli BL21 RIL codon plus (Stratagene) cells and purified using the Ni-NTA agarose affinity resin (Qiagen). Various Akt constructs used in this study are as follows. Akt1-FL: Akt1 (a.a.1–480); Akt2-FL: Akt2 (1–481); Akt3-FL: Akt3 (1–479); Akt1-FL-L2: Akt1 (1–111)-Akt2 (112–151)-Akt1 (150–480); Akt2-FL-L1: Akt2 (1–111)-Akt1 (112–149)-Akt2 (152–481); Akt1-PH-L1: Akt1 (1–149); Akt1-PH-L2: Akt1 (1–111)-Akt2 (112–151); Akt2-PH-L1: Akt2 (1–111)-Akt1 (112–149); Akt2-PH-L2: Akt2 (1–151); Akt3-PH-L2: Akt3 (1–111)-Akt2 (112–151).
Expression of Akt FL proteins in insect cells and protein purification
The donor vector pFLHT was separately constructed to express each Akt protein. The cDNAs of human Akt1 (residues 1–480), murine Akt 2 (residues 1–481) and murine Akt3 (residues 1–479) were amplified from TrueORF Gold cDNA (Origene) using the Phusion HF DNA polymerase (NEB). PCR fragments encoding each form of Akt were respectively cloned next to the N-terminal fused His6 tag through BamHI and HindIII sites into the region controlled by the polyhedrin and p10 promoters of the pFLHT vector. E. coli DH10Bac competent cells were transformed by pFLHT-AKT1, pFLHT-Akt2 and pFLHT-Akt3 and the transformants were selected by a combination of three antibiotics (gentamicin, kanamycin and tetracycline) and cultured for 24 h at 37 °C. The selected bacmids for Akt1, Akt2 and Akt3 were purified using a midi prep kit (QIAGEN). The integrity of the plasmid sequence was verified by DNA sequencing. Each Bacmid (6 µg) was suspended separately in 100 µl of Grace’s media with supplement. Then 18 µl of Cellfectin II (Thermo Fisher) was suspended in 300 µl of Grace’s media with supplement. The Cellfectin II-media mixture was divided into three equal portions and each portion was mixed with each bacmid and incubated for 30 min at room temperature. Sf9 cells were seeded in 6-well plates as 0.9 × 106 cells per well in Grace’s media without supplement. The bacmid cellfectin mixture was transferred into the well and incubated for 5 h at 27 °C. The transfected media was replaced with Grace’s media with supplement and 1% pen/strep. Incubate the cells were incubated for 72 h at 27 °C. P0 virus was harvested after 72 h and P1, P2 and P3 virus were subsequently amplified. After harvesting P3 virus for each protein, one liter of Hifive suspension cells (when the cell density was 4 × 106 cells/ml) were infected using 20 ml P3 virus and incubated at 27 °C in a shaker (120 rpm) for 48 h. The cells were harvested after 48 h by centrifugation 18000g for 10 min. The cells were re-suspended in 100 ml lysis buffer (20 mM Tris-HCl (pH 7.9), 320 mM NaCl, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride (PMSF), 0.5 mM tris(2-carboxyethyl)phosphine (TCEP), 100 µM leupeptin, 1 µM pepstatin A) and lysed by using a homogenizer (EmulsiFlex–C3; Avestin). The lysate was centrifuged at 160000g for 1 h and the clear supernatant was loaded onto a Histrap FF crude column (GE Healthcare) and washed with buffer A (20 mM Tris-HCl (pH 7.9), 320 mM NaCl, 1 mM PMSF, 0.5 mM TCEP, 100 µM leupeptin, 1 µM pepstatin A), eluted with linear gradient with buffer B (20 mM Tris-HCl (pH 7.9), 320 mM NaCl, 1 M imidazole, 1 mM PMSF, 0.5 mM TCEP, 100 µM leupeptin, 1 µM pepstatin A). The identity and integrity of the final protein sample were confirmed by sodium dodecylsulfate polyacrylamide gel electrophoresis. Fractions containing Akt protein were pooled and concentrated using an Amicon Ultra-15 (30,000 Da MWCO) concentrator (Millipore).
Spectrofluorometric measurements
Horiba Flurolog–3 spectrofluorometer was used for all cuvette-based fluorescence measurements. Lipid sensors (typically 500 nM) were added to large unilamellar vesicles with various lipid compositions and the emission spectra of DAN and NR3 were measured with excitation wavelength set at 380 nm and 530 nm, respectively, in a single-photon excitation mode. Vesicle binding of the proteins was monitored in terms of an increase in fluorescence emission intensity (∆F) (at 445 nm for DAN and at 590 nm for NR3). For each protein, ∆F was normalized against the maximal fluorescence increase (∆Fmax) and ∆F/∆Fmax was plotted against wavelength or the PIP3 or PI34P2 concentration in vesicles.
Calibration of lipid Sensors by fluorescence microscopy
All fluorescence microscopy measurements were carried out in a two-photon excitation mode using a custom–built multi–photon, four-channel microscope equipped with two femtosecond–pulsed laser sources (Newport) (Stahelin et al., 2004). In vitro calibration of PIP3 or PI34P2 was performed using POPC/POPE/POPS/PI/cholesterol/PIP3 (or PI34P2) (13–x/40/26/10/10/x: x = 0–3 mole%) or POPC/POPS/PIP3 (or PI34P2) (70–x/30/x: x = 0–3 mole%) giant unilamellar vesicles (GUV). Mole% is defined as 100 x mole fraction. These GUV were mixed with the sensors in the concentration range of 0–500 nM. DAN–labeled and NR3–labeled sensors were two–photon excited at 780 nm and 900 nm, respectively. 480/40 nm and 560/50 nm band pass filters were employed for the blue channel and the green channel, respectively, whereas 620/60 nm and 670/50 nm band pass filters were used for the orange channel and the red channel, respectively. For DAN-labeled sensors, blue channel fluorescence signals derive from membrane-bound sensors only whereas green channel signals are from both membrane-bound and sensors. Likewise, orange channel fluorescence signals derive from membrane-bound sensors only whereas red channel signals are from both membrane–bound and sensors for NR3– labeled sensors. For each PIP3 (or PI34P2) concentration, 10 GUVs were selected for image analysis by Image-Pro Plus (Media Cybernetics, Inc.). For data analysis, the region of interest (the membrane in our case) was selected by setting a threshold intensity (or brightness) value on the basis of the intensity distribution profile of the image. Since the orange channel (or blue channel for DAN-labeled sensors) always gives stronger membrane signals than the red channel (or green channel for DAN-labeled sensors) for NR3-labeled sensors, we first selected the mask from the orange (or blue) channel and superimposed it onto the same image in the red (or green) channel. The estimated membrane region of the vesicle was validated by comparing it with the membrane region in the differential interference contrast image of the vesicle. The total intensity of GUV (FB(total) and FG(total) for DAN–labeled sensors and FO(total) and FR(total) for NR3–labeled sensors) were divided by the total area of the pixels that constitute each GUV to yield the average intensities, FB and FG for DAN–labeled sensors and FO and FR for NR3–labeled sensors, which were then used to prepare the ratiometric calibration curves. For DAN-labeled sensors, non-linear least-squares analysis of the (FB/FG) versus the PIP3 (or PI34P2) concentration ([PIP3]) plot using the equation; FB/FG = (FB/FG)min + (FB/FG)max/(1 + Kd/[PIP3]) yielded Kd, (FB/FG)max, and (FB/FG)min values. Kd, (FB/FG)max, and (FB/FG)min are equilibrium dissociation constant (in mole%), and the maximal and minimal FB/FG values, respectively. The theoretical calibration curve was then constructed using these values and [PIP3] from an unknown sample was calculated using the calibration curve. The same calibration was performed for NR3–labeled sensors.
Sensor calibration and cellular lipid quantification
NIH 3T3 (or other) cells were seeded into 50 mm round glass–bottom plates and grown at 37°C in a humidified atmosphere of 95% air and 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) (Life Technologies) supplemented with 10% (v/v) fatal bovine serum (FBS), 100 U/ml penicillin G, and 100 µg/ml streptomycin sulfate (Life Technologies) and cultured in the plates for about 48 h before lipid quantification. All cell lines were cultured bi-weekly and stocks of cell lines were passaged no more than ten times for use in experiments. For simultaneous PIP3 and PI34P2 quantification in NIH 3T3 (or other) cells, an equimolar mixture of DAN-eMyoX-PH and NR3-eTapp1-cPH was delivered into the cells by microinjection. Typically, 20–30 femtoliter of 0.5–1 µM sensor solution was microinjected into the cell. All microscopy measurements and imaging data analysis were performed as described (Liu et al., 2014; Yoon et al., 2011). NIH 3T3 cells were serum-starved for 16 h and stimulated with 50 ng/ml PDGF-BB. The three– dimensional display of local lipid concentration was obtained using the surf function in MATLAB. The angular profile of photon counts in the cellular PM was calculated by Image–Pro Plus software.
Membrane translocation analysis of Akt
For calculating the relative ratio between the fluorescence intensity of PM (Fm) and the cytosolic area (Fc), the line profile function in Image-Pro Plus software (Media Cybernetics) was used. The detailed calculation method was described as reported. (Cai et al., 2014) Briefly, a line was drawn across the cell image, and the distance-dependent intensity plot with two peaks indicating PM was obtained. If the nucleus region has a strong signal, the line was drawn over the nonnuclear region. Fm was calculated by averaging the two PM peaks whereas Fc was calculated by averaging the intensity of the area between the two peaks. The degree of EE translocation could not be robustly quantified by the same analysis because of a much smaller surface area of EE than that of PM.
Biosensor-based Akt activity assay
ReAktion1 (Ananthanarayanan et al., 2007) and ReAktion2 (Zhou et al, unpublished) FRET biosensors were used to monitor the activation of Akt. These biosensors were composed of the N-terminal CFP, followed by Akt-FL and the C-terminal YFP. Conformational changes caused by phosphorylation of PM-bound Akt bring the donor (CFP) and acceptor (YFP) molecules within the biosensors into close proximity, allowing FRET to occur. The detailed development process of ReAktion2 will be reported elsewhere. The FRET experiments were performed with a custom-modified Olympus FV3000 confocal microscope. The 405 nm laser was used to excite CFP and the 480/40 nm and 530/40 nm filters were used to collect CFP and YFP fluorescence emission intensity (FCFP and FYFP), respectively. The CFP to YFP FRET (FRETR) for ReAktion1 and ReAktion1 was determined in terms of the increase in YFP intensity. To correct for all potential bleed-through effects, all possible combinations of excitation and emission were measured with each laser using CFP and YFP molecules separately expressed in NIH 3T3 cells: e.g., 405-nm excitation of CFP and YFP, respectively, each separately collected with 480/40 nm and 530/40 nm filters. The correction parameters for the bleed-through of CFP (a = 0.42) and YFP (b = 0.2), respectively, into the 530/40 nm filter. The corrected FRET (FRETS) values were then calculated using the following formula: FRETS = FRETR – a FCFP – b FYFP. (Wang et al., 2014) To compensate for the different cellular expression levels of Akt biosensors, FRETS was normalized against donor signals (FCFP) to yield nFRET at a given time. (Wang et al., 2014) FRETS was processed with MATLAB software while nFRET was calculated with Image-ProPlus software.
Single-molecule tracking
Single-molecule imaging was performed using a custom-built total internal reflection fluorescence microscope as described previously. NIH 3T3 cells were plated on the 8-well chambered coverglass (Lab-Tek, Thermo Fisher Scientific) at the density of 1 × 105 for 24 h, Akt1 FL-EGFP (or Akt2 FL-EGFP) and SNAP-tagged eTapp1-PH (or SNAP-tagged eMyox-PH) were co-transfected into cells using the jetPRIME system (Polyplus-transfection) according to the manufacturer’s protocols. The transfected NIH 3T3 cells were serum starved for 16 h and subsequently labeled with SNAP-Cell tetramethylrhodamine (TMR)-Star (New England Biolabs). Labeled cells were washed to remove the free dye, stimulated with 50 ng/ml PDGF-BB, the two protein molecules were simultaneously tracked and analyzed as described. The images were spatially corrected following the algorithm described previously (Koyama-Honda et al., 2005). All particle tracking, data analysis and image processing were carried out with in-house programs written in MATLAB. Co-localization analysis of two molecules was performed with a fixed threshold criterion (i.e., <400 nm) for co-localization (Koyama-Honda et al., 2005). The same size of PM surface was analyzed for each data. The percentage Akt molecules spending a given colocalzation time (>0.2 sec) with eTapp1-PH (or eMyox-PH) on the PM of NIH 3T3 cells was calculated from the total number of Akt molecules and displayed as a histogram. Data were fit into a single exponential decay equation (i.e., P = Po e-kt) to determine the dissociation rate constant (k) values by non-linear least-squares analysis and the half-life values of co-localization were calculated as ln2/k. 50–100 images were analyzed for each data point.
Surface plasmon resonance (SPR) analysis
All SPR measurements were performed at 23°C in 20 mM Tris, pH 7.4, containing 0.16 M NaCl using a lipid-coated L1 chip in the BIACORE X-100 system (GE Healthcare) as described previously. LUVs of POPC/POPS/PI34P2 (or PIP3) (77:20:3) and POPC were used as the active surface and the control surface, respectively. Since SPR measurements focused on determination of the lipid headgroup specificity and the relative membrane affinity of AKT isoforms and their PH domain constructs, we simplified the lipid compositions of LUVs to clearly distinguish their PIP3 and PI34P2 specificity. Sensorgrams were analyzed assuming a Langmuir-type binding between the protein (P) and protein binding sites (M) on vesicles (that is, P + M↔PM). The Req values were plotted against the protein concentrations (Po), and the Kd was established by nonlinear least squares analysis of the binding isotherm using the equation, Req = Rmax/(1 + Kd/Po). For kinetic SPR measurements, the flow rate was maintained at 30 µl/min for both association and dissociation phases.
siRNA knockdown
NIH 3T3 cells were plated on 6-well plate at the density of ~1.5 × 105 for 24 h and 10 pmol siRNAs were transfected into cells using jetPRIME (Polyplus transfection) according to the manufacturer’s protocols. After 48 h transfection, the cells were harvested for Western blot analysis.
Western blot analysis
Transfected cells or treated cells were lysed in the cell lysis buffer (20 mM Tris-HCl, pH 7.5, containing 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% Triton-X, protease inhibitors and phosphatase inhibitors (1 mM Na3VO4, 1 mM NaF, 1 µg/ml leupeptin, 1 mM PMSF, 1.5 mM benzamidine, and 2 µg/ml pepstatin). The same amounts of proteins were loaded onto a polyacrylamide gel to run sodium dodecylsulfate polyacrylamide gel electrophoresis. Proteins were separated and transferred to a polyvinylidene difluoride membrane. The membrane was blocked with 5% (w/v) nonfat milk for 1 h and incubated overnight at 4°C with various antibodies (1:1000 dilution for all antibodies). After the unbound antibodies were removed, the membranes were incubated with the horseradish peroxidase secondary antibody for 1 h at room temperature before development.
Immunofluorescence staining
After NIH 3T3 cells in DMEM were stimulated with 50 ng/ml PDGF-BB for the indicated period, they were washed with the phosphate buffered saline (PBS) for 3 min three time. Cells were fixed with 4% (m/v) paraformaldehyde for 30 min at room temperature. Cells were washed with PBS for 3 min three time and incubated with 0.5% Triton X-100 (v/v) for 20 min at room temperature. Cells were washed with PBS for 10 min three time and blocked with 5% (m/v) bovine serum albumin (BSA) or milk for 30 min at room temperature. Cells were incubated with a primary antibody (all antibodies were diluted 1000-fold in 1% BSA solution) overnight at 4 ℃ or for 2 h at room temperature. Cells were washed with PBS for 10 min three time and incubated with the secondary antibody for 45 min under dark. Cells were washed with PBS for 10 min three and imaged using an Olympus FV3000 laser scanning confocal microscope.
QUANTIFICATION AND STATISTICAL ANALYSIS
All imaging data analysis and image processing were carried out with in-house programs written in MATLAB. The band intensities of western blot were quantified by ImageJ software (Bethesda, MD). Statistical significance was calculated by the Student’s t-test. Significance levels are: * p <0.05 and ** p < 0.01. The number of experiments, the number of total cells analyzed (n), and significance are reported in the figure legends. Sample sizes for cellular imaging and assays were chosen as the minimum number of independent observations required for statistically significant results.
Supplementary Material
HIGHLIGHTS.
PIP3 and PI34P2 after PI3K activation is quantitatively determined in live cells.
PIP3 and PI34P2 have distinctively different spatiotemporal history and dynamics.
PIP3 and PI34P2 induce site- and isoform-specific activation of Akt proteins
ACKNOWLEDGEMENTS
This work was supported by the grants from the National Institutes of Health (R35GM122530 for WC, R21ES028384 for JHM, R56MH107387 for LWG, R01AG016927 and R01CA090764 for NH, and R35CA197622 and R01GM111665 for JZ). JHM was also supported by National Science Foundation grant MCB-1412692.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Ananthanarayanan B, Fosbrink M, Rahdar M, and Zhang J (2007). Live-cell molecular analysis of Akt activation reveals roles for activation loop phosphorylation. J. Biol. Chem 282, 36634–36641. [DOI] [PubMed] [Google Scholar]
- Boucrot E, Ferreira AP, Almeida-Souza L, Debard S, Vallis Y, Howard G, Bertot L, Sauvonnet N, and McMahon HT (2015). Endophilin marks and controls a clathrin-independent endocytic pathway. Nature 517, 460–465. [DOI] [PubMed] [Google Scholar]
- Braccini L, Ciraolo E, Campa CC, Perino A, Longo DL, Tibolla G, Pregnolato M, Cao Y, Tassone B, Damilano F, et al. (2015). PI3K-C2gamma is a Rab5 effector selectively controlling endosomal Akt2 activation downstream of insulin signalling. Nat Commun 6, 7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calleja V, Alcor D, Laguerre M, Park J, Vojnovic B, Hemmings BA, Downward J, Parker PJ, and Larijani B (2007). Intramolecular and intermolecular interactions of protein kinase B define its activation in vivo. PLoS Biol 5, e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calleja V, Laguerre M, Parker PJ, and Larijani B (2009). Role of a novel PH-kinase domain interface in PKB/Akt regulation: structural mechanism for allosteric inhibition. PLoS Biol 7, e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantley LC (2002). The phosphoinositide 3-kinase pathway. Science 296, 1655–1657. [DOI] [PubMed] [Google Scholar]
- Chin YR, and Toker A (2010). The actin-bundling protein palladin is an Akt1-specific substrate that regulates breast cancer cell migration. Mol. Cell 38, 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A, Schwartz AL, Dautry-Varsat A, and Lodish HF (1983). Kinetics of internalization and recycling of transferrin and the transferrin receptor in a human hepatoma cell line. Effect of lysosomotropic agents. J. Biol. Chem 258, 9681–9689. [PubMed] [Google Scholar]
- Clark J, Anderson KE, Juvin V, Smith TS, Karpe F, Wakelam MJ, Stephens LR, and Hawkins PT (2011). Quantification of PtdInsP3 molecular species in cells and tissues by mass spectrometry. Nat. Methods 8, 267–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNitto JP, and Lambright DG (2006). Membrane and juxtamembrane targeting by PH and PTB domains. Biochim. Biophys. Acta 1761, 850–867. [DOI] [PubMed] [Google Scholar]
- Ebner M, Lucic I, Leonard TA, and Yudushkin I (2017). PI(3,4,5)P3 Engagement Restricts Akt Activity to Cellular Membranes. Mol. Cell 65, 416–431 e416. [DOI] [PubMed] [Google Scholar]
- Eramo MJ, and Mitchell CA (2016). Regulation of PtdIns(3,4,5)P3/Akt signalling by inositol polyphosphate 5-phosphatases. Biochem. Soc. Trans 44, 240–252. [DOI] [PubMed] [Google Scholar]
- Franke TF, Kaplan DR, Cantley LC, and Toker A (1997). Direct regulation of the Akt proto-oncogene product by phosphatidylinositol-3,4-bisphosphate. Science 275, 665–668. [DOI] [PubMed] [Google Scholar]
- Franke TF, Yang SI, Chan TO, Datta K, Kazlauskas A, Morrison DK, Kaplan DR, and Tsichlis PN (1995). The protein kinase encoded by the Akt proto-oncogene is a target of the PDGF-activated phosphatidylinositol 3-kinase. Cell 81, 727–736. [DOI] [PubMed] [Google Scholar]
- Fruman DA, and Rommel C (2014). PI3K and cancer: lessons, challenges and opportunities. Nat. Rev. Drug Discov 13, 140–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins PT, and Stephens LR (2015). PI3K signalling in inflammation. Biochim. Biophys. Acta 1851, 882–897. [DOI] [PubMed] [Google Scholar]
- Hawkins PT, and Stephens LR (2016). Emerging evidence of signalling roles for PI(3,4)P2 in Class I and II PI3K-regulated pathways. Biochem. Soc. Trans 44, 307–314. [DOI] [PubMed] [Google Scholar]
- Jethwa N, Chung GH, Lete MG, Alonso A, Byrne RD, Calleja V, and Larijani B (2015). Endomembrane PtdIns(3,4,5)P3 activates the PI3K-Akt pathway. J. Cell Sci 128, 3456–3465. [DOI] [PubMed] [Google Scholar]
- Kielkowska A, Niewczas I, Anderson KE, Durrant TN, Clark J, Stephens LR, and Hawkins PT (2014). A new approach to measuring phosphoinositides in cells by mass spectrometry. Adv Biol Regul 54, 131–141. [DOI] [PubMed] [Google Scholar]
- Kim EK, Tucker DF, Yun SJ, Do KH, Kim MS, Kim JH, Kim CD, Birnbaum MJ, and Bae SS (2008). Linker region of Akt1/protein kinase Balpha mediates platelet-derived growth factor-induced translocation and cell migration. Cell. Signal 20, 2030–2037. [DOI] [PubMed] [Google Scholar]
- Koyama-Honda I, Ritchie K, Fujiwara T, Iino R, Murakoshi H, Kasai RS, and Kusumi A (2005). Fluorescence imaging for monitoring the colocalization of two single molecules in living cells. Biophys. J 88, 2126–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Chew C., Lunardi A, Gulluni F, Ruan DT, Chen M, Salmena L, Nishino M, Papa A, Ng C, Fung J, et al. (2015). In Vivo Role of INPP4B in Tumor and Metastasis Suppression through Regulation of PI3K-AKT Signaling at Endosomes. Cancer Discov 5, 740–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, and Marshall AJ (2015). Phosphatidylinositol (3,4) bisphosphate-specific phosphatases and effector proteins: A distinct branch of PI3K signaling. Cell. Signal 27, 1789–1798. [DOI] [PubMed] [Google Scholar]
- Liu SL, Sheng R, Jung JH, Wang L, Stec E, O’Connor MJ, Song S, Bikkavilli RK, Winn RA, Lee D, et al. (2017). Orthogonal lipid sensors identify transbilayer asymmetry of plasma membrane cholesterol. Nat. Chem. Biol 13, 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SL, Sheng R, O’Connor MJ, Cui Y, Yoon Y, Kurilova S, Lee D, and Cho W (2014). Simultaneous in situ quantification of two cellular lipid pools using orthogonal fluorescent sensors. Angew. Chem. Int. Ed. Engl 53, 14387–14391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Yu J, Yan J, Wei Z, and Zhang M (2011). Structural basis of the myosin X PH1(N)-PH2-PH1(C) tandem as a specific and acute cellular PI(3,4,5)P(3) sensor. Mol Biol Cell 22, 4268–4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek M, Kielkowska A, Chessa T, Anderson KE, Barneda D, Pir P, Nakanishi H, Eguchi S, Koizumi A, Sasaki J, et al. (2017). PTEN Regulates PI(3,4)P2 Signaling Downstream of Class I PI3K. Mol. Cell 68, 566–580 e510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna D, Albanese A, Park WS, and Cho W (2007). Mechanistic basis of differential cellular responses of phosphatidylinositol 3,4-bisphosphate- and phosphatidylinositol 3,4,5-trisphosphate-binding pleckstrin homology domains. J. Biol. Chem 282, 32093–32105. [DOI] [PubMed] [Google Scholar]
- Manning BD, and Cantley LC (2007). AKT/PKB signaling: navigating downstream. Cell 129, 1261–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD, and Toker A (2017). AKT/PKB Signaling: Navigating the Network. Cell 169, 381–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marat AL, Wallroth A, Lo WT, Muller R, Norata GD, Falasca M, Schultz C, and Haucke V (2017). mTORC1 activity repression by late endosomal phosphatidylinositol 3,4-bisphosphate. Science 356, 968–972. [DOI] [PubMed] [Google Scholar]
- Nogueira V, Park Y, Chen CC, Xu PZ, Chen ML, Tonic I, Unterman T, and Hay N (2008). Akt determines replicative senescence and oxidative or oncogenic premature senescence and sensitizes cells to oxidative apoptosis. Cancer Cell 14, 458–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh C, Janakiraman V, Wu WI, Foo CK, Kljavin NM, Chaudhuri S, Stawiski E, Lee B, Lin J, Li H, et al. (2012). Disruption of PH-kinase domain interactions leads to oncogenic activation of AKT in human cancers. Proc Natl Acad Sci U S A 109, 19368–19373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MJ, Sheng R, Silkov A, Jung DJ, Wang ZG, Xin Y, Kim H, Thiagarajan-Rosenkranz P, Song S, Yoon Y, et al. (2016). SH2 Domains Serve as Lipid-Binding Modules for pTyr-Signaling Proteins. Mol. Cell 62, 7–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posor Y, Eichhorn-Gruenig M, Puchkov D, Schoneberg J, Ullrich A, Lampe A, Muller R, Zarbakhsh S, Gulluni F, Hirsch E, et al. (2013). Spatiotemporal control of endocytosis by phosphatidylinositol-3,4-bisphosphate. Nature 499, 233–237. [DOI] [PubMed] [Google Scholar]
- Renard HF, Simunovic M, Lemiere J, Boucrot E, Garcia-Castillo MD, Arumugam S, Chambon V, Lamaze C, Wunder C, Kenworthy AK, et al. (2015). Endophilin-A2 functions in membrane scission in clathrin-independent endocytosis. Nature 517, 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso G, Blaustein M, Pozzi B, Mammi P, and Srebrow A (2015). Akt/PKB: one kinase, many modifications. Biochem. J 468, 203–214. [DOI] [PubMed] [Google Scholar]
- Rodgers SJ, Ferguson DT, Mitchell CA, and Ooms LM (2017). Regulation of PI3K effector signalling in cancer by the phosphoinositide phosphatases. Biosci. Rep 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng R, Chen Y, Yung Gee H., Stec E, Melowic HR, Blatner NR, Tun MP, Kim Y, Kallberg M, Fujiwara TK, et al. (2012). Cholesterol modulates cell signaling and protein networking by specifically interacting with PDZ domain-containing scaffold proteins. Nat Commun 3, 1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe LM, Yuzugullu H, and Zhao JJ (2015). PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat. Rev. Cancer 15, 7–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Stephens L, and Hawkins P (2012). PI3K signalling: the path to discovery and understanding. Nat. Rev. Mol. Cell Biol 13, 195–203. [DOI] [PubMed] [Google Scholar]
- von Kleist L, Stahlschmidt W, Bulut H, Gromova K, Puchkov D, Robertson MJ, MacGregor KA, Tomilin N, Pechstein A, Chau N, et al. (2011). Role of the clathrin terminal domain in regulating coated pit dynamics revealed by small molecule inhibition. Cell 146, 471–484. [DOI] [PubMed] [Google Scholar]
- Wang X, Wang Y, Zhou Y, Hendron E, Mancarella S, Andrake MD, Rothberg BS, Soboloff J, and Gill DL (2014). Distinct Orai-coupling domains in STIM1 and STIM2 define the Orai-activating site. Nat Commun 5, 3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt SA, Kimber WA, Fleming IN, Leslie NR, Downes CP, and Lucocq JM (2004). Detection of novel intracellular agonist responsive pools of phosphatidylinositol 3,4-bisphosphate using the TAPP1 pleckstrin homology domain in immunoelectron microscopy. Biochem. J 377, 653–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y, Lee PJ, Kurilova S, and Cho W (2011). In situ quantitative imaging of cellular lipids using molecular sensors. Nat Chem 3, 868–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


