Abstract
Herein, we evaluated the association of the Toll-like receptor 6 (TLR6) single nucleotide polymorphisms (SNPs) rs3796508 (Val327Met) and rs5743810 (Ser249Pro) with breast cancer (BC) susceptibility in Saudi Arabian women, using in silico analysis. We found no significant differences in genotypic and allelic frequencies for rs3796508 between the BC patients (n = 127) and healthy individuals (n = 116). However, 86% of the BC patients, versus 98% of the healthy controls, carried the rs5743810 Pro allele (OR = 0.103, CI = 0.036–0.293, P = 0.00001). Advanced analysis based on the comparison of the estrogen receptor (ER)-positive and -negative patients with the healthy controls indicated a significant association between rs5743810 allelic frequency and BC risk protection (OR = 0.100, CI = 0.034–0.297, P = 0.00001 for ER+ BC cases; OR = 0.102, CI = 0.033–0.318, P = 0.00001 for ER−BC cases). Furthermore, rs5743810 was associated with BC risk protection at either above or below 48 years of age at diagnosis (OR = 0.101, CI = 0.022–0.455, P = 0.00037 for age ≤48 years; OR = 0.120, CI = 0.028–0.519, P = 0.00087 for age >48 years). Such associations were not found for rs3796508. In silico analysis indicated that these SNPs had neutral effects within the TLR6 structure, confirming the protective role of rs5743810. Our findings therefore suggest a strong association between rs5743810 and protection against BC risk in Saudi Arabian women. Importantly, the rs5743810 Pro allele could be a potential BC diagnostic biomarker in this ethnic population.
Introduction
In recent years, breast cancer (BC) has become the most frequently diagnosed cancer among women worldwide, with approximately 2.5 million cases in 2015 [1]. The lethal disease is also one of the most prevalent cancers among women living in Gulf Cooperation Council countries, including Saudi Arabia [2]. In Saudi Arabia, BC is considered the leading cause of female mortality among all other cancers [3], being more prevalent among young women in comparison to women suffering from BC in Western countries [4]. According to the 2013 cancer incidence report from the Saudi Health Council, BC cases accounted for approximately 16% of all malignances among adults of both sexes and approximately 29% of all malignances among Saudi Arabian females.
The relationship between the dysregulation of innate immunity-related genes and BC development is widely documented in the literature, including that of Toll-like receptors (TLRs)[5–7]. Importantly, the human TLRs are essential components of the innate immunity that provides host protection against infections caused by both viral and bacterial organisms[5]. TLR expression is induced by different types of immune cells, such as dendritic cells and macrophages [8]; however, their expression is also found in non-immune system cells, such as oral mucosal cells and skin keratinocytes [9]. So far, 10 different human TLRs have been recognized, with different subcellular distributions, and play crucial roles in the human innate immune response via inflammatory cytokines [8, 10]. TLR1, TLR2, TLR4, TLR5, TLR6, and TLR10 are present on the cell surface and can identify membrane lipids of microbes. However, the endosomal receptors TLR3, TLR7, TLR8, and TLR9 have the ability to identify host-derived nucleotides and microbial pathogens and trigger inflammatory responses [11]. TLR1 and TLR6 form heterodimers with TLR2, which are crucial in mucosal immune response regulation [12]. The mRNA expression of TLRs is found in many different types of cancers and plays a fundamental role in cancer occurrence and progression through the regulation of cell survival and proliferation. For instance, it was found that TLR1, TLR6, and TLR7 were upregulated in the human non-small-cell lung cancer cell line SPC-A1 [13]. Furthermore, the TLR2 and TLR6 genes were highly expressed in human melanoma cells [14] and in patients with ovarian cancer[14]. The clinical significance of the expression levels of TLR1, TLR2, TLR4, TLR5, TLR6, TLR8, and TLR10 in human renal carcinoma cells, relative to the levels in normal renal cells, has been reported [15].
Notably, genetic factors have been observed to be principal causes of BC development[16]. Genetic variations of single nucleotide polymorphisms (SNPs) normally occur in the human genome and have been comprehensively examined in genetic cancer studies[17]. Recent studies have revealed that SNPs of multiple genes are associated with the initiation of tumorigenesis, including BC risk [17, 18]. In 2015, Cotterchio and colleagues revealed that polymorphism in TLR6 was associated with the risk of pancreatic cancer [19]. Additionally, TLR1 and TLR6 are located within the same gene cluster [20] and polymorphisms in these genes have been reported to increase advanced prostate cancer risks [21]. Previous studies have shown that a variant of TLR6 rs13281615 was associated with increased BC risk in the Chinese population [22]. The TLR6 SNPs rs3796508 and rs5743810 have been reported in different types of diseases, but not in any malignancies. Therefore, the current study aimed to identify the potential association between TLR6 rs3796508 and rs5743810 and increased BC risk by comparing their frequencies in Saudi Arabian women with BC and in healthy women. Additionally, the potential use of these SNPs as diagnostic biomarkers for BC was evaluated.
Results
Basic clinical characteristics of the study subjects
All of the study participants were of Saudi Arabian ethnicity and exclusively female. The selected clinical characteristics of the included population are shown in Table 1. There were no variations in either demographic or other pertinent characteristics between the BC cases and healthy controls; however, there were some clinical differences for other aspects. The BC group consisted of 127 women (median age at diagnosis 48 ± 8.2 years), who were selected from the Oncology Centre at King Faisal Specialist Hospital & Research Center from 2002 to 2004. No significant differences in age and gender distribution were seen between the BC group and the control group. The control group was age-matched to the BC group (i.e., median age 48 years) and comprised 116 healthy women who had neither a family nor a personal history of BC. Of the 127 cases in the BC group, 45 were aged ≤48 years and 82 were aged >48 years. By contrast, in the control group, 62 women were ≤48 years of age and 54 were >48 years of age. In addition, the family histories of BC and health-affecting behaviors (e.g., smoking habit) were found to be similar between the two study groups (Table 1). Other clinical data for those populations, including stage of BC, medications, presence of other diseases, and status of the estrogen receptor (ER+ and ER–), human epidermal growth factor receptor (positive and negative), and progesterone receptor (positive and negative), were collected and are summarized in Table 1.
Table 1. Study population characteristics among Saudi BC cases and healthy controls used for genotyping.
| Variable | Parameter | Cases N (%) |
Controls N (%) |
|---|---|---|---|
| Total persons | - | 127 | 116 |
| Age (Years): Median age (48±8.2) | ≤48 | 45 (35.43%) | 62 (53.49) |
| >48 | 82 (64.57%) | 54 (46.55%) | |
| Estrogen receptor | ER+ | 76 (59.84%) | NA |
| ER- | 49 (38.58%) | NA | |
| Progesterone receptor | PR+ | 71 (55.91%) | NA |
| PR- | 55 (43.31%) | NA | |
| HER Status | HER+ | 49 (38.59%) | NA |
| HER- | 78 (61.42%) | NA | |
| Smoking habits | Smokers | 0 (0%) | 0(0%) |
| No Smokers | 127 (100%) | 116 (100%) |
Abbreviations: BC, breast cancer; NA, not applicable.
Association of TLR6 gene polymorphisms with BC risk in Saudi Arabian female patients
In the current study, the distributions of the two TLR6 SNPs (rs3796508 G/A and rs5743810 C/T) did not deviate significantly from HWE (P > 0.05). The genotypic and allelic distributions of the two TLR6 SNPs among the BC patients and healthy controls are presented in Table 2. Association analysis revealed no significant differences in genotypic and allelic frequencies between the BC group and control group for TLR6 rs3796508 (Table 3). However, the genotypic frequencies of rs3796508 G/A in the BC group were 94%, 6%, and 0% for Val/Val, Val/Met, and Met/Met, respectively, whereas they were 93%, 6%, and 1%, respectively, in the control group. The homozygous “Met/Met” genotype of rs3796508 did not indicate any association with BC cases when compared with the healthy controls (OR = 0.291, CI = 0.012–7.216, P = 0.285; see Table 3). For rs5743810 C/T, the Ser/Pro genotype was present in 28% of the BC patients and in 3% of the control subjects. Interestingly, there were significant differences in the frequency of the Pro allele of this SNP between the two groups, with 86% of BC patients and 98% of healthy controls carrying the allele (OR = 0.103, CI = 0.036–0.293, P = 0.00001; Table 3).
Table 2. Distribution of genotypes and allele frequencies of the TLR6 gene loci among Saudi BC cases and controls.
| Genotype | Cases | HWE P-value |
Controls | HWE P-value |
|---|---|---|---|---|
| rs3796508 | ||||
| Val/Val | 117 | 0.047396* | 102 | 0.852705 |
| Val/Met | 8 | 7 | ||
| Met/Met | 0 | 1 | ||
| rs5743810 | ||||
| Ser/Ser | 0 | 0.711682 | 0 | 0.062743 |
| Ser/Pro | 36 | 4 | ||
| Pro/Pro | 91 | 116 |
Table 3. Genotype and allele frequencies of TLR6 gene polymorphism in Saudi BC cases and controls.
| SNP ID | Genotype | Cases | Controls | OR | (95% CI) | χ2 –Value | P*- Value |
|---|---|---|---|---|---|---|---|
| n = 125 | n = 110 | ||||||
| Val/Val | 117 (0.94) | 102 (0.93) | Ref | ||||
| rs3796508 | Val/Met | 8 (0.06) | 7 (0.06) | 0.996 | 0.349–2.843 | 0.0009 | 0.99453 |
| Met/Met | 0 (0.00) | 1 (0.01) | 0.291 | 0.012–7.216 | 1.14 | 0.28542 | |
| Val/Met +Met/Met | 8 (0.06) | 8 (0.07) | 0.872 | 0.316–2.406 | 0.07 | 0.79099 | |
| Val | 242 (0.97) | 211 (0.96) | Ref | ||||
| Met | 8 (0.03) | 9 (0.04) | 0.775 | 0.294–2.045 | 0.27 | 0.60574 | |
| n = 127 | n = 120 | ||||||
| Ser/Ser | 0 (0.00) | 0 (0.00) | Ref | ||||
| rs5743810 | Ser/Pro | 36 (0.28) | 4 (0.03) | 8.111 | 0.14–461.1 | — | 1 |
| Pro/Pro | 91 (0.72) | 116 (0.97) | 0.785 | 0.015–39.962 | — | 1 | |
| Ser/Pro +Pro/Pro | 127 (1) | 120 (1) | 1.058 | 0.021–53.747 | — | 1 | |
| Ser | 36 (0.14) | 4 (0.02) | Ref | ||||
| Pro | 218 (0.86) | 236 (0.98) | 0.103 | 0.036–0.293 | 25.94 | <0.00001* |
* p <0.05
Correlation of the genotypic distribution of TLR6 gene polymorphisms with clinical parameters of the two study groups
We further evaluated the TLR6 genotypes in relation to the different demographic and clinical variables. Advanced analysis of the genotypic distribution of the SNPs based on the age at BC diagnosis revealed that the frequency of rs3796508 distribution did not appear to be associated with the risk of BC development in both the younger (≤48 years) and older (>48 years) populations. However, the distributions of rs3796508 genotypes in BC patients ≤48 years of age were 91%, 9%, and 0% for Val/Val, Val/Met, and Met/Met, respectively, whereas those in the control patients were 98%, 2%, and 0%, respectively. In addition, the frequencies of the heterozygous “Val/Met” genotype of rs3796508 were similar between the BC and control groups in both age-based subpopulations (i.e., ≤48 years and >48 years) (Table 4). By contrast, rs5743810 showed a significant association with protection against advanced BC risk in the patients who were diagnosed with BC at both ≤48 and >48 years of age. However, based on the allelic frequencies, the Pro allele gave a 9-fold higher protection against the risk of BC than did the Ser allele in Saudi Arabian women in both age-based subpopulations (OR = 0.101, CI = 0.022–0.455, P = 0.00037 for women aged ≤48 years; and OR = 0.120, CI = 0.028–0.519, P = 0.00087 for women aged >48 years) (Table 4).
Table 4. Genotype and allele frequencies of TLR6 SNPs in Saudi BC cases versus controls based on age of diagnostic.
| SNP ID | Genotype | Cases | Controls | OR | (95% CI) | χ2 Value | P*- Value |
|---|---|---|---|---|---|---|---|
| patient’s age ≤ 48 years | |||||||
| rs3796508 | n = 44 | n = 56 | |||||
| Val/Val | 40(0.91) | 55(0.98) | Ref | ||||
| Val/Met | 4(0.09) | 1(0.02) | 5.500 | 0.592–51.090 | 2.77 | 0.09615 | |
| Met/Met | 0(0.00) | 0(0.00) | 1.370 | 0.027–70.519 | — | 1 | |
| Val/Met +Met/Met | 4(0.09) | 1(0.02) | 5.500 | 0.592–51.090 | 2.77 | 09615 | |
| Val | 84(0.95) | 111(0.99) | Ref | ||||
| Met | 4(0.05) | 1(0.01) | 5.286 | 0.580–48.162 | 2.70 | 0.18404 | |
| rs5743810 | n = 45 | n = 55 | |||||
| Ser/Ser | 0(0.00) | 0(0.00) | Ref | ||||
| Ser/Pro | 14(0.31) | 2(0.04) | 5.800 | 0.092–365.479 | — | 1 | |
| Pro/Pro | 31(0.69) | 53(0.96) | 0.589 | 0.011–30.413 | — | 1 | |
| Ser/Pro+Pro/Pro | 45(1) | 55(1) | 0.820 | 0.016–42.132 | — | 1 | |
| Ser | 14(0.16) | 2(0.02) | Ref | ||||
| Pro | 76(0.84) | 108(0.98) | 0.101 | 0.022–0.455 | 12.69 | 0.00037* | |
| SNP ID | Genotype | Cases | Controls | OR | (95% CI) | χ2 Value | P*- Value |
| patient’s age > 48 years | |||||||
| rs3796508 | n = 81 | n = 54 | |||||
| Val/Val | 77 (0.95) | 47 (0.87) | Ref | ||||
| Val/Met | 4 (0.05) | 6 (0.11) | 0.407 | 0.109–1.517 | 1.89 | 0.16922 | |
| Met/Met | 0 (0.00) | 1 (0.02) | 0.204 | 0.008–5.118 | 1.62 | 0.20350 | |
| Val/Met +Met/Met | 4 (0.05) | 7 (0.13) | 0.349 | 0.097–1.256 | 2.79 | 0.09499 | |
| Val | 158 (0.98) | 100 (0.93) | Ref | ||||
| Met | 4 (0.02) | 8 (0.07) | 0.316 | 0.093–1.078 | 3.72 | 0.07084 | |
| rs5743810 | n = 82 | n = 55 | |||||
| Ser/Ser | 0 (0.00) | 0 (0.00) | Ref | ||||
| Ser/Pro | 22 (0.27) | 2 (0.04) | 9.000 | 0.144–560.703 | — | 1 | |
| Pro/Pro | 60 (0.73) | 53 (0.96) | 1.131 | 0.022–57.981 | — | 1 | |
| Ser/Pro+Pro/Pro | 82 (1) | 55 (1) | 1.486 | 0.029–76.027 | — | 1 | |
| Ser | 22 (0.13) | 2 (0.02) | Ref | ||||
| Pro | 142 (0.87) | 108 (0.98) | 0.120 | 0.028–0.519 | 11.08 | 0.00087* | |
* = P<0.05, Ref = Reference allele
Finally, we analyzed the associations of the genotypic and allelic frequency distributions of rs3796508 and rs5743810 with the ER status of the BC patients. In the case of rs3796508, no significant associations were found with either ER+ or ER−status compared with the controls (Table 5), as the genotypic frequencies of this SNP were similar between both groups analyzed. In the BC group, 95%, 5%, and 0% carried the Val/Val, Val/Met, and Met/Met genotypes, respectively, whereas the distributions were 93%, 6%, and 1%, respectively, for the control group. However, based on the ER+ or ER−status, the rs5743810 Pro allele frequency was again more greatly associated with BC risk protection in this ethnic group than was the Ser allele (OR = 0.100, CI = 0.034–0.297, P = 0.00001 for ER+ BC cases; and OR = 0.102, CI = 0.033–0.318, P = 0.00001 for ER−BC cases) (Table 5). Moreover, the distribution frequencies of genotypes Ser/Ser, Ser/Pro, and Pro/Pro were similar in the ER+/ER−subgroups of BC patients compared with the control group (Table 5).
Table 5. Genotype and allele frequencies TLR6 SNPs in Saudi BC cases and controls based on ER+/ER- status.
| SNP ID | Genotype | Cases | Controls | OR | (95% CI) | χ2 Value | P*- Value |
| ER+ | |||||||
| rs3796508 | n = 75 | n = 110 | |||||
| Val/Val | 71 (0.95) | 102 (0.93) | Ref | ||||
| Val/Met | 4 (0.05) | 7 (0.06) | 0.821 | 0.232–2.909 | 0.09 | 0.75954 | |
| Met/Met | 0 (0.00) | 1 (0.01) | 0.478 | 0.019–11.898 | 0.69 | 0.40504 | |
| Val/Met +Met/Met | 4 (0.05) | 8 (0.07) | 0.718 | 0.208–2.477 | 0.28 | 0.59899 | |
| Val | 146 (0.97) | 211 (0.96) | Ref | ||||
| Met | 4 (0.03) | 9 (0.04) | 0.642 | 0.194–2.125 | 0.53 | 0.46507 | |
| rs5743810 | n = 76 | n = 120 | |||||
| Ser/Ser | 0 (0.00) | 0 (0.00) | Ref | ||||
| Ser/Pro | 22 (0.29) | 4 (0.03) | 5.000 | 0.087–286.553 | — | 1 | |
| Pro/Pro | 54 (0.71) | 116 (0.97) | 0.468 | 0.009–23.889 | — | 1 | |
| Ser/Pro+Pro/Pro | 76 (1) | 120 (1) | 0.635 | 0.012–32.331 | — | 1 | |
| Ser | 22 (0.14) | 4 (0.02) | Ref | ||||
| Pro | 130 (0.86) | 236 (0.98) | 0.100 | 0.034–0.297 | 24.65 | <0.00001* | |
| SNP ID | Genotype | Cases | Controls | OR | (95% CI) | χ2 Value | P*- Value |
| ER- | |||||||
| rs3796508 | n = 48 | n = 110 | |||||
| Val/Val | 44 (0.92) | 102 (0.93) | Ref | ||||
| Val/Met | 4 (0.08) | 7 (0.06) | 1.325 | 0.369–4.756 | 0.19 | 0.66555 | |
| Met/Met | 0 (0.00) | 1 (0.01) | 0.768 | 0.031–19.214 | 0.43 | 0.51193 | |
| Val/Met +Met/Met | 4 (0.08) | 8 (0.07) | 1.159 | 0.332–4.051 | 0.05 | 0.81698 | |
| Val | 92 (0.96) | 211 (0.96) | Ref | ||||
| Met | 4 (0.04) | 9 (0.04) | 1.019 | 0.306–3.394 | 0.0009 | 1 | |
| rs5743810 | n = 49 | n = 120 | |||||
| Ser/Ser | 0 (0.00) | 0 (0.00) | Ref | ||||
| Ser/Pro | 14 (0.29) | 4 (0.03) | 3.222 | 0.056–186.825 | — | 1 | |
| Pro/Pro | 35 (0.71) | 116 (0.97) | 0.305 | 0.006–15.635 | — | 1 | |
| Ser/Pro+Pro/Pro | 49 (1) | 120 (1) | 0.411 | 0.008–20.993 | — | 1 | |
| Ser | 14 (0.14) | 4 (0.02) | Ref | ||||
| Pro | 84 (0.86) | 236 (0.98) | 0.102 | 0.033–0.318 | 21.98 | <0.00001* | |
* = P<0.05, Ref = Reference allele
In silico analysis
Different bioinformatics algorithms and software were used to predict the pathogenicity and damaging effects of the rs5743810 (Ser249Pro) and rs3796508 (Val327Met) mutations. The majority of the algorithms considered these two SNPs as being neutral, with harmless effects, and there was no evidence of deleterious events correlated with the mutations. This exclusion of negative effects of the mutations by the in silico analysis supported our findings that these SNPs might have some positive effects (Table 6). Proline is considered to be a unique amino acid with a specific structure, where the side chain is connected to the protein backbone twice, creating a 5-membered nitrogen ring. Therefore, any mutation that introduces a Pro residue at a site where the backbone torsion angles are unable to accommodate such a residue is usually associated with clashes and damaging effects because of the energy changes. Nevertheless, our in silico predictions indicated that there were no damaging effects associated with rs5743810 (Ser249Pro) and s3796508 (Val327Met), confirming that these two SNPs are protective mutations.
Table 6. Predict the pathogenicity and damaging effects of the rs5743810 (Ser249Pro) and rs3796508 (Val327Met) mutations.
| Mutation nucleotide | Protein Change | Mutation taster | Polyphen-2.0 | SIFT | PROVEAN | Mutation Assessor | FATHMM | CONDEL |
|---|---|---|---|---|---|---|---|---|
| c.C>T | p.V327M | (0.99) Harmless polymorphism |
(0.069) Neutral |
(0.02) Damaging |
(-0.716) Neutral |
(2.015) Predicted functional (low-Medium) |
(3.03) Tolerated |
(0.362) Neutral |
| c.A>G | p.S249PRO | (0.99) Harmless polymorphism |
(0.000) Neutral |
(0.56) Neutral |
(1.124) Neutral |
(-1.06) Predicted non-functional (Neutral) |
(2.49) Tolerated |
(0.326) Neutral |
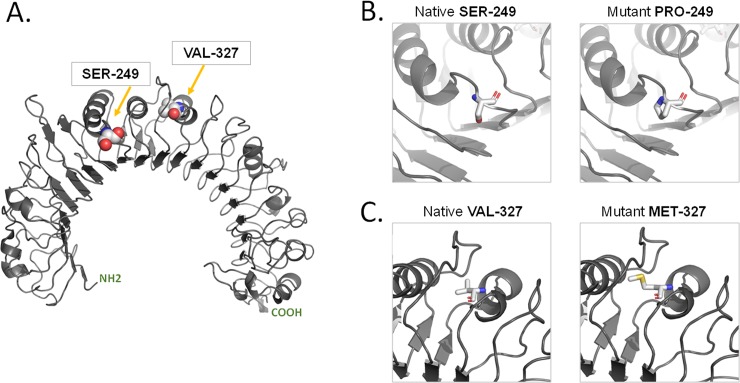
On the basis of the solved crystal structure of the Toll/interleukin-1 receptor domain of TLR6 (PDB 4OM7) from positions 640 to 782, a homology model was constructed using the SWISS-MODEL template library (SMTL version 2017-10-23, PDB release 2017-10-13), which models tertiary and quaternary protein structures using evolutionary information. The best template structure matching the target sequence was selected (3a79.1.B), having 66% sequence identity and a resolution of 2.90 Angstrom. The model revealed that Glu33-Lys573 has an overall curved shape. Using the “mutation” tool in SWISS-MODEL, Ser249 was mutated to Pro249, and Val327 was mutated to Met327 (Fig 1). The original side chain was substituted with the best rotamer with the lowest energy score of the Thr50 amino acid.
Fig 1.
Structural function of TLR6: A) Homology model, showing the location of Ser-249 and Val-327 in spheres representation. B) Native Ser-249 and Mutant Pro-249 in sticks representation. C) Native Val-327 C. Mutant Met-327 in sticks representation. CPK Colours used for illustration.
Discussion
Polymorphisms in different genes are known to be caused by genetic changes in the DNA composition [23]. SNPs may result in DNA genetic variations, which may change the gene-encoded protein function and/or activity by means of amino acid exchange [24]. We hypothesized that the TLR6 gene could play a vital role in innate immune system activation, and that dysregulation of this gene function may result in the development of metastatic BC. It was previously reported that genetic changes in TLRs genes are correlated with increased BC risk [3, 25] and several TLR gene polymorphisms have been identified to be related to increased BC susceptibility [26]. The primary objective of the present study was to investigate the association of TLR6-associated SNPs with increased risk of BC development in Saudi Arabian women. Specifically, the SNPs studied were rs3796508 (979 G>A) and rs5743810 (745 C>T), located on exonic regions of chromosome 4, with valine changed to methionine for rs3796508 (Val327Met) and serine changed to proline (Ser249Pro) for rs5743810. Assessment of rs3796508 revealed no association between its genotypic and allelic frequency distributions with risk of BC in this population. So far, no previous study has reported a clear association between this polymorphism and the development of cancer, including BC. Thus, the rs3796508 polymorphism can affect TLR6 expression in the women suffering from BC without being the main cause of the disease development. However, the significantly higher frequency of the minor Pro allele of rs5743810 indicated that it had a strong association with protection against both BC development and metastatic risks in Saudi Arabian women, both in the youngest and the oldest cases and also in the different types of BC (ER+ or ER–). The rs5743810 SNP is described in the literature as a downstream gene variant, which means that it is located in an exonic region of the gene. This localization means that rs5743810 may have an effect on TLR6 expression and function. The change in amino acid from Ser to Pro results in functional deterioration of TLR6 and predisposes individuals to dysregulation of the innate immune system, which can potentially cause development of the cancer. It is believed that polymorphisms in coding regions are currently relevant to cancer development owing to their roles in gene expression regulation. TLR6 functionally interacts with TLR2 to mediate the cellular response to bacterial lipoproteins and activate the NF-κB pathway and inflammatory events, and consequently, may contribute to tumor development and progression [27–29]. Despite the enhanced expression and activation of TLR6, its function and role in cancer are not clearly understood. Upon TLR2/TLR6 stimulation, the Pro phenotype of the TLR6 rs5743810 SNP was found to be associated with lower secretions of proinflammatory cytokines as well as tumor necrosis factor-alpha and IL-6 (among others), decreasing the risk of BC development, as compared with the Ser or wild-type phenotype responses. A Ser249Pro polymorphism in the extracellular domain of the encoded protein may be associated with BC susceptibility in some populations. Some studies on clinical samples have pointed to TLR6-related tumor progression. Rogers et al. 2013 [30] observed an association between sequence variants in TLR6 and prostate cancer susceptibility among men of African descent[30]. Furthermore, TLR6 expression was also associated with smoking [31, 32].
Our Structure model of TLR-6 illustrate the importance of the in silico evaluation of functional significance and representing how the Ser/Pro change affect ligand induced dimerization. Briefly, Proline is considered to be a unique amino acid with a specific structure, where the side chain is connected to the protein backbone twice, creating a 5-membered nitrogen ring. Therefore, any mutation that introduces a Pro residue at a site where the backbone torsion angles are unable to accommodate such a residue is usually associated with clashes and damaging effects because of the energy changes. Ser/Pro change is near the dimerization interface however this dimerization is not directly interfering with Ser/Pro change. Ser/Pro change is also towards the end of an alpha-helical LRR segment: possibly this could alter the conformation of LRR9 which is involved in ligand binding [Ref 1.]. Nevertheless, our in silico predictions indicated that there were no damaging effects associated with rs5743810 (Ser249Pro) and s3796508 (Val327Met), confirming that these two SNPs are protective mutations. These outcomes may show that the rs5743810 (Ser249Pro) SNP of the TLR6 gene may be distributed differently among diverse cancer types and ethnicities, and can thus be used as a biomarker for the diagnosis of different cancers (rather than a specific marker for BC). However, no data on the functional implications of this SNP on TLR6 expression are currently available.
Conclusions
Novelty and impact
Breast cancer (BC) remains the most prevalent cancer among female worldwide. The lethal disease represents 19.9% of the total cancer-related deaths in Saudi Arabia women and ranked as the first leading cause of deaths. Due to the high rate of the consanguineous marriage in SA, several genetics basis diseases become more prevalence, including BC. Therefore, studying the association of the TLR6 SNPs with BC susceptibility in Saudi Arabian women is highly needed.
The present study clarifies significant roles for TLR6 polymorphic variants in determining BC susceptibility risk among the Saudi Arabian female population, especially that of rs5743810. Additionally, the in silico analysis indicates a protective role for the rs5743810 mutation. However, further investigations are necessary to address the association owing to the limited number of BC samples in the present study. In addition, further independent studies of genetic correlations between TLR6 sequence variants and BC risk in other ethnic populations would be of great interest. Because the exact mechanisms by which TLR6 polymorphisms contribute to cancer pathogenesis are unknown, such elucidation will be a challenge for future research. We believe that these SNPs and their BC risk associations could contribute to their development as biomarkers for the early detection of BC.
Materials and methods
Ethical approval certificate
This case-controlled study was accredited by the Ethics Review Committee at King Faisal Medical City (Ethical approval number 15-089E), and the panel members assented to the use of a consent form. Prior to sample collection, all participating study subjects completed a questionnaire for demographic information, such as the patient’s age at diagnosis, gender, family health history, past medical history, tumor location, tumor characteristics, and reproductive records. The estrogen receptor level was determined via immunohistochemical analysis. All participants in the current investigation agreed to publication of the data generated from this study.
Study design and patient recruitment
The present case-controlled BC study included 127 Saudi Arabian women with histologically confirmed BC between 2011 and 2016 at King Faisal Medical City (Riyadh, Kingdom of Saudi Arabia) prior to any medical treatment. Whole blood specimens were obtained from the BC patients for genotyping were from doctor Abdulrahman Al Naeem at Department of Women's Imaging, King Fahad Medical City, Riyadh, Saudi Arabia. Additionally, blood specimens were collected from 116 healthy women who lacked any clinical signs of BC were from doctor Sana Abdulla Ajaj at Family Medicine Department, College of Medicine, King Saud University, Riyadh, Saudi Arabia. All subjects provided informed written consent to participate in the study and to approve the use of their biological samples for genetic analyses. The design of the study was approved by the local Ethics Committee number 15-089E from King Faisal Medical City, Riyadh and our study don't include any minorsGenomic DNA was extracted from all blood specimens. Both the BC patients and the healthy controls were frequency matched within five years by age and sex. Healthy female volunteers were allowed to participate in the study if they had a normal mammography and no previous cancer history.
DNA extraction
For both groups of women, 3 mL of blood was drawn into EDTA-vacutainer vials (BD Vacutainer Systems, Plymouth, UK). Subsequently, genomic DNA was immediately isolated from 200 μL of the EDTA-anticoagulated blood using a QIAmp DNA Blood Mini Kit (Qiagen, Valencia, CA, USA) in accordance with the manufacturer’s instructions. The DNA samples were preserved at –20°C until used. The isolated genomic DNA concentration was measured with a Nano-Drop 8000 spectrophotometer (Thermo Scientific, Waltham, MA, USA), and the DNA purity was evaluated by measuring the absorbance ratios A260/A280 and A260/A230.
Selected TLR6 polymorphisms and DNA genotyping
Two SNPs in the TLR6 gene were investigated and genotyped using the TaqMan allelic discrimination assay. Three selection criteria for the TLR6 gene polymorphisms were used: (1) their positions, (2) their frequencies and their affiliations with cancer peril, as mentioned formerly, and (3) their correlations with many cancers in various ethnic groups, as reported in various literature reviews. The TLR6 polymorphisms used in the current study—rs3796508 (979 G>A, Val327Met) and rs5743810 (745 C>T, Ser249Pro)—are in the exonic region. It is currently believed that polymorphisms in coding regions are relevant to cancer development owing to their roles in gene expression regulation (S1 Table). Sample genotyping was executed in a 96-well format by applying a QuantStudio 7 Flex Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). In brief, each genotyping reaction was performed in an overall mixture volume of 10 μL containing DNA at a final amount of 15 ng, 5.6 μL of 2× TaqMan Genotyping Master Mix (Applied Biosystems), and 0.2 μL of 40× TaqMan Genotyping SNP Assay reagent (Applied Biosystems).
All primer sequences and probe mixtures were purchased from Applied Biosystems. The efficiencies of the marker and sample genotyping were validated through the performance of both positive and negative controls to confirm the genotyping quality. Genotyping of >10% of randomly selected samples was duplicated to obtain 100% identical results.
In silico analysis
To determine the structural functions along with the effects of the rs5743810 (Ser249Pro) and rs3796508 (Val327Met) mutations, several in silico algorithmic software were used; namely, MutationTaster, PolyPhen-2.0 [33], SIFT[34], PROVEAN [35], MutationAssessor [36], FATHMM,[37] and CONDEL[38]. A homology model of the 3D structure of TLR6 was built using the SWISS-MODEL template library (SMTL version 2017-10-23, PDB release 2017-10-13) [39].
Statistical analysis
Statistical analysis of the data was conducted using the SPSS version 16.0 statistical package (SPSS, Chicago, IL, USA). All probability values (P values) below 0.05 were considered statistically significant. Differences in the allelic and genotypic association of each SNP were tested for Hardy-Weinberg equilibrium (HWE). The odds ratios (ORs) and 95% confidence intervals (CIs) were calculated using the Chi-squared test to compare genetic variations between the BC patients and the healthy participants.
Supporting information
TLR6_HUMAN : NM_006068 NP_006059 p.Val327Met
TLR6_HUMAN : NM_006068 NP_006059 p.Ser249Pro.
(DOCX)
Abbreviations
- BC
breast cancer
- CI
confidence interval
- ER
estrogen receptor
- OR
odds ratio
- SNPs
single nucleotide polymorphisms
- TLR6
Toll-like receptor 6
- TLRs
Toll-like receptors
- HWE
Hardy-Weinberg equilibrium (HWE)
- Pro
Proline
- Ser
Serine
- Val
Valine
- Met
Methionine
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Fitzmaurice C, Allen C, Barber RM, Barregard L, Bhutta ZA, Brenner H, et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-years for 32 Cancer Groups, 1990 to 2015: A Systematic Analysis for the Global Burden of Disease Study. JAMA oncology. 2017;3(4):524–48. Epub 2016/12/06. 10.1001/jamaoncol.2016.5688 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albeshan SM, Mackey MG, Hossain SZ, Alfuraih AA, Brennan PC. Breast Cancer Epidemiology in Gulf Cooperation Council Countries: A Regional and International Comparison. Clinical breast cancer. 2017. Epub 2017/08/07. 10.1016/j.clbc.2017.07.006 . [DOI] [PubMed] [Google Scholar]
- 3.Semlali A, Almutairi M, Parine NR, Al Amri A, Shaik JP, Al Naeem A, et al. No genetic relationship between TLR2 rs4696480, rs3804100, and rs3804099 gene polymorphisms and female breast cancer in Saudi populations. Oncotargets Ther. 2017;10:2325–33. 10.2147/Ott.S121618 ISI:000400265300001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alrashidi AG, Ahmed HG, Alshammeri KJK, Alrashedi SA, BA AL, Alshammari FNM, et al. Knowledge and Perceptions of Common Breast Cancer Risk Factors in Northern Saudi Arabia. Asian Pacific journal of cancer prevention: APJCP. 2017;18(10):2755–61. Epub 2017/10/27. doi: 10.22034/APJCP.2017.18.10.2755 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhatelia K, Singh K, Singh R. TLRs: linking inflammation and breast cancer. Cell Signal. 2014;26(11):2350–7. Epub 2014/08/06. 10.1016/j.cellsig.2014.07.035 . [DOI] [PubMed] [Google Scholar]
- 6.Semlali A, Jalouli M, Parine NR, Al Amri A, Arafah M, Al Naeem A, et al. Toll-like receptor 4 as a predictor of clinical outcomes of estrogen receptor-negative breast cancer in Saudi women. Onco Targets Ther. 2017;10:1207–16. Epub 2017/03/11. 10.2147/OTT.S112165 ; PubMed Central PMCID: PMC5338938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhide MR, Mucha R, Mikula I Jr., Kisova L, Skrabana R, Novak M, et al. Novel mutations in TLR genes cause hyporesponsiveness to Mycobacterium avium subsp. paratuberculosis infection. BMC genetics. 2009;10:21 Epub 2009/05/28. 10.1186/1471-2156-10-21 ; PubMed Central PMCID: PMC2705378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drexler SK, Foxwell BM. The role of toll-like receptors in chronic inflammation. The international journal of biochemistry & cell biology. 2010;42(4):506–18. Epub 2009/10/20. 10.1016/j.biocel.2009.10.009 . [DOI] [PubMed] [Google Scholar]
- 9.Beklen A, Hukkanen M, Richardson R, Konttinen YT. Immunohistochemical localization of Toll-like receptors 1–10 in periodontitis. Oral microbiology and immunology. 2008;23(5):425–31. Epub 2008/09/17. 10.1111/j.1399-302X.2008.00448.x . [DOI] [PubMed] [Google Scholar]
- 10.Basith S, Manavalan B, Yoo TH, Kim SG, Choi S. Roles of toll-like receptors in cancer: a double-edged sword for defense and offense. Archives of pharmacal research. 2012;35(8):1297–316. Epub 2012/09/04. 10.1007/s12272-012-0802-7 . [DOI] [PubMed] [Google Scholar]
- 11.Fang C, Wei X, Wei Y. Mitochondrial DNA in the regulation of innate immune responses. Protein & cell. 2016;7(1):11–6. Epub 2015/10/27. 10.1007/s13238-015-0222-9 ; PubMed Central PMCID: PMC4707157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morgan ME, Koelink PJ, Zheng B, den Brok MH, van de Kant HJ, Verspaget HW, et al. Toll-like receptor 6 stimulation promotes T-helper 1 and 17 responses in gastrointestinal-associated lymphoid tissue and modulates murine experimental colitis. Mucosal immunology. 2014;7(5):1266–77. Epub 2014/03/29. 10.1038/mi.2014.16 ; PubMed Central PMCID: PMC4137742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ke X, Wu M, Lou J, Zhang S, Huang P, Sun R, et al. Activation of Toll-like receptors signaling in non-small cell lung cancer cell line induced by tumor-associated macrophages. Chinese journal of cancer research = Chung-kuo yen cheng yen chiu. 2015;27(2):181–9. Epub 2015/05/06. 10.3978/j.issn.1000-9604.2015.03.07 ; PubMed Central PMCID: PMC4409966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mauldin IS, Wang E, Deacon DH, Olson WC, Bao Y, Slingluff CL Jr. TLR2/6 agonists and interferon-gamma induce human melanoma cells to produce CXCL10. International journal of cancer Journal international du cancer. 2015;137(6):1386–96. Epub 2015/03/15. 10.1002/ijc.29515 ; PubMed Central PMCID: PMC4497924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu H, Xu SS, Cheng QQ, He LM, Li Z. [Expression and clinical significance of Toll-like receptors in human renal carcinoma cell 786–0 and normal renal cell HK-2]. Zhonghua yi xue za zhi. 2011;91(2):129–31. Epub 2011/03/23. . [PubMed] [Google Scholar]
- 16.Korak T, Ergul E, Uren N, Altinok D, Simsek T, Utkan NZ, et al. RAD51 (rs1801320) gene polymorphism and breast cancer risk in Turkish population. Int J Clin Exp Patho. 2017;10(2):2181–6. ISI:000395739000157. [Google Scholar]
- 17.Jang JP, Baek IC, Choi EJ, Kim TG. Multiplex Genotyping of Cytokine Gene SNPs Using Fluorescence Bead Array. PloS one. 2015;10(2). ARTN e0118008 10.1371/journal.pone.0118008 ISI:000350322700092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hein A, Rack B, Li L, Ekici AB, Reis A, Lux MP, et al. Genetic Breast Cancer Susceptibility Variants and Prognosis in the Prospectively Randomized SUCCESS A Study. Geburtsh Frauenheilk. 2017;77(6):651–9. 10.1055/s-0042-113189 ISI:000404556200017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cotterchio M, Lowcock E, Bider-Canfield Z, Lemire M, Greenwood C, Gallinger S, et al. Association between Variants in Atopy-Related Immunologic Candidate Genes and Pancreatic Cancer Risk. PloS one. 2015;10(5). ARTN e0125273 10.1371/journal.pone.0125273 ISI:000354049700060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kutikhin AG. Association of polymorphisms in TLR genes and in genes of the Toll-like receptor signaling pathway with cancer risk. Hum Immunol. 2011;72(11):1095–116. 10.1016/j.humimm.2011.07.307 ISI:000296546100019. [DOI] [PubMed] [Google Scholar]
- 21.Kazma R, Mefford JA, Cheng I, Plummer SJ, Levin AM, Rybicki BA, et al. Association of the Innate Immunity and Inflammation Pathway with Advanced Prostate Cancer Risk. PloS one. 2012;7(12). ARTN e51680 10.1371/journal.pone.0051680 ISI:000312386800063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan M, Ji SM, Liaw CS, Yap YS, Law HY, Yoon CS, et al. Association of common genetic variants with breast cancer risk and clinicopathological characteristics in a Chinese population. Breast cancer research and treatment. 2012;136(1):209–20. 10.1007/s10549-012-2234-y ISI:000309940300020. [DOI] [PubMed] [Google Scholar]
- 23.Gupta AK, Cherman AM, Tyring SK. Viral and nonviral uses of imiquimod: a review. Journal of cutaneous medicine and surgery. 2004;8(5):338–52. Epub 2005/05/04. 10.1177/120347540400800504 . [DOI] [PubMed] [Google Scholar]
- 24.Yates CM, Sternberg MJ. The effects of non-synonymous single nucleotide polymorphisms (nsSNPs) on protein-protein interactions. Journal of molecular biology. 2013;425(21):3949–63. Epub 2013/07/23. 10.1016/j.jmb.2013.07.012 . [DOI] [PubMed] [Google Scholar]
- 25.Resler AJ, Malone KE, Johnson LG, Malkki M, Petersdorf EW, McKnight B, et al. Genetic variation in TLR or NFkappaB pathways and the risk of breast cancer: a case-control study. BMC cancer. 2013;13:219 Epub 2013/05/03. 10.1186/1471-2407-13-219 ; PubMed Central PMCID: PMC3651307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu S, Wang X, Shi Y, Han L, Zhao Z, Zhao C, et al. Toll-like receptor gene polymorphisms and susceptibility to Epstein-Barr virus-associated and -negative gastric carcinoma in Northern China. Saudi journal of gastroenterology: official journal of the Saudi Gastroenterology Association. 2015;21(2):95–103. Epub 2015/04/07. 10.4103/1319-3767.153832 ; PubMed Central PMCID: PMC4392582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong J, Cho IH, Kwak KI, Suh EC, Seo J, Min HJ, et al. Microglial Toll-like receptor 2 contributes to kainic acid-induced glial activation and hippocampal neuronal cell death. The Journal of biological chemistry. 2010;285(50):39447–57. Epub 2010/10/07. 10.1074/jbc.M110.132522 ; PubMed Central PMCID: PMC2998094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hennessy EJ, Parker AE, O'Neill LA. Targeting Toll-like receptors: emerging therapeutics? Nature reviews Drug discovery. 2010;9(4):293–307. Epub 2010/04/10. 10.1038/nrd3203 . [DOI] [PubMed] [Google Scholar]
- 29.Park SJ, Youn HS. Isoliquiritigenin suppresses the Toll-interleukin-1 receptor domain-containing adapter inducing interferon-beta (TRIF)-dependent signaling pathway of Toll-like receptors by targeting TBK1. Journal of agricultural and food chemistry. 2010;58(8):4701–5. Epub 2010/04/02. 10.1021/jf100484r . [DOI] [PubMed] [Google Scholar]
- 30.Rogers EN, Jones DZ, Kidd NC, Yeyeodu S, Brock G, Ragin C, et al. Toll-like receptor-associated sequence variants and prostate cancer risk among men of African descent. Genes Immun. 2013;14(6):347–55. 10.1038/gene.2013.22 ISI:000324099900001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohailan M, Alanazi M, Rouabhia M, Alamri A, Parine NR, Alhadheq A, et al. Effect of smoking on the genetic makeup of toll-like receptors 2 and 6. Onco Targets Ther. 2016;9:7187–98. Epub 2016/12/07. 10.2147/OTT.S109650 ; PubMed Central PMCID: PMC5123654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Semlali A, Witoled C, Alanazi M, Rouabhia M. Whole cigarette smoke increased the expression of TLRs, HBDs, and proinflammory cytokines by human gingival epithelial cells through different signaling pathways. PloS one. 2012;7(12):e52614 Epub 2013/01/10. 10.1371/journal.pone.0052614 ; PubMed Central PMCID: PMC3532503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nature methods. 2010;7(4):248–9. 10.1038/nmeth0410-248 ; PubMed Central PMCID: PMC2855889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature protocols. 2009;4(7):1073–81. 10.1038/nprot.2009.86 . [DOI] [PubMed] [Google Scholar]
- 35.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PloS one. 2012;7(10):e46688 10.1371/journal.pone.0046688 ; PubMed Central PMCID: PMC3466303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic acids research. 2011;39(17):e118 10.1093/nar/gkr407 ; PubMed Central PMCID: PMC3177186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shihab HA, Gough J, Cooper DN, Stenson PD, Barker GL, Edwards KJ, et al. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Human mutation. 2013;34(1):57–65. 10.1002/humu.22225 ; PubMed Central PMCID: PMC3558800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gonzalez-Perez A, Lopez-Bigas N. Improving the Assessment of the Outcome of Nonsynonymous SNVs with a Consensus Deleteriousness Score, Condel. Am J Hum Genet. 2011;88(4):440–9. 10.1016/j.ajhg.2011.03.004 WOS:000289664300004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic acids research. 2014;42(Web Server issue):W252–8. 10.1093/nar/gku340 ; PubMed Central PMCID: PMC4086089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TLR6_HUMAN : NM_006068 NP_006059 p.Val327Met
TLR6_HUMAN : NM_006068 NP_006059 p.Ser249Pro.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.