Abstract
Background:
Although increased height has been associated with osteosarcoma risk in previous epidemiologic studies, the relative contribution of stature during different developmental time-points remains unclear. Furthermore, how genetic determinants of height impact osteosarcoma etiology remains unexplored. Genetic variants associated with stature in previous genome-wide association studies may be biomarkers of osteosarcoma risk.
Methods:
We tested the associations between osteosarcoma risk and polygenic scores for adult height (416 variants), childhood height, (six variants), and birth length (five variants) in 864 osteosarcoma patients and 1879 controls of European ancestry.
Results:
Each standard deviation increase in the polygenic score for adult height, corresponding to a 1.7-cm increase in stature, was associated with a 1.10-fold increase in risk of osteosarcoma (95% CI: 1.01–1.19, P= 0.027). Each standard deviation increase in the polygenic score for childhood height, corresponding to a 0.5-cm increase in stature, was associated with a 1.10-fold increase in risk of osteosarcoma (95% CI: 1.01–1.20, P= 0.023). The polygenic score for birth length was not associated with osteosarcoma risk (P=0.11). When adult and childhood height scores were modeled together, they were independently associated with osteosarcoma risk (P=0.037 and 0.043, respectively). An eQTL for CILP2 (cartilage intermediate layer protein 2), rs8103992, was significantly associated with osteosarcoma risk after adjustment for multiple comparisons (OR=1.35, 95% CI: 1.16–1.56, P=7.93×10−5, Padjusted=0.034).
Conclusions:
Genetic propensity to taller adult and childhood height attainments contributed independently to osteosarcoma risk in our data. These results suggest that the biological pathways affecting normal bone growth may be involved in osteosarcoma etiology.
Keywords: Osteosarcoma, height, growth, Mendelian Randomization, polygenic risk score
Keywords: Pediatric Cancer, Osteosarcoma, Height, Mendelian Randomization, Polygenic Score
Precis:
Polygenic scores representing genetic propensity to taller adult and childhood statures were each associated with elevated osteosarcoma risk. These results suggest that the biological pathways affecting normal bone growth may be involved in osteosarcoma etiology.
INTRODUCTION
Osteosarcoma is the most commonly diagnosed primary malignant bone tumor, with peak incidence occurring during adolescence1. Osteosarcomas frequently arise at sites of rapid bone growth, especially the metaphyses of long bones. These aggressive tumors occur most frequently around the time of puberty, after which risk declines substantially1. Correspondingly, osteosarcoma diagnosis tends to occur at younger ages in females than in males, reflecting their earlier age of peak height velocity2, 3. Although average age at osteosarcoma diagnosis is younger in females, the overall incidence of osteosarcoma is lower in females than in males, possibly related to population-level sex-differences in height attainment4, 5. These epidemiologic observations suggest that factors involved in osteoblast proliferation during normal bone growth may also be involved in osteosarcoma formation.
A subset of osteosarcoma diagnoses is attributable to heritable cancer predisposition syndromes6–9, with 3.8% of cases estimated to carry known or likely Li-Fraumeni syndrome-associated mutations10, and 7% of patients experiencing secondary malignancies suggestive of hereditary cancer syndromes11. Epidemiologic research exploring the role of common genetic variation in predisposition to osteosarcoma has suggested a role for variants in the DNA repair12, growth hormone3 and telomere maintenance pathways13 as potential risk factors. However, only a single genome-wide association study (GWAS) of osteosarcoma risk has been published14. This GWAS reported two osteosarcoma risk loci at 6p21.3 and 2p25.2, but the biologic mechanisms underlying these associations remain unknown.
Aside from male sex, the factor most consistently associated with increased osteosarcoma risk is above-average height1, 4, 15. In 1967, Fraumeni observed that children with osteosarcoma were significantly taller at the time of diagnosis than a hospital-based control group and suggested that “the origin of at least some of these tumors is a function of skeletal growth rates during childhood and adolescence”16. Subsequent studies have generally replicated these findings, observing that individuals in the 90th percentile of height are at increased risk1 and that height at diagnosis or one year prior are both greater in cases than in the general population17, 18. However, other studies have been less convincing, observing that osteosarcoma patients had shorter birth length19, that age- and sex-specific height percentiles were not associated with osteosarcoma risk20, that only patients diagnosed during adolescence had elevated heights21, and that cases showed no consistent pattern of differences in growth 22.
Recent GWAS investigating the genomic architecture of human height have identified hundreds of variants associated with stature. A recent meta-analysis of GWAS data from more than 250,000 individuals identified 423 independent loci explaining one-fifth of the heritability for adult height attainment, implicating several pathways relevant to cancer (e.g. WNT/β-catenin and Hedgehog signaling)23. Another GWAS meta-analysis of nearly 20,000 subjects has identified six loci associated with childhood height attainment prior to the onset of pubertal growth acceleration (i.e. the “take-off phase”), measured at age 10 in girls and age 12 in boys24. This same consortium identified additional loci associated with birth length, including several that overlapped with genes implicated in adult height attainment25. Given that osteoblast proliferation in the context of both normal bone growth and osteosarcomagenesis may rely on shared pro-growth pathways, we sought to determine if genetic determinants of birth length, childhood height or adult height play roles in osteosarcoma etiology.
We evaluated the individual and combined (i.e. as polygenic scores) effects of 416 SNPs associated with adult height attainment, six SNPs associated with childhood height attainment, and five SNPs associated with birth length in 864 osteosarcoma cases and 1879 controls of European ancestry from two independent case-control studies. We further assessed whether there is causal evidence of height affecting osteosarcoma risk using a Mendelian randomization framework. Finally, we investigated whether genetic associations were enriched among certain growth-related pathways, or are modified by subject sex or clinical presentation to seek further insight into the etiology and progression of this aggressive malignancy.
MATERIALS AND METHODS
California osteosarcoma case-control study:
The study was approved by the Institutional Review Boards at the UC, Berkeley and the California Department of Public Health (CDPH). The CDPH Genetic Diseases Screening Branch obtains newborn blood samples from all neonates born within the state for the purpose of disease screening. Remaining bloodspots have been archived at −20°C since 1982 and are available for approved research. We linked statewide birth records (1982–2009) to cancer diagnosis data from the California Cancer Registry (1988–2011). Included in this analysis were 207 osteosarcoma cases and 696 controls born in California from 1982–2009. Cases were diagnosed with osteosarcoma before age 20, per CCR record. Controls were matched on birth year, sex, and maternal self-reported race/ethnicity. Detailed characteristics of these subjects have been reported previously26 and appear in Supplementary Table 1.
DNA extraction from California newborn bloodspots:
A one-third portion of a 12-mm dried bloodspot was partitioned into three uniform segments and placed in a 2-mL microcentrifuge tube prior to the extraction, carried out using the QIAamp DNA Investigator Kit (Qiagen). 280μL of Buffer ATL and 20μL of Proteinase K were added to each sample. Samples were vortexed and incubated in a dry-bath shaker at 900 rpm and 56°C for one hour. After incubation, samples were briefly centrifuged and the lysate solution was transferred to a new 2 mL microcentrifuge tube, while the solid remnants were discarded. 1 μL of 1 ng/μL carrier RNA was added to the lysate then briefly vortexed. Samples were placed in the Qiagen Qiacube automated work station for DNA isolation, yielding a purified DNA sample in ATE buffer.
Genotyping and quality control of California case and control specimens:
DNA specimens were assigned to genotyping plates using blocked randomization according to case-control status, reported ethnicity, and sex. DNA was genotyped on the Affymetrix Axiom Latino Array. DNA samples were genotyped on an Affymetrix TITAN system, and raw image files were processed with Affymetrix Genetools.
Duplicate samples (n=34) had average genotype concordance >99%. Call-rate filtering for SNPs and samples was performed iteratively as follows: SNPs with call-rates <92% were removed, then samples with call-rates <95% were removed, then SNPs with call-rates <97% were removed, then samples with call-rates <97% were removed. Any SNP displaying significant departure from Hardy-Weinberg equilibrium P<1.0×10−5 among European-ancestry controls was excluded. Samples with mismatched reported versus genotyped sex were excluded. We performed identity-by-descent analyses in PLINK and excluded one member of any sample pair that had an identity-by-descent proportion >0.1827. Using genome-wide SNP data from 1184 HapMap Phase 3 samples, we performed principal component analysis using unlinked autosomal biallelic SNPs with allele frequency>0.05 and removed from analysis any sample showing evidence of non-European ancestry (>3 SDs from mean CEU values on PCs 1–3) (Supplementary Figures 1a-b).
NCI/Geisinger osteosarcoma dataset:
A second osteosarcoma case-control dataset was built from dbGaP study accession phs000734.v1.p1 (A Genome-wide Association Study (GWAS) of Risk for Osteosarcoma) and phs000381.v1.p1 (eMERGE Geisinger eGenomic Medicine MyCode Project Controls). Specimens were genotyped on the Illumina OmniExpress array and underwent quality-control filtering as previously described13. Briefly, SNPs with call rates <0.98 were removed from analyses. Following removal of poorly performing SNPs, subjects with genotyping call rates < 0.97 were removed. Ancestry-informative principal components were calculated using Eigenstrat and HapMap reference samples and mean values of the first five principal components were calculated among HapMap CEU samples27. Subjects that fell more than 3 SDs from the mean CEPH values on PCs 1–3 were excluded from further analyses (Supplementary Figure 1c-d). Because the NCI/Geisinger cases were selected in a clinic-based manner and include subjects from hospitals in Italy and Spain, we performed additional PC analyses in these subjects to investigate more cryptic intra-European ancestral differences using Human Genome Diversity Panel (HGDP) reference samples (Supplementary Figure 1e-f).
Following removal of subjects with non-European ancestry, SNPs with Hardy–Weinberg equilibrium P < 1.0×10−5 among controls were removed. Osteosarcoma cases and controls from the California and NCI/Geisinger datasets were compared, and both duplicated and cryptically-related samples were excluded from the NCI/Geisinger dataset (IBD>0.18). A total of 657 non-overlapping European-ancestry cases and 1183 controls were included in the final NCI/Geisinger dataset. These osteosarcoma patients were predominantly children and adolescents (age <21), although individual-level age data were unavailable and the original publication did not restrict to a specific age range. These patients are a subset of those included in a previous GWAS14.
Genotype imputation:
Haplotype phasing was performed with SHAPEIT v2.79028 and whole-genome imputation was carried out using the Minimac3 software29 with 64,976 human haplotypes from the 2016 release of the Haplotype Reference Consortium used as the imputation reference panel30. SNPs with imputation quality (info) scores less than 0.60 or posterior probabilities less than 0.90 were excluded to remove poorly-imputed SNPs, as previously done for the same Axiom array design31. Imputation quality metrics for SNPs included in the polygenic scores are presented in Supplementary Table 2, with info scores ranging from 0.76–1.0.
Construction of polygenic scores and statistical analyses:
We estimated the association between osteosarcoma risk and weighted polygenic height scores [for adult height attainment (PHSadult), childhood height attainment (PHSchild), and birth length (PHSbirth)], with adjustment for five ancestry-informative principal components, sex, and birth year (California dataset only) using logistic regression, then combining results for the California dataset and the NCI/Geisinger dataset using fixed-effects meta-analysis.
To construct the PHSadult, we selected the most statistically significant SNP from each of the 423 independent height-associated loci reported by Wood et al23. We further screened these 423 variants and ensured that no two SNPs had a pairwise R2≥0.10. The PHSadult for each individual was calculated as the number of effect alleles (i.e. the allele associated with taller stature) at each SNP multiplied by the weight for that SNP (i.e., the beta value of standardized height per effect allele reported in previous GWAS regression analyses23) summed across all height SNPs. Among 423 height-associated SNPs, 416 were successfully genotyped or imputed in our osteosarcoma case-control data. To determine the validity of the PHSadult, we assessed its association with adult height among 1183 NCI/Geisinger control subjects ages 21+, with adjustment for five ancestry-informative principal components and sex. Although other large GWAS of height have been performed, the next largest was published by the same group and all subjects in those analyses are a subset of those included in Wood, et al. 32.
The PHSchild was built using six independent SNPs that were associated with height at age 10 in girls and 12 in boys at genome-wide statistical significance in Cousminer, et al24. The PHSbirth was built using five independent SNPs that were associated with birth length at P<1.0×10−6 and were successfully replicated in an independent dataset at P<0.05 (one variant) or were located within 25kb of a SNP associated with adult height in a prior GWAS (four variants), as reported by van der Valk, et al.25. Unfortunately, childhood height and birth length data were unavailable in either the California or the NCI/Geisinger datasets.
None of the height-associated SNPs were located within 1Mb of a previously-identified osteosarcoma risk locus on chromosome 2. However, one adult height SNP, rs12214804, was located 152kb away from a previously-identified osteosarcoma risk SNP on chromosome 6 (rs1906953), with an R2 of 0.046 in European-ancestry HapMap subjects. We therefore performed a sensitivity analysis by excluding rs12214804 from the PHSadult and re-calculating its association with osteosarcoma risk.
PHSbirth and PHSchild were not correlated with each other (R=−0.0026, P=0.89), but each was positively correlated with the PHSadult (R=0.15, P=2.2×10−15 and R=0.15, P=1.4×10−12, respectively) due to linkage disequilibrium (LD) between SNPs comprising the models. When we constructed a new reduced PHSadult based on 411 total SNPs (after excluding five SNPs linked to those used in constructing the PHSbirth variable), correlation between the two polygenic height scores disappeared (R=0.0049, P=0.74). Similarly, when we constructed a new reduced PHSadult based on 410 total SNPs (after excluding six SNPs linked to those used in constructing the PHSchild variable), correlation between the two polygenic height scores disappeared (R=−0.012, P=0.53). Given the observed correlations between the PHS variables, their associations with osteosarcoma risk are presented without correction for multiple comparisons.
In order to study the combined effect of multiple polygenic height scores, we performed association analyses with both PHSchild and PHSadult as predictors (using the reduced PHSadult based on 410 total SNPs) and osteosarcoma as the outcome using a logistic regression model, adjusting for five ancestry-informative principal components, birth year (for the California dataset only), and sex.
Prior GWAS analyses of height reported beta estimates as standardized height rather than height in centimeters (cm) per effect allele. For purposes of interpreting effect sizes, a conversion was made to approximate the height in cm corresponding to each standard deviation of polygenic height score. We calculated this by multiplying the standard deviation of each polygenic height score (for standardized height) by the standard deviation of population height in cm, averaged between boys aged 12 and girls aged 10 for childhood height, and between men and women aged 20 years or older for adult height in the United States33.
For sex-stratified analyses, we examined association signals separately in males and females and tested for heterogeneity of effect using Cochran’s Q test. We also conducted case-only analyses, assessing whether polygenic scores for height were associated with differences in clinical presentation, including age at diagnosis, tumor location, and presence of metastasis. Clinical data were available for only the California cases and are coded as previously described26.
Mendelian randomization analyses
We used an inverse-variance weighted Mendelian randomization approach to assess the causal association between height and osteosarcoma risk using summarized data34, repeated for each of the three height measures. We also performed sensitivity analyses to address potential issues of invalid Mendelian randomization assumptions using the MR-Egger regression method to assess potential directional pleiotropy35 and a weighted-median approach which provides a consistent estimate when up to 50% of the SNPs used are invalid instruments36.
Pathway analysis of height-associated SNPs:
For each of the 423 SNPs independently associated with adult height attainment in Wood et al.23, we identified the nearest protein-coding gene in GRCh37. These RefSeq gene names were then queried in two pathway analysis databases: the Kyoto Encyclopedia of Genes and Genomes (KEGG) and the Protein ANalysis THrough Evolutionary Relationships (PANTHER) Classification System. After identifying highly-represented biological pathways within the height-associated gene list (KEGG P-value ≤0.001; PANTHER gene count ≥10), we recalculated the PHSadult, this time reducing the number of SNPs contributing to the model to only those residing within each respective pathway. In this way, we aimed to determine if any specific biological processes contributing to height attainment were driving the association between PHSadult and osteosarcoma risk.
Single SNP association analyses:
Single-locus association statistics for imputed and directly-genotyped SNPs were calculated using logistic regression in SNPTESTv2, using an allelic additive model and probabilistic genotype dosages37. The effect of individual SNPs on osteosarcoma risk was calculated with adjustment for the first five principal components from Eigenstrat. Analyses were carried out separately in the California osteosarcoma case-control dataset and the NCI/Geisinger case-control dataset. Fixed-effects meta-analysis was carried out using the program META38. In addition to SNPs used to create the polygenic height scores, we also tested four SNPs related to other adolescent anthropometric traits, including: childhood height in males, pubertal growth, pubertal growth in females, and late pubertal growth24. To assess statistical significance of associations in the single-SNP analyses, a Bonferroni correction was applied for 431 total tests (416 successfully imputed adult height attainment SNPs, six childhood height attainment SNPs, four related adolescent anthropometric trait SNPs, and five birth length SNPs), resulting in a significance threshold of 1.2×10−4.
RESULTS
After removing samples with genotyping call-rates below 97%, individuals showing evidence of non-European ancestry and cryptically-related individuals, a total of 864 osteosarcoma cases and 1879 controls remained for association analyses. This included 207 osteosarcoma cases and 696 controls from the California dataset and 657 non-overlapping osteosarcoma cases and 1183 controls from the NCI/Geisinger dataset. Among the 423 SNPs previously reported to be independently associated with adult height attainment23, 416 were successfully genotyped or imputed in the California and NCI/Geisinger datasets and used to construct the weighted polygenic height score for adult height attainment (PHSadult), with 410 SNPs used to construct the reduced PHSadult score that is uncorrelated with childhood height. Among the six SNPs reported to be associated with childhood height attainment prior to the pubertal growth spurt, and five SNPs reported to be associated with birth length, all were successfully genotyped or imputed in both datasets and used to construct weighted polygenic height scores (PHSchild and PHSbirth, respectively).
The PHSadult was strongly positively correlated with adult height attainment in both men (R=0.33, P=3.2×10−20) and women (R=0.37, P=1.04×10−17) among 1183 NCI/Geisinger control subjects with adult height information available, indicating that it is likely to serve as a robust predictor for adult height attainment. In multivariable logistic regression analyses, each standard deviation increase in the PHSadult was associated with a 1.10-fold increased risk of osteosarcoma (95% CI: 1.01–1.19, P= 0.027) (Table 1). Based on the distribution of PHSadult and a conversion factor using reported height statistics of adults in the US population33, this result suggests that a 1.7-cm increase in adult height attainment confers an approximately 10% increased risk of osteosarcoma. Sensitivity analyses excluding rs12214804, a variant in weak LD with a previously-identified osteosarcoma risk locus, showed no meaningful attenuation of the PHSadult association (P-value changed from 0.027 to 0.032), suggesting that the osteosarcoma risk variant is not driving the observed association with PHSadult.
Table 1.
Odds ratios and 95% confidence intervals for osteosarcoma risk associated with polygenic height scores
| Height trait(s) modeled by polygenic score | SNPs in polygenic score(s) | OR (95% CI)a | P-value(s)b |
|---|---|---|---|
| Birth | 5 | 1.07 (0.99–1.17) | 0.11 |
| Childhood | 6 | 1.10 (1.01–1.20) | 0.023 |
| Adult | 416 | 1.10 (1.01–1.19) | 0.027 |
| Adultreducedc | 410 | 1.09 (1.00–1.19) | 0.048 |
| Adultreducedc & Childhood | 410 & 6 | 1.09 (1.00–1.19) & 1.10 (1.01–1.20) | 0.048 & 0.023 |
OR represents the osteosarcoma risk associated with one standard deviation increase in polygenic height score, corresponding to approximately 1.7 cm of adult height, 0.5 cm of childhood height, and 0.14 cm of birth length, estimated by multiplying the standard deviation of each polygenic height score (calculated using beta values of standardized height from prior GWAS regression analyses) by the standard deviation of population height in centimeters averaged between men and women aged 20 years and over for adult height, between boys aged 12 and girls aged 10 for childhood height, and between male and female infants from birth to two months for birth length, reported from the National Health and Nutrition Examination Survey data on the U.S. population in 2007–2010
P-values are unadjusted for multiple testing
Adult polygenic height score excluding SNPs that are in linkage disequilibrium with childhood height SNPs
In analyses of childhood height attainment, each standard deviation increase in the PHSchild was associated with a 1.10-fold increased risk of osteosarcoma (95% CI: 1.01–1.20, P= 0.023) (Table 1). Based on the distribution of PHSchild and a conversion factor using reported height statistics of children in the US population, this result suggests that a 0.5-cm increase in childhood height attainment confers an approximately 10% increased risk of osteosarcoma. When PHSchild and the reduced PHSadult were modeled together, both polygenic height scores retained statistical significance, suggesting that childhood and adult height attainment are independently associated with risk of osteosarcoma (Pchild=0.023 and Padult=0.048, respectively) (Table 1). No interaction on the log-additive scale was detected between PHSchild and PHSadult (P=0.98).
In analyses of birth length, each standard deviation increase in the PHSbirth was not significantly associated with osteosarcoma risk, although the direction of the association is consistent with that of childhood and adult height (OR=1.07, 95% CI: 0.99–1.17, P=0.11).
No differences on PCs 1–3 were observed between cases and controls when compared using t-tests (P>0.10). However, 12 cases from the NCI/Geisinger dataset clustered with HGDP French and Italian subjects, without overlapping controls (Supplementary Figure 1e-f). Sensitivity analyses excluding these cases did not result in meaningful change for PHSadult or PHSchild (P=0.023 and 0.029, respectively), suggesting that ancestry differences are unlikely to drive the associations between height-related variants and osteosarcoma risk.
Analyses of the PHSchild and PHSadult variables did not reveal any significant interactions between either variable with subject sex (Pinteraction=0.78 and 0.50, respectively), although the effect estimate was slightly higher for PHSadult in male subjects (Supplementary Table 3). In case-only analyses of the California subjects, where clinical data were available, we did not observe differences in either the PHSchild or PHSadult according to the presence of metastatic disease, tumor stage, differentiation, size, location (long bone of the arm or leg versus another site), or patient’s age at diagnosis.
Both inverse-variance weighted and the weighted median Mendelian randomization estimates of the causal effect of height on osteosarcoma risk are consistent with those obtained by regressing the polygenic height scores on osteosarcoma when scaled to approximate the unit of height corresponding to one standard deviation increase in polygenic height score (Supplementary Table 4). However, the Mendelian randomization estimates were not significant (P>0.05), except for the adult height inverse-variance weighted estimate (P=0.029). No directional pleiotropy was detected for any of the height measures (MR Egger intercept P>0.05) (Supplementary Table 4).
Querying the 423 genes associated with adult height attainment against the KEGG database identified two over-represented biological pathways: “Hedgehog signaling” (eight genes) and “Wnt signaling” (12 genes). The PANTHER database identified five gene ontology pathways containing 10 or more members, including “Gonadotropin-releasing hormone receptor” (18 genes), “Wnt signaling” (13 genes), “CCKR signaling map” (10 genes), “Inflammation mediated by chemokine and cytokine signaling” (10 genes), and “Integrin signaling” (10 genes) (Supplementary Table 5). We calculated seven new PHSadult, each comprised of only those SNPs belonging to genes in the corresponding pathway. Of these seven pathways, only the PHSadult based on 10 SNPs involved in the “CCKR signaling map” pathway was associated with risk of osteosarcoma (P= 0.027) (Table 2). Although the direction of effect again indicated that variants associated with taller height increased risk of osteosarcoma, none of the 10 SNPs comprising the model was itself associated with osteosarcoma risk at P<0.05 (Table 3).
Table 2.
Odds ratios and 95% confidence intervals for osteosarcoma risk associated with polygenic scores of seven highly-represented biological pathways within the height-associated gene list
| Pathway modeled by polygenic score | Pathway analysis database | SNPs in polygenic score(s) | OR (95% CI)a | P-value(s)b |
|---|---|---|---|---|
| Hedgehog | KEGG | 8 | 1.02 (0.93–1.11) | 0.70 |
| WNT | KEGG | 12 | 0.99 (0.91–1.08) | 0.88 |
| Gonadotropin-releasing hormone receptor | PANTHER | 18 | 1.03 (0.95–1.13) | 0.44 |
| WNT | PANTHER | 13 | 1.00 (0.91–1.08) | 0.92 |
| CCKR/gastrin | PANTHER | 10 | 1.10 (1.01–1.20) | 0.027 |
| Inflammation | PANTHER | 10 | 1.07 (0.98–1.16) | 0.12 |
| Integrin | PANTHER | 10 | 1.01 (0.93–1.10) | 0.79 |
OR represents the osteosarcoma risk associated with one standard deviation increase in polygenic height score for variants belonging to each biologic pathway identified from the height-associated gene list.
P-values are unadjusted for multiple testing
Table 3.
SNPs previously associated with adult height and located in genes belonging to the “CCKR signaling map” pathway and their associations with osteosarcoma risk.
| SNP | Chr:BPa | Gene | Alleleb | EAF | P-value | OR (95% CI)c |
|---|---|---|---|---|---|---|
| rs6746356 | 2:174815898 | SP3 | A/C | 0.75 | 0.37 | 1.06 (0.93–1.22) |
| rs833152 | 2:183219101 | PDE1A | C/A | 0.42 | 0.24 | 1.07 (0.95–1.21) |
| rs3915129 | 3:4728104 | CTNNB1 | G/T | 0.48 | 0.68 | 1.03 (0.91–1.16) |
| rs2633761 | 3:41243742 | ITPR1 | A/G | 0.47 | 0.053 | 0.89 (0.79–1.00) |
| rs9291926 | 5:67599656 | PIK3R1 | T/G | 0.48 | 0.51 | 1.04 (0.92–1.18) |
| rs6894139 | 5:88327782 | MEF2C | T/G | 0.52 | 0.12 | 1.10 (0.97–1.24) |
| rs10995319 | 10:52762887 | PRKG1 | T/C | 0.77 | 0.96 | 1.00 (0.87–1.16) |
| rs12435366 | 14:35838389 | NFKBIA | C/T | 0.76 | 0.18 | 1.10 (0.96–1.27) |
| rs8069300 | 17:11984232 | MAP2K4 | G/C | 0.48 | 0.57 | 1.04 (0.92–1.17) |
| rs2117563 | 17:73368985 | GRB2 | G/A | 0.83 | 0.91 | 0.99 (0.84–1.16) |
Positions are GRCh37/hg19
Effect allele, listed first, is the allele associated with taller stature in Wood, et al.
Odds ratio corresponds to the effect of each additional copy of the effect allele, previously associated with adult height, on risk of osteosarcoma.
Single SNP analyses identified 22 variants that were nominally associated with osteosarcoma risk at P<0.05, one of which was part of the PHSchild (Supplementary Table 6) and 21 of which were part of the PHSadult (Supplementary Table 7). One of the variants associated with adult height, rs8103992, was significantly associated with osteosarcoma risk after Bonferroni correction for 431 tests (OR=1.35, 95% CI: 1.16–1.56, P=7.93×10−5, Padjusted=0.034).
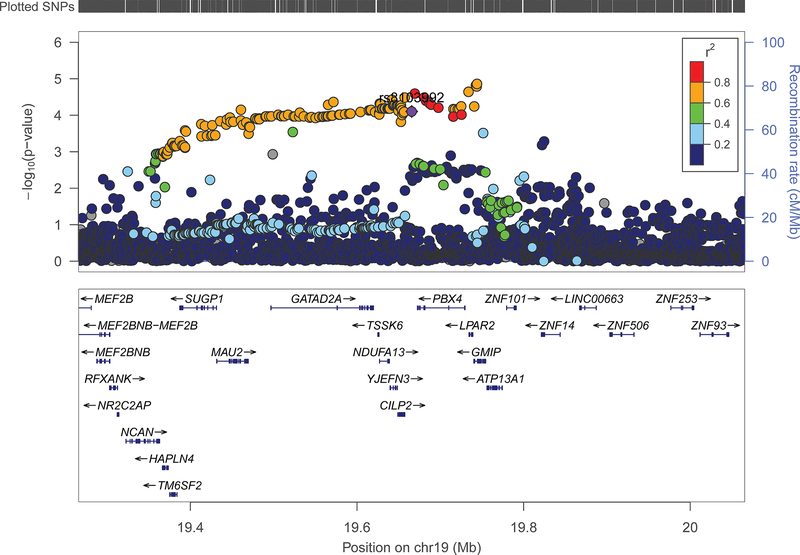
Located in an intergenic region on chromosome 19, rs8103992 is a reported expression quantitative trait locus (eQTL) for the adjacent gene CILP2 (cartilage intermediate layer protein 2) and the downstream gene LPAR239. We assessed whether rs8103992 was located within any regulatory loci using the Epigenome Browser and found that it overlapped a putative enhancer in osteoblast cells, as supported by the peaks for DNase I hypersensitivity, H3K27 acetylation, and H3K4 monomethylation in osteoblast cells40.The risk allele (A) is predicted to maintain a transcription factor binding site for HNF4 and FOXA2, as well as the chimeric Ewing sarcoma fusion gene product EWSR1-FLI141-44. Assessing all common SNPs (MAF>0.01) in an 800kb window centered on rs8103992 revealed a cluster of SNPs located about 75kb away that were in high LD (R2>0.60), which were slightly more significantly associated with osteosarcoma risk in our case-control data (Figure 1). The lead SNP in the region, rs11878202 (OR=1.35, 95% CI: 1.18–1.55, P=1.38×10−5), is located in an intron of GMIP and is itself an eQTL for several genes, including: CILP2, LPAR2, PBX4, and GMIP.
Figure 1: Association of SNPs with risk of osteosarcoma in an extended region of chromosome 19p13.11 previously associated with adult height attainment.
Genotypes include both “on array” SNPs and additional SNPs imputed as described in the Methods. A recent GWAS of adult height attainment identified rs8103992 (purple diamond shape) as the lead SNP in this region23 and we used it as one of 416 SNPs comprising our polygenic height score. Other SNPs are displayed by color, showing their extent of genetic linkage with rs8103992. The SNP most significantly associated with risk of osteosarcoma, rs11878202, is located in an intron of GMIP. Recombination rate, genetic position, and the locations of nearby genes are indicated.
DISCUSSION
Our results indicate that a genetic predisposition to taller stature is a risk factor for osteosarcoma. Using 416 SNPs associated with adult height attainment in a recent GWAS of more than 250,000 subjects, we constructed a polygenic height score that was significantly associated with both adult height and osteosarcoma risk. Our data suggest a 1.10-fold increase in osteosarcoma risk per standard deviation in polygenic height score, which corresponds to an approximately 1.7-cm increase in stature. This can be re-scaled to make direct comparisons with a previous Mendelian randomization study that reported effects sizes per 10-cm increase in genetically-predicted height45. While they observed an approximately 10% increased risk of lung cancer, 20% increased risk of breast cancer, and 60% increased risk of colorectal cancer associated with each 10-cm increase in genetically-predicted adult height, our re-scaled estimate is equivalent to a 75% increase in osteosarcoma risk per 10-cm increase in adult height.
In addition to the observed association between osteosarcoma risk and a genetic predisposition to taller adult stature, we also observed a significant association with a genetic predisposition to taller childhood stature prior to the take-off phase of the pubertal growth spurt. Of note, when the polygenic score for childhood and adult height attainment were modeled together, both scores retained statistical significance, suggesting that childhood and adult height attainment are independently associated with risk of osteosarcoma. This addresses a question that previous observational studies have been poorly-suited to answer due to the high correlation between childhood and adult height. Because our childhood and adult height variables are based on genetic polymorphisms rather than physical measurements, there was no correlation between these variables after removing SNPs in linkage disequilibrium.
The independent effects of childhood height and adult height observed in our data appear plausible given that up to 20% of adult stature is obtained after childhood during the pubertal growth spurt46. Because childhood height captures pre-pubertal growth, and adult height additionally captures pubertal growth, it is possible that our observed associations reflect the effects of pre-pubertal and pubertal growth on osteosarcoma risk. This is perhaps logical, considering that a majority of osteosarcoma patients are diagnosed before they reach adult height, many even before puberty4.
In single-SNP analyses, only rs8103992 was significantly associated with osteosarcoma risk after Bonferroni correction. This SNP, previously associated with adult height attainment, is a known eQTL for the nearby genes CILP2 and LPAR239. Interestingly, the risk (A) allele is associated with taller stature and reportedly preserves a transcription factor binding motif for the chimeric Ewing sarcoma fusion gene product EWSR1-FLI141, 42, 47, suggesting that this SNP may promote bone growth in both healthy and malignant contexts. Although SNPs in CILP2 have been previously associated with adult height attainment and cholesterol levels23, 48, they have not been reported to influence anthropometric traits in children. However, a recent epigenome-wide study of body composition in preschool children identified CpGs in CILP2 that were significantly associated with both BMI and fat free mass index49. Thus, the relationship between genetic and epigenetic variation in CILP2, body composition in children and adults, and future occurrence of disease merits further attention in longitudinal studies.
We utilized two approaches to assess the relationship between height and osteosarcoma risk in this study; 1) estimating the association between the polygenic score for height and osteosarcoma risk, and 2) estimating the causal effect of height on osteosarcoma risk using Mendelian randomization. Although the two approaches address similar questions of identifying the link between height and osteosarcoma, the estimates obtained from the two approaches have different interpretations50. The former, an estimate of osteosarcoma risk associated with height-related genetic variants, aims more generally to determine common biological mechanisms between the two phenotypes while the latter, an estimate of osteosarcoma risk due to height, requires that additional assumptions be satisfied for the estimate to be valid. The Mendelian randomization assumptions are 1) the SNPs used as instrumental variables are associated with the phenotype of interest (i.e. height), 2) the SNPs are not associated with confounders that influence both height and osteosarcoma risk, and 3) the SNPs are associated with osteosarcoma only through their effects on height51.
The first Mendelian randomization assumption is likely valid for the SNPs used for adult height, as the PHSadult was highly correlated with height in the Geisinger controls and explained 4.9% of the variation in adult height attainment after controlling for sex (F-statistic = 140.9). We did not have childhood height or birth length data available to assess the performance of the PHSchild or PHSbirth, and the proportion of variation explained by these variants was generally not reported in the prior GWAS24, 25. However, among the five SNPs used for the PHSbirth, one had a reported proportion of variance explained of 0.05%25, suggesting that the five-SNP birth length polygenic score is likely not a strong instrumental variable under a Mendelian randomization framework. By carefully excluding individuals with non-European ancestry, balancing cases and controls by ancestry, and adjusting for ancestry-informative principal components in all analyses, inflation of test statistics due to population stratification has likely been controlled, addressing a potential violation of the second assumption. Pleiotropy, wherein a SNP impacts osteosarcoma risk through mechanisms beyond its effect on height, may contribute to the association between stature and osteosarcoma risk. Although MR-Egger regression analyses revealed no evidence of directional pleiotropy35, the method is still susceptible to bias if multiple variants are associated with the same confounder. Furthermore, while we observed estimates of similar direction and magnitude between the polygenic score association approach and the Mendelian randomization approach, only the inverse-variance weighted approach for adult height reached significance (P<0.05), suggesting that caution is warranted in making a causal interpretation between height and osteosarcoma risk. Therefore, we interpret our results more generally that genetic predisposition to taller stature is a risk factor for osteosarcoma.
Prior literature suggests that the greater incidence of osteosarcoma in males is attributable to population-level sex-differences in height attainment4, 5. Associations with the polygenic scores did not significantly differ between males and females, suggesting that the osteosarcoma risk attributable to genetic determinants of height is not sex-specific and that a 1.7 cm increase in adult height confers approximately the same magnitude of osteosarcoma risk in men as in women. However, given that men achieve taller average stature than women, this also implies that the absolute risk of osteosarcoma will be higher in men than in women despite the risk associated with a per-unit increase in genetic height being approximately the same. It is also possible that the per-unit increase in height has a greater effect on osteosarcoma risk in males than females, as previously suggested1, but that this effect is not captured by the genetic determinants of height investigated in our study.
Whether genetic variation associated with increased height impacts risk of osteosarcoma via upregulation of shared pro-growth pathways or via independent mechanisms is not clear, but evidence suggests that these phenotypes have overlapping genetic determinants. Our pathway-based analyses did not identify a subset of height-associated SNPs that were clearly driving the observed association between the PHSadult and osteosarcoma risk, although a 10-SNP model limited to variants involved in the “CCKR signaling map” pathway was associated with risk. This was intriguing because CCKR signaling is the process which regulates gastrin expression, and gastrin over-expression has been shown to accelerate bone turnover rates in murine models52.
Overall, our results align well with a pleiotropic hypothesis wherein osteoblast proliferation is regulated by shared mechanisms in the contexts of both normal bone growth and osteosarcomagenesis. Since height is not a classically modifiable risk factor, our findings primarily serve to elucidate the genetic and biological factors linking height and cancer risk rather than to guide specific public health recommendations. In conclusion, we observe that genetic predisposition to taller childhood and adult stature contribute independently to osteosarcoma risk, suggesting that the biological pathways affecting normal bone growth are also involved in osteosarcoma etiology.
Supplementary Material
ACKNOWLEDGEMENTS
The biospecimens and/or data used in this study were obtained from the California Biobank Program (SIS request number 550). The California Department of Public Health is not responsible for the results or conclusions drawn by the authors of this publication. The collection of cancer incidence data used in this study was supported by the California Department of Public Health pursuant to California Health and Safety Code Section 103885; Centers for Disease Control and Prevention’s (CDC) National Program of Cancer Registries, under cooperative agreement 5NU58DP003862–04/DP003862; the National Cancer Institute’s Surveillance, Epidemiology and End Results Program under contract HHSN261201000140C awarded to the Cancer Prevention Institute of California, contract HHSN261201000035C awarded to the University of Southern California, and contract HHSN261201000034C awarded to the Public Health Institute. The ideas and opinions expressed herein are those of the author(s) and do not necessarily reflect the opinions of the State of California, Department of Public Health, the National Cancer Institute, and the Centers for Disease Control and Prevention or their Contractors and Subcontractors.”
Datasets used for the analyses described in this manuscript were obtained from dbGaP study accession phs000734.v1.p1 [A Genome-wide Association Study (GWAS) of Risk for Osteosarcoma] and phs000381.v1.p1 [eMERGE Geisinger eGenomic Medicine (GeM) - MyCode Project Controls].Samples and data in this study were provided by the Geisinger MyCode® Project. Funding for the MyCode® Project was provided by a grant from Commonwealth of Pennsylvania and the Clinic Research Fund of Geisinger Clinic. Funding support for the genotyping of the MyCode® cohort was provided by a Geisinger Clinic operating funds and an award from the Clinic Research Fund.
Funding: This work was supported by the National Institutes of Health T32 CA151022–06 (C.Z.), R01CA155461 (J.L.W.), and by ‘A’ Awards from The Alex’s Lemonade Stand Foundation (A.J.dS., K.M.W.).
Footnotes
Conflicts of Interest: None
Author contributions:
Acquisition of data: Helen M. Hansen, Julio Gonzalez-Maya, Ivan V. Smirnov, Joseph L. Wiemels, and Kyle Walsh
Conceptualization and design: Chenan Zhang, Catherine Metayer, Joseph L. Wiemels, and Kyle Walsh
Analysis and interpretation of results: Chenan Zhang, Adam J. de Smith, Qingyi Wei, William C. Eward, Joseph L. Wiemels, and Kyle Walsh
Writing-original draft: Chenan Zhang and Kyle Walsh
Writing-review and editing: Chenan Zhang, Libby M. Morimoto, Adam J. de Smith, Helen M. Hansen, Julio Gonzalez-Maya, Alyson A. Endicott, Ivan V. Smirnov, Catherine Metayer, Qingyi Wei, William C. Eward, Joseph L. Wiemels, and Kyle M. Walsh
REFERENCES
- 1.Mirabello L, Pfeiffer R, Murphy G, et al. Height at diagnosis and birth-weight as risk factors for osteosarcoma. Cancer Causes Control. 2011;22: 899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Savage SA, Mirabello L. Using epidemiology and genomics to understand osteosarcoma etiology. Sarcoma. 2011;2011: 548151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Musselman JR, Bergemann TL, Ross JA, et al. Case-parent analysis of variation in pubertal hormone genes and pediatric osteosarcoma: a Children’s Oncology Group (COG) study. Int J Mol Epidemiol Genet. 2012;3: 286–293. [PMC free article] [PubMed] [Google Scholar]
- 4.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer. 2009;115: 1531–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duong LM, Richardson LC. Descriptive epidemiology of malignant primary osteosarcoma using population-based registries, United States, 1999–2008. J Registry Manag. 2013;40: 59–64. [PMC free article] [PubMed] [Google Scholar]
- 6.Wang LL, Gannavarapu A, Kozinetz CA, et al. Association between osteosarcoma and deleterious mutations in the RECQL4 gene in Rothmund-Thomson syndrome. J Natl Cancer Inst. 2003;95: 669–674. [DOI] [PubMed] [Google Scholar]
- 7.Chauveinc L, Mosseri V, Quintana E, et al. Osteosarcoma following retinoblastoma: age at onset and latency period. Ophthalmic Genet. 2001;22: 77–88. [DOI] [PubMed] [Google Scholar]
- 8.Wunder JS, Gokgoz N, Parkes R, et al. TP53 mutations and outcome in osteosarcoma: a prospective, multicenter study. J Clin Oncol. 2005;23: 1483–1490. [DOI] [PubMed] [Google Scholar]
- 9.Lipton JM, Federman N, Khabbaze Y, et al. Osteogenic sarcoma associated with Diamond-Blackfan anemia: a report from the Diamond-Blackfan Anemia Registry. J Pediatr Hematol Oncol. 2001;23: 39–44. [DOI] [PubMed] [Google Scholar]
- 10.Mirabello L, Yeager M, Mai PL, et al. Germline TP53 variants and susceptibility to osteosarcoma. J Natl Cancer Inst. 2015;107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hauben EI, Arends J, Vandenbroucke JP, van Asperen CJ, Van Marck E, Hogendoorn PC. Multiple primary malignancies in osteosarcoma patients. Incidence and predictive value of osteosarcoma subtype for cancer syndromes related with osteosarcoma. Eur J Hum Genet. 2003;11: 611–618. [DOI] [PubMed] [Google Scholar]
- 12.Mirabello L, Yu K, Berndt SI, et al. A comprehensive candidate gene approach identifies genetic variation associated with osteosarcoma. BMC Cancer. 2011;11: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walsh KM, Whitehead TP, de Smith AJ, et al. Common genetic variants associated with telomere length confer risk for neuroblastoma and other childhood cancers. Carcinogenesis. 2016;37: 576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Savage SA, Mirabello L, Wang Z, et al. Genome-wide association study identifies two susceptibility loci for osteosarcoma. Nat Genet. 2013;45: 799–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arora RS, Kontopantelis E, Alston RD, Eden TO, Geraci M, Birch JM. Relationship between height at diagnosis and bone tumours in young people: a meta-analysis. Cancer Causes Control. 2011;22: 681–688. [DOI] [PubMed] [Google Scholar]
- 16.Fraumeni JF Jr. Stature and malignant tumors of bone in childhood and adolescence. Cancer. 1967;20: 967–973. [DOI] [PubMed] [Google Scholar]
- 17.Cotterill SJ, Wright CM, Pearce MS, Craft AW, Group UMBTW. Stature of young people with malignant bone tumors. Pediatr Blood Cancer. 2004;42: 59–63. [DOI] [PubMed] [Google Scholar]
- 18.Gelberg KH, Fitzgerald EF, Hwang S, Dubrow R. Growth and development and other risk factors for osteosarcoma in children and young adults. Int J Epidemiol. 1997;26: 272–278. [DOI] [PubMed] [Google Scholar]
- 19.Operskalski EA, Preston-Martin S, Henderson BE, Visscher BR. A case-control study of osteosarcoma in young persons. Am J Epidemiol. 1987;126: 118–126. [DOI] [PubMed] [Google Scholar]
- 20.Troisi R, Masters MN, Joshipura K, Douglass C, Cole BF, Hoover RN. Perinatal factors, growth and development, and osteosarcoma risk. Br J Cancer. 2006;95: 1603–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Longhi A, Pasini A, Cicognani A, et al. Height as a risk factor for osteosarcoma. J Pediatr Hematol Oncol. 2005;27: 314–318. [DOI] [PubMed] [Google Scholar]
- 22.Buckley JD, Pendergrass TW, Buckley CM, et al. Epidemiology of osteosarcoma and Ewing’s sarcoma in childhood: a study of 305 cases by the Children’s Cancer Group. Cancer. 1998;83: 1440–1448. [DOI] [PubMed] [Google Scholar]
- 23.Wood AR, Esko T, Yang J, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet. 2014;46: 1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cousminer DL, Berry DJ, Timpson NJ, et al. Genome-wide association and longitudinal analyses reveal genetic loci linking pubertal height growth, pubertal timing and childhood adiposity. Hum Mol Genet. 2013;22: 2735–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Valk RJ, Kreiner-Moller E, Kooijman MN, et al. A novel common variant in DCST2 is associated with length in early life and height in adulthood. Hum Mol Genet. 2015;24: 1155–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Endicott AA, Morimoto LM, Kline CN, Wiemels JL, Metayer C, Walsh KM. Perinatal factors associated with clinical presentation of osteosarcoma in children and adolescents. Pediatr Blood Cancer. 2017;64. [DOI] [PubMed] [Google Scholar]
- 27.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Connell J, Gurdasani D, Delaneau O, et al. A general approach for haplotype phasing across the full spectrum of relatedness. PLoS Genet. 2014;10: e1004234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das S, Forer L, Schonherr S, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48: 1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCarthy S, Das S, Kretzschmar W, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48: 1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wiemels JL, Walsh KM, de Smith AJ, et al. GWAS in childhood acute lymphoblastic leukemia reveals novel genetic associations at chromosomes 17q12 and 8q24.21. Nat Commun. 2018;9: 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lango Allen H, Estrada K, Lettre G, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467: 832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fryar CD, Gu Q, Ogden CL. Anthropometric reference data for children and adults: United States, 2007–2010. Vital Health Stat 11. 2012: 1–48. [PubMed] [Google Scholar]
- 34.Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37: 658–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44: 512–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol. 2016;40: 304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11: 499–511. [DOI] [PubMed] [Google Scholar]
- 38.Liu JZ, Tozzi F, Waterworth DM, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010;42: 436–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Consortium GT, Laboratory DA, Coordinating Center -Analysis Working G, et al. Genetic effects on gene expression across human tissues. Nature. 2017;550: 204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou X, Li D, Zhang B, et al. Epigenomic annotation of genetic variants using the Roadmap Epigenome Browser. Nat Biotechnol. 2015;33: 345–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bryne JC, Valen E, Tang MH, et al. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008;36: D102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pique-Regi R, Degner JF, Pai AA, Gaffney DJ, Gilad Y, Pritchard JK. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res. 2011;21: 447–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsunoda T, Takagi T. Estimating transcription factor bindability on DNA. Bioinformatics. 1999;15: 622–630. [DOI] [PubMed] [Google Scholar]
- 44.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40: D930–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khankari NK, Shu XO, Wen W, et al. Association between Adult Height and Risk of Colorectal, Lung, and Prostate Cancer: Results from Meta-analyses of Prospective Studies and Mendelian Randomization Analyses. PLoS Med. 2016;13: e1002118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luo ZC, Karlberg J. Critical growth phases for adult shortness. Am J Epidemiol. 2000;152: 125–131. [DOI] [PubMed] [Google Scholar]
- 47.Boyle AP, Hong EL, Hariharan M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22: 1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kathiresan S, Melander O, Guiducci C, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40: 189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rzehak P, Covic M, Saffery R, et al. DNA-Methylation and Body Composition in Preschool Children: Epigenome-Wide-Analysis in the European Childhood Obesity Project (CHOP)-Study. Sci Rep. 2017;7: 14349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson JR, Minelli C, Del Greco MF. Mendelian Randomization using Public Data from Genetic Consortia. Int J Biostat. 2016;12. [DOI] [PubMed] [Google Scholar]
- 51.Didelez V, Sheehan N. Mendelian randomization as an instrumental variable approach to causal inference. Stat Methods Med Res. 2007;16: 309–330. [DOI] [PubMed] [Google Scholar]
- 52.Krishnamra N, Limlomwongse L. Acute and long-term effects of gastrin on muscle and bone calcium uptake in rats. J Nutr Sci Vitaminol (Tokyo). 1995;41: 687–697. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.