Abstract
Objective:
Uterine carcinosarcoma (UCS) is a rare and aggressive form of uterine cancer. It is biphasic, exhibiting histological features of both malignant epithelial (carcinoma) and mesenchymal (sarcoma) elements, reflected in ambiguity in accepted treatment guidelines. We sought to study the genomic and transcriptomic profiles of these elements individually to gain further insights into the development of these tumors.
Methods:
We macro-dissected carcinomatous, sarcomatous, and normal tissues from formalin fixed paraffin embedded uterine samples of 10 UCS patients. Single nucleotide polymorphism microarrays, targeted DNA sequencing and whole-transcriptome RNA-sequencing were performed. Somatic chromosomal alterations (SCAs), point mutation and gene expression profiles were compared between carcinomatous and sarcomatous components.
Results:
In addition to TP53, other recurrently mutated genes harboring putative driver or loss-offunction mutations included PTEN, FBXW7, FGFR2, KRAS, PIK3CA and CTNNB1, genes known to be involved in UCS. Intra-patient somatic mutation and SCA profiles were highly similar between paired carcinoma and sarcoma samples. An epithelial-mesenchymal transition (EMT) signature tended to differentiate components, with EMT-like status more common in advancedstage patients exhibiting higher inter-component SCA heterogeneity.
Conclusions:
From DNA analysis, our results indicate a monoclonal disease origin for this cohort. Yet expression-derived EMT statuses of the carcinomatous and sarcomatous components were often discrepant, and advanced cases displayed greater genomic heterogeneity. Therefore, separately-profiled components of UCS tumors may better inform disease progression or potential.
Keywords: uterine cancer, carcinosarcoma; genomics, heterogeneity, biphasic, haplotype
Introduction
Uterine carcinosarcoma (UCS), also referred to as Malignant Mixed Müllerian Tumors (MMMT) of the uterus, is a malignant neoplasm of the female genital tract. As implied, the neoplasm shows histological features of both epithelial elements (adenocarcinoma) and mesenchymal elements (sarcoma). Overall, MMMT patients exhibit 5-year survival of between 35% and 65% for earlier stage and 10% or less for late stages [1]. Although UCS is a relatively rare malignancy, representing less than 5% of all uterine cancers, it is known for its aggressive clinical course and accounts for a disproportionate number (15%) of all uterine cancer deaths [2]. This is due to several factors, including late stage at presentation (10% of patients will have metastatic disease and 60% will have extrauterine disease at time of diagnosis) [3], and limited clinical trials investigating optimal treatment approaches. Therapeutic approaches developed uniquely for UCS are inadequate and borrowing insights from other cancers has proved challenging. Indeed, UCS was traditionally classified as a sarcoma and treatments were thus directed as such. In 2009, the International Federation of Gynecology and Obstetrics introduced a new staging system for uterine sarcomas that made UCS a variant of endometrial carcinoma [4], based on greater similarity in clinical course and risk factors, compared to sarcoma. This recent change reflects a relatively shallow appreciation of the complexity of UCS, motivating additional research toward understanding its pathology, progression and tumor development, and how these relate to clinical outcome.
The unique biphasic nature of carcinosarcomas has given rise to a body of work to better understand its origins. Three theories are generally accepted as plausible [5, 6]. The collision theory indicates that the tumors are biclonal, arising from separate cells that later merge. The conversion theory states that a single cell undergoes metaplastic differentiation. Finally, the combination theory captures aspects of the previous two, i.e. that a common precursor differentiates bi-directionally before merging. Most molecular and histopathological evidence is supportive of the conversion theory for a majority of tumors [6]. Wada et. al. assessed the X inactivation patterns in 25 carcinosarcoma cases. They found similar patterns of X chromosome inactivation in 19 out of 25 cases, suggesting monoclonal (conversion or combination) derivation [7]. Other studies have shown consistent patterns of deletions, preserved across carcinoma and sarcoma components, also suggesting monoclonal origin [8–10]. Schulten et al. [11] showed 2 of 3 cases had similar karyotypic abnormalities with additional ones in the sarcomatous components. They also demonstrated the presence of frequent 8q gains in both components of carcinosarcoma and de novo endometrial carcinoma, suggesting a monoclonal origin with additional genetic aberrations in areas having undergone sarcomatous transformation. While most clinical, histological, and molecular evidence supports the conversion theory, and it is now generally assumed that the neoplasm is derived from sarcomatous differentiation of high-grade carcinoma [12, 13], there are examples of a bona fide bi-clonal origin of the tumor (collision theory) for some UCS patients. Wada, for example, found 3 patients exhibiting X-inactivation patterns indicating a collision origin [7]. From these observations, it appears there exists heterogeneity in modes of UCS development, with a monoclonal origin as the more common mode. Further investigations are required to better characterize their relative frequencies and understand their clinical relevance.
McConechy et al, performed component-separated analyses, reporting DNA mutations in 27 genes of 13 patients and reported mutation patterns that were consistent with a monoclonal theory [13]. Zhao et al performed component-separated whole-exome sequencing analyses on 6 uterine and ovarian carcinosarcoma patients and indicated a similar conclusion [14]. The Cancer Genome Atlas for UCS performed multi-platform molecular characterizations from unseparated, mixedcomponent cancer samples, studying both components of UCS via bulk analyses [15]. While this comprehensive effort sheds light on the genomics and transcriptomics of this cancer relative to other sites, such a design is not geared toward resolution of molecular origins and development of the disease.
Unavailable from deep DNA point mutations assessments of McConechy et al [13] and Zhao et al [14], and the multiple-platform approach of TCGA is a more complete picture of the landscape of somatic chromosomal alterations (SCAs) leading to acquired allelic imbalance in each tissue component, as well as a corresponding analysis of gene expression profiles. Here we sought to address this void, obtaining separated carcinomatous and sarcomatous tissues from 10 patients diagnosed with homologous UCS (see Table 1). Separately in both tissue types in each patient, using paired adjacent normal tissue as a contrast, we performed genome-wide SCA and targeted point mutation profiling, using SNP DNA microarrays and deep next-generation sequencing (NGS). We also performed whole-transcriptome (NGS) expression profiling in these tissues (RNAseq), attempting to relate expression patterns to underlying genomic features.
Table 1.
Patient clinical overview
| Subject | Age at diagnosis (yrs) |
Stage at diagnosis |
Response | Carcinoma EMT classification |
Sarcoma EMT classification |
|---|---|---|---|---|---|
| 0317 | 58 | IIB | Complete response | epithelial | EMT |
| 0609 | 89 | IIA | Complete response | EMT | EMT |
| 0619 | 64 | IB | Complete response | epithelial | epithelial |
| 0622 | 68 | IIIA | Complete response | epithelial | EMT |
| 0724 | 39 | IVB | Progressive disease | --- | --- |
| 0819 | 71 | IIIB | NA | --- | EMT |
| 0829 | 61 | IA | Complete response | epithelial | epithelial |
| 0909 | 65 | IA | Complete response | --- | epithelial |
| 1104 | 74 | IIIA | Complete response | EMT | EMT |
Materials and Methods
Sample collection
Samples from 10 patients diagnosed with UCS were collected at Avera McKennan Hospital & University Health Center. This study was approved under the Avera McKennan Institutional Review Board (#2015.020). Subjects were consented to the Gynecologic Specimen Bank (GSB), which includes specimen collection and genetic/genomic evaluation. We were also allowed to use deidentified samples from deceased patients as per the Avera Institutional Review Board (approval date April 24, 2015). The three components (carcinoma, sarcoma, and normal) were identified from corresponding pathology slides by a pathologist (RS); histologic classifications are provided in Supplementary Table 1. For this study to reduce additional sources of variation, patients were selected based on a confirmed homologous uterine carcinosarcoma determination. The tissue samples were collected during surgery (resection) and were preserved in formalin fixed paraffin embedded (FFPE) blocks. The slide was overlaid onto the FFPE block and macro-dissection of an FFPE core using a punch biopsy tool (three - 1mm punches or one – 3mm punch) was performed. DNA and RNA were extracted using the Qiagen AllPrep DNA/RNA FFPE kit (Valencia, CA, USA) as per manufacturer’s protocol.
SNP arrays
Single nucleotide polymorphism (SNP) data was generated using the Illumina Infinium® OmniExpressExome-8 v 1.3 BeadChip array (Illumina Inc., San Diego, CA, USA). Extracted DNA was subjected to the Infinium HD FFPE DNA Restoration protocol prior to genotyping (Illumina Inc., San Diego, CA). Infinium processing was carried out following the manufacturer’s instructions. Raw intensity files were analyzed in Illumina Genome Studio Genotyping Module software (v2011.1). Cluster locations for genotype calling were imported from a vendor supplied cluster file (HumanOmniExpressExome-8v1–2_A.egt).
DNA sequencing, alignment and mutation calling
Targeted deep sequencing across 174 amplicons within 26 cancer genes was performed to assess somatic variation using the Illumina TruSight Tumor 26 Sample Preparation Kit (Illumina Inc., San Diego, CA). DNA samples were subjected to FFPE quality control quantitative PCR (qPCR) to determine suitability for PCR amplification relative to a non-FFPE reference sample. Amplicon based DNA sequencing libraries were pooled at a normalized concentration (4 nmol/L) and quantified via qPCR utilizing the KAPA Library Quantfication Kit (KAPA Biosystems, Woburn, MA) prior to sequencing on an Illumina MiSeq to a mean depth of 6000x. FASTQ files were generated in MiSeq Reporter software v1.0.0 (Illumina Inc., San Diego, CA). To call somatic mutations, we applied the Illumina Amplicon-DS Somatic Variant Caller. Calls in the tumor samples were deemed potential germline variants and removed if variant reads existed in the paired-normal sample. Tumor variants with lower than 2% allele frequencies were excluded. Further, sequencing regions harboring mutations were visually inspected across samples using the Integrative Genomics Viewer to identify potential false positive mutations due to systematic alignment issues and the potential presence of mutant reads in the project’s normal samples [16]. Finally, to focus on potentially significant mutations, we report and interpret only those mutations that are annotated in the COSMIC (Catalog of Somatic Mutations in Cancer) database.
RNA sequencing, alignment and analysis
Whole transcriptome sequencing was performed to quantify gene expression. Each FFPE RNA sample was assessed for degradation on an RNA 6000 Nano chip with a 2100 BioAnalyzer (Agilent; Santa Clara, CA). The average concentration and RNA integrity score (RIN) of the sample set averaged 678.5ng/ul and 2.24 respectively. Sequencing libraries were prepared using the TruSeq Stranded Total RNA Library Prep Kit with Ribo-Zero Gold (Illumina, Inc; San Diego, CA) following the low sample procedure. Fragmentation was reduced to zero to accommodate the FFPE nature of the samples. The concentration of each library was determined by qPCR utilizing the KAPA Library Quantification Kit for Next Generation Sequencing (KAPA Biosystems; Woburn, MA) prior to sequencing. Sequencing-by-synthesis (SBS) was performed on a HiSeq2500 utilizing v4 chemistry with paired-end 101-bp reads and a 6-bp index read resulting in approximately 4.2 billion paired-end (or total) reads. De-multiplexing of the raw sequence data and FASTQ generation was carried out using bcl2fastq (v2.17) where each sample averaged 138.4 million paired-end reads. Sequence reads were aligned to the GENCODE version 19 transcriptome using STAR [17], resulting in 75 million reads per sample for downstream analyses. RSEM [18] was utilized for gene expression quantification. We removed six samples (two normal, three carcinomatous, and one sarcomatous samples) from our analyses because their alignment rates were less than 30%. We then applied EBSeq to detect any differentially expressed genes between carcinomas and sarcomas.
Detection of chromosomal alterations
To profile SCAs, we applied a haplotype-aware hidden Markov model (HMM), hapLOH, designed to detect acquired allelic imbalance, including those at low mutant cell fractions or exhibited in tumor samples of low cellularity [19]. To do so we performed standard SNP genotype quality control procedures and estimated haplotypes using MACH [20] in the normal samples. To increase phasing accuracy, we integrated samples from other studies with the same array platform. One patient was removed because of a failed genotyping correlation QC step that verifies that paired carcinomas and sarcomas came from the same individual. Thus in our genomic analyses, we have nine patients (27 samples) in total. We then applied hapLOH to each carcinomatous, sarcomatous, and normal sample with two aberrant states, summarizing SCAs as those with maximum posterior probabilities from the HMM exceeding 0.95 and boundaries established where posterior probabilities fell below 0.5. We took the SCA calls that were made for each sample of a patient and then tested if these events existed in the other samples of the same individual using the following algorithm. We applied a binomial test to assess whether there was higher than expected phasing (utilizing the estimated germline haplotype) within the A/B alleles having allele frequencies greater than the expected 0.5 (for a heterozygous site) within called event regions derived from other samples of the same patient. In addition, we statistically tested whether the excessive haplotype in each sample, is the under-represented haplotype (over the same genomic region) of other samples from the same patient. These regions where opposite haplotypes are over-represented are deemed to be separate mutation events which we term mirrored AI.
Downstream comparisons of the RNA-seq, DNA somatic mutation and SCA calls, between tumor components and among patients, as well as generation of graphical displays, were performed using R (www.r-project.org).
Inter-component heterogeneity
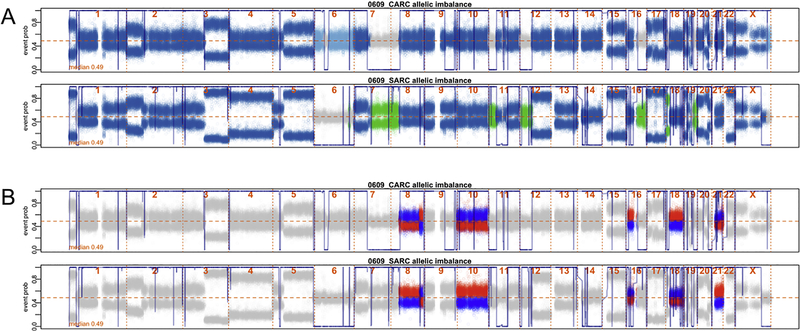
We assessed inter-component heterogeneity by quantifying differences in genomic SCAs between UCS component samples within each patient. First, regions of the genome that exhibited component-specific AI, were identified (see the light-blue and green regions of Figure 1A). Next, for regions where both components exhibited AI, we identified regions where paired samples had different over-represented haplotypes, signifying putative component-specific events (see the blue and red regions of Figure 1B). We identified these component-specific regions by first taking event regions called by hapLOH, and then applying a binomial test checking for inconsistency of the over-represented allele at each heterozygous site between the paired samples over that region. Our inter-component heterogeneity measure is then the proportion of heterozygous sites that are in component-specific events compared to the total number of event heterozygous sites.
Figure 1. Identification of UCS somatic chromosomal alterations (SCAs) that are shared, carcinoma-specific and sarcoma-specific.
(A) B-allele frequencies across the genome of the carcinoma (top) and sarcoma (bottom) components of a UCS tumor from subject 0609. The fluctuating blue line across the plot represents a posterior probability of SCAs across each sample’s genome. Dark blue dots represent the B-allele frequencies at markers in the genome where both the carcinoma and sarcoma components exhibit SCAs. Light blue represents portions of the genome that have carcinoma-specific SCAs (see chr 6). Similarly, green represents portions of the genome that have sarcoma-specific SCAs (see chr 7q). Both the carcinoma-specific and sarcoma-specific SCAs are characterized as private SCAs in downstream analyses. (B) The blue and red markers for each plot correspond to the same maternal and paternal haplotypes for the pair of samples described in (A), identifying regions of the genome that are discrepant in their overrepresented haplotypes. This suggests that events at these regions of the genome are independent; thus, events in these regions are re-classified from shared to private SCAs.
Results
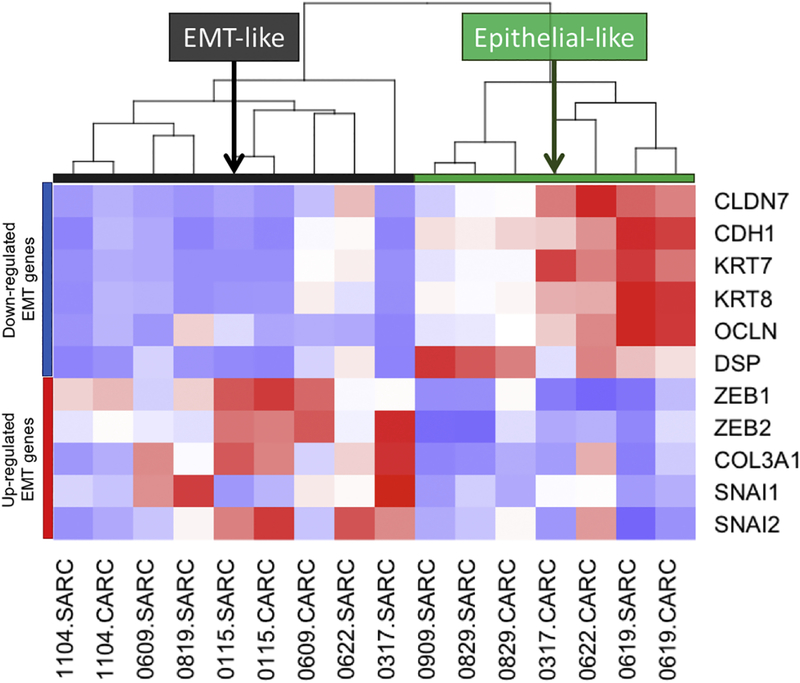
Gene expression patterns between the carcinoma and sarcoma samples revealed few differences (Supplementary Figure 1), where gene set analyses of differentially expressed genes failed to identify any component-specific aberrant cancer pathways. An epithelial-to-mesenchymal (EMT) phenotype has previously been associated with UCS, and more specifically with higher-degrees of sarcomatous cells in bulk UCS tumor analyses [21]; thus, we investigated the EMT statuses in our separate component samples. Through clustering of samples based on the expression of known EMT genes (6-down regulated and 5 up-regulated genes [15]), nine samples were identified as EMT-like and seven as epithelial-like (Figure 2). EMT-like statuses were more common in the sarcomas (5 of 8) versus carcinomas (2 of 6), although not at statistical significance. For 7 patients where both components were successfully profiled, 5 component-pairs resulted in common EMTlike statuses. For 2 component-pairs from patients 0317 and 0622, sample classification resulted in discrepant EMT-like statuses, where the carcinoma components were epithelial-like and the sarcomatous components were EMT-like.
Figure 2. Gene expression pattern in UCS component samples based on EMT signature genes.
Samples are segregated into two groups based on the unsupervised clustering of gene expression values of EMT-associated genes.
All described mutations have previously been reported in the COSMIC database. A total of 37 somatic mutations were identified from the 26 cancer-gene sequencing panel, with one to three mutations being called per sample (see Supplementary Table 2). Mutations in TP53 were the most prevalent, detected in 12 of 18 (67%) samples from 6 of 9 patients (Figure 3). Other recurrently mutated genes identified include PTEN (6/18; 33%), FBXW7 (4/18; 22%), FGFR2 (4/18; 22%), KRAS (4/18; 22%), PIK3CA (3/18; 17%) and CTNNB1 (2/18; 11%) – all of which have been previously identified in gynecologic carcinosarcomas [13, 15, 22] (Figure 3). Our sample size is too modest to robustly compare specific gene mutation frequencies with previous findings. Mutations identified in the sarcomas were identical to those identified in their carcinoma counterparts with the exception of one mutation identified (18 of 19). A highly recurrent COSMIC PIK3CA E542K mutation was unique to subject 0609’s sarcoma component. Other highly recurrent COSMIC and putative driver mutations were identified in PIK3CA (E545K), KRAS (G12A and G12S), CTNBB1 (D32A) and TP53 (R175H and R273C), identified in both UCS component samples of 5 subjects.
Figure 3. An integrated view of genomic AI, mutations and gene expression for UCS carcinoma and sarcoma components.
Each column represents a UCS sample. The samples are sorted by EMT status first (as determined by RNA-seq clustering of EMT-specific genes) and then by decreasing amounts of private SCAs (which is a putative measure of UCS heterogeneity equal to the proportion of the genome that is aberrant in the corresponding sample but not in its UCS component counterpart).
A mean of 89% of the genomic SCA regions identified per UCS tumor were shared between paired carcinoma and sarcoma samples, with 5.7% and 5.8% being unique to the carcinoma and sarcoma components respectively. The total load of genomic SCA per sample is illustrated in the top column bar of Figure 3. The amount of component-shared SCA is in light-gray and the component-specific SCA is illustrated in black, which is a measure of inter-component heterogeneity. EMT-like status was associated with advanced stage upon diagnosis in each of the patients (Fisher’s test, p = 0.02; comparing stage I UCS versus stages II-IV). EMT status was also associated with higher degrees of inter-component heterogeneity based on the amounts of inter-component SCA differences (Wilcoxon rank sum, p = 0.03).
Discussion
Modern statistical genomic approaches allow inexpensive, genome-wide SCA profiling of subclonal mutations. These techniques, which leverage germline haplotype information to disentangle recurrent mutations from those that may have arisen earlier, had not been previously applied to the distinct sarcoma and carcinoma components of UCS. We sought to contextualize these discoveries in the landscape of mutations discovered from targeted DNA and whole-transcriptome RNA surveys.
Consistent with recent mutation-based studies, both carcinomatous and sarcomatous components exhibited similar molecular profiles, suggesting that both components in UCS originated from a single clone. Where a mutation difference existed within components of a single patient, the sarcoma component harbored the additional mutation (a highly recurrent PIK3CA E542K mutation), hinting that in this case, the sarcoma may have evolved from the carcinoma. Where inter-component heterogeneity based on SCA profiling exists, quantification of genomic differences may provide insights into the progression of UCS and may be clinically informative. Our results imply that higher degrees of SCA heterogeneity reflect more advanced disease.
In addition, gene expression classification of EMT-status may provide similar insights into tumor progression. Of importance, we observed that gene expression based classification of EMT status of the carcinomatous and sarcomatous components are often discrepant; thus, in a mixed-cell population, profiling of the UCS tumor in bulk may fail to identify an EMT-like status, indicating that component-based profiling may be required in such a scenario.
There is no widely accepted grading system for UCS. Clinical staging is the most important prognostic factor for patients with UCS and is used to guide treatment decisions. Treatment approaches are based on the extent of the primary tumor and whether distant metastatic disease (stage IV) is documented. Surgery is recommended for patients with no evidence of metastatic disease, often combined with systemic chemotherapy. The treatment of women with metastatic UCS is essentially palliative, although surgery may be performed to relieve local symptoms if considered beneficial; chemotherapy is often employed but has only limited efficacy [23]. For women with extrauterine disease limited to the peritoneum, surgical cytoreduction is recommended but local recurrence is common. Currently, patient evaluations do not consider molecular features of UCS such as EMT status, although it is well accepted in cancer research that EMT status is a measure of metastatic potential. Larger studies would be required to assess a relationship with outcomes and thus any future utility of this as a clinical marker.
In summary, our study separately profiled of the carcinomatous and sarcomatous components of UCS separately and quantified the inter-component heterogeneity and suggests that this comparison be clinically informative. Our data strongly support a model of the carcinomatous and sarcomatous components arising from a common precursor or perhaps one of the components arising from the other at late stage. The size of our gene panel does not allow a more detailed dendrogram analysis. That there is some divergence of mutational and SCA profile suggests that the two components remain separate and do not transdifferentiate after divergence. The combination of DNA somatic mutation, gene expression, and allelic imbalance data examined here contribute to our collective expectations regarding the heterogeneous nature of UCS.
Supplementary Material
Highlights.
UCS genomic profiling of distinct carcinoma and sarcoma components reveals deep similarities and implies monoclonal origin.
An EMT-like gene expression signature was observed in samples from advanced stage UCS patients.
Higher levels of genomic inter-component heterogeneity was associated with greater predicted metastatic potential.
Component-specific profiling may provide a means of refining clinical staging for UCS patients.
Acknowledgements
The authors wish to acknowledge NIH grants P30 CA016672 and R01 HG005859.
Footnotes
Conflict of Interest
The authors do not declare any conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.George EM, Herzog TJ, Neugut AI, Lu YS, Burke WM, Lewin SN, Hershman DL, Wright JD: Carcinosarcoma of the ovary: natural history, patterns of treatment, and outcome. Gynecol Oncol 2013, 131:42–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arend R, Doneza JA, Wright JD: Uterine carcinosarcoma. Curr Opin Oncol 2011, 23:531–536. [DOI] [PubMed] [Google Scholar]
- 3.Gonzalez Bosquet J, Terstriep SA, Cliby WA, Brown-Jones M, Kaur JS, Podratz KC, Keeney GL: The impact of multi-modal therapy on survival for uterine carcinosarcomas. Gynecol Oncol 2010, 116:419–423. [DOI] [PubMed] [Google Scholar]
- 4.Page BR, Pappas L, Cooke EW, Gaffney DK: Does the FIGO 2009 endometrial cancer staging system more accurately correlate with clinical outcome in different histologies? Revised staging, endometrial cancer, histology. Int J Gynecol Cancer 2012, 22:593–598. [DOI] [PubMed] [Google Scholar]
- 5.Kanthan R, Senger JL: Uterine carcinosarcomas (malignant mixed mullerian tumours): a review with special emphasis on the controversies in management. Obstet Gynecol Int 2011, 2011:470795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kernochan LE, Garcia RL: Carcinosarcomas (malignant mixed Mullerian tumor) of the uterus: advances in elucidation of biologic and clinical characteristics. J Natl Compr Canc Netw 2009, 7:550–556; quiz 557. [DOI] [PubMed] [Google Scholar]
- 7.Wada H, Enomoto T, Fujita M, Yoshino K, Nakashima R, Kurachi H, Haba T, Wakasa K, Shroyer KR, Tsujimoto M, et al. : Molecular evidence that most but not all carcinosarcomas of the uterus are combination tumors. Cancer Res 1997, 57:5379–5385. [PubMed] [Google Scholar]
- 8.Abeln EC, Smit VT, Wessels JW, de Leeuw WJ, Cornelisse CJ, Fleuren GJ: Molecular genetic evidence for the conversion hypothesis of the origin of malignant mixed mullerian tumours. J Pathol 1997, 183:424–431. [DOI] [PubMed] [Google Scholar]
- 9.Fujii H, Yoshida M, Gong ZX, Matsumoto T, Hamano Y, Fukunaga M, Hruban RH, Gabrielson E, Shirai T: Frequent genetic heterogeneity in the clonal evolution of gynecological carcinosarcoma and its influence on phenotypic diversity. Cancer Res 2000, 60:114–120. [PubMed] [Google Scholar]
- 10.Jin Z, Ogata S, Tamura G, Katayama Y, Fukase M, Yajima M, Motoyama T: Carcinosarcomas (malignant mullerian mixed tumors) of the uterus and ovary: a genetic study with special reference to histogenesis. Int J Gynecol Pathol 2003, 22:368–373. [DOI] [PubMed] [Google Scholar]
- 11.Schulten HJ, Gunawan B, Enders C, Donhuijsen K, Emons G, Fuzesi L: Overrepresentation of 8q in carcinosarcomas and endometrial adenocarcinomas. Am J Clin Pathol 2004, 122:546–551. [DOI] [PubMed] [Google Scholar]
- 12.McCluggage WG: Uterine carcinosarcomas (malignant mixed Mullerian tumors) are metaplastic carcinomas. Int J Gynecol Cancer 2002, 12:687–690. [DOI] [PubMed] [Google Scholar]
- 13.McConechy MK, Hoang LN, Chui MH, Senz J, Yang W, Rozenberg N, Mackenzie R, McAlpine JN, Huntsman DG, Clarke BA, et al. : In-depth molecular profiling of the biphasic components of uterine carcinosarcomas. J Pathol Clin Res 2015, 1:173–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao S, Bellone S, Lopez S, Thakral D, Schwab C, English DP, Black J, Cocco E, Choi J, Zammataro L, et al. : Mutational landscape of uterine and ovarian carcinosarcomas implicates histone genes in epithelial-mesenchymal transition. Proc Natl Acad Sci U S A 2016, 113:12238–12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cherniack AD, Shen H, Walter V, Stewart C, Murray BA, Bowlby R, Hu X, Ling S, Soslow RA, Broaddus RR, et al. : Integrated Molecular Characterization of Uterine Carcinosarcoma. Cancer Cell 2017, 31:411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP: Integrative genomics viewer. Nat Biotechnol 2011, 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR: STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li B, Dewey CN: RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011, 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vattathil S, Scheet P: Haplotype-based profiling of subtle allelic imbalance with SNP arrays. Genome Res 2013, 23:152–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR: MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol 2010, 34:816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pang A, Carbini M, Moreira AL, Maki RG: Carcinosarcomas and Related Cancers: Tumors Caught in the Act of Epithelial-Mesenchymal Transition. J Clin Oncol 2018, 36:210–216. [DOI] [PubMed] [Google Scholar]
- 22.Jones S, Stransky N, McCord CL, Cerami E, Lagowski J, Kelly D, Angiuoli SV, Sausen M, Kann L, Shukla M, et al. : Genomic analyses of gynaecologic carcinosarcomas reveal frequent mutations in chromatin remodelling genes. Nat Commun 2014, 5:5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berton-Rigaud D, Devouassoux-Shisheboran M, Ledermann JA, Leitao MM, Powell MA, Poveda A, Beale P, Glasspool RM, Creutzberg CL, Harter P, et al. : Gynecologic Cancer InterGroup (GCIG) consensus review for uterine and ovarian carcinosarcoma. Int J Gynecol Cancer 2014, 24:S55–60. [DOI] [PubMed] [Google Scholar]
- 24. Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BM, Haag JD, Gould MN, Stewart RM, Kendziorski C: EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 2013, 29:1035–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.