Abstract
Early-stage estrogen receptor-positive (ER+) breast cancer (BCa) is the most common type of BCa in the United States. One critical question with these tumors is identifying which patients will receive added benefit from adjuvant chemotherapy. Nuclear pleomorphism (variance in nuclear shape and morphology) is an important constituent of breast grading schemes, and in ER+ cases, the grade is highly correlated with disease outcome. This study aimed to investigate whether quantitative computer-extracted image features of nuclear shape and orientation on digitized images of hematoxylin-stained and eosinstained tissue of lymph node-negative (LN−), ER+ BCa could help stratify patients into discrete (<10 years short-term vs. >10 years long-term survival) outcome groups independent of standard clinical and pathological parameters. We considered a tissue microarray (TMA) cohort of 276 ER+, LN− patients comprising 150 patients with long-term and 126 patients with short-term overall survival, wherein 177 randomly chosen cases formed the modeling set, and 99 remaining cases the test set. Segmentation of individual nuclei was performed using multiresolution watershed; subsequently, 615 features relating to nuclear shape/texture and orientation disorder were extracted from each TMA spot. The Wilcoxon’s rank-sum test identified the 15 most prognostic quantitative histomorphometric features within the modeling set. These features were then subsequently combined via a linear discriminant analysis classifier and evaluated on the test set to assign a probability of long-term vs. short-term disease-specific survival. In univariate survival analysis, patients identified by the image classifier as high risk had significantly poorer survival outcome: hazard ratio (95% confident interval) = 2.91(1.23–6.92), p = 0.02786. Multivariate analysis controlling for T-stage, histology grade, and nuclear grade showed the classifier to be independently predictive of poorer survival: hazard ratio (95% confident interval) = 3.17(0.33–30.46), p = 0.01039. Our results suggest that quantitative histomorphometric features of nuclear shape and orientation are strongly and independently predictive of patient survival in ER+, LN− BCa.
Introduction
Breast cancer is the most frequently diagnosed cancer in women and is also the second leading cause of cancer related deaths in women in the United States. Hormone receptor-positive breast cancers (estrogen receptor-positive (ER+) and/or progesterone receptor (PR+)) make up almost 70–75% of invasive breast cancers [1]. In these cases, axillary lymph node (LN) status is the most important prognostic predictor of survival [2]. While it is standard clinical practice to offer adjuvant therapy to patients who have evidence of metastatic disease in axillary lymph nodes, clinical trials have demonstrated that LN− patients may benefit as well [3]. However, the overall effect is small and likely only a small proportion of ER+, LN-negative (LN−) patients have measurable benefit from adjuvant chemotherapy. Specific prognosis and treatment options (choice of hormonal therapy alone or hormonal + chemotherapy) are typically determined by molecular assays such as Oncotype DX [4], in which measurement of mRNA expression of a set of genes, dominated by estrogen signaling genes and proliferation genes, can identify tumors likely to do well with hormonal therapy alone and have no further benefit from chemotherapy. However, these assays tend to be expensive and tissue destructive. Interestingly, histological tumor grade has also been shown to be correlated with disease recurrence and patient outcome in LN−, ER+ breast cancer [5]. Unfortunately, breast cancer grading is subjective and hence suffers from low inter-observer concordance [6, 7], especially with respect to grade 2 tumors.
With the digitization of pathology slides, there has been substantial interest from many groups in developing and applying histomorphometric image analysis approaches for quantitative characterization and classification of tissue images of cancers [8]. A number of these approaches have focused on breast cancer grading, risk stratification, and outcome prediction. Basavanhally et al. [9] showed that the global arrangement of nuclei, as quantified by graph features, is useful in stratifying breast cancer patients into low and high risk of recurrence groups. Tambasco et al. [10] reported that the morphologic complexity of nuclear architecture in the epithelium was predictive of disease-specific survival for patients with invasive breast cancer. Beck et al. [11] showed that nuclear morphologic features within the stroma was significantly associated with survival in breast cancer. Yuan and colleagues [12] showed that quantitative measurements of the extent and density of lymphocytic infiltration was predictive of risk of recurrence following endocrine therapy in ER+ breast cancers. More recently, Romo-Bucheli et al. [13, 14] showed that quantitative estimation of mitotic activity and tubular formation in ER+ breast cancer histology images via machine learning and image analysis approaches was strongly correlated with the corresponding Oncotype DX risk categories.
Nuclear pleomorphism, a term employed in cancer grading, refers to the degree of variability in shape, size, and morphology of cell nuclei. For example, Lu et al. [15] found that the local cellular diversity in terms of nuclear shape, size, and texture are prognostic in the context of oral cavity squamous cell carcinomas. In addition to variability in size and shape, some researchers have observed differences in nuclear polarity between more and less aggressive breast cancers. For instance, in certain more aggressive cancers, the nuclei tend to be vesicular, often have prominent nucleoli, and occasionally manifest in the form of large and aberrant presentations. It stands to reason then that computer-extracted measurements relating to differential nuclear polarity may provide a measurable surrogate for the underlying biological aggressiveness of a tumor. Dalla et al. [16] showed that nuclear polarity was helpful in distinguishing grades of bladder cancer. Kararizou et al. [17] found that the mean value of nuclear orientation was associated with the degree of malignancy of human gliomas. Noy et al. [18] reported that the nuclear orientation was highly correlated to the recurrence of meningiomas. In prostate cancer, Lee et al. [19] found that the extent of disorder in nuclear orientation was strongly associated with likelihood of biochemical recurrence following surgery. In the context of breast cancer diagnosis, Herrera-Espiñeira et al. [20] found that computerized measurements of nuclear shape, orientation, and texture disorder could discriminate benign and malignant breast lesions with an accuracy of over 95% from hematoxylin and eosin (H&E) images alone.
In this work, we seek to evaluate whether computer extracted measurement of nuclear orientation combined with features relating to nuclear shape and texture are predictive of aggressiveness and prognosis in early-stage ER+, LN− breast cancers. A set of 177 patients was used as the modeling set and their corresponding H&E stained tissue microarray (TMA) images were employed for feature discovery and classifier construction. Following the application of a nuclear segmentation algorithms on the TMA images, a total of 615 nuclear shape, texture, and nuclear orientation features were extracted from the regions corresponding to the segmented nuclei. The top discriminative features that were most predictive of overall survival were identified via a five-fold crossvalidation based feature selection method on the modeling set. These features were then used to train an image-based machine learning classifier of overall survival. We then applied the image classifier to an independent test set (n = 99) to predict overall survival. Finally, we analyzed if the predicted labels generated by the image classifier were independently correlated with overall patient outcome.
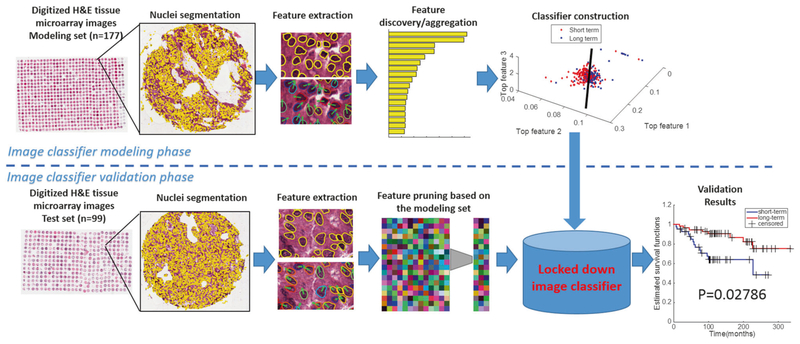
Figure 1 illustrates the overall workflow for the construction and evaluation of the image-based classifier.
Fig. 1.
Overall workflow of construction and validation of image classifier
Materials and methods
Patient selection
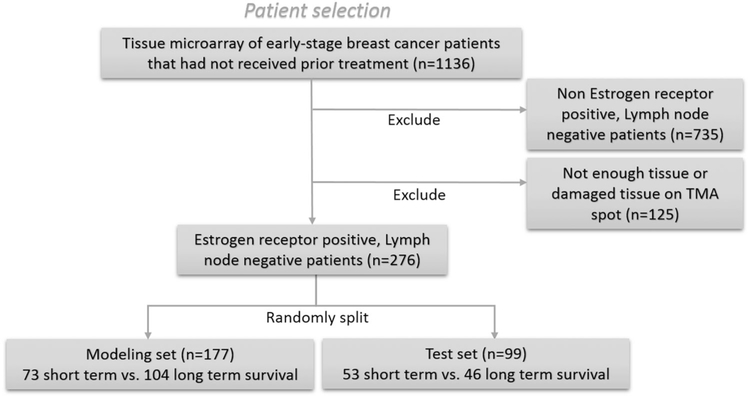
With approval from the Human Investigation Committee at Yale University (protocol #9505008219), patients with early-stage breast cancer (diagnosed with invasive ductal carcinoma) were serially retrospectively collected from the Yale Pathology archives. With approval from the Institutional Review Board of Yale University (“Digital whole slide imaging for the detection of breast cancer and metastasis in lymph nodes”), patients with early-stage breast cancer were identified. Two 2 mm punches from each tumor were obtained to construct the array. All early-stage breast cancer patients with ER+ LN− breast cancer with a sufficient amount of tissue, that is, >60% of the area, available in the digitized TMA were selected for our study. These inclusion criteria resulted in a total of 276 patients being identified with corresponding tissue images for analysis. The TMA cores were divided into two groups corresponding to patients who passed away after the hormonal therapy within 10 years (short-term survival) and those who passed away after the treatment >10 years (long-term survival). The total number of non-censored long-term survival patients was 65 out of 150. A set of 177 randomly chosen cases (73 short-term vs. 104 long-term survival) were used as a modeling set to train the classifier. The remaining 99 cases (53 short-term vs. 46 long-term survival) were used as a test set for independent validation. Table 1 summarizes the modeling set and test set. All TMA cores were digitized at ×40 magnification (0.25 μm/pixel spatial resolution) using a digital whole-slide scanner (Aperio Scanscope XT) (Fig. 2).
Table 1.
Summary of clinical and pathological features of the studied early-stage ER+, LN− breast cancer cohorts
| All data sets, N (%) | Modeling set, N (%) | Test set, N (%) | P value* | |
|---|---|---|---|---|
| No. of patients | 276 | 177 | 99 | |
| Age | 59.0±11.9 | 60.4±11.2 | 56.5±12.2 | ϒ0.0105 |
| Race | <0.0001 | |||
| White | 259 (93.8) | 175 (98.9) | 84 (84.9) | |
| Other | 17 (6.2) | 2(1.1) | 15 (15.1) | |
| Histology grade | 1 | |||
| Well/moderately differentiated | 39 (60.9) | 1 (50.0) | 38 (61.3) | |
| Poorly differentiated | 25 (39.1) | 1 (50.0) | 24 (38.7) | |
| Histology grade | ||||
| Well differentiated | 8 (12.5) | 1 (50.0) | 7 (11.3) | 0.2361 |
| Moderately/poorly differentiated | 56 (87.5) | 1 (50.0) | 55 (88.7) | |
| Nuclear grade | 0.0056 | |||
| 1/2 | 197 (89.9) | 132 (86.3) | 65 (98.5) | |
| 3 | 22 (10.1) | 21 (13.7) | 1 (1.5) | |
| Nuclear grade | 0.4091 | |||
| 1 | 60 (27.4) | 39 (25.5) | 21 (31.8) | |
| 2/3 | 159 (72.6) | 114 (74.5) | 45 (68.2) | |
| T-stage | <0.0001 | |||
| T1 | 248 (89.9) | 172 (97.2) | 76 (76.8) | |
| T2 | 28 (10.1) | 5 (2.8) | 23 (23.2) | |
| Her2 status | 0.0336 | |||
| Positive | 94 (34.4) | 69 (39.0) | 25 (26.0) | |
| Negative | 179 (65.6) | 108 (61.0) | 71 (74.0) | |
| Chemotherapy | <0.0001 | |||
| Yes | 25 (9.1) | 0 (0.0) | 25 (25.3) | |
| No | 251 (90.9) | 177 (100.0) | 74 (74.7) | |
| Survival status | 0.0588 | |||
| Long term (≥10 years) | 150 (54.4) | 104 (58.8) | 46 (46.5) | |
| Short term (<10 years) | 126 (45.6) | 73 (41.2) | 53 (54.5) |
P values were calculated from two-sided Fisher’s exact test.
P value was calculated from Welch two sample t test. P values are computed between the modeling and test set. Values in bold are statistically significant, p < 0.05
Fig. 2.
Inclusion and exclusion criteria for patient selection for the modeling and test sets
Image analysis
Nuclear detection and segmentation
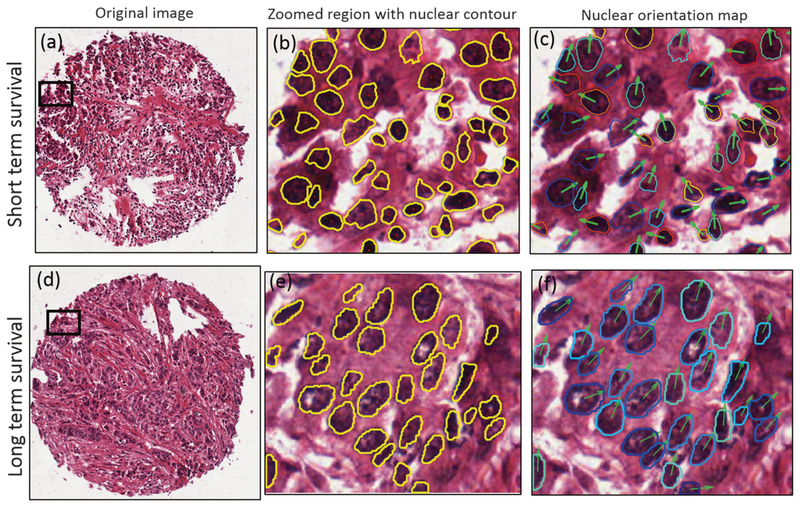
Individual nuclei were automatically segmented from the background using a watershed-based nuclear segmentation method [21–23] at ×40 magnification (0.25 μm/pixel resolution). The automated segmentation takes into account the intensity/gradient data for each nucleus so that cell boundaries can be extracted for subsequent feature extraction (Figs. 1 and 4).
Fig. 4.
Representative H&E-stained TMA spot images for a short-term survival and d long-term survival patient. b, e Zoomed region with pre-segmented nuclear contour, from which the nuclear shape and orientation measurement are extracted. c, f The nuclear orientation map, in which the nuclear contours with different colors indicate different nuclear orientations; the nuclear orientation is also demonstrated by arrows. The long-term survival patient, shown in f, tends to have uniform distribution of nuclear shape (similar in nuclear shape/size in e than that in b) and nuclear orientation (similar colors/orientation in f than that in c), compared to that of short-term survival patient
Feature extraction
Two different categories of quantitative histomorphometric features were extracted. The features are related to nuclear shape/texture and local nuclear orientation disorder as described below:
Nuclear orientation disorder [19] (13 descriptors): This class of features includes a set of 13 local nuclear orientation-based features which were extracted in an attempt to quantify the disorder of nuclear orientations in local regions [24] (Figs. 1 and 4c, f show examples).
Nuclear shape/texture (45 descriptors): This set of features were focused on quantifying nuclear morphology using different measurements including: area to perimeter ratio, smoothness of boundary, invariant moments, Fourier descriptors of boundary, area, major/minor axis of the best fit eclipse, mean intensity, intensity range, and so on. These nuclear shape/texture features are intended to capture the disorder of nuclear shape and texture, attributes that have been previously shown to be implicated in tumor aggressiveness [25].
First-order summary statistic descriptors (mean, median, standard deviation, skewness, and kurtosis) were calculated for each TMA spot for both nuclear shape/texture, and nuclear orientation (with six configurations of local cell clusters) features, resulting in a total of 615 features for each spot. Additional mathematical and technical details pertaining to the approach are described in the Supplementary material section.
Feature selection
Three different feature selection methods, (1) minimum redundancy maximum relevance [26], (2) Wilcoxon’s rank-sum test, and (3) Random Forest [27], were used to identify the best features that maximally distinguished the two classes (short-term and long-term survival) within the modeling set. The approaches were implemented in conjunction with a five-fold crossvalidation scheme and run over 100 iterations. The most frequently selected features were identified and were quantitatively evaluated using box and whisker plots to compare feature expression between the short-term and long-term survival patients. We limited the number of features for inclusion in the machine learning classifier to 15 in order to avoid model overfitting and the curse of dimensionality problem, an issue relating to the generalizability of the classifier when there are too many features with respect to the number of training exemplars.
Classifier construction
Four different machine learning classifiers, linear discriminant analysis, quadratic discriminant analysis, support vector machine, and Random Forest classifier were implemented in conjunction with the top selected features provided by the feature selection methods. The machine learning classifiers were evaluated across 100 iterations of five-fold cross-validation within the modeling set. We then identified the top-performing combination of feature selection scheme and machine learning classifier (from among a total of 12 different combinations) based on the area under the receiver operating characteristic curve (AUC) obtained across all images within the modeling set. This classifier was then locked down as the one to be validated on the test set.
Survival analysis
The survival analysis was performed on the test dataset only. Each test set image was assigned a probability of survival by the locked down machine learning classifier. For AUC computation, a threshold was then applied to this probability at different levels from 0 to 1 with an increment of 0.01. Thus, each test instance was assigned to either the long-term or short-term survival categories for each threshold. The predicted labels for each classifier were then compared to the ground truth labels (actual follow-up outcome information) to evaluate the performance of the classifier.
Associations between the binary image classifier results and the other categorical clinical and pathologic variables were determined by two-sided Fisher’s exact tests. Welch’s unequal variance t test was used for age. Overall survival was defined as the time interval between the date of hormonal therapy and the date of death. The Kaplan–Meier product-limit method was used to estimate empirical survival probabilities as illustrated by Kaplan–Meier curves. Log-rank tests were applied to examine survival differences, indicating the significance of a categorical variable being prognostic for a survival endpoint. Multivariate Cox proportional hazard models were also used to investigate the independent prognostic ability of variables after accounting for the other clinical and pathologic variables. Hazard ratios, associated 95% confidence intervals, and P values were reported. All tests were two sided, with the significance level set at 0.05.
Results
Clinicopathological features of the patient cohorts
Clinical and pathologic features of the modeling and test sets are provided in Table 1. Patients were primarily in their late 50s, and the majority of patients (>84%) were white. Around 9% were treated with chemotherapy and about 10% of the tumors were graded higher than T1. Approximately 60% of the tumors were well or moderately differentiated. The modeling set contains 73 short-term survival patients (41.2%) and the test set contains 53 short-term survival patients (54.5%), with a threshold of 10 years applied on the patient followed up date until death.
Discriminative features
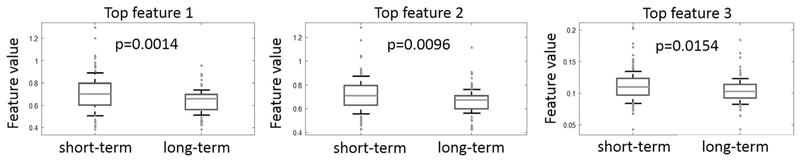
In the modeling set, the nuclear orientation feature dominates the top 15 discriminative features that were used to construct the image classifier (12 out of 15 features were nuclear orientation-related features, for more details refer to Supplementary materials Table S1). The nuclear orientation features aim to measure the disorder in the orientation of adjacent nuclei. Figure 3 shows the top three discriminative features identified within the modeling set. These top three features are all related to the nuclear orientations, which are range of tensor information_measure1 with two different cell cluster graphs density (a = 0.42 and a = 0.43), and standard deviation of tensor information_measure1_a = 0.42. These features reflecting the degree of disorder in terms of local nuclear orientation.
Fig. 3.
Top three discriminative features identified within the modeling set (1 = Orientation: range tensor information_measure1_a = 0.42; 2 = Orientation:range tensor information_measure1_a = 0.43; 3 = Orientation:standard deviation tensor information_measure1_a = 0.42). They are entropy-related measurement capering the degree of disorder of nuclear orientation locally (for more feature explanation please refer to the Supplementary Materials Appendix). Higher feature values reflect more disorder in orientations of adjacent nuclei. The p values were calculated via the paired Student’s t test
The nuclear segmentation and corresponding nuclear orientation feature maps are shown in Fig. 4, for representative long-term and short-term survival cases. The first to the fourth columns show H&E-stained images of tumors from short-term and long-term survival patient groups, the segmented nuclear contours, the nuclear orientation feature maps, and zoomed regions, respectively. The third column in Fig. 4 illustrates the differences of nuclear orientation feature map between short-term and long-term survival patients. For the short-term survival patients, there is a greater variation in nuclear orientation, as evidenced by the arrows and pre-segmented nuclear contour colors (Fig. 4c, in which different nuclear contour colors represent different nuclear orientations). By contrast, for the patients in the long-term survival group, the nuclear orientations appear to be more uniform locally (Fig. 4f).
Classifier evaluation
The performance of the 12 combinations of feature selection and classifier schemes, with respect to four different feature families, in terms of classification performance on the modeling set are summarized in Table 2. Note that since the combination of Wilcoxon’s rank-sum test and linear discriminant analysis yielded the best performance in distinguishing two patient groups on the modeling set (AUC =0.67 ± 0.03), we settled on a combination of these two analytical tools for constructing the locked down image classifier. The LDA classifier yielded an AUC of 0.64 in differentiating the long-term vs. short-term survival patients on the independent test set. By applying a threshold of 0.5 to the output of the image classifier, a corresponding accuracy = 65%, specificity = 73.91%, and sensitivity =57.41% was obtained.
Table 2.
Performance of four different classifiers with three different feature selection methods in the modeling set
| Classifier | Feature selection | AUC | Accuracy | Specificity | Sensitivity |
|---|---|---|---|---|---|
| LDA | WRST | 0.67 ±0.03 | 0.70 ±0.02 | 0.81 ±0.11 | 0.53 ±0.07 |
| MRMR | 0.64 ±0.03 | 0.67 ± 0.02 | 0.76 ±0.11 | 0.55 ± 0.07 | |
| RF | 0.63 ± 0.03 | 0.66 ±0.02 | 0.80±0.15 | 0.45 ± 0.09 | |
| QDA | WRST | 0.54 ±0.04 | 0.63 ± 0.02 | 0.88 ±0.14 | 0.27 ± 0.08 |
| MRMR | 0.56 ±0.03 | 0.64 ±0.02 | 0.86 ±0.11 | 0.31 ±0.07 | |
| RF | 0.54 ±0.03 | 0.62 ±0.02 | 0.89 ±0.13 | 0.24 ± 0.08 | |
| RF | WRST | 0.63 ± 0.03 | 0.61 ±0.03 | 0.73 ±0.04 | 0.45 ± 0.04 |
| MRMR | 0.66 ±0.04 | 0.63 ± 0.03 | 0.77 ± 0.06 | 0.44 ±0.04 | |
| RF | 0.63 ± 0.03 | 0.62 ±0.03 | 0.72 ±0.05 | 0.46 ± 0.04 | |
| SVM | WRST | 0.67 ±0.02 | 0.64 ±0.02 | 0.85 ±0.06 | 0.33 ±0.03 |
| MRMR | 0.66 ±0.04 | 0.63 ± 0.03 | 0.86 ±0.07 | 0.28 ±0.04 | |
| RF | 0.67 ±0.03 | 0.61 ±0.02 | 0.83 ±0.05 | 0.30 ±0.04 |
AUC area under the receptor operating curve, LDA/QDA linear/quadratic discriminant analysis, SVM support vector machine, RF Random Forest classifier, MRMR minimum redundancy, maximum relevance feature selection method, WRST Wilcoxon’s rank-sum test, RF Random Forest feature selection method
The best performance in each metric/column is shown in bold
Survival analysis
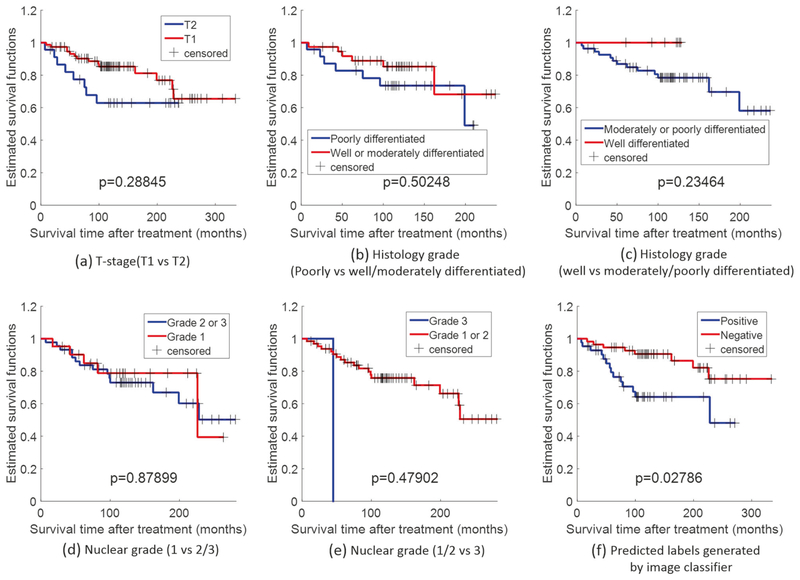
Patients were followed for an average of 129 months (range 7–334 months). Table 3 provides results of univariate logrank survival analysis for the major clinical/pathologic variables and for the image classifiers on the test set. Positive patients obtained by the image classifier (i.e., patients identified by the image classifier to have poor prognosis) had statistically significantly worse survival in univariate analysis (Table 3). As seen in Table 3, among the other major clinical and pathological variables, the binary label generated by the image classifier was a predictive feature that significantly correlated to the survival outcome with follow-up (hazard ratio = 2.91, 95% confident interval = 2.91(1.23–6.92), p = 0.02786). Kaplan–Meier survival curves for the image classifier are presented in Fig. 5. Multivariate survival analysis controlling for the effects of the major prognostic variables is presented in Table 4. The predicted labels obtained by the image classifier still correlated with worse survival independent of the other variables (hazard ratio = 3.17, 95% confident interval =0.33–30.46, p = 0. 01039).
Table 3.
Univariate log-rank analysis for overall survival on the test set
| Variable | Disease-specific survival, HR (95% CI) | P value |
|---|---|---|
| Age | ||
| ≤55 vs. >55 | 1.40 (0.60–3.29) | 0.57664 |
| Race | ||
| Nonwhite vs. white | 0.28 (0.08–0.98) | 0.09464 |
| T-stage | ||
| T1 vs. T2 | 1.94 (0.72–5.20) | 0.28845 |
| Histology grade | ||
| Well/moderately differentiated vs. poorly differentiated | 1.74 (0.56–5.39) | 0.50248 |
| Histology grade | ||
| Well differentiated vs. moderately/poorly differentiated | 3.50 (0.74–16.58) | 0.23464 |
| Nuclear grade | ||
| 1 vs. 2/3 | 1.23 (0.46–3.28) | 0.87899 |
| 1/2 vs. 3 | 74.02 (0.80–6854.43) | 0.47902 |
| Tubule formation | ||
| 1 vs. 2/3 | 1.82 (0.66–5.03) | 0.36692 |
| 1/2 vs. 3 | 1.68 (0.37–7.65) | 0.77774 |
| Nuclear pleomorphism | ||
| 1 vs. 2/3 | 2.29 (0.16–33.57) | 0.93666 |
| 1/2 vs. 3 | 0.30 (0.06–1.68) | 0.35288 |
| Mitotic count | ||
| 1 vs. 2/3 | 74.02 (0.80–6854.43) | 0.47902 |
| Her2 status | ||
| Positive vs. negative | 1.72 (0.65–4.52) | 0.39566 |
| Chemotherapy | ||
| Yes vs. no | 0.55 (0.22–1.40) | 0.30872 |
| Image classifier | ||
| Positive vs. negative | 2.91 (1.23–6.92) | 0.02786 |
CI confidence interval, HR Mantel–Haenszel hazard ratio
Values in bold are statistically significant, p < 0.05
Fig. 5.
Kaplan–Meier survival curve of a T-stage (T1 vs. T2), b histology grade (poorly vs. well/moderately differentiated), c histology grade (well vs. moderately/poorly differentiated), d nuclear grade (1 vs. 2/3), e nuclear grade (1/2 vs. 3), and f predicted labels generated bythe image classifier trained on the test set for predicting overall survival
Table 4.
Multivariate Cox proportional hazard model analysis controlling for major prognostic variables on the test set
| Variable | P value | Hazard ratio (95% Confidence interval) |
|---|---|---|
| T-stage | ||
| T1 vs. T2 | 0.18069 | 1.84 (0.56–6.13) |
| Histology grade | ||
| Well differentiated vs. moderately/poorly differentiated | 0.31457 | 1.58 (0.64–3.90) |
| Nuclear grade | ||
| 1 vs. 2/3 | 0.59577 | 1.32 (0.77–2.27) |
| Image classifier | ||
| Positive vs. negative | 0.01039 | 3.17 (0.33–30.46) |
Discussion
One of the most difficult current dilemmas for clinicians surrounds the identification of patients who will benefit most from chemotherapy in the ER+, LN− setting. There are significant risks associated with chemotherapy, and the need to identify patients who are unlikely to benefit from it is paramount. Standard clinical and histopathologic features alone cannot reliably answer this question. Currently, molecular tests such as Oncotype DX, Mammaprint, and PAM50 (and many others) are routinely performed in the clinic. These tests are all tissue destructive and costly. However, utilization of quantitative histomorphology may represent a reliable alternative to these molecular methods. Given the histologic features of any tumor represent the phenotype of all of the genomic alterations that occur, it is reasonable to infer that the extraction of features by quantitative histomorphology may shed important light upon the underlying biological pathways that drive tumorigenesis and can, in turn, predict the most appropriate therapy.
In this work, we evaluated digitized H&E-stained histology images corresponding to ER+, LN− breast cancers patients, and creating a prognostic classifier for predicting patient overall survival. Specifically, an image analysis-based classifier was constructed that utilizes computer-extracted quantitative image features relating to the nuclear orientation, shape, and texture from digitized H&E-stained slides with breast cancer microarray tumor sections. Our aim was to evaluate the correlation of nuclear shape and orientation disorder with prognosis in early-stage ER+, LN− breast cancers.
In the modeling set, features relating to heterogeneity of nuclear orientation using second-order statistics were found to be overexpressed in short-term survival patients (worse prognosis tumors) compared to the ones with better prognosis, that is, long-term survival patients. This is intuitive since in the aggressive tumors, tissue structures become poorly differentiated as a result of rapid disorganized cell growth, encouraging the nuclei in these structures to form highly irregular organizational patterns. While nuclear orientation and shape disorder has been shown to be important for urothelial cancers including bladder and prostate [16, 19, 28], our study is the first to quantitatively assess the role of disorder in nuclear polarity in predicting short-term vs. long-term survival in ER+, LN− breast cancers.
In addition to nuclear orientation disorder, we also looked at measurements relating to nuclear shape and texture (5 first-order statistics on 45 features = 225). The three most prognostic features were the intensity range of the nuclear surface, standard deviation of the Fractal Dimension [29] of the nuclear boundary, and range of nuclear solidity. The discriminating nuclear shape and texture features also suggest a trend that the higher range/variance of the nuclear shape and texture are associated with a worse prognostic outcome (an explanation of the discriminating features along with corresponding boxplots is shown in the Supplementary materials appendix). Variance in nuclear shape and orientation are hallmarks of nuclear pleomorphism in the context of breast cancer and computer-extracted features of nuclear shape and texture appear to be capturing this aspect of breast cancer grading. These findings corroborate the studies of Basavanhally et al. [30] and Tambasco et al. [10] who both reported that global nuclear architecture features were correlated to cancer grade and patient survival. Our findings are also in alignment with those of Beck et al. [11], who found that appearance and shape measurements of stromal nuclei were important for predicting survival in breast cancer patients, independent of molecular subtype.
Our study did have its limitations since it leveraged TMAs, which notably contain a much smaller snapshot of the overall tumor characteristics as compared to whole-slide biopsies. In the future, we intend to extend this approach to whole-slide images as they may provide a more comprehensive view of the tumors. Also, since the patient image data originated from a single facility as opposed to multiple facilities, typical confounding variables such as tissue staining quality differences and differing patient population characteristics were not considered. This batch effect could potentially affect the image and feature analysis steps and will need to be rigorously investigated in future work [31]. Another limitation was that we did not explicitly distinguish cancer cells from tumor-infiltrating lymphocytes (TILs). The partition of cells into cancer and TILs could potentially have enabled the development of an even more sophisticated classifier by considering the individual contributions of the tumor as well as its habitat and associated immune response [32].
In summary, this study models and validates the independent prognostic value of a computerized H&E image based classifier of nuclear morphology in a cohort of 276 early-stage ER+, LN− breast cancer patients. The separation of early-stage ER+ and LN− breast cancer into short-term and long-term survival cohorts holds special prognostic significance as prior knowledge of the risk of immediate recurrence (<10 years) will help in treatment planning. We believe this is a critical first step in being able to develop a predictive image-based classifier to identify which LN−, ER+ breast cancer patients stand to gain additional benefit from adjuvant chemotherapy.
Supplementary Material
Acknowledgements
We thank Autumn Watson BA for her expert technical assistance with the TMA construction. We also thank Donna M Posey for her wonderful assistance with clerical support and spreadsheet data management for the various aspects of this study. We also thank Tyler Smallman for his assistance with the digital image scanning and file management. Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award numbers 1U24CA199374–01, R01CA202752–01A1, R01CA208236–01A1, R21CA179327–01, R21CA195152–01, the National Institute of Diabetes and Digestive and Kidney Diseases under award number R01DK098503–02, National Center for Research Resources under award number 1 C06 RR12463–01, the DOD Prostate Cancer Synergistic Idea Development Award (PC120857); the DOD Lung Cancer Idea Development New Investigator Award (LC130463), the DOD Prostate Cancer Idea Development Award; the DOD Peer Reviewed Cancer Research Program W81XWH-16–1–0329, the Case Comprehensive Cancer Center Pilot Grant, VelaSano Grant from the Cleveland Clinic, the Wallace H Coulter Foundation Program in the Department of Biomedical Engineering at Case Western Reserve University. Dr Lu is partially supported by the National Natural Science Foundation of China (Grant Nos. 61401263, 61672333, 61672334, 61673251, 61502615, 61573232, 61501287). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Compliance with ethical standards
Conflict of interest Dr Madabhushi is an equity holder in Elucid Bioimaging and in Inspirata Inc. He is also a scientific advisory consultant for Inspirata Inc. and also sits on its scientific advisory board. He is also an equity holder in Inspirata Inc. Additionally his technology has been licensed to Elucid Bioimaging and Inspirata Inc. He is also involved in an NIH U24 grant with PathCore Inc. Dr. Ganesan is an equity holder in Inspirata Inc., is on its scientific advisory board, and holds patents licenced to Inspirata Inc. He has served as a consultant for Novartis and Roche. Dr Rimm is a consultant or advisor to Astra Zeneca, Agendia, Agilent, Bethyl Labs, Biocept, BMS, Cepheid, Cell Signaling Technology, Cepheid, Merck, OptraScan, Perkin Elmer, and Ultivue and Astra Zeneca, Cepheid, Navigate/Novartis, Gilead Sciences, Ultivue, and Perkin Elmer fund research in his lab. Dr. Lu, Dr. Romo-Bucheli, Dr. Janowczyk, Dr. Gilmore, and Mr. Wang declare no competing financial interests.
Electronic supplementary material The online version of this article (https://doi.org/10.1038/s41374-018-0095-7) contains supplementary material, which is available to authorized users.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. [DOI] [PubMed] [Google Scholar]
- 2.Tsai J, Bertoni D, Hernandez-Boussard T, et al. Lymph Node ratio analysis after neoadjuvant chemotherapy is prognostic in hormone receptor-positive and triple-negative breast cancer. Ann Surg Oncol. 2016;23:3310–6. [DOI] [PubMed] [Google Scholar]
- 3.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG). Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet. 2011;378:771–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han HS, Magliocco AM. Molecular testing and the pathologist’s role in clinical trials of breast cancer. Clin Breast Cancer. 2016;16:166–79. [DOI] [PubMed] [Google Scholar]
- 5.Khoury T, Huang X, Chen X, et al. Comprehensive histologic scoring to maximize the predictability of pathology-generated equation of breast cancer Oncotype DX Recurrence Score. Appl Immunohistochem Mol Morphol. 2016;24:703–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elmore JG, Longton GM, Carney PA, et al. Diagnostic concordance among pathologists interpreting breast biopsy specimens. JAMA. 2015;313:1122–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyer JS,Alvarez C, et al. , for the Cooperative Breast Cancer Tissue Resource. Breast carcinoma malignancy grading by Bloom–Richardson system vs proliferation index: reproducibility of grade and advantages of proliferation index. Mod Pathol. 2005;18:1067–78. [DOI] [PubMed] [Google Scholar]
- 8.Gurcan MN, Boucheron LE, Can A, et al. Histopathological image analysis: a review. IEEE Rev Biomed Eng. 2009;2:147–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Basavanhally A, Jun Xu, Madabhushi A, et al. Computer-aided prognosis of ER+ breast cancer histopathology and correlating survival outcome with Oncotype DX assay. IEEE. 2009;851–854. 10.1109/ISBI.2009.5193186. [DOI] [Google Scholar]
- 10.Tambasco M, Eliasziw M, Magliocco AM. Morphologic complexity of epithelial architecture for predicting invasive breast cancer survival. J Transl Med. 2010;8:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beck AH, Sangoi AR, Leung S, et al. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci Transl Med. 2011;3:108ra113. [DOI] [PubMed] [Google Scholar]
- 12.Heindl A, Sestak I, Naidoo K, et al. Relevance of spatial heterogeneity of immune infiltration for predicting risk of recurrence after endocrine therapy of ER+ breast cancer. J Natl Cancer Inst. 2018;110 10.1093/jnci/djx137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romo-Bucheli D, Janowczyk A, Gilmore H, et al. A deep learning based strategy for identifying and associating mitotic activity with gene expression derived risk categories in estrogen receptor positive breast cancers: Automated Mitotic Index Predicts Breast Cancer Risk. Cytometry A. 2017. 10.1002/cyto.a.23065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Romo-Bucheli D, Janowczyk A, Gilmore H, et al. Automated tubule nuclei quantification and correlation with Oncotype DX risk categories in ER+ breast cancer whole slide images. Sci Rep. 2016;6:32706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu C, Lewis JS, Dupont WD, et al. An oral cavity squamous cell carcinoma quantitative histomorphometric-based image classifier of nuclear morphology can risk stratify patients for disease specific survival. Mod Pathol. 2017. 10.1038/modpathol.2017.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalla Palma P, Mauri MF, Forti S, et al. Grading in superficial papillary bladder carcinoma, with an emphasis on nuclear orientation. Anal Quant Cytol Histol. 1996;18:305–8. [PubMed] [Google Scholar]
- 17.Kararizou EG, Davaki PT, Davou RG, et al. Morphometric assessments for the prognostic evaluation of human gliomas. Acta Cytol. 2011;55:203–4. [DOI] [PubMed] [Google Scholar]
- 18.Noy S, Vlodavsky E, Klorin G, et al. Computerized morphometry as an aid in distinguishing recurrent versus nonrecurrent meningiomas. Anal Quant Cytol Histol. 2011;33:141–50. [PubMed] [Google Scholar]
- 19.Lee G, Ali S, Veltri R, et al. Cell orientation entropy (COrE): predicting biochemical recurrence from prostate cancer tissue microarrays. Med Image Comput Comput Assist Interv. 2013;16: 396–403. [DOI] [PubMed] [Google Scholar]
- 20.Herrera-Espiñeira C, Marcos-Muñoz C, López-Cuervo JE. Diagnosis of breast cancer by measuring nuclear disorder using planar graphs. Anal Quant Cytopathol Histopathol. 1997;19:519–23. [PubMed] [Google Scholar]
- 21.Lu C, Mahmood M, Jha N, et al. A robust automatic nuclei segmentation technique for quantitative histopathological image analysis. Anal Quant Cytopathol Histopathol. 2012;34:296–308. [PubMed] [Google Scholar]
- 22.Veta M, van Diest PJ, Kornegoor R, et al. Automatic nuclei segmentation in H&E stained breast cancer histopathology images. PLoS ONE. 2013;8:e70221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu C, Xu H, Xu J, et al. Multi-pass adaptive voting for nuclei detection in histopathological images. Sci Rep. 2016;6:33985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ali S, Veltri R, Epstein JA, et al. Cell cluster graph for prediction of biochemical recurrence in prostate cancer patients from tissue microarrays. Proc. SPIE 2013;8676:8676–8676–11. 10.1117/12.2008695. [DOI] [Google Scholar]
- 25.Lee G, Veltri RW, Zhu G, et al. Nuclear shape and architecture in benign fields predict biochemical recurrence in prostate cancer patients following radical prostatectomy: preliminary findings. Eur Urol Focus. 2016. 10.1016/j.euf.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng H, Long F, Ding C. Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans Pattern Anal Mach Intell. 2005;27: 1226–38. [DOI] [PubMed] [Google Scholar]
- 27.Menze BH, Kelm BM, Masuch R, et al. A comparison of random forest and its Gini importance with standard chemometric methods for the feature selection and classification of spectral data. BMC Bioinform. 2009;10:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castellini P, Montironi MA, Zizzi A, et al. Recurrent papillary urothelial neoplasm of low malignant potential. Subtle architectural disorder detected by quantitative analysis in DAXX immunostained tissue sections. Hum Pathol. 2014;45:745–52. [DOI] [PubMed] [Google Scholar]
- 29.Bose P, Brockton NT, Guggisberg K, et al. Fractal analysis of nuclear histology integrates tumor and stromal features into a single prognostic factor of the oral cancer microenvironment. BMC Cancer. 2015;15 10.1186/s12885-015-1380-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Basavanhally A, Ganesan S, Feldman M, et al. Multi-field-of-view framework for distinguishing tumor grade in ER+ breast cancer from entire histopathology slides. IEEE Trans Biomed Eng. 2013;60:2089–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leo P, Lee G, Shih NNC, et al. Evaluating stability of histomorphometric features across scanner and staining variations: prostate cancer diagnosis from whole slide images. J Med Imag (Bellingham, WA). 2016;3:047502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bense RD, Sotiriou C, Piccart-Gebhart MJ, et al. Relevance of tumor-infiltrating immune cell composition and functionality for disease outcome in breast cancer. J Natl Cancer Inst. 2017;109: djw192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.