Abstract
Purpose:
To compare the prognostic accuracy of gene expression profiling (GEP) combined with PRAME status versus the clinical Tumor-Node-Metastasis (TNM) staging in patients with uveal melanoma (UM).
Design:
Retrospective cohort study.
Methods:
The study included 240 consecutive patients with UM. Tumors were assessed for GEP status (Class 1 or Class 2) using a validated 15-gene assay, and FRAME expression status using quantitative PCR. TNM staging was according to the American Joint Committee on Cancer (AJCC) 8th edition. Statistical analysis included univariate and multivariate Cox proportional hazard models. Metastasis was the primary endpoint.
Results:
GEP was Class 1 in 128 (53.3%) cases, and Class 2 in 112 (46.7%) cases. PRAME status was negative in 157 (65.4%) cases and positive in 83 (34.6%) cases. TNM was stage I in 26 (10.8%) cases, IIA in 67 (27.9%) cases, IIB in 50 (20.8%) cases, IIIA in 59 (24.6%) cases and IIIB in 38 (15.8%) cases. Metastatic disease was detected in 59 (24.6%) cases after median follow-up of 29 months (mean 42 months; range 1–195 months). Variables associated with metastasis included (in order of decreasing significance): GEP class (P=1.5 × 10−8), largest basal tumor diameter (P=2.5 × 10−6), PRAME status (P=2.6 × 10−6), and TNM stage (P=3.7 × 10−6). The prognostic accuracy of an optimized 3-category GEP/PRAME model (P = 8.6 × 10−14) was superior to an optimized TNM model (P = 1.3 × 10−5).
Conclusions:
In UM, molecular prognostic testing using GEP and FRAME provides prognostic accuracy that is superior to TNM staging.
INTRODUCTION
Uveal melanoma (UM) is the most common primary malignancy of the eye and leads to fatal metastasis in up to half of patients.1 Despite ongoing improvements in the diagnosis and management of UM, survival rates have not improved as a result of micrometastasis occurring prior to treatment of the primary tumor.2,3 Consequently, preemptive treatment of micrometastatic disease in the adjuvant setting may be required to improve the survival rate in UM. Indeed, there are an increasing number of clinical trials designed to evaluate adjuvant therapy in patients with high risk UM.4,5 However, in order to utilize adjuvant therapy most effectively, an accurate method is needed to distinguish high risk patients who may benefit from adjuvant therapy from low risk patients who do not require such therapy.
The American Joint Committee on Cancer (AJCC) Tumor-Node-Metastasis (TNM) staging system has been used for a variety of cancers to stratify patients according to metastatic risk.6 The TNM system divides solid tumor types into 4 stages based on the assumption that cancer progresses temporally from primary tumor to local invasion, regional lymphatic extension, and distant metastasis.7 However, anatomic staging systems such as the TNM are now being re-examined in light of evolving knowledge, such as new prognostic cancer biomarkers.7–9 Further, there is growing recognition that many cancers do not progress in the stepwise manner stipulated by the TNM formula, such as the lack of lymph node dissemination (the “N” component of the TNM) in UM. Additionally, the inherent complexity of the TNM methodology limits its precision, reproducibility, and ease of use in the clinical setting.10,11
As an alternative, molecular prognostic testing based on gene expression profiling (GEP) has been shown to yield superior prognostic accuracy compared to clinical, histopathologic and chromosomal features.12–15 An optimized GEP test for routine clinical use has been developed using a 15-gene array on a microfluidics quantitative PCR platform, allowing accurate analysis of very small needle biopsy samples.12 The test uses a machine learning algorithm and an annotated training set to assign tumor samples to Class 1 (low metastatic risk) versus Class 2 (high risk),16 and it is the only such test for UM to be validated in a prospective, multicenter study.12 Further, Class 1 tumors have been shown to harbor mutations in the translation elongation factor EIF1AX and the splicing factor SF3B1, whereas Class 2 tumors are strongly associated with mutations in the tumor suppressor gene BAP1.17–20 More recently, the cancer-testis antigen PRAME (Preferentially Expressed Antigen in Melanoma) was found to represent an independent biomarker providing an additional layer of prognostic precision to the Class 1/Class 2 GEP system.21–23 The presence of PRAME mRNA, which can be assessed from the sample biopsy sample as the GEP, is associated with increased metastatic risk in both Class 1 and Class 2 UMs, although the optimal use of FRAME status as a complement to the GEP has not been established.
In this study, we hypothesized that the prognostic accuracy of the GEP/PRAME molecular prognostic system is non-inferior to the AJCC 8th edition TNM staging system for UM. To test this hypothesis, we compared GEP and FRAME to the TNM clinical staging system in 240 patients with UM treated by a single surgeon. Further, we optimized a method for combining GEP and PRAME into a simple 3-category prognostic system.
METHODS
Clinical Data Collection
This retrospective cohort study was approved by the Institutional Review Board of the University of Miami School of Medicine. Patient information was accessed with proper informed consent and in accordance with the Health Insurance Portability and Accountability Act (HIPAA). The study included 240 patients with primary UMs arising from the choroid and/or ciliary body from the ocular oncology practice of JWH. The ocular pathology laboratory routinely provided cytologic verification of fine needle biopsy samples in patients treated with plaque radiotherapy, and by histopathologic analysis in those treated with enucleation. Patients with primary iris melanomas and those who presented with metastasis were excluded. Collected data included age at diagnosis, sex, largest basal diameter (LBD), tumor thickness, ciliary body involvement, extraocular extension, node status, primary treatment modality, first detection of metastasis, date and cause of death, and date of last follow up. LBD was measured using ultrasonography and indirect ophthalmoscopy, and the larger value of the two was used. Extraocular extension was assessed by ultrasonography in patients undergoing plaque radiotherapy and by histopathologic analysis in patients undergoing enucleation. Tumors were staged according to the AJCC 8th edition TNM staging manual.24 Since most tumors were treated by I-125 plaque radiotherapy, where the histopathologic classification could not be applied, we used only the clinical classification for all patients. GEP class status (Class 1 versus Class 2) was determined with a prospectively validated 15-gene expression profile available as the DecisionDX-UM™ test.25 The GEP test also sub-classifies Class 1 tumors into Class 1A (low metastatic risk) and Class 1B (intermediate risk).26 However, this sub-classification was not used here since PRAME was used to sub-classify both Class 1 and Class 2 tumors, as described in the Results. RNA expression of PRAME was determined by quantitative PCR and categorized as PRAME+ or PRAME-, as previously described.21
Statistical Analysis
Progression free survival (PFS) was measured as the time interval between diagnosis of UM and first detection of metastatic disease. In patients who did not develop metastasis, survival was censored at last follow up. Kaplan-Meier survival curves were used to analyze associations between prognostic factors and PFS. Differences in PFS among prognostic groups were analyzed for statistical significance using the log rank test. Prognostic variables were evaluated using Cox proportional hazards regression (using both simultaneous and stepwise methods). Statistical analysis was performed with MedCalc software (version 18; Ostend, Belgium).
RESULTS
Among 240 consecutive patients diagnosed with UM arising from the choroid and/or ciliary body (Table 1), primary treatment consisted of I-125 plaque radiotherapy in 165 (68.8%) cases, enucleation in 74 (30.8%) cases, and observation in 1 case (0.4%). Tumor sample was obtained by fine needle aspiration biopsy in 166 (69.2%) and by post-enucleation needle biopsy in 74 (30.8%). GEP was Class 1 in 128 (53.3%) cases and Class 2 in 112 (46.7%) cases. PRAME was positive in 83 (34.6%) cases, including 38 (15.8%) Class 1 cases and 45 (18.8%) Class 2 cases. After a median follow up of 29 months (mean 42 months; range 1–195 months), metastasis was detected in 59 (24.6%) cases (Supplemental Material at AJO.com).
Table 1.
Summary of clinicopathologic and molecular features in 240 patients with uveal melanoma
| Variable | Summary Data (N=240) |
|---|---|
| Age at diagnosis (years) | |
| Mean | 62.4 |
| Median | 64 (14 to 93) |
| Sex | |
| Female | 121 (50.4%) |
| Male | 119 (49.6%) |
| Largest basal diameter (mm) | |
| Mean | 14.6 |
| Median (range) | 15.0 (3 to 24) |
| No. of tumors with LBD < 12 | 60 (25.0%) |
| No. of tumors with LBD ≥ 12 | 180 (75.0%) |
| Thickness (mm) | |
| Mean | 6.9 |
| Median (range) | 6.4 (1.2 to 16.4) |
| Ciliary body involvement | |
| Yes | 104 (43.3%) |
| No | 136 (56.7%) |
| Extraocular extension | |
| Yes | 17 (7.1%) |
| No | 51 (21.3%) |
| Unable to be assessed | 172 (72.0%) |
| Gene expression profile | |
| Class 1 | 128 (53.3%) |
| Class 2 | 112 (46.7%) |
| PRAME status | |
| PRAME (−) | 157 (65.4%) |
| PRAME (+) | 83 (34.6%) |
| Gene expression profile and PRAME status | |
| Class 1 PRAME (−) | 90 (37.5%) |
| Class 1 PRAME (+) | 38 (15.8%) |
| Class 2 PRAME (−) | 67 (27.9%) |
| Class 2 PRAME (+) | 45 (18.8%) |
| TNM stage | |
| I | 26 (10.8%) |
| IIA | 67 (27.9%) |
| IIB | 50 (20.8%) |
| IIIA | 59 (24.6%) |
| IIIB | 38 (15.8%) |
| Metastasis | |
| Yes | 59 (24.6%) |
| No | 181 (75.4%) |
| Last status | |
| Alive without metastasis | 175 (72.9%) |
| Alive with metastasis | 23 (9.6%) |
| Melanoma specific mortality | 36 (15.0%) |
| Non-melanoma specific mortality | 6 (2.5%) |
| Treatment | |
| Observation | 1 (0.4%) |
| Plaque brachytherapy | 165 (68.8%) |
| Enucleation | 74 (30.8%) |
| Follow-up (months) | |
| Mean | 42 |
| Median (Range) | 29 (1 to 195) |
Abbreviations: TNM, Tumor-Node-Metastasis; PRAME, preferentially expressed antigen in melanoma.
First, we used univariate Cox proportional hazards analysis to identify variables that were significantly associated with metastasis. For this initial step, we did not discretize continuous variables (age, LBD, thickness) in order to avoid arbitrary cutoff intervals. The factors demonstrating the strongest association with metastasis included: GEP class (P = 1.5 × 10−8), LBD (P = 2.5 × 10−6), PRAME status (P = 2.6 × 10−6), and TNM stage (P = 3.7 × 10−6) (Table 2). We then used multivariate Cox proportional hazards analysis to identify prognostic variables that provided significant independent prognostic information. These variables included: GEP (P = 2.8 × 10−6), PRAME (P = 2.3 × 10−4) and TNM (P = 7.1 × 10−4) (Table 3). We then analyzed the TNM clinical variables independently. This multivariate analysis revealed that LBD was the only TNM clinical variable that contributed prognostic information that was independent of GEP and PRAME (Table 3).
Table 2.
Univariate Cox proportional hazards analysis of clinicopathologic and molecular prognostic variables in 240 patients with uveal melanoma
| Covariate | Regression Coefficient, β (SE) | Wald Statistic | P-value | Hazard Coefficient, Exp(b) (95% CI) |
|---|---|---|---|---|
| Gene expression profile | 1.8230 (0.3218) | 32.10 | 1.5 × 10−8 | 6.1907 (3.2948 to 11.632) |
| Largest basal diameter | 0.1595 (0.0339) | 22.21 | 2.5 × 10−6 | 1.1729 (1.0976 to 1.2534) |
| PRAME status | 1.2500 (0.2660) | 22.09 | 2.6 × 10−6 | 3.4902 (2.0724 to 5.8781) |
| TNM stage | 0.5384 (0.1163) | 21.42 | 3.7 × 10−6 | 1.7132 (1.3639 to 2.1520) |
| Tumor thickness | 0.1168 (0.0341) | 11.78 | 6.0 × 10−4 | 1.1239 (1.0514 to 1.2015) |
| Ciliary body involvement | 0.8256 (0.2756) | 8.97 | 0.0027 | 2.2832 (1.3302 to 3.9188) |
| Male gender | 0.6345 (0.2694) | 5.54 | 0.019 | 1.8860 (1.1124 to 3.1978) |
| Age | 0.01806 (0.00987) | 3.35 | 0.067 | 1.0182 (0.9987 to 1.0381) |
| Extraocular extension | 0.4271 (0.3731) | 1.31 | 0.25 | 1.5328 (0.7378 to 3.1848) |
Abbreviations: SE, standard error; CI, confidence interval; TNM, Tumor-Node-Metastasis; PRAME, Preferentially Expressed Antigen in Melanoma
Table 3.
Multivariate Cox proportional hazards analysis of clinicopathologic and molecular prognostic variables in 240 patients with uveal melanoma
| Clinical variables incorporated into TNM stage | ||||
| Covariate | Regression Coefficient, β (SE) | Wald Statistic | P-value | Hazard Coefficient, Exp(b) (95% CI) |
| Gene expression profile | 1.5594 (0.3329) | 21.95 | 2.8 × 10−6 | 4.7559 (2.4768 to 9.1319) |
| PRAME status | 10061(0.2732) | 13.56 | 2.3 ×10−4 | 2.7349 (1.6009 to 4.6720) |
| TNM stage | 0.4136 (0.1221) | 11.47 | 7.1 × 10−4 | 1.5122 (1.1903 to 1.9213) |
| Male gendera | ||||
| Clinical variables analyzed separately | ||||
| Covariate | Regression Coefficient, β (SE) | Wald Statistic | P-value | Hazard Coefficient, Exp(b) (95% CI) |
| Gene expression profile | 1.6787 (0.3276) | 26.26 | 3.0 × 10−7 | 5.3484 (2.8195 to 10.1833) |
| Largest basal diameter | 0.1340 (0.0391) | 11.76 | 6.0 × 10−4 | 1.1434 (1.0591 to 1.2344) |
| PRAME status | 0.7989 (0.2784) | 8.23 | 0.0041 | 2.2230 (1.2881 to 3.8364) |
| Tumor thicknessa | ||||
| Ciliary body involvementa | ||||
| Male gendera | ||||
Excluded by the Cox multivariate model due to lack of significant independent prognostic value
Abbreviations: SE, standard error; CI, confidence interval; PRAME, preferentially expressed antigen in melanoma; TNM, Tumor-Node-Metastasis
Next, we directly compared the ability of the TNM versus GEP/PRAME to stratify metastatic risk. To optimize the TNM, we used Kaplan-Meier analysis to perform pairwise comparisons between TNM stages (except stage IV) to identify and combine prognostically redundant categories (Figure 1, Top left). There was no significant difference between stages I versus IIA, I versus IIB, I versus IIIA, IIA versus IIB, IIB versus IIIA, or IIIA versus IIIB (Figure 1, Middle left). Consequently, stages IIA + IIB and stages IIIA + IIIB were combined to form three categories with modestly improved statistical significance (P = 1.3 × 10−5) (Figure 1, Bottom left). A similar procedure was performed for GEP/PRAME (Figure 1, Top right). The four GEP/PRAME categories provided statistically significant separation between survival curves (P = 3.3 × 10−13), that was superior to TNM survival curves (P = 6.1 × 10−5). All GEP/PRAME categories were non-redundant except Class 1PRAME+ and Class 2PRAME-, which were then combined (Figure 1, Middle right) to form three GEP/PRAME categories: Class 1PRAME-, Class 1PRAME+ or Class 2PRAME-, and Class 2PRAME+ (Figure 1, Bottom right). The prognostic accuracy of the optimized GEP/PRAME categories (P = 8.6 × 10−14) was superior to the optimized TNM categories (P = 1.3 × 10−5). At every follow up point, the GEP+PRAME model maintained a greater separation between metastatic risk groups than did the TNM. For patients without metastasis after ≥5 years follow-up (n=45), a false positive “high risk” result would have been given in 16 (35.6%) patients using the TNM, compared to 3 (6.7%) using GEP/PRAME. To further investigate this tendency for increased false positives with the TNM, we performed a sub-analysis of tumors with LBD ≥12 mm (n=180). The TNM classified all of these larger tumors as being at increased metastatic risk (Figure 2, Left), whereas GEP/PRAME identified 52/180 (29%) of these tumors as having low metastatic risk, only 3 (6%) of which gave rise to metastasis (Figure 2, Right).
FIGURE 1.
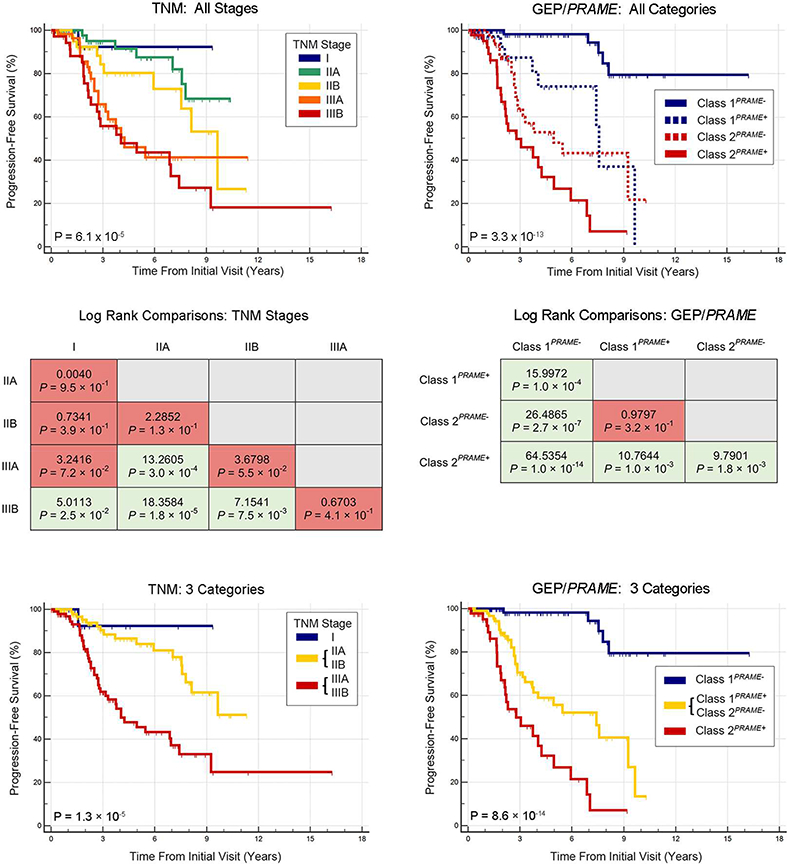
Kaplan-Meier survival analysis in 240 patients with uveal melanoma using Tumor-Node-Metastasis staging and gene expression profiling/PRAME classification. (Top left) Survival curves are shown using all Tumor-Node-Metastasis stages except stage IV. (Middle left) Comparisons of log-rank statistics are shown between Tumor-Node-Metastasis stages. (Bottom left) Survival curves are shown using optimized Tumor-Node-Metastasis categories. (Top right) Survival curves are shown using all gene expression profiling/PRAME categories. (Middle right) Comparisons of log-rank statistics are shown between gene expression profiling/PRAME categories. (Bottom right) Survival curves are shown using optimized gene expression profiling/PRAME categories.
FIGURE 2.
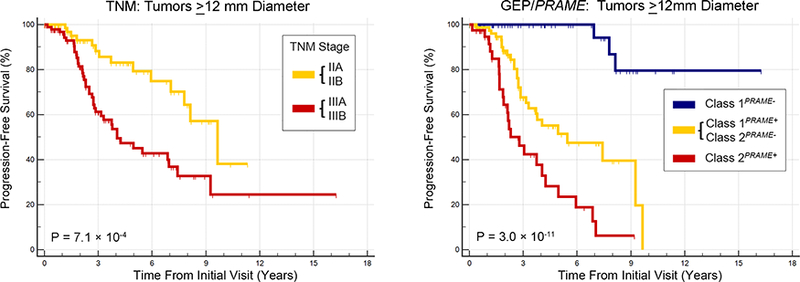
Kaplan-Meier survival analysis in 180 patients with uveal melanoma with largest basal diameter ≥12 mm. (Left) Survival curves are shown using optimized Tumor-Node-Metastasis categories. (Right) Survival curves are shown using gene expression profiling/PRAME categories.
DISCUSSION
In this study, we confirmed that GEP, PRAME and TNM stage were each prognostic of metastasis in UM. Individually, GEP and PRAME both demonstrated prognostic accuracy that was superior to the TNM staging system. Combining GEP and PRAME into a 3-category model further enhanced the prognostic accuracy of this molecular classification system.
There are several limitations to the use of the TNM system for prognostication in UM. First, the TNM assigns increased metastatic risk to all UMs with LBD > 12 mm, yet almost a third of these “large tumors” have low metastatic risk based on their molecular profile. This could result in false positive classification of UMs as having a high metastatic risk, leading to over-management of such patients. Second, several variables used in the clinical “T” stage provide redundant prognostic information. For example, increased tumor thickness is related to increased LBD and ciliary body involvement. This may explain why the prognostic accuracy of LBD alone was similar to the entire clinical TNM staging system in this study and others.10 Third, the assessment of “T” variables, such as measuring tumor dimensions and determining ciliary body involvement, is not standardized and may vary from center to center.27,28. Fourth, some variables that were included in the TNM system for UM to conform to the standard TNM template are of little or no value in UM. For example, it is usually possible to assess extraocular tumor extension only in eyes treated by enucleation (which is performed in a minority of cases). Nodal involvement - the “N” component - does not occur in UM, thereby rendering this dimension of the TNM system irrelevant. Perhaps most importantly, the dependence of TNM staging on anatomic and morphologic features fails to accommodate new scientific understanding of cancer behavior, including powerful molecular prognostic biomarkers such as GEP and PRAME.29
We acknowledge several limitations of this study. First, this was a single center retrospective study, whereas we prefer prospective multi-center validation of prognostic markers.12 Second, while the sample size was adequate for the intended purpose of this study, we would prefer a larger number of subjects to provide statistical power for detailed sub-analyses (such as the role of LBD in the GEP/PRAME prognostic system). Third, the median follow-up was relatively short (29 months), whereas we would prefer longer follow-up to minimize effects of lead time bias. However, a sub-analysis of patients with at least 5 years follow-up yielded results that were consistent with the overall findings. These limitations will each be addressed by the Collaborative Ocular Oncology Group Study Number 2 (COOG2), an ongoing prospective, multi-center clinical study funded by the National Cancer Institute. This study will also formally compare the new GEP/PRAME model described here to the existing Class 1A/1B/2 system, as well as mutations in BAP1, SF3B1 and EIF1AX and chromosomal copy number changes.
In conclusion, this study confirmed that both GEP and PRAME were individually superior to TNM in predicting a patient’s risk of developing metastasis from UM. Moreover, GEP could be combined with PRAME to create an even more efficient and simplified 3-category molecular prognostic model. These findings continue to support the superior prognostic accuracy of these molecular biomarkers over anatomic features. Despite the deficiencies of the TNM system for personalized management of individual patients, it continues to be valuable for grouping patients with similar extent of disease into discrete “bins” for purposes of clinical, epidemiologic and health policy research.
Supplementary Material
ACKNOWLEDGEMENTS/DISCLOSURES
a. Funding/Support: J. William Harbour was supported by the National Institutes of Health (Bethesda, MD) R01 CA125970, the Alcon Research Institute (Fort Worth, TX), Research to Prevent Blindness, Inc. Senior Investigator Award (New York, NY), and a generous gift from Dr. Mark J. Daily. The Bascom Palmer Eye Institute received funding from National Institutes of Health (Bethesda, MD) Core Grant P30EY014801. Department of Defense (Washington, DC) 329 Grant #W81XWH-13–1-0048, and a Research to Prevent Blindness Unrestricted Grant (New York, NY).
b. Financial Disclosures: J. William Harbour is the inventor of intellectual property used in the study and receives royalties from its commercialization. He is a paid consultant for Castle Biosciences, licensee of this intellectual property. Scott D. Walter served on an advisory board for Castle Biosciences. The following authors have no financial disclosures: Louis Cai, Manuel Paez-Escamilla, Bercin Tarlan, Christina L.Decatur, and Barbara M. Perez. All authors attest that they meet the current ICMJE criteria for authorship.
c. Other Acknowledgements: The authors wish to thank William J. Feuer, M.S. (Bascom Palmer Eye Institute, Miami, FL) for statistical support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Ramaiya KJ, Harbour JW. Current management of uveal melanoma. Expert Review of Ophthalmology. 2007;2(6):939–946. [Google Scholar]
- 2.Singh AD, Topham A. Survival rates with uveal melanoma in the United States: 1973–1997. Ophthalmology. 2003;110(5):962–965. [DOI] [PubMed] [Google Scholar]
- 3.Eskelin S, Pyrhonen S, Summanen P, Hahka-Kemppinen M, Kivela T. Tumor doubling times in metastatic malignant melanoma of the uvea - Tumor progression before and after treatment. Ophthalmology. 2000; 107(8): 1443–1449. [DOI] [PubMed] [Google Scholar]
- 4.Valsecchi ME, Orloff M, Sato R, et al. Adjuvant sunitinib in high-risk patients with uveal melanoma: comparison with institutional controls. Ophthalmology. 2017. [DOI] [PubMed] [Google Scholar]
- 5.Bol KF, van den Bosch T, Schreibelt G, et al. Adjuvant dendritic cell vaccination in high-risk uveal melanoma. Ophthalmology. 2016;123(10):2265–2267. [DOI] [PubMed] [Google Scholar]
- 6.AJCC cancer staging manual. New York, NY: Springer Science+Business Media; 2016. [Google Scholar]
- 7.Burke HB. Outcome prediction and the future of the TNM staging system. J Natl Cancer Inst. 2004;96(19):1408–1409. [DOI] [PubMed] [Google Scholar]
- 8.Quirke P, Williams GT, Ectors N, Ensari A, Piard F, Nagtegaal I. The future of the TNM staging system in colorectal cancer: time for a debate? Lancet Oncol. 2007;8(7):651–657. [DOI] [PubMed] [Google Scholar]
- 9.Jouffret L, Turrini O, Ewald J, Moutardier V, Iovanna JL, Delpero JR. Long-term survivors after pancreatectomy for cancer: the TNM classification is outdated. ANZ J Surg. 2015;85(11):860–864. [DOI] [PubMed] [Google Scholar]
- 10.Skinner CC, Augsburger JJ, Augsburger BD, Correa ZM. Comparison of Alternative Tumor Size Classifications for Posterior Uveal Melanomas. Invest Ophthalmol Vis Sci. 2017;58(9):3335–3342. [DOI] [PubMed] [Google Scholar]
- 11.Harbour JW, Augsburger JJ, Char DH. Author reply: To PMID Ophthalmology. 2013;120(7):e51. [DOI] [PubMed] [Google Scholar]
- 12.Onken MD, Worley LA, Char DH, et al. Collaborative Ocular Oncology Group report number 1: prospective validation of a multi-gene prognostic assay in uveal melanoma. Ophthalmology. 2012;119(8):1596–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Correa ZM, Augsburger JJ. Independent prognostic significance of gene expression profile class and largest basal diameter of posterior uveal melanomas. Am J Ophthalmol. 2016;162:20–27 e21. [DOI] [PubMed] [Google Scholar]
- 14.Petrausch U, Martus P, Tonnies H, et al. Significance of gene expression analysis in uveal melanoma in comparison to standard risk factors for risk assessment of subsequent metastases. Eye. 2007;22(8):997–1007. [DOI] [PubMed] [Google Scholar]
- 15.Worley LA, Onken MD, Person E, et al. Transcriptomic versus chromosomal prognostic markers and clinical outcome in uveal melanoma. Clin Cancer Res. 2007;13(5):1466–1471. [DOI] [PubMed] [Google Scholar]
- 16.Onken MD, Worley LA, Tuscan MD, Harbour JW. An accurate, clinically feasible multi-gene expression assay for predicting metastasis in uveal melanoma. J Mol Diagn. 2010;12(4):461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harbour JW, Onken MD, Roberson ED, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330(6009):1410–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harbour JW, Roberson ED, Anbunathan H, Onken MD, Worley LA, Bowcock AM. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nature genetics. 2013;45(2):133–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Decatur CL, Ong E, Garg N, et al. Driver mutations in uveal melanoma: associations with gene expression profile and patient outcomes. JAMA Ophthalmol. 2016;134(7):728–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin M, Masshofer L, Temming P, et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nature genetics. 2013;45:933–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Field MG, Decatur CL, Kurtenbach S, et al. PRAME as an independent biomarker for metastasis in uveal melanoma. Clin Cancer Res. 2016;22(5):1234–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Field MG, Durante MA, Decatur CL, et al. Epigenetic reprogramming and aberrant expression of PRAME are associated with increased metastatic risk in Class 1 and Class 2 uveal melanomas. Oncotarget. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reichstein D New concepts in the molecular understanding of uveal melanoma. Curr Opin Ophthalmol. 2017;28(3):219–227. [DOI] [PubMed] [Google Scholar]
- 24.Edge SB. AJCC cancer staging manual. 8th ed. New York: Springer; 2017. [Google Scholar]
- 25.Harbour JW, Chen R. The DecisionDx-UM Gene Expression Profile Test Provides Risk Stratification and Individualized Patient Care in Uveal Melanoma. PLoS Curr. 2013;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plasseraud KM, Wilkinson JK, Oelschlager KM, et al. Gene expression profiling in uveal melanoma: technical reliability and correlation of molecular class with pathologic characteristics. Diagn Pathol. 2017;12(1):59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harbour JW, Augsburger JJ, Char DH. Gene expression profiling versus TNM classification. Ophthalmology. 2013;120(7):e52–53. [DOI] [PubMed] [Google Scholar]
- 28.Kujala E, Damato B, Coupland SE, et al. Staging of ciliary body and choroidal melanomas based on anatomic extent. J Clin Oncol. 2013;31 (22):2825–2831. [DOI] [PubMed] [Google Scholar]
- 29.Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer. 2005;5(11):845–856. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


