Abstract
The molecular heterogeneity of clear cell renal carcinoma (ccRCC) makes prediction of disease progression and therapeutic response difficult. Thus, this report investigates the functional significance, mechanisms of action, and clinical utility of miR-182-5p and metastasis associated lung adenocarcinoma transcript 1 (MALAT1/NEAT2), a long non-coding RNA (lncRNA), in the regulation of kidney cancer using human kidney cancer tissues as well as in vitro and in vivo model systems. Profiling of miR-182-5p and MALAT-1 in human renal cancer cells and clinical specimens was done by quantitative real-time PCR (qPCR). The biological significance was determined by series of in vitro and in vivo experiments. The interaction between miR-182-5p and MALAT-1 was investigated using luciferase reporter assays. In addition, the effects of miR-182-5p overexpression and MALAT-1 downregulation on cell cycle progression were assessed in ccRCC cells. The data indicates that miR-182-5p is downregulated in ccRCC; the mechanism being CpG hypermethylation as observed from 5-Aza CdR treatment that decreased promoter methylation and expression of key methylation regulatory genes like DNMT1, DNMT3a and DNMT3b. Overexpression of miR-182-5p inhibited cell proliferation, colony formation, apoptosis and led to G2/M-phase cell cycle arrest by directly targeting MALAT-1. Downregulation of MALAT-1 led to upregulation of p53, downregulation of CDC20, AURKA, drivers of the cell cycle mitotic phase. Transient knockdown of MALAT-1 mimicked the effects of miR-182-5p overexpression. Finally, overexpression of miR-182-5p decreased tumor growth in mice, compared to controls; thus, demonstrating its anti-tumor effect in vivo.
Keywords: Clear cell renal carcinoma, miR-182-5p, MALAT-1, CDC20, AURKA, p53
Introduction
Renal cell carcinoma (RCC) is a leading cause of death accounting for nearly 14,000 (~2.4%) deaths in the United States in 2017, with clear cell carcinoma (ccRCC) being the most common histological subtype (1). Although early stage ccRCC is curable by surgery, there are still many patients who develop recurrence or localized tumor with distant metastasis. Compared to other cancers, there are few tumor biomarkers for renal cancer (2). It is therefore necessary, to improve our understanding of ccRCC pathogenesis and identify new biomarkers for development of new diagnostic and therapeutic approaches.
Micro RNAs (miRNA) are small, non-protein coding molecules that play an important role in regulation of gene expression by inhibition of their target genes. They are found to be involved in controlling several tumor associated processes like cell proliferation, apoptosis, migration, invasion and are frequently dysregulated in tumor tissues (3,4). With regards to miR-182 function, there are contradictory reports of it being associated with invasive and /or metastatic potential (5–7). While in glioblastoma, osteosarcoma and clear cell renal carcinoma, miR-182 plays a tumor suppressive role (8–11). Overall these studies suggest cell-dependent role of miR-182 across different cancer types.
Long noncoding RNAs (lncRNAs) are a class of RNA with more than 200 nucleotides, and may act as regulators for tumor suppressive miRNAs. Metastasis-associated lung adenocarcinoma transcript 1 (MALAT-1) is a highly conserved nuclear lncRNA among mammals. Its overexpression enhances the tumorigenic potential of renal cancer cells (12–15) and knockdown of MALAT-1 is known to induce G2/M phase arrest in many types of cancer (16–18). Previous studies have shown that lncRNAs may interact with miRNAs and modulate each other’s expression (19). MALAT-1 expression in some cancers is suppressed by miRNAs. For example, miR-205 and miR-200 suppress MALAT-1 in ccRCC (13,20).
Previously we performed a preliminary miRNA microarray screening in renal cancer cell lines compared to a non-malignant HK2 cell (21–25). miR-182 was found to be among the significantly downregulated miRNAs in cancer cells compared to HK2. The present study was undertaken to define the role of miR-182 in ccRCC and investigate its relationship with long non-coding RNA MALAT-1. The study, for the first time, reveals a link between these two ncRNAs, and demonstrates a novel regulation of MALAT-1 expression by miR-182-5p. Finally, we show the tumor suppressor role of miR-182-5p and its involvement in regulating the proteins that control mitotic progression in ccRCC.
Materials and Methods
Human specimens and cells
Normal and cancerous renal tissues were obtained from 53 representative FFPE (Formaldehyde fixed paraffin embedded) tissue blocks of radical nephrectomy specimens from the Pathology Department of the Veterans Affairs (VA) Medical center of San Francisco. The blocks were from kidney cancer patients who underwent surgery at the VA Medical center between 1980–2009. Written informed consent was obtained from all patients and the study was in accordance with institutional guidelines (IRB approval no: 16-18555). Sections (4μm) of the blocks were prepared, H&E stained, and slides were reviewed by a board-certified pathologist to mark the normal and cancer areas. Subsequently, 12μm slides were made from the blocks and microdissection was performed using the marked H&E stained slides as a template.
Human renal cell carcinoma cell lines ACHN (ATCC # CRL-1611), Caki-1 (ATCC # HTB-46), 786-O (ATCC # CRL-1932) and a non-malignant renal cell line HK-2 (ATCC # CRL-2190) were obtained from the American Type Culture Collection (ATCC) (Manassas, VA) and grown under recommended conditions. These human-derived cell lines were validated by DNA short tandem repeat analysis by ATCC. Cell line experiments were performed within 5–6 months of their procurement/resuscitation. ACHN cells were cultured in MEM media, Caki-1 in McCoy’s 5A (modified) media and 786-O in RPMI media, each supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. Immortalized non-transformed HK-2 cells were maintained in keratinocyte serum free media supplemented with prequalified human recombinant epidermal growth factor 1–53 (EGF 1–53) and bovine pituitary extract (BPE) (GIBCO/Invitrogen, Carlsbad, CA, USA). All cell lines were maintained in an incubator with a humidified atmosphere of 95% air and 5% CO2 at 37°C.
RNA isolation and quantitative RT-PCR
Total RNA was extracted from cells and human tissue samples using a miRNeasy kit and miRNeasy FFPE kit (Qiagen, Germany) respectively according to manufacturer’s instructions. Assays to quantify mature miRNA and mRNAs were performed using TaqMan microRNA (miR-182-5p, RNU48) and gene expression assays (MALAT-1, HPRT1, DNMT1, DNMT3a, DNMT3b) (Applied Biosystems Inc., Foster City, CA, USA). Human Hypoxanthine Phosphoribosyltransferase 1 (HPRT1) and RNU48 were used as endogenous controls.
Real-time reverse transcription-polymerase chain reaction (RT-PCR) was carried out using Quant Studio7 PCR system and Taqman universal PCR master mix as per manufacturer’s instructions. Validation of the downregulated genes from RT2 profiler was done using RT-PCR with specific primers and SYBR Green (Applied Biosystems). The primer sequences for AURKA, BMI1, FLT1, CDC20, CCND2, SKP2, VEGF-C are provided in supplemental table 1.
The expression levels for miR-182-5p and MALAT-1 in tumor tissues were compared with paired adjacent normal tissues. The expression levels of miRNA/RNA were calculated as the amount of target miRNA/RNA relative to that of RNU48/HPRT1 control to normalize the initial input of total RNA. Also, we used mRNA and miRNA expression data from The Cancer Genome Atlas (TCGA) now available at the genomic data commons (GDC) portal (https://gdc-portal.nci.nih.gov/).
Protein extraction and Western blot analysis
miR-182-5p mimic and ASO-MALAT-1 transfected cells along with corresponding control were harvested after 72hrs and subjected to lysis using NP-40 (Thermo Scientific). Protein concentration was measured using the BCA protein assay (Thermo Scientific). Western blots were performed using NuPAGE 4-12% Bis-Tris protein gels (Invitrogen). For gel electrophoresis, the iBlot 2 dry blotting system (Invitrogen) and transferred onto plyvinylidene fluoride (PVDF) transfer membranes. Membranes were incubated with Odyssey blocking buffer (LI-COR) prior to incubation with primary antibodies overnight at 4°C. Goat anti-rabbit IgG (H+L) 800 CW or goat anti-mouse (H+L) 680RD was applied for 45 minutes at room temperature (1:15000, LI-COR) before washing with PBS with Tween 20. An Odyssey Infrared Imaging System Scanner was used to generate immunoblot images and LI-COR Odyssey software (LI-COR Biosciences) was utilized for band quantification. The following primary antibodies were used- Aurora A/AURKA (Cell Signaling, 14475T), CDC20 (GeneTex, GTX111137), p53 (Cell signaling #2524), Beta-actin (Cell Signaling #3700, #4970).
Transient transfection
To induce miR182-5p overexpression, cells were transfected with miRVana miR-182-5p mimic (Thermo Fisher Scientific) using Lipofectamine RNAi Max (Thermo Fisher Scientific). To verify the transfection efficiency of miRNA mimic, miRVana miRNA mimic negative control #1 (Thermo Fisher Scientific) was included in each transfection experiment.
For MALAT-1, we used antisense oligonucleotides (ASO) from IDT (Integrated DNA technologies). Cells were reverse transfected using Lipofectamine RNAi Max (Thermo Fisher Scientific). For reverse transfection, cells were plated in growth media without antibiotics and the transfection reagents were added on the same day. All transfection experiments were carried out according to manufacturer’s protocol and for 72hrs.
For p53 overexpression, cells were transfected with plasmid DNA (pcDNA3 p53 wild type (WT)), gift from David Meek, Addgene plasmid #69003) (26) using Lipofectamine 3000 (Thermo FisherScientific). To verify the transfection efficiency, corresponding control plasmid DNA (pcDNA3 empty expression vector) was included in each transfection experiment.
5Aza-CdR treatment of cells and RNA extraction
For demethylation studies, cells were treated daily with 5-AZA-Deoxycitydine (5Aza-CdR) (Sigma-Aldrich) for 72hrs at concentration of 20uM (27). Total RNA was isolated using a miRNeasy mini kit (Qiagen). And the levels of miRNA were determined. The TCGA database was used to check the methylation status of miR-182 promoter in both normal and tumor samples. The Wanderer, a web based tool that offers convenient and fast access to gene-centered TCGA methylation data (28) was used to determine the difference across the samples. The adjusted p-values were generated through Benjamini and Hochberg adjustment (False Discovery rate) for multiple testing.
In order to confirm the methyaltion status of the miR-182 promoter in renal cancer cell lines we extracted DNA from these cells using DNeasy tissue kit (Qiagen) and modified with sodium bisulphite using EZ DNA methylation-Gold kit (Zymo Research, Orange, CA, USA) according to the manufacturer’s instructions. Bisulfite-treated DNA was analyzed by methylation specific quatitative polymerase chain reaction (MS-qPCR) with primer pairs for unmethylated and methylated regulatory region of miR-182-5p. MS-qPCR was performed as described earlier (29). For each sample, the percent of methylation was calculated by the difference of Ct in methylated sample (Ct-M) and Ct in unmethylated sample (Ct-U). The sequences of the primers are given in supplementary table 1.
Cell viability and clonogenicity assays
Cell viability for both miR-182-5p overexpression and MALAT-1 knockdown were determined at 24, 48, 72 hours by using the CellTiter 96® AqueousOne Solution Cell Proliferation Assay kit (Promega), according to the manufacturer’s protocol. For clonogenicity assay, 48 hours post-transfection, cells were counted, seeded at low density (250 cells/plate) and allowed to grow until visible colonies appeared. Then, cells were stained with Geimsa and colonies were counted.
Cell cycle analysis
Cells were transfected with miRNA or negative control and ASO-MALAT-1 or corresponding negative control and harvested after 72hrs, washed and fixed in cold 70% ethanol overnight at −20°C. Cell pellets were stained with PI/RNase staining buffer (BD Pharmingen). All analyses were performed in triplicate and 10,000 gated events/sample were counted.
Apoptosis assay
Cells were transfected with miRNA or negative control and corresponding negative control and harvested after 72hrs. Cells were washed in cold PBS, suspended in 1X binding buffer and stained with Annexin V-FITC and 7AAD viability dye (Annexin V-FITC/7 AAD kit, Beckman Coulter). After 15 minutes of incubation at room temperature in dark, cells were washed and analyzed using BD FACSVerse (BD Pharmingen).
Luciferase reporter assay
To evaluate whether miR-182-5p regulates MALAT-1, we looked for potential binding target sites that were annotated in at least one of the four databases (TargetScan, RNA22, miRanda and Pictar) using miRWalk. We found 3 potential binding sites for miR-182-5p in the MALAT-1 sequence (leftmost position of predicted site-2979, 5065, 5702).
Luciferase reporter vectors were constructed by ligation with annealed custom MALAT-1 oligonucleotides containing putative target binding sites and corresponding nontarget /mutant sites and inserted into the pmiR-GLO reporter vector (Promega, Madison, WI). miR-182-5p mimic and negative control transfected ACHN and Caki-1 cells were co-transfected with 1ng of the pmir-GLO vector with the putative MALAT-1 binding site and luciferase activity was measured 48 hours after transfection using the Dual Luciferase Reporter Assay system (Promega). Relative luciferase activity was calculated by normalizing to the Renilla luminescence.
In vivo intratumoral delivery of miR-182-5p
All animal care was in accordance with the guidelines of SFVAMC and the study was approved by the San Francisco VA IACUC (Approval number: 16-004). The therapeutic potential of miR-182-5p was examined by local administration in established tumors using a renal cancer xenograft nude mouse model as previously described (22,30). Nude mice (5-weeks old, River laboratories) (n=10) were injected with 0.8 x 106 ACHN cells (in 100ul volume) subcutaneously in the right flanks. Once palpable tumors developed, caliper measurements were taken once a week and tumor volumes were calculated as {(width)2 * length}/2 {x2y/2, where width (x) < length (y)}. Synthetic miR-182-5p mimic /miR-Con (6.25ug each) complexed with 1.6ul siPORTamine transfection reagent (Ambion, Austin, TX) in 50ul of PBS was injected intratumorally every 4 days. Experiment was terminated two days after the last treatment (day 28) and tumor volumes were measured.
RT2 profiler PCR array analysis
Expression profiling of 84 genes involved in cancer related pathways was performed using human RT2 profiler PCR array PAHS-33z (Qiagen) based on SYBR-Green real time PCR. Three biological replicates were prepared from miRNA and control transfections and the isolated RNA samples from ACHN and Caki1 treated under the same conditions were pooled. RNA was extracted using a miRNA isolation kit (Qiagen), cDNA was synthesized from the RNA pool isolated, using RT2 first strand kit (Qiagen) following the manufacturer’s instructions. SYBR-Green real-time PCR was performed and relative quantification changes in expression were measured by obtaining the threshold cycle and normalizing to the average of two housekeeping genes, HPRT1 and glyceraldehyde 3-phosphate dehydrogenase(GAPDH). Data from the array was analyzed with SABiosciences software by observed Ct values and determining relative fold change as compared with housekeeping genes.
Pathway analysis
We performed functional analysis using the EnrichR platform (31,32). This web based software allows pathway evaluation from its extensive gene set libraries including Gene ontology and various pathway analysis libraries like Kyoto Encyclopedia of Genes and Genomes pathway (KEGG), Reactome pathway, Wikipathway, Panther and Biocarta. Gene set enrichment and pathway analysis in this study was performed with the p-value adjustment by Z-score permutation.
Statistical analysis
All data was derived from at least two or three independent experiments. Statistical analysis was performed using GraphPad Prism 7 and MedCalc. All quantified data was presented as mean ± SE of at least triplicate samples and three experiments performed at different times or as indicated. Statistical significance was assessed by Student's t-test and p-values<0.05 were considered significant. The TCGA data cohort was analyzed using the Mann-Whitney test to compare reads per million between normal and adjacent tumor samples. Receiver operating curves (ROC) were calculated to determine the potential of miR-182-5p to discriminate between malignant and non-malignant samples.
Results
Expression of miR-182-5p in ccRCC
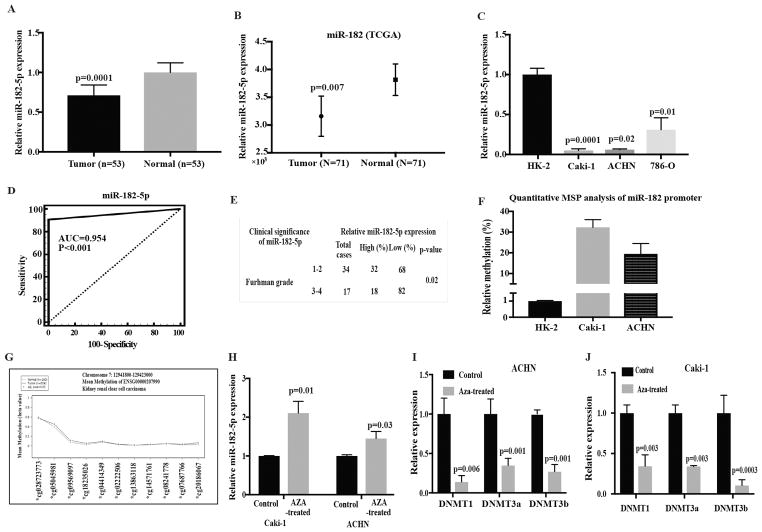
The level of miR-182-5p expression in human ccRCC was measured in 53 tumor and paired normal tissues. miR-182-5p was significantly down-regulated in tumor tissue samples compared to adjacent normal tissues (~79.24%, P-value =0.0001, Fig. 1A). The TCGA data portal was also used to validate the differences in miR-182-5p expression between tumor and adjacent normal tissues using Mann Whitney test from Rank sums test. miR-182-5p levels were significantly lower in tumors compared to normal tissue (p=0.007) (Fig. 1B). We also measured the levels miR-182-5p in renal cancer cell lines ACHN, 786-O, Caki-1 and found them to be lower when compared to HK2 cells. The levels of miR182-5p were significantly low in 786-O, ACHN and Caki-1 (P-value<0.05, Fig. 1C). Thus, the cell line data agrees with the tissue data showing that miR-182-5p is downregulated in ccRCC.
Figure 1.
miR182-5p is downregulated in renal carcinoma. A miR-182-5p expression levels in clear cell renal carcinoma specimens relative to matched adjacent normal as assessed by real-time PCR. B Box-plot of miR-182-5p expression using TCGA (Normal, n= 71; Adjacent cancer tissue, n= 71) database. P-value calculated by Mann-Whitney two-tailed test, Bar = mean± SE. C Relative miR-182-5p expression levels in renal cancer cell lines and an immortalized non-malignant cell line (HK2) as assessed by RT-PCR. Data normalized to RNU48 control. D Statistical assessment of miR-182-5p as a ccRCC biomarker. E Chi-square test showing correlation of miR-182-5p with Fuhrman grade. F Quantitaive MSP analysis of methylation status of miR-182 promoter in renal cancer cell lines. G Average methylation profiles for kidney renal cell carcinoma (KIRC) at the hsa-miR-182-5p gene locus using KIRC dataset (see Methods section). Adjusted p-value calculated by Wilcoxon signed rank test. H miR-182-5p expression level in 5-Aza-CdR treated and untreated RCC cell lines. I and J Expression levels of DNMT1, DNMT3a and DNMT3b in ACHN and Caki-1 cells treated with 5-Aza-CdR. P-value calculated by Student t-test. Bar = mean± SE.
In view of these results, we performed Receiver operating curve (ROC) analyses to examine the diagnostic potential of miR-182-5p in ccRCC. This analysis showed that miR-182-5p expression can be a single significant parameter to discriminate between normal and tumor tissues with an AUC of 0.954 (95% CI: 0.895–0.985, p<0.001) (Fig. 1D). We also determined the correlation between miR-182-5p expression and Fuhrman grade. Chi squared analysis revealed that decreased miR-182-5p expression was correlated with an increase in Fuhrman grade (low to high) (Fig 1E). The clinical demographics of the patient population are summarized in supplementary Table 2. Based on this data, miR-182-5p has significant potential to be a diagnostic biomarker for ccRCC, though this needs to be validated with a larger sample cohort.
Previous studies with melanoma, ovarian, hepatocellular and renal cell carcinoma have identified a distinctive genomic site rich in methylated CpG islands, located upstream of the miR-182 coding sequence (33,34) (Fig. S1). Thus, we analyzed the methylation status of this genomic region in cell lines compared to non-malignant HK2. We observed hypermethylation of miR-182in both Caki-1 and ACHN cells compared to normal HK2 cells (Fig. 1F). Further, we checked the methylation status of miR-182 promoter in TCGA data cohort and observed similar trend with tumor tissues being hypermethylated compared to normal tissues (Fig. 1G).
To clarify the role of epigenetics in miR-182-5p silencing in Caki-1 and ACHN cells we treated these cells with demethylating agent, 5-Aza-CdR and observed that the expression level of miR-182 was increased after 72 hours of treatment indicating possible epigenetic regulation (Fig. 1H). We also investigated the status of methylation regulatory genes such as DNMTl, DNMT3a and DNMT3b after 5-Aza-CdR treatment and found significant decrease in the expression of these genes compared to the cells treated with the vehicle (DMSO) control (Fig. 1I–J).
miR-182-5p overexpression suppresses tumorigenicity in vitro in renal cancer cell lines
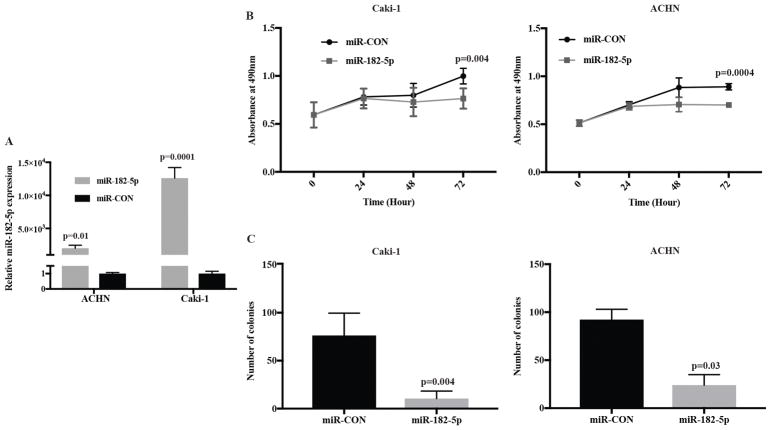
Next, we evaluated the tumor suppressive potential of miR-182-5p in vitro by overexpressing it in ACHN and Caki-1 cells using miR-182-5p mimic (Fig. 2A). miR-182-5p overexpression reduced cell viability and clonogenicity in both ACHN and Caki-1 cells compared to controls (Fig 2B and C). These observations suggest that miR-182-5p overexpression suppresses the tumorigenesis of ccRCC cell lines.
Figure 2.
miR-182-5p over-expression inhibiting ccRCC cell proliferation and colony formation. A Relative expression level of miR-182-5p after transfection with miR-182-5p mimic or control. B Cell viability was determined by MTS assay in Caki-1 and ACHN cells transfected with miR-182-5p mimic and miR-CON. C Colony formation assay with Caki-1, ACHN cells. Results shown are representative of three independent experiments. P-value calculated by Student t-test. Bar = mean± SE.
miR-182-5p overexpression induces apoptosis and affects the metaphase progression of ccRCC cell lines
Apoptosis analyses were performed with ACHN and Caki-1 cells transfected with negative control/ miR-182-5p. It was observed that the average apoptotic fractions (early apoptotic + late apoptotic) were significantly increased with a concomitant decrease in viable cell populations upon miR-182-5p overexpression compared to negative control transfected cells (Fig. 3A). These analyses suggest that miR-182-5p affects apoptotic pathways in ccRCC and plays a pro-apoptotic role.
Figure 3.
miR-182-5p functions as a tumor suppressor gene both in vitro and in vivo. A Flow cytometric analysis of Annexin-V-FITC -7AAD stained Caki-1 and ACHN cells transfected with miR-CON/ miR-182-5p. B Cell cycle analysis showing G2/M arrest in Caki-1 and ACHN cells. C Tumor volume following intra-tumoral injection of control or miR-182-5p mimic into established tumors. Data represents mean of each group and error bars are SEM. Day 0 is start of injection and tumor growth was followed for 28 days and each mouse received total of 9 injections. Representative pictures of mice injected with either control or miR-182-5p mimic (mean ± SE,p-value calculated by student’s t-test).
We also checked the cell cycle status of ACHN and Caki-1 cell lines after transfection with miR-182-5p mimic. The G2/M phase of the cell cycle was significantly increased in both ACHN (17% vs 23% p=0.01) and Caki-1(11% vs 15%, p=0.003) as compared to the control (Fig. 3B).
Intratumoral delivery of miR-182-5p leads to tumor regression in ccRCC nude mouse xenografts
ACHN cells were injected into nude mice and maintained until solid, palpable tumors were formed (Fig. 3C). Control miRNA or miR-182-5p mimics were injected intratumorally at periodic intervals of 3-days and tumor growth was monitored weekly. Interestingly, we observed significant tumor growth inhibition in mice injected with miR-182-5p mimics supporting the tumor suppressive effects of this miRNA in ccRCC (Fig. 3C). The average tumor volume in controls was 754.5 mm3 compared to 153.96 mm3 in mice that received miR-182-5p mimic at the end of the experiment. These results confirm the tumor suppressive effect of miR-182-5p in a RCC xenograft model. The statistical significance of each time-point was determined by student’s t-test.
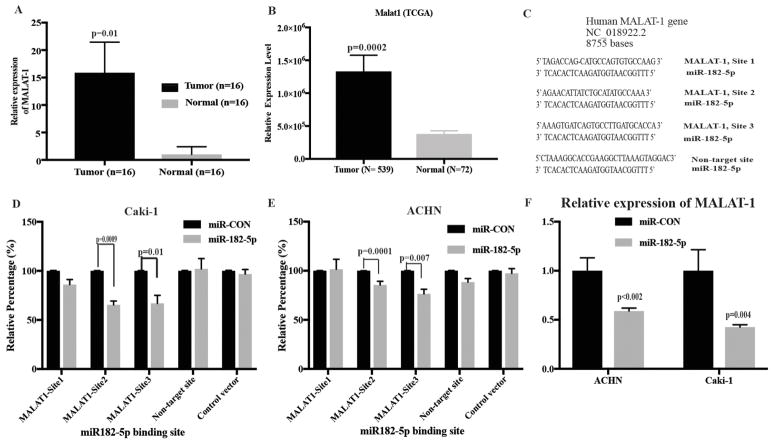
miR-182-5p targets MALAT-1 and decreases its expression in cell lines
Based on our sample cohort and the TCGA database, we observed that MALAT-1 expression is significantly higher in ccRCC compared to normal tissues (Fig. 4A and B). It has been reported that MALAT-1 can modulate cell proliferation through regulating cell cycle progression. Therefore, we hypothesized that miR-182-5p may be involved in controlling ccRCC cell growth by silencing MALAT-1 gene expression. We then used computational algorithms to identify miR-182-5p potential binding sites in the MALAT-1 sequence and identified three potential sites (Fig. 4C). To examine potential miR-182-5p/MALAT-1 interaction experimentally, we performed luciferase reporter assays. Assays with target and non-target MALAT-1 binding sites for miR-182-5p showed that there was decreased luciferase activity with MALAT-1 site 2 and 3 (site 2, p<0.05 and site 3, p<0.05) upon miR-182-5p overexpression while a non-target site had no effect on luciferase activity (Fig. 4D and E). We also found that overexpression of miR-182-5p decreased the levels of MALAT-1 in both ACHN and Caki-1 cells (Fig. 4F).
Figure 4.
MALAT-1 is a direct target of miR-182-5p and upregulated in renal carcinoma. A Quantitative RT-PCR analysis of MALAT-1 in 16 pairs of matched laser micro-dissected tissue samples. B Bar-graph of MALAT-1 expression using TCGA (Normal, n= 72; cancer tissue, n= 539) database. P-value calculated by Mann-Whitney two-tailed test, Bar = mean± SE C Predicted miR-182-5p binding sites in MALAT-1 sequence. D and E Luciferase reporter assays with either wild type, non-target MALAT-1 or luciferase control constructs co-transfected with miR-CON/miR-182-5p in Caki-1 and ACHN cells respectively. F miR-182-5p over-expression caused a significant decrease in MALAT-1 levels compared to control in both the cell lines. P-value calculated by Student t-test. Bar = mean± SE.
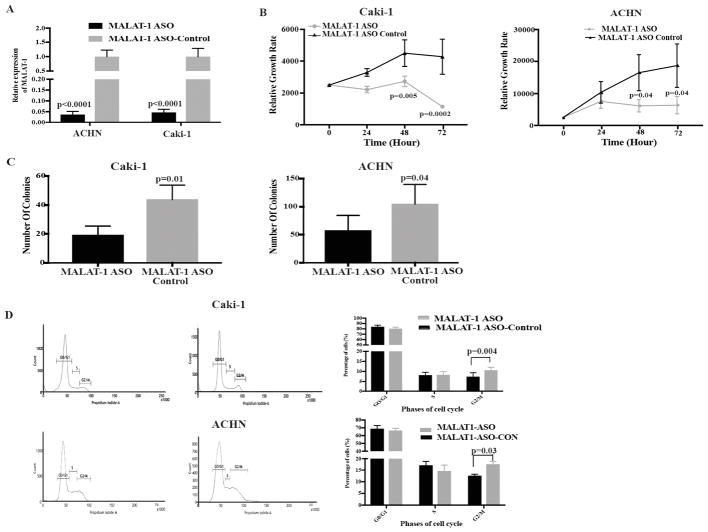
Functional effect of MALAT-1 knockdown
From the above data, we hypothesized that miR-182-5p functions as a tumor suppressor miRNA by downregulating MALAT-1 expression. To check this, we performed MALAT-1 knockdown through ASO-mediated transient transfection in ACHN and Caki-1 cells (Fig. 5A) followed by functional assays. We found that knockdown of MALAT-1 decreased cell and colony formation in both ACHN and Caki-1 cells (Fig. 5B and C).
Figure 5.
MALAT-1 knockdown mimics the effect of miR-182-5p overexpression. A Relative expression of MALAT-1 after transient transfection with MALAT-1 anti-sense oligonucleotide. B Proliferation of Caki-1 and ACHN after anti-MALAT-1 transfection was significantly reduced compared to control. C Anti-MALAT-1 significantly reduces the colony forming ability of both Caki-1 and ACHN. D MALAT-1 knockdown leads to G2/M arrest in both the cell lines. P-value calculated by Student t-test. Bar = mean± SE.
We also examined the effect of MALAT-1 knockdown on cell cycle progression. We found that both ACHN (12.5% vs 17.6%, p-value= 0.03) and Caki-1 cells (7.3% vs 10.5%, p-value= 0.004) showed G2/M arrest after MALAT-1 knockdown compared to controls (Fig. 5D).
miR-182-5p regulates cancer progression through downstream targets of MALAT-1
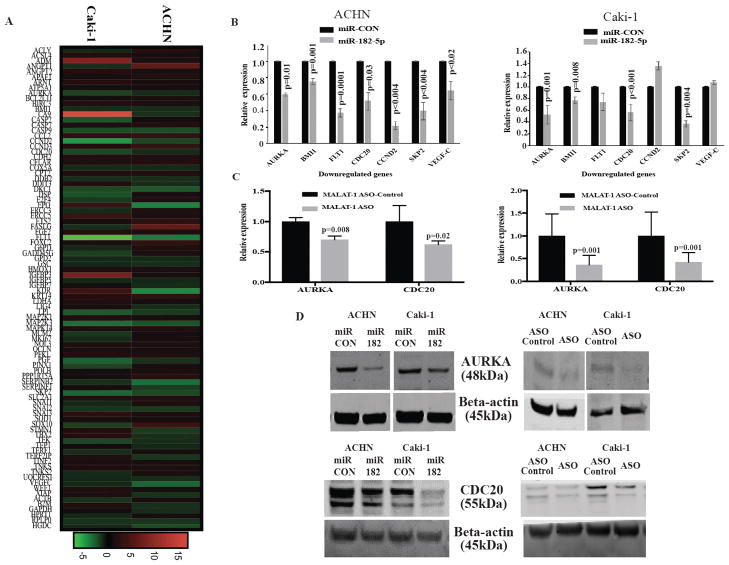
To determine how the overexpression of miR-182-5p influences G2/M progression of the cell cycle through MALAT-1, we next did RT2 qPCR-based array profiling in miR-182-5p overexpressing cells and compared them with negative control. The results showed differences in expression levels for several genes related to apoptosis, cell cycle and angiogenesis in both ACHN and Caki-1 cells (Fig. 6A). miR-182-5p overexpression significantly decreased cell cycle related genes such as -CDC20, AURKA, SKP2 and BMI1. To confirm whether the downregulated genes are related to the cell cycle and its associated pathways, we performed pathway enrichment analysis with the EnrichR pathway analysis tool. The downregulated genes belong to different pathways, including regulation of mitotic cell cycle pathway (Reactome) / cell cycle (KEGG) (Fig. S2). Thus, our cell cycle analyses using flow cytometry corresponded to the qPCR array results. We confirmed the gene expression results from RT2 profiler array by qPCR. Seven genes were selected from 21 that were downregulated in both cell lines (Fig.6B). AURKA and CDC20 protein levels were decreased after overexpression of miR-182-5p in cell lines (Fig. 6D). Knock down of MALAT-1 had similar effects on levels of AURKA and CDC20 suggesting that miR-182-5p exerts its anti-proliferative effect through MALAT-1 (Fig 6C and D).
Figure 6.
miR-182-5p over-expression associated with mitotic progression of the cell cycle. A Heatmap generated from 84 key genes involved in cancer related pathways display the fold cahge for each gene in both Caki-1 and ACHN cells transfected with miR182-5p. Column represent individual cell line and each row represents individual assayed gene. Green represents downregulation of the gene and, red represents upregulation of gene . B qPCR analyses for some of the altered genes after miR-182-5p transfection in both Caki-1 and ACHN cells. C Decreased expression of AURKA and CDC20 in Caki-1 and ACHN cells after MALAT-1 ASO transfection at mRNA level. D Confirmation of decreased expression of AURKA and CDC20 at protein level in Caki-1 and ACHN cells after transfection with miR-182-5p mimic and MALAT-1 ASO transfection compared with respective controls. P-value calculated by Student t-test. Bar = mean± SE.
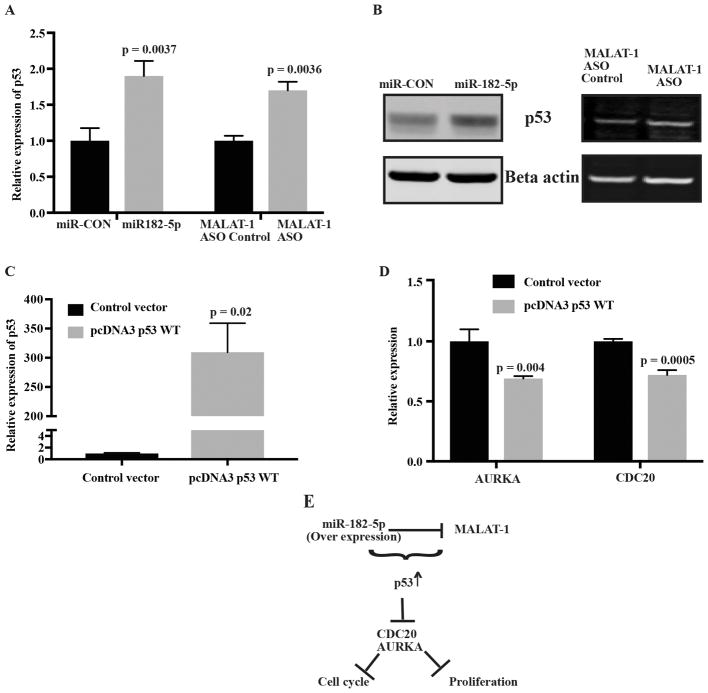
p53 a downstream effector of miR-182-5p overexpression
p53, a downstream target of MALAT-1 is known to downregulate CDC20 (35) and AURKA(36). We looked at changes in the levels of p53 after miR-182-5p overexpression and MALAT-1 knockdown. The levels of p53 increased significantly both at the mRNA and protein levels (Fig 7A and 7B). To confirm whether p53 is involved in cell cycle regulation, we overexpressed p53 in Caki-1 cells (Fig 7C). Overexpression of p53 had no effect on the levels of MALAT-1 and miR-182-5p suggesting that there is no feedback loop amongst them. On the other hand, the levels of CDC 20 and AURKA decreased significantly in p53 overexpressing cells confirming involvement of p53 (Fig 7D). These results show that G2/M arrest is associated with p53 upregulation caused by miR-182-5p overexpression and MALAT-1 downregulation (Fig 7E).
Figure 7.
p53 is a key regulator of miR-182-5p mediated changes. Increased expression of p53 at A mRNA level, B protein level after miR-182-5p over expression and MALAT-1 ASO knockdown in Caki-1 cells. C Relative expression level of p53 after transfection with p53 WT or empty vector. D Relative expression level of CDC20 and AURKA in p53 overexpressed cells. E Schematic representation of miR-182-5p mediated changes in MALAT-1 related downstream pathways through p53.
Discussion
Emerging studies have confirmed the important role of miRNAs in ccRCC tumorigenesis and other cancers. Human miR-182-5p, located at chromosome 7q32 region, is transcribed from a miR-183 family cluster. Contradictory reports describe that miR-182 possesses oncogenic function in prostate, melanoma and ovarian cancer (5–7) while exhibiting tumor-suppressive activity in other cancers such as, lung adenocarcinomas, posterior uveal melanoma and renal carcinoma (10,11,37,38). These studies suggest that miR-182 function is cell context based. Thus, the cell context-dependent network of miR-182-5p regulated genes may determine its biological function in a specific cancer.
Here, in this study, we investigated the status and functional role of miR-182 in ccRCC. In addition, we demonstrated a novel interaction of miR-182-5p with long non-coding RNA MALAT-1. We observed that miR-182 expression was significantly downregulated in human ccRCC clinical samples compared to their matched normal couterparts consistent with the previous reports (10,39,40). A common cause of loss of tumor suppressor miRNAs in human cancer is the silencing of their primary transcripts by CpG island promoter hypermethylation (41). Our in silico analysis in TCGA database showed hypermethylation of miR-182-5p in ccRCC patients. Thus, we considered the possibility that miR-182-5p expression was decreased in ccRCC patients due to promoter hyper-methylation. miR-182 has been reported to be hypermethylated in various cancers (10,33,34). Our results revealed that CpG methylation was significantly higher in renal cancer cells as compared to normal renal cells. Further, a demethylating agent Aza-CdR treatment has been reported to rescue the expression of hypermethylated miRNAs in cancers(42). Our results indicated that treatment with Aza-CdR restored miR-182-5p expression and decreased the expression of key methylation regulating genes such as DNMT1, DNMT3a and DNMT3b in ccRCC cell lines. Thus, confirming that CpG island hyper-methylation is responsible for suppressed miR-182-5p expression in ccRCC.
miRNAs possess several features that make them attractive candidates as new cancer diagnostic and prognostic biomarkers and miRNA signatures have been reported to be useful tools for early diagnosis of cancer (43,44). In this study, we found that miR-182-5p has potential to be a diagnostic biomarker for renal cancer. Further, we observed a tumor suppressor role of miR-182-5p in ccRCC indicated by reduced cell viability, proliferation, colony formation, increased apoptosis and G2/M arrest. In addition, miR-182-5p had tumor suppressor effects in in vivo mouse model, highlighting the fact that restoration of miR-182-5p expression may be an attractive therapeutic modality in ccRCC.
MALAT-1 is generally upregulated in several types of cancer and is considered to be an oncogene (45). We observed MALAT-1 to be overexpressed and its attenuation caused G2/M cell cycle arrest in ccRCC. MALAT-1 has been implicated in cancer cell cycle progression (16,17,46). miR-182 mediated downregulation of MALAT-1, this interaction resulted in suppression of cell cycle promoting genes CDC20 and AURKA, and upregulation of the cell cycle regulator p53. Consistent with our finding, silencing of both genes (CDC20, AURKA) has been known to induce G2/M arrest in various cancers (35,47), whereas, induction of p53 has been reported to induce cell cycle arrest, cellular senescence or apoptosis (48) and is known to suppress the expression of both CDC20 and AURKA (36,49). p53 protein has been shown to activate the transcription of subset of miRNAs including miR-182-5p in neuroblastoma (50). Contrary to this report, in our study there was no change in levels of miR-182 and MALAT-1. However, a decrease in CDC20 and AURKA was observed at transcription level suggesting that p53 is a downstream effector of miR-182-5p/MALAT-1 interaction in ccRCC. These results highlight a novel molecular mechanism of miR-182 interaction with MALAT-1 leading to regulation of cell cycle pathway genes thereby regulating ccRCC progression.
To summarize, we provide evidence that miR-182 is silenced by CpG hypermethylation and may possess diagnostic potential in ccRCC. We also elucidated the tumor suppressor role of miR-182-5p via regulation of MALAT-1 and downstream pathway genes in ccRCC. We have demonstrated for the first time miR-182-5p/MALAT-1 interaction that represents a new mechanism underlying the development of ccRCC. This interaction may provide a better therapeutic opportunity to target ccRCC.
Supplementary Material
Implications.
This is the first study that offers new insight into role of miR-182-5p/MALAT-1 interaction on inhibition of ccRCC progression.
Acknowledgments
We thank Dr. Roger Erickson for his support and assistance with the preparation of the manuscript.
Grant support
This study was supported by the Department of Veterans Affairs VA Merit Review 101 BX001123 and the National Institutes of Health / National Cancer Institute RO1CA199694, RO1CA196848. Dr Dahiya is the recipient of a Senior Research Career Scientist award (#IK6BX004473) from the Department of Veterans Affairs.
Footnotes
Conflict of interest There are no conflicts of interest.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Linehan WM. Genetic basis of kidney cancer: role of genomics for the development of disease-based therapeutics. Genome Res United States. 2012;22:2089–100. doi: 10.1101/gr.131110.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fendler A, Jung K. MicroRNAs as new diagnostic and prognostic biomarkers in urological tumors. Crit Rev Oncog United States. 2013;18:289–302. doi: 10.1615/critrevoncog.2013007176. [DOI] [PubMed] [Google Scholar]
- 4.Fendler A, Stephan C, Yousef GM, Jung K. MicroRNAs as Regulators of Signal Transduction in Urological Tumors. Clin Chem. 2011;57:954–968. doi: 10.1373/clinchem.2010.157727. [DOI] [PubMed] [Google Scholar]
- 5.Wallis CJD, Gordanpour A, Bendavid JS, Sugar L, Nam RK, Seth A. MiR-182 Is Associated with Growth, Migration and Invasion in Prostate Cancer via Suppression of FOXO1. J Cancer Sydney: Ivyspring International Publisher. 2015;6:1295–305. doi: 10.7150/jca.13176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez-Diaz S, et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc Natl Acad Sci U S A United States. 2009;106:1814–9. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu X, Ayub B, Liu Z, Serna VA, Qiang W, Liu Y, et al. Anti-miR182 reduces ovarian cancer burden, invasion, and metastasis: an in vivo study in orthotopic xenografts of nude mice. Mol Cancer Ther United States. 2014;13:1729–39. doi: 10.1158/1535-7163.MCT-13-0982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kouri FM, Hurley LA, Daniel WL, Day ES, Hua Y, Hao L, et al. miR-182 integrates apoptosis, growth, and differentiation programs in glioblastoma. Genes Dev United States. 2015;29:732–45. doi: 10.1101/gad.257394.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu J, Lv G, Zhou S, Zhou Y, Nie B, Duan H, et al. The Downregulation of MiR-182 Is Associated with the Growth and Invasion of Osteosarcoma Cells through the Regulation of TIAM1 Expression. In: Coleman WB, editor. PLoS One Public Library of Science. Vol. 10. 2015. p. e0121175. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Xu X, Wu J, Li S, Hu Z, Xu X, Zhu Y, et al. Downregulation of microRNA-182-5p contributes to renal cell carcinoma proliferation via activating the AKT/FOXO3a signaling pathway. Mol Cancer. 2014;13:109. doi: 10.1186/1476-4598-13-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Li H, Cui L, Feng J, Fan Q. MicroRNA-182 suppresses clear cell renal cell carcinoma migration and invasion by targeting IGF1R. Neoplasma Slovakia. 2016;63:717–25. doi: 10.4149/neo_2016_508. [DOI] [PubMed] [Google Scholar]
- 12.Zhang H, Yang F, Chen S-J, Che J, Zheng J. Upregulation of long non-coding RNA MALAT1 correlates with tumor progression and poor prognosis in clear cell renal cell carcinoma. Tumor Biol. 2015;36:2947–55. doi: 10.1007/s13277-014-2925-6. [DOI] [PubMed] [Google Scholar]
- 13.Hirata H, Hinoda Y, Shahryari V, Deng G, Nakajima K, Tabatabai ZL, et al. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res. 2015;75:1322–31. doi: 10.1158/0008-5472.CAN-14-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuiper RP. Upregulation of the transcription factor TFEB in t(6;11)(p21;q13)-positive renal cell carcinomas due to promoter substitution. Hum Mol Genet. 2003;12:1661–9. doi: 10.1093/hmg/ddg178. [DOI] [PubMed] [Google Scholar]
- 15.Davis IJ, Hsi B-L, Arroyo JD, Vargas SO, Yeh YA, Motyckova G, et al. Cloning of an Alpha-TFEB fusion in renal tumors harboring the t(6;11)(p21;q13) chromosome translocation. Proc Natl Acad Sci. 2003;100:6051–6. doi: 10.1073/pnas.0931430100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X, et al. Long Noncoding RNA MALAT1 Controls Cell Cycle Progression by Regulating the Expression of Oncogenic Transcription Factor B-MYB. In: Clurman BE, editor. PLoS Genet Public Library of Science. Vol. 9. 2013. p. e1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu L, Wu Y, Tan D, Meng H, Wang K, Bai Y, et al. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J Exp {&} Clin Cancer Res. 2015;34:7. doi: 10.1186/s13046-015-0123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Tang X, Shi M, Wen C, Shen B. MiR-216a decreases MALAT1 expression, induces G2/M arrest and apoptosis in pancreatic cancer cells. Biochem Biophys Res Commun. 2017;483:816–22. doi: 10.1016/j.bbrc.2016.12.167. [DOI] [PubMed] [Google Scholar]
- 19.Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res England. 2010;38:5366–83. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao H, Tang K, Liu P, Chen K, Hu J, Zhen J, et al. LncRNA MALAT1 functions as a competing endogenous RNA to regulate ZEB2 expression by sponging miR-200s in clear cell kidney carcinoma. Oncotarget. 2015;6:38005–15. doi: 10.18632/oncotarget.5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dasgupta P, Kulkarni P, Majid S, Varahram S, Hashimoto Y, Bhat NS, et al. MicroRNA-203 inhibits long noncoding RNA HOTAIR and regulates tumorigenesis through epithelial-to-mesenchymal transition pathway in renal cell carcinoma. Mol Cancer Ther. 2018 doi: 10.1158/1535-7163.MCT-17-0925. molcanther.0925.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Majid S, Saini S, Dar AA, Hirata H, Shahryari V, Tanaka Y, et al. MicroRNA-205 inhibits Src-mediated oncogenic pathways in renal cancer. Cancer Res. 2011:71. doi: 10.1158/0008-5472.CAN-10-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saini S, Yamamura S, Majid S, Shahryari V, Hirata H, Tanaka Y, et al. MicroRNA-708 induces apoptosis and suppresses tumorigenicity in renal cancer cells. Cancer Res. 2011;71:6208–19. doi: 10.1158/0008-5472.CAN-11-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamamura S, Saini S, Majid S, Hirata H, Ueno K, Chang I, et al. Microrna-34a suppresses malignant transformation by targeting c-myc transcriptional complexes in human renal cell carcinoma. Carcinogenesis. 2012;33:294–300. doi: 10.1093/carcin/bgr286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ueno K, Hirata H, Shahryari V, Chen Y, Zaman MS, Singh K, et al. Tumour suppressor microRNA-584 directly targets oncogene Rock-1 and decreases invasion ability in human clear cell renal cell carcinoma. Br J Cancer. 2011;104:308–15. doi: 10.1038/sj.bjc.6606028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loughery J, Cox M, Smith LM, Meek DW. Critical role for p53-serine 15 phosphorylation in stimulating transactivation at p53-responsive promoters. Nucleic Acids Res. 2014;42:7666–80. doi: 10.1093/nar/gku501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu R-J, He W, Wang X-B, Li L, Zhao H-B, Liu X-Y, et al. DNMT1-maintained hypermethylation of Krüppel-like factor 5 involves in the progression of clear cell renal cell carcinoma. Cell Death & Dis The Author(s) 2017;8:e2952. doi: 10.1038/cddis.2017.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Díez-Villanueva A, Mallona I, Peinado MA. Wanderer, an interactive viewer to explore DNA methylation and gene expression data in human cancer. Epigenetics {&} Chromatin. 2015;8:22. doi: 10.1186/s13072-015-0014-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nip H, Dar AA, Saini S, Colden M, Varahram S, Chowdhary H, et al. Oncogenic microRNA-4534 regulates PTEN pathway in prostate cancer. Oncotarget. 2016;7:68371–84. doi: 10.18632/oncotarget.12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trang P, Medina PP, Wiggins JF, Ruffino L, Kelnar K, Omotola M, et al. Regression of murine lung tumors by the let-7 microRNA. Oncogene Macmillan Publishers Limited. 2010;29:1580–7. doi: 10.1038/onc.2009.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics England. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res England. 2016;44:W90–7. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marzec-Kotarska B, Cybulski M, Kotarski JC, Ronowicz A, Tarkowski R, Polak G, et al. Molecular bases of aberrant miR-182 expression in ovarian cancer. Genes Chromosom Cancer. 2016;55:877–89. doi: 10.1002/gcc.22387. [DOI] [PubMed] [Google Scholar]
- 34.Anwar SL, Krech T, Hasemeier B, Schipper E, Schweitzer N, Vogel A, Kreipe H, Buurman R, Skawran BLU. hsa-mir-183 is frequently methylated and related to poor survival in human hepatocellular carcinoma. World J Gastroenterol. 2017;23:1568–75. doi: 10.3748/wjg.v23.i9.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kidokoro T, Tanikawa C, Furukawa Y, Katagiri T, Nakamura Y, Matsuda K. CDC20, a potential cancer therapeutic target, is negatively regulated by p53. Oncogene Nature Publishing Group. 2007;27:1562–71. doi: 10.1038/sj.onc.1210799. [DOI] [PubMed] [Google Scholar]
- 36.Sasai K, Treekitkarnmongkol W, Kai K, Katayama H, Sen S. Functional Significance of Aurora Kinases–p53 Protein Family Interactions in Cancer. Front Oncol. 2016:247. doi: 10.3389/fonc.2016.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan D, Dong X, Da Chen X, Yao S, Wang L, Wang J, et al. Role of MicroRNA-182 in Posterior Uveal Melanoma: Regulation of Tumor Development through MITF, BCL2 and Cyclin D2. In: Gonzalez P, editor. PLoS One. Vol. 7. 2012. p. e40967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun Y, Fang R, Li C, Li L, Li F, Ye X, et al. Hsa-mir-182 suppresses lung tumorigenesis through down regulation of RGS17 expression in vitro. Biochem Biophys Res Commun. 2010:396. doi: 10.1016/j.bbrc.2010.04.127. [DOI] [PubMed] [Google Scholar]
- 39.Fu J-H, Yang S, Nan C-J, Zhou C-C, Lu D-Q, Li S, et al. MiR-182 affects renal cancer cell proliferation, apoptosis, and invasion by regulating PI3K/AKT/mTOR signaling pathway. Eur Rev Med Pharmacol Sci. 2018;22:351–7. doi: 10.26355/eurrev_201801_14179. [DOI] [PubMed] [Google Scholar]
- 40.Wang YQ, Guo RD, Guo RM, Sheng W, Yin LR. MicroRNA-182 promotes cell growth, invasion, and chemoresistance by targeting programmed cell death 4 (PDCD4) in human ovarian carcinomas. J Cell Biochem. 2013:114. doi: 10.1002/jcb.24488. [DOI] [PubMed] [Google Scholar]
- 41.Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell United States. 2006;9:435–43. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 42.Majid S, Dar AA, Ahmad AE, Hirata H, Kawakami K, Shahryari V, et al. BTG3 tumor suppressor gene promoter demethylation, histone modification and cell cycle arrest by genistein in renal cancer. Carcinogenesis. 2009;30:662–70. doi: 10.1093/carcin/bgp042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: A new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010:2087–92. doi: 10.1111/j.1349-7006.2010.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heinzelmann J, Henning B, Sanjmyatav J, Posorski N, Steiner T, Wunderlich H, et al. Specific miRNA signatures are associated with metastasis and poor prognosis in clear cell renal cell carcinoma. World J Urol. 2011;29:367–73. doi: 10.1007/s00345-010-0633-4. [DOI] [PubMed] [Google Scholar]
- 45.Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res United States. 2013;73:1180–9. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X, Li M, Wang Z, Han S, Tang X, Ge Y, et al. Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. J Biol Chem United States. 2015;290:3925–35. doi: 10.1074/jbc.M114.596866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang A, Gao K, Chu L, Zhang R, Yang J, Zheng J. Aurora kinases: novel therapy targets in cancers. Oncotarget. 2017;8:23937–54. doi: 10.18632/oncotarget.14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoe KK, Verma CS, Lane DP. Drugging the p53 pathway: understanding the route to clinical efficacy. Nat Rev Drug Discov. 2014;13:217–36. doi: 10.1038/nrd4236. [DOI] [PubMed] [Google Scholar]
- 49.Banerjee T, Nath S, Roychoudhury S. Nucleic Acids Res. Vol. 37. Oxford University Press; 2009. DNA damage induced p53 downregulates Cdc20 by direct binding to its promoter causing chromatin remodeling; pp. 2688–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rihani A, Goethem A, Van Ongenaert M, De Brouwer S, Volders P-J, Agarwal S, et al. Genome wide expression profiling of p53 regulated miRNAs in neuroblastoma. Sci reports. 2015;12(5):9027. doi: 10.1038/srep09027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.