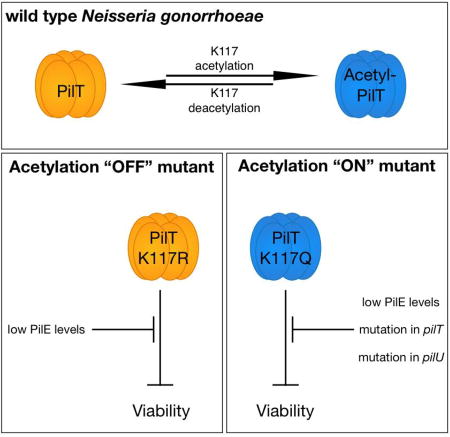
Summary
Post-translational acetylation is a common protein modification in bacteria. It was recently reported that Neisseria gonorrhoeae acetylates the Type IV pilus retraction motor, PilT. Here, we show recombinant PilT can be acetylated in vitro and acetylation does not affect PilT ultrastructure. To investigate the function of PilT acetylation, we mutated an acetylated lysine, K117, to mimic its acetylated or unacetylated forms. These mutations were not tolerated by wild-type Neisseria gonorrhoeae, but they were tolerated by Neisseria gonorrhoeae carrying an inducible pilE when grown without inducer. We identified additional mutations in pilT and pilU that suppress the lethality of K117 mutations. To investigate the link between PilE and PilT acetylation, we found the lack of PilE decreases PilT acetylation levels and increases the amount of PilT associated with the inner membrane. Finally, we found no difference between wild-type and mutant cells in transformation efficiency, suggesting neither mutation inhibits Type IV pilus retraction. Mutant cells, however, form microcolonies morphologically distinct from wt cells. We conclude that interfering with the acetylation status of PilT K117 greatly reduces N. gonorrhoeae viability, and mutations in pilT, pilU, and pilE can overcome this lethality. We discuss the implications of these findings in the context of Type IV pilus retraction regulation.
Keywords: protein acetylation, Neisseria gonorrhoeae acetylome, PilT acetylation, PilU, Type IV pilus
Graphical abstract
Introduction
Post-translational acetylation of proteins is an important determinant of eukaryotic cell activity. Acetylation of a protein can affect its catalytic activity, interactions with other cellular components, or assembly of large protein complexes (Choudhary et al., 2009; Choudhary et al., 2014). Protein acetylation is emerging as a common regulatory theme in prokaryotes. Protein “acetylomes” have been described for Escherichia coli and Salmonella enterica, and recently for Neisseria gonorrhoeae (Ngo) (Wang et al., 2010; Castano-Cerezo et al., 2014; Post et al., 2017). The acetylation of bacterial proteins is known to influence metabolic flux (Wang et al., 2010; Castano-Cerezo et al., 2014; Ouidir et al., 2016); less well understood is the effect of protein acetylation on the function of nonmetabolic proteins.
The Ngo acetylome includes PilT, a nonmetabolic protein that participates in the biogenesis of the Type IV pilus (Tfp), a multicomponent structure expressed by many prokaryotes (reviewed in Giltner et al., 2012). In most bacteria, the Tfp complex consists of an extracellular retractile filament anchored to the cell envelope through assembly components. The filament, a helical polymer of PilE subunits and minor proteins, extends and retracts through the coordinated action of the Tfp biogenesis machinery at the interface of the periplasm and inner-membrane.
The biochemical steps in Tfp assembly and retraction have not been defined, nor have the signal(s) and mechanism(s) that coordinate the switch between fiber assembly and retraction. Studies show that ATP hydrolysis by the cytosolic PilF protein energizes PilE insertion into the base of the fiber through interactions with inner membrane components PilM and PilG (Carbonnelle et al., 2006; McLaughlin et al., 2012; Georgiadou et al., 2012a; Takhar et al., 2013; Chang et al., 2016; Mancl et al., 2016; McCallum et al., 2017). ATP hydrolysis by the hexameric PilT motor causes the filament to retract, presumably by the assembly mechanism in reverse, i.e. interactions of PilT with PilG and PilM disassemble PilE subunits from the base of the pilus (McLaughlin et al., 2012; Georgiadou et al., 2012b; Chang et al., 2016; McCallum et al., 2017). Cryo-electron tomography studies suggest PilF and PilT occupy the same space at the cytosolic face of the Tfp assembly complex, implying occupancy by one or the other of these ATPases determines fiber extension or retraction (Gold et al., 2015; Chang et al., 2016).
In this study, we show biochemically that Ngo PilT is acetylated. We provide genetic evidence that Ngo does not tolerate amino acid substitutions in the acetylatable lysine at position 117 (K117). Suppressing PilE expression, and secondary mutations in pilT and pilU, a Tfp-associated gene co-transcribed with pilT, overcome the reduced viability of pilTK117 mutants. Finally, mutations in PilT K117 do not affect Tfp retraction, per se, but affect the social behavior of Ngo cells. We conclude that blocking the ability of Ngo to toggle between PilT K117 acetylation ON and OFF states is deleterious to the cell.
Results
PilT is acetylated by Ngo
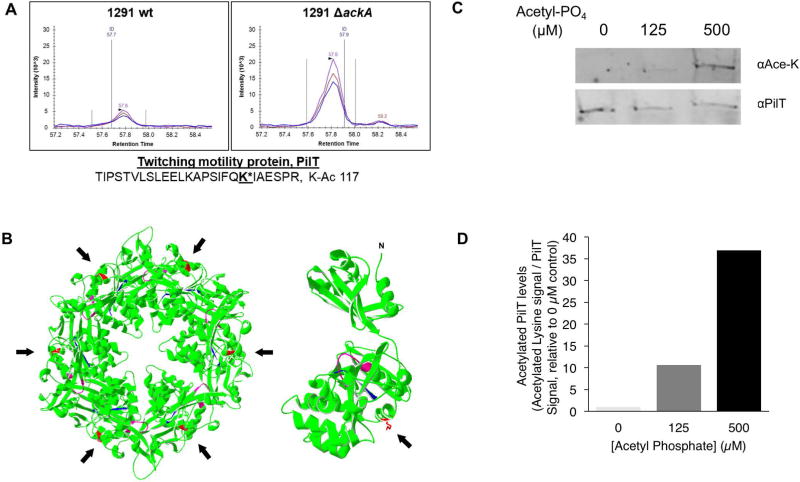
All Neisseria gonorrhoeae strains examined to date acetylate their proteins post-translationally (Fig S1). The Ngo acetylome includes PilT, which comprises the hexameric motor that retracts Type IV pili fibers (Post et al., 2017). Several lysines in PilT are acetylated, the most abundant acetylated residue being lysine 117 (K117). Moreover, the level of acetylated K117 is linked to expression of ackA, a gene involved in the non-enzymatic protein acetylation pathway (Fig. 1A). The acetylated PilT K117 peptide was consistently observed 3.2-fold more frequently in Ngo 1291 ΔackA compared to wt 1291. In light of these findings, we sought to determine the importance of PilT K117 acetylation in Tfp retraction.
Fig 1. PilT is acetylated at K117 in vivo and in vitro.
(A) MS1 ion chromatograms of the precursor ions for M (blue), M+1 (purple), and M+2 (red) for the PilT K117 acetylated peptide isolated from 1291 wt (left) or 1291 ΔackA (right) cells. (B) Predicted structure of the Ngo PilT hexamer (left) and monomer (right) with the K117 side chain indicated by the arrow. Walker box A, magenta; Walker box B, cyan; K117 acetylation site, red. Arrowheads indicate the position of the acetylated lysine K117 in the predicted structures. (C) Immunoblot of purified recombinant PilT after incubation with varying concentrations of acetyl-phosphate (AcP) and immunoprecipitation with anti-PilT antibodies. Representative of 4 independent experiments. (D) Densitometry of panel C demonstrating in vitro acetylated PilT levels.
K117 is 15 amino acids upstream of the first residue of Walker box A, the catalytic domain of the PilT ATPase (Fig. 1B). Modeling of the Ngo PilT monomer maps K117 to a region linking the N- and C-terminal globular domains. The K117 side chain is surface exposed, and distal to the PilT catalytic domain. In the modeled hexamer, K117 does not lie at subunit interfaces; rather, it extends from the exterior rim of the disc and is spatially separated from the internally located catalytic domains.
Purified recombinant PilT (rPilT) assembles into native hexamers (Hockenberry et al., 2016). To determine whether PilT can be acetylated in vitro, an equal number of rPilT hexamers was incubated with increasing concentrations of acetyl-phosphate (AcP), and the products were separated on a non-denaturing gel and immunoblotted simultaneously with antibodies to PilT and acetylated lysine (Ace-K). Results indicate the PilT hexamer is acetylated in vitro in an AcP concentration dependent manner (Fig. 1C). The simultaneous probing of the filter with αPilT and αAce-K antibodies may have resulted in variation in PilT signals, with the two antibodies interfering with each other for binding. Acetylation did not alter the migration properties of the PilT hexamer, indicating it did not alter subunit-subunit interactions.
Ngo does not tolerate mutations in the PilT acetylation site K117
To understand the effect of PilT acetylation on protein function, we attempted to substitute K117 with an amino acid that either mimics an acetylated residue (K117Q) or cannot be acetylated (K117R and K117A). Mutagenesis was performed in E. coli; the constructs were marked with a Kanamycin resistance (KanR) cassette and all plasmid constructs were verified by sequencing before they were transformed into Ngo MS11. A mutant pilTL201C-Kan construct tolerated by Ngo was used as a control. Four independent transformation experiments were performed for each plasmid. Constructs with mutant acetylation sites repeatedly transformed Ngo at a 100-fold lower frequency than the control plasmid containing Ngo pilTL201C (Table 1).
Table 1.
Transformation frequency wt and ipilE strains using pilTK117 mutant DNAs and secondary mutations in transformants.
| PilT mutation | Genotype of recipient |
Transformation frequency (approximate) |
# Transformations performed |
# KanR transformants sequenced |
# KanR transformants with targeted pilT mutation |
# KanR transformants with secondary mutations in pilT, pilT2 or pilU |
|---|---|---|---|---|---|---|
| L201C | wt | ~10−5 | 1 | 10 | 8 | 0 |
| K117Q | wt | ~10−7 | 4 | 42 | 5 | 5 |
| K117R | wt | ~10−7 | 4 | 42 | 0 | 0 |
| K117A | wt | ~10−7 | 4 | 42 | 0 | 0 |
| K117Q | ipilE | ~10−6 | 1 | 10 | 6 | 0 |
| K117R | ipilE | ~10−6 | 1 | 10 | 2 | 0 |
Forty-two KanR clones from each of the K117Q, K117R and K117A transformations were sequenced at the pilT locus. All K117R and K117A clones and 37 K117Q clones were wt at the pilT locus (Table 1). Of the other 5 K117Q clones, 4 had an in-frame 12-basepair deletion upstream of K117Q. In this context, it should be noted that one of the deleted codons encodes an acetylatable lysine, K110 (Fig S2). These 4 clones are likely siblings as they were isolated from the same transformation reaction. Computational modeling of this PilT variant shows the mutation shortens the unstructured region linking the N- and C-terminal domains and reorients the K117 side chain so that it is no longer surface exposed (Fig S2A, B). The fifth clone, obtained from a separate K117Q transformation, had a single base insertion in pilU, a Tfp-associated gene co-transcribed with pilT. The insertion occurred between Walker box A and B, and would result in a truncated and presumably catalytically inactive PilU (Fig. S2C). By contrast, 8/10 pilTL201C transformants selected for sequencing had the targeted mutation in pilT and no secondary mutations in either pilT or pilU. These results suggest that secondary mutations in pilT and pilU can alleviate the toxicity of the mutations at the PilT K117 acetylation site.
Suppression of pilE expression alleviates the toxicity of the pilT K117 mutations
It is unclear why Ngo does not tolerate mutations in PilT K117. We tested whether PilE influences the viability of the pilTK117 mutants. As mentioned, PilT is proposed to cause the Tfp fiber to retract by disassembling PilE subunits from the base of the fiber, and the disassembled subunits are held in the inner membrane until they are reassembled into the fiber by the PilF assemblase (McLaughlin et al., 2012; Chang et al., 2016). In this model, PilE inner membrane levels are balanced by Tfp assembly and retraction. We hypothesized that PilT K117 mutant proteins cause a detrimental accumulation of PilE through aberrant retraction signaling. To test this hypothesis, we constructed a strain in which pilE is transcriptionally fused to the IPTG-inducible lac promoter (ipilE). In the absence of IPTG, ipilE produced negligible amounts of PilE (Fig. S3). ipilE cells were transformed with pilTK117Q and pilTK117R DNA during a brief exposure to IPTG and plated on Kanamycin-containing GCB agar without IPTG. Under these conditions, ipilE cells transformed at a 10-fold higher frequency than wt cells (Table 1).
Ten KanR transformants from each of the pilTK117Q and pilTK117R transformation experiments were analyzed further. Six pilTK117Q and 2 pilTK117R transformants had the targeted mutation in pilT with no secondary mutations in pilT, pilU, pilF, or pilT2 (which is hypothesized to encode an alternative Tfp motor (Kurre et al., 2012)).
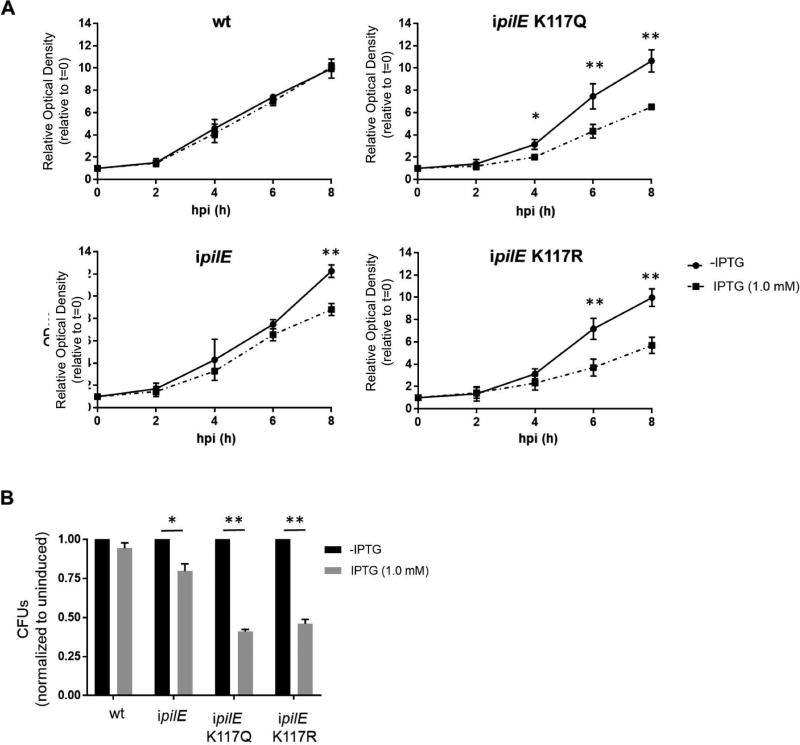
We further examined one ipilE pilTK117Q and one ipilE pilTK117R transformant with the targeted pilT mutation and no secondary mutations in pilT, pilU, pilT2 or pilF. We determined whether induction of pilE expression would affect the growth of these clones. Wt, ipilE, ipilE pilTK117Q and ipilE pilTK117R were grown to log phase in broth, IPTG was added to half of each culture and the optical density (OD600) of the cultures was monitored over time. Wt grew equally well in the presence and absence of IPTG (Fig 2A). ipilE grew at similar rates in the presence and absence of IPTG until 8 hours post-induction (hpi), when the density of the induced culture was visibly lower. The reason for this late stage growth difference is unclear. It is possible that prolonged IPTG induction led to a detrimental accumulation of PilE, as has been observed when PilE is overexpressed in E. coli. In contrast, IPTG induction slowed the growth of ipilE pilTK117Q and ipilE pilTK117R soon after 4 hpi. Viable counts of the 8-hour cultures confirmed the OD600 readings; CFUs of the induced ipilE pilTK117Q and ipilE pilTK117R cultures were ~60% lower than those of uninduced cultures (Fig 2B). Taken together, these results strongly suggest that the presence of PilE is detrimental to the growth of the pilTK117Q and pilTK117R mutants, and shutting down pilE expression can suppress the lethality of these mutations.
Fig 2. Growth of ipilE pilTK117Q and ipilE pilTK117R is inhibited when pilE is induced.
5.0 × 108 wt, ipilE, ipilE pilTK117Q and ipilE pilTK117R cells were inoculated into supplemented GCB in the presence or absence of 1.0 mM IPTG and incubated for 8 h. (A) OD600 of cultures. Average of 3 independent experiments. (B) Viable counts of cultures grown in the presence or absence of IPTG at 8 hours after IPTG induction. Cultures at 8 hpi were diluted and plated on agar without IPTG. Values are CFU counts of cultures grown in the presence of IPTG normalized to those grown in the absence of IPTG. Average of 3 independent experiments. Student’s unpaired t-test; (*) p>0.05, (**) p>0.01.
Secondary mutations in pilT and pilU alleviate the toxicity of the pilT K117 mutations
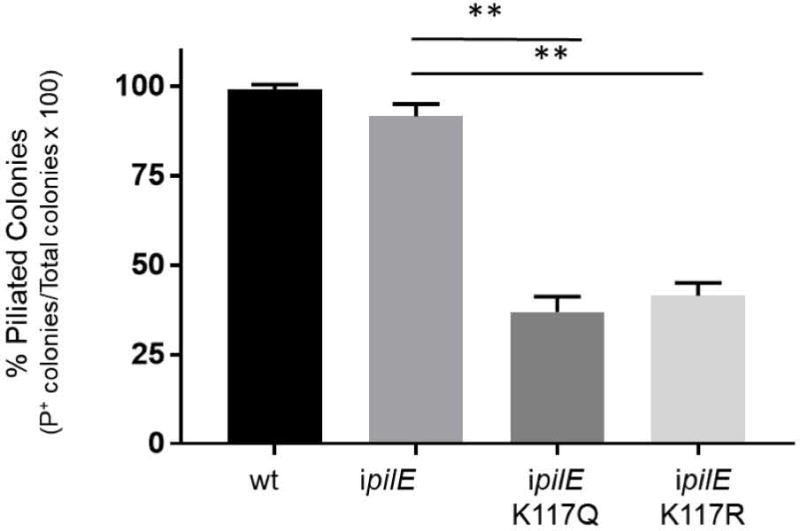
IPTG induction of pilE expression did not completely inhibit the growth of ipilE pilTK117Q and ipilE pilTK117R (Fig 2A, B). We examined these mutants for compensatory mutations that allowed them to grow in the presence of PilE. Wt, ipilE, ipilE pilTK117Q and ipilE pilTK117R cells were grown in broth with IPTG for 8 hours and plated on agar with or without IPTG. Colonies on IPTG plates were analyzed by several methods. First, the piliation status of the colonies was assessed by microscopy. Approximately 10% of ipilE colonies on IPTG plates were nonpiliated, a value comparable to wt Ngo (Fig. 3). By contrast, 60% of ipilE pilTK117Q and ipilE pilTK117R colonies on IPTG plates were nonpiliated. All nonpiliated ipilE pilTK117Q and ipilE pilTK117R colonies on IPTG plates had the targeted mutations in pilT; their pilU and ipilE loci (lac promoter and pilE ORF) were unaltered. The mechanism(s) underlying the nonpiliated phenotype in these isolates remains to be determined.
Fig 3. Piliation state of ipilE pilTK117Q and ipilE pilTK117R colonies on IPTG agar.
The percentage of piliated cells in wt, ipilE, ipilEK117Q and ipilEK117R cultures grown for 8 hours in broth containing IPTG. Average of 2 independent experiments. (*) p<0.05, (**) p<0.01; Student’s unpaired t-test.
We next focused on the mutants that maintained a piliated phenotype upon growth on IPTG plates. The pilT locus of 48 piliated ipilE pilTK117Q and ipilE pilTK117R colonies on IPTG plates was sequenced. pilT had reverted to wt in 33/48 ipilE pilTK117Q clones and 48/48 ipilE pilTK117R clones. In contrast, all ipilE pilTK117Q and ipilE pilTK117R piliated colonies on control plates without IPTG had the targeted mutation in pilT. This indicates there is great selective pressure on Ngo cells to maintain the PilT K117 acetylation site.
In the above experiments, 15 piliated pilTK117Q colonies on IPTG agar retained the targeted mutation in pilT. We determined whether they had secondary mutations in pilE, pilF, pilU, pilT2, pilM, or pilG. Results show that the lac promoter and pilE ORF in all 15 clones were unaltered. One clone had a tyrosine (Y) to histidine (H) substitution and a single base insertion at the midpoint of pilU that would prematurely terminate translation (Fig. S4). This corroborates our previous observation that a secondary mutation in pilU allowed Ngo to tolerate the K117Q mutation (Fig. S2). In the other 14 clones, pilU, pilF, pilT2, pilG, and pilM were unaltered (ORF and 100 bp of upstream and downstream sequence). The mutations that alleviate the toxicity of the PilTK117Q mutation have not yet been determined; nevertheless, these findings indicate that blocking the ability of Ngo to toggle the acetylation status of PilT K117 is detrimental to the cell.
PilE presence promotes PilT acetylation and inhibits PilT membrane localization
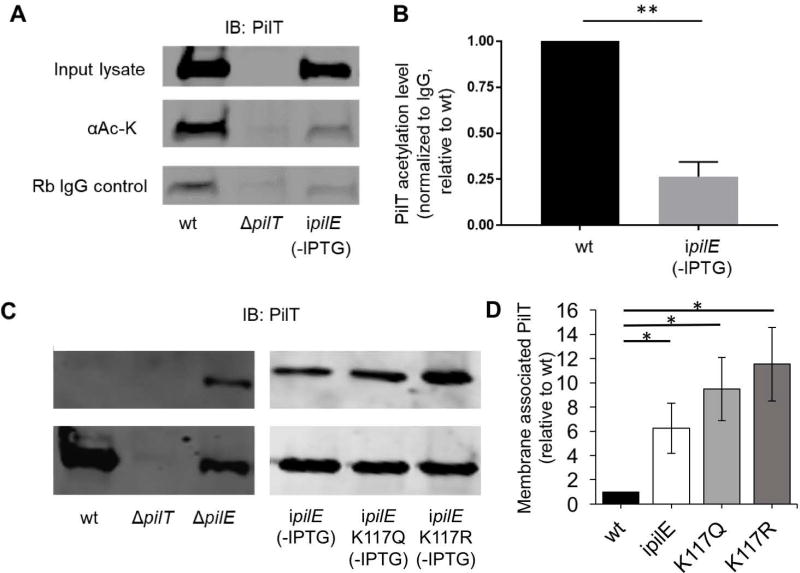
We showed that the presence of PilE affects the viability of pilTK117 mutants. We therefore asked whether PilT acetylation is sensitive to PilE levels, by determining the level of acetylated PilT in wt, ΔpilT, and ipilE. Cells were grown in broth without IPTG for 4 hours, and their lysates were immunoprecipitated with a monoclonal antibody specific for acetylated lysine residues or a control isotype antibody, and immunoblotted with antibodies to PilT. Wt cells have approximately 5-fold more acetylated PilT than cells without PilE (Fig 4A, B), strongly suggesting PilT acetylation is linked to cellular PilE levels.
Fig 4. The level of acetylated PilT is linked to the presence of PilE.
(A) Level of acetylated PilT in whole cell lysates of wt, ΔpilT, and ipilE (uninduced). Lysates were immunoprecipitated with acetylated lysine (Ac-K) or isotype control (Rb IgG) antibodies, separated by 15% SDS-PAGE and immunoblotted with PilT antibodies. A representative immunoblot is shown. (B) Graph of acetylated PilT signals from wt and ipilE cells from 3 independent immunoblots, normalized to the IgG control signal. (C) PilT levels in total membrane and cytosolic/periplasmic fractions of wt, ΔpilT, ΔpilE, and uninduced ipilE, ipilE pilTK117Q, and ipilE pilTK117R cells. Fractions were separated on a 15% denaturing acrylamide gel and immunoblotted with PilT antibodies. Representative of 3 independent experiments. (D) Densitometry analysis of western blots from panel C. PilT levels from membrane fraction were divided by PilT levels in the soluble fraction and are expressed relative to Ngo. (*) p<0.05, (**) P<0.01; Student’s unpaired t-test.
We also examined the subcellular location of PilT in the presence and absence of PilE. A previous report shows PilT is in the cytosol, associated with the inner membranes in wt Ngo (Brossay et al., 1994). Wt, ΔpilT, ΔpilE, ipilE, ipilE pilTK117Q, and ipilE pilTK117R cells were grown in broth without IPTG and their soluble (cytosolic/periplasmic) and membrane (inner and outer membrane) fractions were immunoblotted with antibodies to PilT. Antibodies to GAPDH and H.8 were used to monitor the purity of the cytosolic and membrane fractions, respectively (Fig. S5). In wt cells, PilT was located primarily in the soluble (cytosolic/periplasmic) fraction (Fig 4C). However in ΔpilE, uninduced ipilE, ipilE pilTK117Q, and ipilE pilTK117R cells, PilT was found in both the soluble and membrane fractions (Fig 4C). Although the differences between strains were not statistically significant, ipilE pilTK117Q and ipilE pilTK117R cells consistently contained more membrane-associated PilT than ipilE cells; and ipilE pilTK117R consistently contained the highest levels of membrane-associated PilT (Fig 4D). Attempts to perform these experiments with the ipilE strains under IPTG inducing conditions were unsuccessful because of the large number of bacteria required for the assay and the reduced growth rate of the mutants when pilE is expressed.
PilT acetylation status is a determinant of Ngo social behavior but not genetic competence
Finally, we determined whether the K117Q and K117R mutations affected PilT function, using DNA transformation and microcolony formation as readouts. The growth defect of ipilE pilTK117Q and ipilE pilTK117R was visible beginning 4 hours after IPTG induction (Fig 2A). We therefore determined their transformation frequency at 3 hours post-induction, using genomic DNA from a Rifampicin resistant (RifR) Ngo strain (Table S1). As expected, wt transformed at the same frequency in the presence and absence of IPTG; and ipilE, ipilE pilTK117Q and ipilE pilTK117R were not transformable in the absence of IPTG (data not shown). In the presence of IPTG, ipilE, ipilE pilTK117Q and ipilE pilTK117R transformed at wt frequencies. Thus, mutations in the PilT acetylation site do not affect Ngo genetic competence, and, by implication, its ability to retract Tfp.
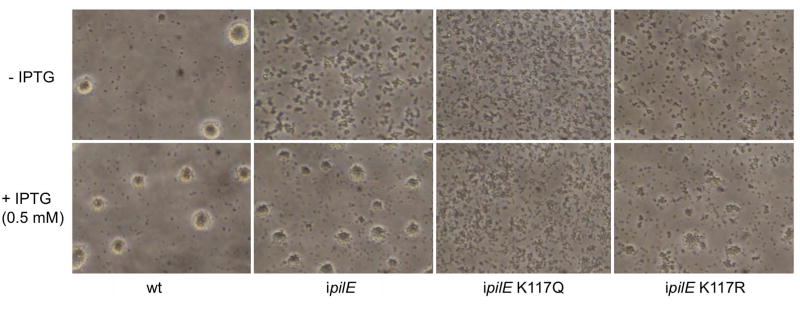
Ngo cells crawl together and organize into spherical microcolonies. This process requires the Tfp retraction. pilTL201C cells expressing an attenuated PilT ATPase motor form aberrantly shaped microcolonies and flatter biofilms (Hockenberry et al., 2016). We determined whether the K117Q and K117R mutations affect microcolony formation. Wt, ipilE, ipilE pilTK117Q and ipilE pilTK117R were examined microscopically after 3 hours of growth in the presence and absence of IPTG (Fig 5). As expected, wt cells aggregated into microcolonies with typically spherical shapes in the presence and absence of IPTG, while ipilE cells formed microcolonies only in the presence of IPTG. ipilE pilTK117Q formed small aggregates in the presence and absence of IPTG. ipilE pilTK117R cells exhibited a different behavior. In the presence of IPTG, they formed microcolonies that were flatter and smaller in diameter than wt structures. The PilT acetylation mutants could not be assessed for biofilm formation because they lost viability in the presence of IPTG. Taken together, these experiments indicate that the two mutations in the K117 acetylation site affects Ngo social behavior in different ways but have no effect on DNA transformation.
Fig 5. ipilE pilTK117Q and ipilE pilTK117R form morphologically distinct microcolonies.
Wt, ipilE, ipilE pilTK117Q, and ipilE pilTK117R were inoculated into microtiter dishes (5.0 × 108) containing supplemented GCB with IPTG (0.5 mM) or without IPTG, incubated for 3 h, and imaged by phase contrast microscopy. Representative of 3 independent experiments.
Discussion
In this report, we provided biochemical evidence that PilT, the Type IV pilus (Tfp) retraction motor protein of Ngo, is acetylated in vitro and in vivo (Fig. 1). We presented genetic evidence that Ngo does not tolerate mutations in the PilT K117 acetylation site that mimics either a constitutively acetylated (K117Q) or unacetylaed (K117R and K117A) residue (Table 1). Using an IPTG-inducible pilE strain, we showed the presence of PilE significantly slows the growth of pilTK117Q and pilTK117R mutants (Fig. 2A).
When pilT acetylation mutants were grown in the presence of PilE, the only viable isolates had a wt pilT, inhibited pilE expression, or had a secondary mutation in pilT or pilU (Table 1, Figs. 3, S1, S3). The compensating mutation in pilT was an in-frame deletion upstream of K117 that removes 4 amino acids, including an acetylatable lysine, K110. Structure prediction of this protein suggests the mutation shifts the K117 sidechain from a surface-exposed to buried position. A more complete structural analysis of this suppressor mutant can disentangle the mechanism in which this mutation abrogates the K117Q mediated lethality.
Frameshift mutations in pilU also abrogated the lethality of the PilTK117Q mutation (Figs. S1, S3). How these mutations alleviate the lethality of PilT K117 mutation is unclear. Only one suppressor mutation renders PilU presumably catalytically inactive. Both, however, result in the deletion of two acetylatable lysines within the C-terminal domain (Figs. S1, S3). PilU was long speculated to play a role in Tfp retraction due its co-transcription with and structural similarity to PilT. Ngo and Neisseria meningitidis mutants deleted of pilU show differences in microcolony formation and infection dynamics compared to wt, further implicating a role for PilU in Tfp retraction (Park et al., 2002; Eriksson et al., 2012). Our findings lend support to this idea. As PilU is also acetylated (Post et al., 2017), it is tempting to speculate that Tfp retraction dynamics are coordinated by differential acetylation of PilT and PilU. The suppressor mutants isolated in this study will be valuable tools for understanding the role of acetylation in PilT function, as well as the function of PilU in Tfp retraction.
Why do the pilTK117Q, pilTK117R and pilTK117A mutants have reduced viability? As mentioned, the model of Tfp retraction invokes the idea that PilE subunits released during fiber retraction are membrane associated. Our results (Fig. 4) are consistent with the notion that PilT K117 mutant proteins cause a detrimental accumulation of PilE subunits. Following this logic, a ΔpilF mutant, which cannot assemble Tfp fibers, or a ΔpilD mutant, which cannot process pre-pilin into its mature form, should also accumulate PilE subunits in the inner membrane and intoxicate Ngo; indeed, pilE transcription is downregulated in both ΔpilF and ΔpilD cells (Dietrich et al., 2009). This supports the idea that Ngo can only tolerate low levels of intracellular PilE in the absence of export.
In cells lacking PilE, more PilT molecules are unacetylated and associated with the membrane (Fig 4), suggesting unacetylated PilT preferentially associates with the membrane. The slightly higher levels of membrane-associated PilT observed in the uninduced ipilE pilTK117R mutant compared to uninduced ipilE and ipilE pilTK117Q (Fig 4C, D) supports this idea. However, PilT remains membrane-associated at high levels in uninduced ipilE pilTK117Q (Fig 4C, D). Together, these observations suggest that the acetylation status of K117 contributes to, but is not the sole determinant of, PilT membrane association. The observation that PilT is acetylated at multiple sites and that one K117Q suppressor mutation has deleted an acetylatable lysine (Fig S2), suggests cooperativity between acetylated lysines across the PilT motor. A detailed molecular analysis of other PilT acetylation sites will resolve this issue.
Mutations in the PilT acetylation site do not affect Tfp retraction, per se, as judged by the normal transformation frequency of mutant cells (Table 1). However, the mutations do affect the social behavior of cells. Wt bacteria crawl together to form microcolonies containing hundreds of cells; in turn, microcolonies crawl towards each other and fuse into larger microcolonies and eventually biofilms. ipilE pilTK117Q cells cluster together into small aggregates in the presence and absence of IPTG, indicating this behavior is independent of PilE (Fig 5). ipilE pilTK117R cells have a different phenotype: they form small aggregates that are similar in shape to microcolonies, but the structures are smaller and flatter. This suggests that Tfp retraction is not the sole determinant of microcolony formation, and that PilT acetylation plays a role in regulating this process.
Our findings linking PilE levels with PilT acetylation (Fig. 4) recall the study in Pseudomonas aeruginosa showing the PilS/R two-component system senses and responds to the levels of membrane-associated PilA, the PilE homologue (Kilmury and Burrows, 2016). PilS interacts directly with PilA, inducing a signal cascade that suppresses pilA expression. When PilA levels are low, PilS does not initiate the cascade, and pilA expression is derepressed. It is possible that in Ngo a similar system senses PilE levels and regulates PilT acetylation and its association with the Tfp assembly complex. In this model, a reduction in membrane-associated PilE, such as occurs during pilus assembly, would trigger PilT deacetylation and association with the membrane. In turn, membrane bound PilT promotes pilus depolymerization and an increase in PilE in the membrane.
For Neisseria, Tfp retraction-mediated motility is the sole mode of locomotion. Tfp fibers are peritrichous in the gonococcus. Ngo crawls over surfaces by a tug-of-war mechanism in which the direction of movement is determined by the sum of vectoral force from all retracting pili at a given time (Marathe et al., 2014; Zaburdaev et al., 2014; Maier and Wong, 2015). A bacterium crawls with directional persistence, implying there is coordination among the multiple Tfp around the cell body (Marathe et al., 2014). Neisseria does not have orthologues of the P. aeruginosa and Myxococcus xanthus Tfp-associated chemotaxis genes, suggesting it uses a different mechanism to control directional movement. In E. coli, acetylation of a flagellar component CheY promotes motor switching and directional motility (Fraiberg et al., 2015). It is reasonable to speculate that the acetylation status of PilT affects its association with Tfp assembly complexes, ultimately coordinating Tfp retraction events across the cell envelope and directional movement of the cell.
In conclusion, blocking the ability of Ngo to toggle between acetylated and unacetylated states of PilT is detrimental to its viability. Our data support a model in which acetylation of PilT influences its cellular location, and perturbing this pathway dysregulates bacterial social behavior. Further investigation into the mechanisms that confer tolerance to the pilT acetylation site mutations and the role of other PilT acetylation sites will shed light on the mechanism of pilus retraction.
Materials and methods
Bacterial strains, growth conditions, and infection studies
For mass spectroscopy analysis, N. gonorrhoeae strain 1291 (Apicella, 1974) and its corresponding acetate kinase (ackA) mutant, 1291 ΔackA, were grown on GC agar plates (Becton Dickinson, Franklin Lakes, NJ) supplemented with 1% IsoVitaleX (Becton Dickinson) and 50 µg/mL kanamycin, when needed, at 37°C with 5% CO2. Bacteria for the acetylome experiments were grown overnight in 500 mL cultures of GC broth supplemented with 1% IsoVitaleX and Kellogg’s supplement at 37°C, rotating at 150 rpm. Three biological replicates were grown for each strain.
For all other experiments, Neisseria gonorrhoeae (Ngo) MS11 and MS11ΔpilT (Dietrich et al., 2009) were used throughout this study. Genomic DNA from Ngo C9.10 (Long et al., 2001) was transformed into Ngo MS11 to construct the IPTG-inducible pilE (ipilE) Erythromycin resistant strain used in this study. All strains were grown on Gonococcal Broth (GCB) agar plates containing Kellogg’s supplements I and II and Vancomycin, Colistin and Nalidixic Acid (VCN) (BD BBL), or in liquid GCB containing Kellogg’s supplements I and II. All cultures were grown at 37°C with 5% CO2. Escherichia coli strains DH5α and BL21 were grown in Luria Broth at 37°C.
Sample Preparation and Proteolytic Digestion
Samples were prepared and analyzed as previously described (Post et al., 2017). Briefly, cell pellets from 1291wt or 1291 ΔackA were washed with PBS, and re-suspended and denatured in a solution of 6 M urea, 100 mM Tris, 75 mM NaCl containing the deacetylase inhibitors 1 µM tricostatin A and 3 mM nicotinamide. Protein lysates generated by microtip sonication, reduced with 20 mM dithiothreitol (37°C for 30 min), and subsequently alkylated with 40 mM iodoacetamide (30 min at RT in the dark). Proteolytic trypsin digestion was performed overnight at 37°C using an enzyme:substrate ratio (wt/wt) of 1:50. Samples were desalted using HLB Oasis SPE cartridges (Waters Corp., Milford, MA).
Affinity purification of lysine-acetylated peptides and mass spectrometric analyses. The polyclonal anti-acetyl-lysine agarose bound antibody conjugate from ImmuneChem (ICP0380-100) (Burnaby, Canada) was prewashed and incubated with 1 mg digested protein lysate in NET buffer, overnight at 4°C, on a rotating platform. Beads were washed, and enriched acetyl peptides were eluted and de-salted using C-18 ZipTips prior to mass spectrometric analyses. All samples were analyzed by reverse-phase HPLC-ESI-MS/MS using an Eksigent UltraPlus nano-LC 2D HPLC system (Dublin, CA) connected to an orthogonal quadrupole time-of-flight (QqTOF) TripleTOF 5600 mass spectrometer (SCIEX, Framingham, MA), as previously described (Post et al., 2017).
Bioinformatic database searches
All mass spectrometric data was searched, as previously described (Post et al., 2017), using ProteinPilot 4.5 (rev. 1656) with the Paragon algorithm 4.5.0.0, 1654 (SCIEX) (Shilov et al., 2007) using a N. gonorrhoeae strain FA 1090 custom database downloaded from the Kyoto Encyclopedia of Genes and Genomes (KEGG) website (http://www.genome.jp/kegg/). The Proteomics System Performance Evaluation Pipeline (PSPEP) tool was used to generate the FDR analyses using a concatenated forward and reverse decoy database to search the data (Shilov et al., 2007). For identification of acetylated peptides, we required a peptide confidence cut-off threshold of 99%.
Quantitative Skyline MS1 Filtering and Data Analysis
MS1 chromatogram-based quantitation was performed in Skyline 2.6, an open source software project (http://proteome.gs.washington.edu/software/skyline) (MacLean et al., 2010) using the MS1 ion intensity chromatogram processing algorithm MS1 Filtering (Schilling et al., 2012), as previously described (Schilling et al., 2015). Briefly, all raw files acquired in data-dependent acquisition mode were directly imported into Skyline 2.6, and MS1 precursor ions were extracted for all peptides present in the comprehensive MS/MS spectral libraries that had been generated from corresponding database searches. Quantitative analysis for acetyl-enriched samples was based on extracted ion chromatograms (XICs) and the resulting precursor ion peak areas for M, M+1, and M+2; final quantitative comparisons were typically based on only the highest ranked precursor ion, which was then compared between different conditions.
All peak areas obtained from the extracted precursor ion chromatograms using MS1 Filtering were exported from Skyline into Excel spreadsheets using the statistical program ‘MS1Probe’, (http://skytools.maccosslab.org). For each biological replicate and strain, the peak areas for individual K-acetyl peptides were assembled into a data array and the peak area ratios between the two strains (1291ackA/1291wt) were calculated. Significance was assessed using two-tailed Student’s t-test requiring p-values<0.05. Acetylation sites were designated as AckA-dependent if there was at least a two-fold change in the relative peak areas between the ΔackA mutant and the wt strains with a significance of p<0.05 in two or more biological replicates. The overall average ratio for each candidate peptide was generated by log-transforming the individual biological replicate ratios, generating an overall ratio, and finally converting back to a natural ratio.
PilT structure prediction
Structure predictions of Ngo PilT were performed by Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre/) and based on the structure of Pseudomonas aeruginosa PilT (PDB model 3JVU). Resultant files were examined using Swiss PDBViewer (v4.1).
Cloning, overexpression, purification, and in vitro acetylation of PilT
PilT over-expression and purification was performed as previously described (Hockenberry et al., 2016). Purified PilT hexamers (final concentration 1.25 µM of PilT) were incubated with 0, 125, or 500 mM acetyl-phosphate (Sigma) in acetylation buffer (Tris HCl (50 mM, pH 7.5), NaCl (150 mM), EDTA (1 mM) (Choudhary et al., 2014). Reactions were incubated for 3 hours at 37°C. Samples were separated by electrophoresis in an 8% non-denaturing polyacrylamide gel. The separated proteins were transferred to a nitrocellulose membrane (0.45 µm, GE Healthcare Biosciences) using the Trans-Blot SD Semi-Dry Transfer Cell (Life Technologies). The membrane was blocked in Tris-buffered saline containing 0.1% Tween-20 (TBST) and non-fat dry milk (5% w/v) for 1 h at room temperature, then probed with αPilT and αAceK (Cell Signaling Technologies) for 1 h at room temperature. The membrane was washed and probed with α-rabbit (LICOR) and α-mouse diluted in 5% milk/TBST for 1 h at room temperature. Blots were imaged on the LICOR Odyssey Infrared Imaging System and analyzed using ImageJ.
Immunoprecipitation of acetylated PilT
5 × 108 cfu of wt, ΔpilT, and ipilE were resuspended in 0.5 mL RIPA buffer and incubated for 10 minutes at room temperature. The lysate was centrifuged at high speed to remove cellular debris. The whole cell lysate was incubated with non-specific αIgG and Protein A agarose beads for 2 h. The lysate was centrifuged for 30 minutes at 3000 xg. αAcetylated lysine (Cell Signaling Technologies) or nonspecific IgG control antibody (Santa Cruz) and Protein A agarose beads were added to the precleared lysate and incubated overnight at 4°C. After 16–20 hours, the samples were centrifuged for 30 minutes at 3000 xg. The supernates were banked and the pelleted agarose beads were washed 5 times with RIPA buffter. 25 µL of 1× Laemmeli buffer was added to the beads and samples were boiled for 10 min. 20 µL of the samples were separated by SDS-PAGE (15% acrylamide) and immunoblotted for PilT and PilE (SM1) as described above. Blots were imaged on the LICOR Odyssey Infrared Imaging System and analyzed by densitometry using ImageJ.
Cellular fractionation
5.0 × 108 cfu of the appropriate strains were separated into soluble and membrane fractions by ultracentrifugation as previously described. Detection of PilT in these fractions was determined by Western blotting as described above. Because the cytosolic purity control runs at the same molecular weight as PilT (37 kDa), parallel membranes were immunoblotted with the H.8 (generous gift from Sanjay Ram) antibody and αGAPDH (ThermoFisher, GA1R) were used as membrane and cytosolic fraction purity controls, respectively.
Construction of the Ngo K117Q, K117R and K117A mutants
The K117Q, K117R and K117A alleles were inserted into the endogenous pilT locus using by allelic replacement, as described (Hockenberry et al., 2016a). Briefly, the Ngo pilTU operon was amplified using primers 106F+R and the K117Q or K117R mutations were introduced into pilT using overlap extension PCR so as not to interrupt the pilU expression. Mutagenized pilTU DNAs were inserted into the pUC19-Kan-pilU3’ at the SmaI/BamHI site to create pUC19-pilTK117QU::Kan, pUC19-pilTK117RU::Kan and pUC19-pilTK117AU::Kan (Fig. S6) . All constructs were confirmed by sequencing (Elim Biopharmicueticals). The plasmids were transformed into Ngo MS11 and transformants were plated on GCB agar containing Kanamycin (50 mg/L). The pilT locus of Kanamycin resistant transformants was sequenced to confirm it had the intended mutation.
Introduction of K117Q and K117R into ipilE
The K117Q and K117R mutations were transferred into the ipilE strain by growing the strain for 3 h in 1.0 mM IPTG prior to transformation with pUC19-pilTK117QU::Kan or pUC19-pilTK117RU::Kan plasmids. Transformants were selected on GCB agar containing Kanamycin (50 mg/L), and without IPTG. The pilT locus of Kanamycin resistant transformants was sequenced to confirm it contained the intended mutation.
Screening for secondary mutations in ipilE K117Q and ipilE K117R
ipilE K117Q and ipilE K117R cells were grown for 8 h in supplemented GCB broth containing IPTG (1.0 mM), then plated on GCB agar containing IPTG (1.0 mM). Piliated colonies were passed three times on GCB agar containing IPTG (1.0 mM), to isolate colonies that maintained the piliated phenotype. After the third passage, a colony from each clone was suspended in 50 µL GCB + 30% glycerol and stored at −80°C for future analysis.
DNA Transformation
Assays were performed as previously described. Briefly, bacteria were grown for 3 h in GCB broth containing IPTG (1.0 mM). 1.0 × 107 cfu were incubated with 1.0 µg of Ngo MS11 RifR genomic DNA in GCB broth containing IPTG (1.0 mM) and MgSO4 (5.0 mM) for 20 min at 37°C, 5% CO2. The cells were transferred to 900 µL pre-warmed GCB containing Kellogg’s supplement I and II and incubated for 2 h at 37°C, 5% CO2. The cells were harvested and spread onto supplemented GCB or supplemented GCB containing Rifampicin (50 µg/L) agar plates. Transformation frequency was calculated as the number of RifR cfu / total cfu / µg RifR genomic DNA. Values from 3 independent experiments were averaged.
Microcolony formation
Bacteria were harvested from GCB agar plates grown for 16 h and resuspended in supplemented GCB broth. 5.0 × 107 cfu of each strain were inoculated into a well of a 12 well dish (Falcon) containing 1.0 mL of pre-equilibrated supplemented GCB +/− IPTG (0.5 mM). Images of the bacteria were acquired after 3 h incubation at 37°C, 5% CO2 using a camera mounted to an inverted microscope (Nikon).
Supplementary Material
Abbreviated Summary.
Cells can control protein activity through post-translational modification. Here, we show by genetic analysis that the viability of pathogen Neisseria gonorrhoeae is dramatically reduced when its PilT protein is prevented from switching between acetylated and deacetylated states. Loss of viability is overcome when secondary mutations occur in genes encoding proteins known or suspected to interact with PilT at the membrane. These findings demonstrate that acetylation is an important determinant of PilT function, and imply that PilT influences membrane dynamics.
Acknowledgments
We are grateful for the stimulating discussions with B. Fane, M Rendón, WJ Kim, and MC Ma during these studies. M. Rendón gave this manuscript a thorough review. We are thankful to Hank Seifert for the gift of the inducible pilE strain and Sanjay Ram for the H.8 antibody. This work was supported by the National Institutes of Health grant R01AI107966 awarded to MS and A108255 to DMBP and MAA.
Footnotes
Author Contributions: Conception and design of the study: AH, DP, MA, MS; Acquisition, analysis, and interpretation of the data: AH, DP, KR, MA, MS; Writing the manuscript: AH, DP, KR, MA, MS.
References
- Apicella MA. Antigenically Distinct Populations of Neisseria gonorrhoeaei Isolation and Characterization of the Responsible Determinants. J Infect Dis. 1974;130:619–625. doi: 10.1093/infdis/130.6.619. [DOI] [PubMed] [Google Scholar]
- Brossay L, Paradis G, Fox R, Koomey M, Hébert J. Identification, localization, and distribution of the PilT protein in Neisseria gonorrhoeae. Infect Immun. 1994;62:2302–8. doi: 10.1128/iai.62.6.2302-2308.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonnelle E, Helaine S, Nassif X, Pelicic V. A systematic genetic analysis in Neisseria meningitidis defines the Pil proteins required for assembly, functionality, stabilization and export of type IV pili. Mol Microbiol. 2006;61:1510–1522. doi: 10.1111/j.1365-2958.2006.05341.x. [DOI] [PubMed] [Google Scholar]
- Castano-Cerezo S, Bernal V, Post H, Fuhrer T, Cappadona S, Sanchez-Diaz NC, et al. Protein acetylation affects acetate metabolism, motility and acid stress response in Escherichia coli. Mol Syst Biol. 2014;10:762–762. doi: 10.15252/msb.20145227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y-W, Rettberg LA, Treuner-Lange A, Iwasa J, Sogaard-Andersen L, Jensen GJ. Architecture of the type IVa pilus machine. Science (80-) 2016;351 doi: 10.1126/science.aad2001. aad2001-aad2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, et al. Lysine Acetylation Targets Protein Complexes and Co-Regulates Major Cellular Functions. Science (80-) 2009;325 doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat Rev Mol Cell Biol. 2014;15 doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- Dietrich M, Mollenkopf H, So M, Friedrich A. Pilin regulation in the pilT mutant of Neisseria gonorrhoeae strain MS11. FEMS Microbiol Lett. 2009;296:248–56. doi: 10.1111/j.1574-6968.2009.01647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson J, Eriksson OS, Jonsson A-B. Loss of meningococcal PilU delays microcolony formation and attenuates virulence in vivo. Infect Immun. 2012;80:2538–47. doi: 10.1128/IAI.06354-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraiberg M, Afanzar O, Cassidy CK, Gabashvili A, Schulten K, Levin Y, Eisenbach M. CheY’s acetylation sites responsible for generating clockwise flagellar rotation in E scherichia coli. Mol Microbiol. 2015;95:231–244. doi: 10.1111/mmi.12858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiadou M, Castagnini M, Karimova G, Ladant D, Pelicic V. Large-scale study of the interactions between proteins involved in type IV pilus biology in Neisseria meningitidis: characterization of a subcomplex involved in pilus assembly. Mol Microbiol. 2012a;84:857–73. doi: 10.1111/j.1365-2958.2012.08062.x. [DOI] [PubMed] [Google Scholar]
- Giltner CL, Nguyen Y, Burrows LL. Type IV Pilin Proteins: Versatile Molecular Modules. Microbiol Mol Biol Rev. 2012;76:740–772. doi: 10.1128/MMBR.00035-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold VAM, Salzer R, Averhoff B, Kühlbrandt W, Averhoff B, Friedrich A, et al. Structure of a type IV pilus machinery in the open and closed state. Elife. 2015;4:385–393. doi: 10.7554/eLife.07380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockenberry AM, Hutchens DM, Agellon A, So M. Attenuation of the Type IV Pilus Retraction Motor Influences Neisseria gonorrhoeae Social and Infection Behavior. MBio. 2016a;7:e01994–16. doi: 10.1128/mBio.01994-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmury SLN, Burrows LL. Type IV pilins regulate their own expression via direct intramembrane interactions with the sensor kinase PilS. Proc Natl Acad Sci U S A. 2016;113:6017–22. doi: 10.1073/pnas.1512947113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurre R, Höne A, Clausen M, Meel C, Maier B. PilT2 enhances the speed of gonococcal type IV pilus retraction and of twitching motility. Mol Microbiol. 2012;86:857–65. doi: 10.1111/mmi.12022. [DOI] [PubMed] [Google Scholar]
- Long CD, Hayes SF, Putten JP, van Harvey HA, Apicella MA, Seifert HS. Modulation of gonococcal piliation by regulatable transcription of pilE. J Bacteriol. 2001;183:1600–9. doi: 10.1128/JB.183.5.1600-1609.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier B, Wong GCL. How Bacteria Use Type IV Pili Machinery on Surfaces. Trends Microbiol. 2015 doi: 10.1016/j.tim.2015.09.002. [DOI] [PubMed] [Google Scholar]
- Mancl JM, Black WP, Robinson H, Yang Z, Schubot FD. Crystal Structure of a Type IV Pilus Assembly ATPase: Insights into the Molecular Mechanism of PilB from Thermus thermophilus. Structure. 2016;24:1886–1897. doi: 10.1016/j.str.2016.08.010. [DOI] [PubMed] [Google Scholar]
- Marathe R, Meel C, Schmidt NC, Dewenter L, Kurre R, Greune L, et al. Bacterial twitching motility is coordinated by a two-dimensional tug-of-war with directional memory. Nat Commun. 2014;5:630–633. doi: 10.1038/ncomms4759. [DOI] [PubMed] [Google Scholar]
- McCallum M, Tammam S, Khan A, Burrows LL, Howell PL. The molecular mechanism of the type IVa pilus motors. Nat Commun. 2017;8:15091. doi: 10.1038/ncomms15091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin LS, Haft RJF, Forest KT. Structural insights into the Type II secretion nanomachine. Curr Opin Struct Biol. 2012;22:208–16. doi: 10.1016/j.sbi.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouidir T, Kentache T, Hardouin J. Protein lysine acetylation in bacteria: Current state of the art. Proteomics. 2016;16:301–309. doi: 10.1002/pmic.201500258. [DOI] [PubMed] [Google Scholar]
- Park H-SM, Wolfgang M, Koomey M. Modification of type IV pilus-associated epithelial cell adherence and multicellular behavior by the PilU protein of Neisseria gonorrhoeae. Infect Immun. 2002;70:3891–903. doi: 10.1128/IAI.70.7.3891-3903.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post DMB, Schilling B, Reinders LM, D’Souza AK, Ketterer MR, Kiel SJ, et al. Identification and characterization of AckA-dependent protein acetylation in Neisseria gonorrhoeae. PLoS One. 2017;12:e0179621. doi: 10.1371/journal.pone.0179621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling B, Christensen D, Davis R, Sahu AK, Hu LI, Walker-Peddakotla A, et al. Protein acetylation dynamics in response to carbon overflow in Escherichia coli. Mol Microbiol. 2015;98:847–863. doi: 10.1111/mmi.13161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling B, Rardin MJ, MacLean BX, Zawadzka AM, Frewen BE, Cusack MP, et al. Platform-independent and Label-free Quantitation of Proteomic Data Using MS1 Extracted Ion Chromatograms in Skyline. Mol Cell Proteomics. 2012;11:202–214. doi: 10.1074/mcp.M112.017707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilov IV, Seymour SL, Patel AA, Loboda A, Tang WH, Keating SP, et al. The Paragon Algorithm, a Next Generation Search Engine That Uses Sequence Temperature Values and Feature Probabilities to Identify Peptides from Tandem Mass Spectra. Mol Cell Proteomics. 2007;6:1638–1655. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- Takhar HK, Kemp K, Kim M, Howell PL, Burrows LL. The platform protein is essential for type IV pilus biogenesis. J Biol Chem. 2013;288:9721–8. doi: 10.1074/jbc.M113.453506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–7. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaburdaev V, Biais N, Schmiedeberg M, Eriksson J, Jonsson A-B, Sheetz MP, Weitz DA. Uncovering the Mechanism of Trapping and Cell Orientation during Neisseria gonorrhoeae Twitching Motility. Biophys J. 2014;107:1523–1531. doi: 10.1016/j.bpj.2014.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.