Abstract
Background
Foetal RHD genotyping can be predicted by real-time polymerase chain reaction (qPCR) using cell-free foetal DNA extracted from maternal plasma. The object of this study was to determine the diagnostic accuracy and feasibility of non-invasive RHD foetal genotyping, using a commercial multiple-exon assay, as a guide to appropriate administration of targeted antenatal immunoprophylaxis.
Material and methods
Cell-free foetal DNA was extracted from plasma of RhD-negative women between 11–30 weeks of pregnancy. The foetal RHD genotype was determined non-invasively by qPCR amplification of exons 5, 7 and 10 of the RHD gene using the Free DNA Fetal Kit® RhD. Results were compared with serological RhD cord blood typing at birth. The analysis of diagnostic accuracy was restricted to the period (24–28+6 weeks) during which foetal genotyping is usually performed for targeted antenatal immunoprophylaxis.
Results
RHD foetal genotyping was performed on 367 plasma samples (24–28+6 weeks). Neonatal RhD phenotype results were available for 284 pregnancies. Foetal RHD status was inconclusive in 9/284 (3.2%) samples, including four cases with RhD maternal variants. Two false-positive results were registered. The sensitivity was 100% and the specificity was 97.5% (95% CI: 94.0–100). The diagnostic accuracy was 99.3% (95% CI: 98.3–100), decreasing to 96.1% (95% CI: 93.9–98.4) when the inconclusive results were included. The negative and positive predictive values were 100% (95% CI: 100–100) and 99.0% (95% CI: 97.6–100), respectively. There was one false-negative result in a sample collected at 18 weeks. After inclusion of samples at early gestational age (<23+6 week), sensitivity and accuracy were 99.6% (95% CI: 98.7–100) and 95.5% (95% CI: 93.3–97.8), respectively.
Discussion
This study demonstrates that foetal RHD detection on maternal plasma using a commercial multiple-exon assay is a reliable and accurate tool to predict foetal RhD phenotype. It can be a safe guide for the appropriate administration of targeted prenatal immunoprophylaxis.
Keywords: haemolytic disease of the foetus and the newborn (HDFN), RHD genotyping, immunoprophylaxis, prenatal diagnosis
Introduction
Haemolytic disease of the foetus and the newborn (HDFN) has been the main cause of neonatal and perinatal morbidity and mortality for many decades1,2. The impact of this disease in economically advanced countries has been greatly reduced by the existence of surveillance and prevention programmes. Until the 1960s HDFN affected about 7,000 neonates per year with a mortality of 1.5/1,000 births. The introduction of postnatal anti-D immunoglobulin (RhIg) (late 1960s) drastically decreased the risk of anti-D alloimmunisation, such that the current incidence of RhD HDFN is 0.01–0.03% and the mortality rate is lower than 2/10,000 births3–6.
National guidelines from scientific societies and health institutions7–13 strongly recommend that all RhD-negative women are routinely offered RhIg post-partum, during antenatal care (at 28 weeks) and following any potentially sensitising event in which foeto-maternal haemorrhage may have occurred. Current policy and legal practice is that women should be given appropriate information about RhIg - its benefits to foetal health (in current and future pregnancies) and potentially adverse events - in order to give a conscious consent.
The rate of RhD maternal-foetal incompatibility depends on the prevalence of RhD-negative and RhD-positive phenotypes, which is linked to RHD haplotype frequencies. In a predominantly white population about 40% of RhD-negative women carry a RhD-negative foetus14–16. Therefore, during a pregnancy, 40% of RhD-negative women receive unnecessary administration of one or more RhIg, prepared from pooled human plasma and, even if current preparations are safe, they are exposed to a risk of infection from viral or prion contamination17–21. Furthermore, there are ethical concerns about the source of hyperimmune plasma, its world-wide shortage and the wastage of an expensive product. Haemovigilance reports registered incidents involving neglected, inappropriate and/or unnecessary administration of RhIg: in 2016 SHOT reported 333 adverse events out of 409 reports (81.4%) related to omission or late administration of RhIg (2 anti-D immunisation) and 69/409 inappropriate administrations22.
The likely future direction of prevention of RhD HDFN lies in defining the RHD fetal genotype from cell-free foetal DNA (cffDNA) in maternal plasma5. The discovery of circulating foetal DNA in maternal plasma allowed invasive procedures, associated with the risk of miscarriage, transplacental haemorrhage and alloimmmunisation stimulus, to be abandoned23. Large-scale studies demonstrated the feasibility of real-time polymerase chain reaction (PCR)-based screening for foetal RHD to guide targeted antenatal immunoprophylaxis24–29, restricting the administration of this immunoprophylaxis to RhD-negative women who carry a RhD-positive foetus. In 2010 and 2011 Denmark and the Netherlands, respectively, implemented nationwide antenatal screening for foetal RHD genotyping; regional availability of prenatal RHD screening is also reported in Sweden, Belgium, United Kingdom, Czech Republic, France and Germany30,31.
At present, in Italy, a routine protocol for non-invasive foetal RHD genotyping does not exist: this test is performed by a limited number of specialised Transfusion Service laboratories for anti-D alloimmunised patients only or by private laboratories, which are not to be trusted. In 2015 the Regional Blood Centre of the Region of Emilia-Romagna (Italy) supported a project including two phases: the first had the aim of determining the diagnostic accuracy and feasibility of non-invasive foetal genotyping at different gestational ages, comparing results with serological RhD typing on cord blood; the second phase planned to introduce RHD foetal genotyping into the antenatal screening programme in Emilia-Romagna. We report here the results of the first phase of the study.
Materials and methods
Five Regional Immunohaematology and Transfusion Services participated in the first phase of the project: Bologna (S.Orsola-Malpighi Polyclinic and Maggiore Hospital), Ferrara, Imola and Rimini.
RhD-negative pregnant women, with RhD-positive partners or partners of unknown RhD phenotype, presenting in hospitals at different gestational ages for antenatal immunohaematological tests (first trimester screening, third trimester - around 28 weeks) or invasive diagnostic procedures, were asked to take part in the study. Local physicians obtained written informed consent for genetic tests and for personal information/biological samples, and completed a report with pregnancy data. Approval was obtained from the Ethical Committee of the coordinating centre (S. Orsola-Malpighi Polyclinic - Bologna).
EDTA-anticoagulated blood (6–9 mL) was drawn and shipped, at room temperature within 48 hours of collection, to the Advanced Immunohaematology Laboratory in S. Orsola-Malpighi Polyclinic in Bologna. Samples for which informed consent was absent or incomplete, haemolysed samples and samples received more than 48 hours after collection were not accepted.
The compliant samples were double-centrifuged, aliquoted and stored at −30 °C until further processing. Two aliquots of maternal blood were frozen at −30 °C, as a source of maternal DNA for analysing potential maternal RhD variants.
Foetal DNA was treated twice for DNA extraction, using manual extraction by microcolumn: the QIAamp® DNA DSP Blood Mini Kit (Qiagen, Hilden, Germany) was used initially and then replaced by the QIAamp® DSP DNA Virus Kit (Qiagen), which is more specific for small DNA fragments32. Given the high percentage of inconclusive results and low threshold cycle (Ct) in controls, we finally performed extraction with the QIAamp® Circulating Nucleic Acid Kit (Qiagen). Foetal DNA was extracted from 1 mL plasma samples containing 5 μL of diluted maize DNA (as an extraction/amplification control), according to the manufacturer’s instructions.
Using the Free DNA Fetal Kit® RhD (Institut de Biotechnologies Jacques-Boy, Reims, France) and LightCycler® 480 Probes Master (Roche, Rotkreuz, Switzerland), we carried out, in duplicate, real-time PCR analysis for RHD exons 5, 7 and 10 on cffDNA isolated from maternal plasma, generating six test results for each sample. Three controls were added for each series of extractions: RhD-negative and RhD-positive plasma controls provided with the kit, a blank control with extraction water instead of plasma, and nuclease-free water used for PCR mix (single amplification).
All PCR tests were performed with the Dx Real-time System (Biorad, Hercules, CA, USA) applying the amplification conditions indicated by the manufacturer. Traceability of all phases of the analysis protocol (plasma storage, DNA extraction, PCR amplification) with manual transfer of plasma/DNA was guaranteed by the supervision and signatures of two operators on dedicated check-lists.
The manufacturer’s instructions suggested the following Ct values as references for the acceptance of batch and sample results: Ct values between 35 and 41 for each target in samples, Ct values <39 for the positive control, Ct values ≤ 37 for the maize DNA IVR2 exon for both samples and controls. These values were applied during the validation of the method. Subsequently, in accordance with scientific consultant suggestions, Ct value ranges of each target (RHD exons 5, 7 and 10 in samples and in positive control, maize DNA) were determined for each lot of probes (Table I). Ct values <40 for exon 5 and <41 for exons 7 and 10 were interpreted as a positive signal.
Table I.
Cut-off Ct values applied in the typing algorithm.
| Target | Range | |
|---|---|---|
| Positive samples | Exon 5 | 34.33–39.33 |
| Exon 7 | 35.84–40.80 | |
| Exon 10 | 35.54–40.78 | |
| Positive controls | Exon 5 | 34.24–36.18 |
| Exon 7 | 35.95–37.81 | |
| Exon 10 | 35.34–37.96 | |
| Maize DNA | 34.42–35.64 | |
Ct: Cycle threshold
Results were validated only if: (i) no amplification curve was observed for each target of the RhD-negative control, blank control and PCR water (to exclude contamination); (ii) the Ct values for exons 5, 7 and 10 of the RhD-positive control were, respectively, 35.21±0.97, 36.88±0.93 and 36.65±1.31; and (iii) the maize DNA IVR2 exon was amplified with a Ct range of 35.03±0.61, confirming the efficacy of DNA extraction and the absence of PCR inhibition for each sample/control.
The genotype was reported as RHD negative when all RHD PCR reactions were negative (6/6) and the RHD genotype was considered positive when at least 5/6 PCR reactions were positive. RHD PCR-positive reactions ≤4/6 were reported as inconclusive. Discrepant or inconclusive results were repeated on a new plasma aliquot to identify a technical error, a different sensibility of probes with low cffDNA concentrations or a coding/not coding RhD variant.
A RHD PCR-positive reaction with Ct values below 35 or lower than the Ct of the positive control for each target, in one or more RHD exons, suggested the presence of a silent RHD gene in the maternal genome33 and invalidated the foetal RHD genotype, which was considered as RHD positive. Afterwards serological RhD phenotyping, antiglobulin RhD test and PCR-single strand polymorphism (SSP) molecular typing were performed on the maternal sample to identify or exclude a RhD variant.
Predicted RhD phenotype from the non-invasively determined foetal RHD genotype was confirmed by serological RhD cord blood typing at birth, performed locally following validated procedures.
Results
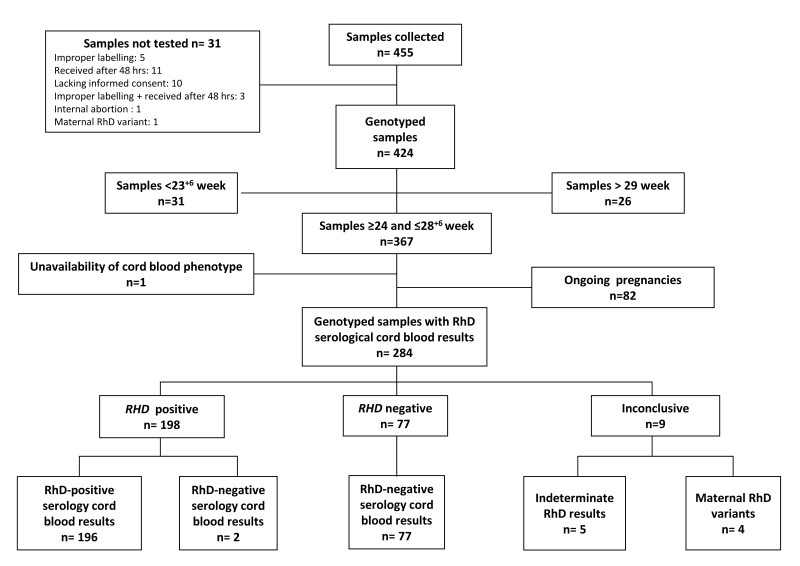
From February 2016 to January 2018, 455 RhD-negative pregnant women were recruited; 31/455 collected samples were not tested (Figure 1). One of the aims of the study was to determine the diagnostic accuracy of non-invasive foetal genotyping at different gestational ages, however, we collected a small number of samples from women in the first trimester of pregnancy. Therefore, we restricted the analysis of diagnostic accuracy to the period of 24–28+6 weeks, during which foetal genotyping is usually performed for targeted antenatal RhIg administration. Out of 424 genotyped samples, 31 collected before 23+6 weeks and 26 after 29 weeks were excluded; one sample was excluded because no cord blood phenotype was obtained owing to a stillbirth (Figure 1). The foetal RHD genotype and the RhD serological cord blood phenotype were available for 284 pregnancies; 82 pregnancies are still ongoing.
Figure 1.
Flow chart summarising the results of non-invasive foetal RHD genotyping.
With regards to ethnicity, the majority of the women were Caucasian (273 samples, 96.1%), nine (3.1%) were African, one (0.3%) was Asian (India), and one (0.3%) came from Central America. It should be noted that these rates do not represent the proportion of racial mixture in the region (31%)33. In fact, because of language difficulties, participation in the project by foreign women was low (Table II). The antibody screening performed at the beginning of third trimester of pregnancy was negative in 260/284 pregnant women and positive in 24/284 owing to passive anti-D from previous RhIg, administered after chorionic villous sampling in the first trimester. Post-natal screening, performed 6 months after delivery, was negative in all tested women (248/284).
Table II.
Population characteristics.
| Maternal age (years) mean (range) | 33.8±4.8 |
|
| |
| Ethnicity | |
| Caucasian | 273 |
|
| |
| African | 9 |
|
| |
| Asian | 1 (India) |
|
| |
| Central America | 1 (Cuba) |
|
| |
| Parity | |
| Nulliparous | 118 |
|
| |
| Multiparous | 166 |
|
| |
| Twin pregnancies | 6 |
|
| |
| Prenatal antibody screening | |
| Negative | 260/284 |
|
| |
| Positive* | 24/284 |
|
| |
| Postnatal antibody screening** | |
| Negative | 248/284 |
|
| |
| Positive | 0/284 |
Anti-D from previous immunoprophylaxis, administered in first trimester;
Performed 6 months after delivery; 36/284 women have not yet been screened, because they gave birth less than 6 months ago.
As a result of a validation process, which overcame the problems that had surfaced with the QIAamp® DNA DSP Blood Mini Kit and QIAamp® DSP DNA Virus Kit (Qiagen), the foetal DNA isolation is now performed with QIAamp® Circulating Nucleic Acid Kit (Qiagen), more specific than the previous ones for extraction of small DNA fragments of human origin34. Table III shows details about the efficiency of DNA recovery of the extraction kits.
Table III.
Typing results.
| Extraction kit | Typed samples | Conclusive results with one measurement | Conclusive results with two measurements | Conclusive results with three or more measurements | Inconclusive results | False-positive | False-negative |
|---|---|---|---|---|---|---|---|
| QIAamp DNA DSP Blood Mini Kit | 46 N | 38 | 4 | 2 | 2 | 0 | 0 |
| QIAamp DSP DNA Virus Kit | 209 N+2 I | 170 | 19 | 2 | 20 | 1 | 1 |
| QIAamp Circulating Nucleic Acid Kit | 96 N+16 I | 104 | 4 | 0 | 4 | 1 | 0 |
N: new samples; I: sample with inconclusive results obtained with other extraction kits.
In 198/284 samples a RHD-positive test result (69.7%) and in 77/284 samples (27.1%) a RHD-negative test result were obtained. In two cases the foetuses have been reported as RHD positive, whereas the RhD cord blood serology showed the neonates to be RhD negative. In the first sample, the suspicious of a mistake during one of the manual steps of the typing process prompted us to introduce the check-lists; the second sample was probably mistaken by contamination. No pseudogenes or non-coding variant genes were identified. There was one false-negative result (RhD-positive foetus with a RHD-negative maternal cffDNA result) in a sample taken at 18 weeks of gestation.
Genotyping results were inconclusive in 9/284 (3.2%) samples; in five cases the reaction pattern was variable in subsequent repetitions and not interpretable according to our criteria: RhD phenotyping at birth confirmed one as RhD negative and four as RhD positive. In 4/11 samples we could not identify the RHD fetal genotype because of maternal RhD variants (Table IV). In two samples 6/6 positive signals with low Ct values were obtained, while RhD serological typing was repeatedly negative. PCR-SSP analysis on maternal DNA predicted the presence of RhD variants with very low antigen expression (RHD*01W.29 and RHD*11). In two pregnancies the reaction pattern was a single amplification signal only with the exon 10 probe and low Ct values: the antiglobulin RhD typing was negative and the PCR-SSP analysis revealed the presence of hybrid genes, associated with a RhD-negative phenotype, in which RHD exons 5 and 7 are substituted by the corresponding RHCE.
Table IV.
Maternal RhD variants.
| ID | Week of gestation | Rh phenotype | RhD AG test | Ct exon 5 | Ct exon 7 | Ct exon 10 | Maternal RHD genotype | Predicted maternal RhD phenotype |
|---|---|---|---|---|---|---|---|---|
| 227/2016 | 28 | Ccee | Positive | 28.07 | 29.42 | 29.03 | RHD*01W.29 (RHD*weak D type 29) | CcDuee |
| 230/2016 | 28 | Ccee | Positive | 32.47 | 34.15 | 33.60 | RHD*11 (RHD*weak D type 11) | CcDuee |
| 245/2016 | 29 | Ccee | Negative | N.A. | N.A. | 33.42 | RHD-CE(2–9)-D | Ccdee |
| 248/2016 | 28 | *Ccee | Negative | N.A. | N.A. | 31.76 | RHD-CE(3–7)-D | Ccdee |
N.A.: not amplified; AG: anti-globulin;
partial C antigen associated with d(C)ces aplotype.
The sensitivity of the non-invasive foetal RHD genotyping was 100% and the specificity was 97.5% (95% CI: 94.0–100). The diagnostic accuracy was 99.3% (95% CI: 98.3–100), decreasing to 96.1% (95% CI 93.9–98.4) when the inconclusive results were included. The negative and positive predictive values were 100% (95% CI: 100–100) and 99.0% (95% CI: 97.6–100) respectively.
Discussion
The determination of foetal genotyping on circulating cffDNA in maternal plasma transformed prenatal cares in all respects35. About 20 years ago, Lo and co-workers36 demonstrated the presence of male foetal DNA sequences in maternal plasma, where it can be detected as early as at 5 weeks of gestation; however, the paucity of cffDNA and its coexistence with maternal DNA were limitations to its diagnostic use, especially at early gestational ages. The diagnostic application of non-invasive RHD foetal genotyping, previously defined at 16 weeks by Lo et al.36, was subsequently antedated to 11–14 weeks37.
cffDNA from maternal plasma is usually isolated manually with commercially available kits, but automation of the process allows its application on a large scale5,38 with good recovery of foetal DNA. Finning et al.19 demonstrated that a high-throughput technique using robotic isolation of DNA from maternal plasma could reduce the false-negative rate to 0.2% in the third trimester, although it remained 3.5% at 11–13 weeks37. Recently Moise et al.39 reported a 0.32% false-negative rate in first trimester samples analysed with a mass spectrometry platform, opening new perspectives in testing methods. Different, specific and highly sensitive methodologies for the detection of cffDNA were successfully applied for diagnostic purposes from 11–12 weeks of gestation37,40–49, exploiting the presence/absence of the RHD gene amplification and assuming a negative RHD maternal genotype (the RHD gene is deleted in Caucasians and 19% of Afro-Americans) corresponding to the RhD-negative phenotype. A recent meta-analysis by Zhu et al.50, performed on 41 publications and including ~11,000 samples, reported an overall diagnostic accuracy of 98.5% (99% in the first trimester) and a negative predictive value of 98%.
The clinical application was initially restricted to anti-D immunised women at high risk of HDFN5,51,52 and was then extended to non-immunised RhD-negative women for the appropriate administration of targeted prenatal RhIg. Since the first large-scale feasibility studies24,25, many authors have demonstrated the accuracy of foetal RHD genotyping, also at early gestational age37,47,48, and have evaluated strategies to implement it (Table V). Besides nations in which, nationally or regionally, non-invasive foetal RHD genotyping is part of routine antenatal screening, it is under evaluation in other European countries.
Table V.
Published studies on non-invasive RHD foetal genotyping performed with real-time polymerase chain reaction.
| Reference (First Author, year, ref) | Country | Samples (n) | Gestational week | RHD exons | Controls | False-positive results | False-negative results | Inconclusive results | Sensitivity (%) | Specificity (%) | Accuracy (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Rouillac-Le Sciellour C, 2004, 53 | France | 893 | 7–40 | 7, 10 | None | 99.5 | |||||
| van der Schoot CE, 2006, 24 | The Netherlands | 1,257 | 30* | 7 | None | 0 | 3 | 0 | 99.6 | ||
| Rouillac-Le Sciellour C, 2007, 54 | France | 300 | 10–34 | 7, 10 | Maize DNA (T) | 2 | 100 | 99.9 | 99.3 | ||
| Finning K, 2008, 19 | UK | 1,869 | 8–38 | 5, 7 |
CCR5 (T) SRY (F) |
14 (0.8%) | 3 (0.2%) | 56 (3%) | 99.7§ | 94§ | 99.7§ |
| Minon JM, 2008, 55 | Belgium | 545 | 10–38 | 4, 5, 10 |
CCR5 (T) SRY (F) |
1 | 100 | 99.8 | |||
| Muller SP, 2008, 25 | Germany | 1,113 | 6–32 | 5, 7 | β-globin (T) | NR | NR | NR | 99.7/99.8# | 99.2/98.1# | 99.5/99.2# |
| Hyland CA, 2009, 56 | Australia | 140 | 12–40 | 4, 5, 10 |
CCR5 (T) SRY RASSF1A (F) |
0 | 100 | 96.4 | |||
| Wang XD, 2009, 57 | China | 78 | 14–40 | β-globin (T) SRY, STR’s (F) |
3 | 0 | 3 | 100 | 94.9 | ||
| Morano, D 2010, 58 | Italy | 11 | 14–22 | 4, 5, 10 |
GAPD (T) SRY (F) |
1 | 1 | 3 | 54 | ||
| Mohammed NB, 2010, 59 | Pakistan | 21 | 20–39 | 5 | β-globin (T) | 3 | 1 | 62.3 | 62.5 | ||
| Chinen PA, 2010, 60 | Brazil | 102 | 7–36 | 7, 10 | β-globin (T) | 2 | 0 | 100 | |||
| Scheffer PG, 2011, 34 | The Netherlands | 212 | 7–38 | 5, 7 | Albumin (T) SRY, biallelic polymorphisms (F) |
0 | 0 | 100 | |||
| Akolekar R, 2011, 37 | UK | 586 | 11–14 | 5, 7 | CCR5 (T) | 6 (3.5%) | 84 (14.3%) | 98.2 | 100 | 98.8 | |
| Clausen FB, 2012, 61 | Denmark | 2,312 | 25* | 5, 7/5, 10/5, 10 ** |
CCR5 SOD GAPDH (T) |
0.26% (6) | 0.087% (2) | 3.2% (74) | 99.9 | 99.3 | 99.6 |
| Daniels G, 2012, 26 | UK | 4,876 | 7–24 | 5, 7 | NR | 17 | 3 | >99.2& | |||
| De Haas M, 2012, 62 | The Netherlands | 6,941 | NR | 5, 7 | None | 0.2% | 56 (1.1%) | 12 (0.2%) | >99.6@ | ||
| Grande M, 2013, 63 | Spain | 284 | 24–26 | 5, 7 (+6, 10) | β-actin (T) SRY (F) |
1 | 100 | 99 | 99.6 | ||
| Macher HC, 2012, 64 | Spain | 1,012 | 10–28 | 5, 7 | SRY (F) | 3 | 100 | 98.6 | 99.3 | ||
| Oliveira J, 2011, 31 | Portugal | 19 | 22–37 | 7, 10 |
SRY (F) GLO (T) |
1 (5.9%) | 80 | 100 | 67 | 94 | |
| Sbarsi I, 2012, 65 | Italy | 31 | NR | 4, 5, 10 |
CCR5 (T) SRY (F) |
100 | 100 | 100 | |||
| Wikman AT, 2012, 28 | Sweden | 4,118 | 8–40 | 4 | GAPDH (T) | 14 (1.1%) | 23 (1.1) | 14 | 98.9$ | ||
| Dovč-Drnovšek T, 2013, 66 | Slovenia | 153 | 7–38 | 5, 7, 10 (+ int 4) |
ALB (T) SRY (F), |
100 | 100 | 100 | |||
| Tiblad E, 2013, 67 | Sweden | 8,374 | 29* | 4 | GAPDH (T) | 98.9 | 98.9 | ||||
| Chitty LS, 2014, 48 | UK | 2,288 | 11–23 | 5, 7 |
CCR5 (T) SRY (F) |
18 | 19 | 393 (8%) | 99.34 | 94.91 | 91.24 |
| Schmidt LC, 2014, 68 | Brasil | 55 | 12–29 | 5, 7 | SRY (F) | 1 | 5 (9.1%) | 100 | 100 | 100 | |
| De Haas M, 2016, 29 | The Netherlands | 25,789 | 27* | 5, 7 | None | 225 (0.57%) | 9 (0.03%) | 0.21% | 99.94 | 97.74 | 99.09 |
| Vivanti A, 2016, 49 | France | 416 | 10–14 | 10 | Mouse GALT gene (T) |
7 | 0 | 9 (2.2%) | 100 | 95.2 | 98.3 |
| Trucco Boggione C, 2017, 69 | Argentina | 298 | 19–28 | 4, 5, 7, 10 | SRY (F) | 1 | 8 | 100 | 98.8 | ||
| Geifman-Holtzaman O, 2006, 70 | Meta-analysis | 3,261 (37 articles) | 8–42 | 95.4 | 98.6 | 91.4*** | |||||
| Zhu Y, 2014, 50 | Meta-analysis | 11,129 (41 studies) | 99 | 98 | 98.5 (99% in first trimester) |
The foetus was categorised as RhD positive if RHD predicted phenotype was RhD positive, RHD variant or inconclusive;
extraction by spin-column method/magnetic tips;
the sensitivity was 99.3% at 10 weeks and 100% after 22 weeks;
specificity was >99.8 at 11 weeks (3 false negatives out 4,011 samples);
Estimated from reported false-negative results <0.25%;
mean;
multicentre study;
(90.8 % in the first trimester, 85.0% in the second trimester, 85.3% in the third trimester); CCR5: C-C chemokine receptor type 5; GAPDH: glycerldehyde 3-phosphate dehydrogenase; SOD: superoxidase dismutase; (T): total cell-free DNA control; (F): foetal cell-free DNA control; NR: not reported.
In Italy the first experience58 of foetal RHD genotyping was conducted in 2010, when cffDNA was tested by real-time PCR and RhD foetal status was successfully determined. This was followed by the Pavia experience65 in 2012, validating the local protocol and demonstrating feasibility and 100% accuracy.
The project supported by the Regional Blood Centre of Emilia-Romagna gave us the opportunity to evaluate the diagnostic accuracy of a commercial test for non-invasive foetal RHD genotyping and to introduce it into the routine antenatal protocol for screening RhD-negative pregnant women.
The validation of the process was complex because of the difficulties encountered with foetal DNA extraction, which is a critical step25. In fact, the quantity of foetal nucleic acids is minimal in comparison with the background of maternal DNA and, despite the high sensitivity and specificity of testing methods, they are prone to false-negative results. False-negative results could cause the lack of anti-D administration and the risk of alloimmunisation. In our study we had one false negative result in an 18-week sample: RhD-positive typing of the neonate was confirmed on a new sample from the newly born baby. After exclusion of an operating mistake, the retrospective analysis showed that the typing session (in which extraction was performed with the QIAamp® DSP DNA Virus Kit) had RhD positive control Ct values at the higher limits, although in the defined range. The genotyping on a second maternal plasma aliquot, extracted with the QIAamp® Circulating Nucleic Acid Kit, confirmed the positive serological result. The QIAamp® DSP DNA Virus Kit extraction kit is one of three tested during this study and, although the best in an International Workshop32, it showed a low specificity in foetal DNA recovery, as well as a failure to meet defined Ct values for amplification and extraction controls. These performances forced us to repeat the test even more than once (Table III), but the results were often only clarified when using the QIAamp® Circulating Nucleic Acid Kit, which revealed greater efficiency in foetal DNA recovery and showed a higher conclusive result rate at first analysis. Therefore, in these samples, two of the main causes of false-negative results are concurrent: the efficiency of foetal DNA extraction63 and the early gestational age, when the quantity of foetal DNA is minimal37,48.
The maize DNA provided in the Free DNA Fetal Kit®RhD is considered an adequate control for both DNA extraction and PCR amplification71 and the lack of a positive control for cffDNA is considered acceptable if foetal genotyping is used for screening. Relying on our experience and with the aim of avoiding false-negative results, we will implement a second analysis at least 4 weeks later, as recommended54,72, to confirm negative results obtained at an early gestational age, as an alternative to a cffDNA control37,54.
The inconclusive result rate was 3.2%, as expected with amplification of more targeted RHD exons. In fact, this strategy could increase indeterminate results, conceivably because of low foetal DNA recovery (unrelated to gestational age) in RhD-positive samples and different primer affinity, and because of contamination or non-specific amplification in RhD-negative samples, but at the same time it reduces false results, compared to analysis of single exons. Even if there are experiences demonstrating the reliability of single-exon assays (with a low false-negative rate)24,28,49,67,70 and their ease of application in routine analysis, we chose the opportunity, offered by a commercial CE-marked kit, to explore more RHD exons simultaneously and to identify maternal and/or foetal RHD variants that may not be recognisable by the conventional serological techniques or may cause maternal alloimmunisation, respectively. In a complex and polymorphic gene such as RHD, the amplification/not amplification of a single exon (5, 7, 10 or others) is informative only for the single exon analysed and it could deceive, giving only a partial view of the gene status. In fact, the disadvantage of amplifying only one RHD exon is the risk of false-positive results, if the target is a non-coding exon (i.e. exon 7 in RHDψ), or false-negative results, if the target is substituted by a RHCE coding sequence (i.e. exon 5 in RHD*DVI).
On the other hand, the availability of primers that amplify three pivotal exons of RHD (exons 5, 7 and 10) allows the gene to be studied from a different point of view and with more accuracy. After the first decade of experience, the Special Non-invasive Advances in Fetal and Neonatal Evaluation Network73 recommended the use of specific primers for exons 5 and 7 to overcome the complexity of RHD. Exon 5 is involved in many RHD variants and hybrid genes: for example, it does not amplify in RHDψ (coding for the RhD-negative phenotype in 67% of black Africans) and RHD*6(RHD*DVI) (the most common RHD variant in Caucasians)29,74 alleles. The analysis of exon 5 alone gives a correct negative result in the first case, but a false negative result in the second one: in the latter case it would mean no RhIg administration in a RhD-incompatible pregnancy and the risk of anti-D alloimmunisation. Exon 7 has many D-specific nucleotides and is absent only in a minority of RHD variants (such as RHD-CE-Ds)54,74, while it is amplified in the RHDψ allele giving a false-positive result, which would mean inappropriate RhIg administration. However, if exons 5 and 7 are analysed together, a discrepant amplification result between them suggests a variant, induces a more detailed study and guides appropriate management of pregnancy. Finally, as in many hybrid RHD-CE-D genes (some of which associated with weak or partial antigen expression) exons 5 and 7 are both replaced by the corresponding RHCE exons, the inclusion of exon 10 confirms or rules out the presence of the RHD gene, depending on positive or negative results; in fact, exon 10 is one of the most preserved RHD exons (although it is a non-coding exon and it could give false-positive results if tested alone)75 and warrants an high specificity of the test.
This compound of primers is useful in RHD foetal genotyping of pregnant African women, in whom the frequency of RhD variants is higher (up to 6–7%) than in Caucasians (0.2–1%)14. In fact one of the RHD maternal variants (RHD*CE(3–7)-D) was identified in an African woman associated with a d(C)ces allele and partial C; the other hybrid gene (RHD-CE(2–9)-D), associated with a RhD-negative phenotype and RHCE*Ce or RHCE*cE alleles in Europeans, was found in a Caucasian woman.
About one decade ago, Rouillac-Le Sciellour et al.54 performed preliminary studies with the Free DNA Fetal Kit® RhD (with primers for RHD exons 7 and 10) and reported high sensitivity and specificity rates (100% and >99% respectively); however, in this article the authors affirmed that “the first generation kit is not suitable for correct genotyping of foetuses from women carrying a RHDψ pseudogene in their genome”. In fact, the kit could not discriminate the presence of RHDψ in a foetus from a normal coding RHD gene because exons 7 and 10 were amplified in both cases: if RHDψ was present, this would mean inappropriate immunoprophylaxis or wrong evaluation of HDFN risk in the case of an alloimmunised pregnant woman. The subsequent implementation of exon 5 primers is helpful for the identification of RhD variants.
The low number of samples collected at an early gestational age is a weakness of our investigation and we, therefore, excluded such samples from the statistical analysis of the study. Nevertheless, after the inclusion of samples taken at an early gestational age (<23+6 week), the sensitivity and accuracy of the test were 99.6% (95% CI: 98.7–100) and 95.5% (95% CI: 93.3–97.8) respectively. These results encourage the performance of further studies to confirm the reliability of this protocol also in the first trimester.
The high sensitivity (100%) and diagnostic accuracy (99.3%) rates, obtained in the period during which foetal genotyping is usually performed for targeted antenatal RhIg administration (24–28+6 weeks), show that fetal RHD genotyping is a reliable test for managing RhD-negative pregnancies and a powerful diagnostic tool in prenatal care, if appropriate strategies are applied. Beyond economic and ethical advantages, we believe that it is a way to prevent anti-D alloimmunisation and HDFN with an integrated and multidisciplinary management of pregnancy: it deflects attention from RhIg to pregnant women and their babies and improves safety, without increasing the risk of immunisation or foetal disease.
Conclusions
The findings of this study demonstrate that foetal RHD detection on maternal plasma using a commercial multiple-exon assay is a reliable and accurate tool to predict foetal RhD phenotype, with acceptable rates of false-negative and false-positive results. It can be a safe guide for the appropriate administration of targeted prenatal RhIg, avoiding unnecessary exposure to immunoprophylaxis. The presence of three RHD exon primers increases the sensibility and provides the opportunity to identify RhD variants in mixed ethnic populations. The validation of the process allows its introduction into routine clinical practice in the Region.
Footnotes
Funding and resources
This work was financially supported by a grant from the Regional Blood Centre of the Region of Emilia-Romagna (Italy): “Introduction of non-invasive RHD fetal genotyping form maternal plasma as a guide for appropriate administration of anti-D immunoglobulin for maternal alloimmunisation and HDFN prevention” (PG0060400-Piano Sangue e Plasma 2013–15).
Authorship contributions
CC, PF, AG, GL, SN and SM contributed to the sample collection. SP and LR performed the foetal genotyping. SM and LR collected the data and wrote the manuscript. VR revised the draft paper.
The Authors declare no conflicts of interest.
References
- 1.de Haas M, Thurik FF, Koelewijn JM, van der Schoot CE. Haemolytic disease of the fetus and newborn. Vox Sang. 2015;109:99–113. doi: 10.1111/vox.12265. [DOI] [PubMed] [Google Scholar]
- 2.Urbaniak SJ, Greiss MA. RhD haemolytic disease of the fetus and the newborn. Blood Rev. 2000;14:44–61. doi: 10.1054/blre.1999.0123. [DOI] [PubMed] [Google Scholar]
- 3.Crowther C, Middleton P. Anti-D administration after childbirth for preventing Rhesus alloimmunisation. Cochrane Database Syst Rev. 2000;2:CD000021. doi: 10.1002/14651858.CD000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crowther CA, Keirse MJ. Anti-D administration in pregnancy for preventing Rhesus alloimmunisation. Cochrane Database Syst Rev. 2000;2:CD000020. doi: 10.1002/14651858.CD000020. [DOI] [PubMed] [Google Scholar]
- 5.de Haas M, Finning K, Massey E, Roberts DJ. Anti-D prophylaxis: past, present and future. Transfus Med. 2014;24:1–7. doi: 10.1111/tme.12099. [DOI] [PubMed] [Google Scholar]
- 6.National Institute for Health and Clinical Excellence. Review of NICE technology appraisal guidance. London UK: Aug, 2008. [Accessed on 06/02/2018]. Routine antenatal anti-D prophylaxis for women who are rhesus D negative. Available at: https://www.nice.org.uk/guidance/ta156/resources/routine-antenatal-antid-prophylaxis-for-women-who-are-rhesus-d-negative-pdf-82598318102725. [Google Scholar]
- 7.Qureshi E, Massey D, Kirwan T, et al. BCSH guideline for the use of anti-D immunoglobulin for the prevention of haemolytic disease of the fetus and newborn. Transfus Med. 2014;24:8–20. doi: 10.1111/tme.12091. [DOI] [PubMed] [Google Scholar]
- 8.Bennardello F, Coluzzi S, Curciarello G, et al. Raccomandazioni per la prevenzione ed il trattamento della Malattia Emolitica del Feto e del Neonato. Società Italiana di Medicina Trasfusionale e Immunoematologia. 2014. [Accessed on 06/02/2018]. Available at: http://www.simti.it/linee_guida.aspx?ok=1.
- 9.Royal Australian and New Zealand College of Obstetricians and Gynecologists. Guidelines for the use of Rh (D) immunoglobulin (Anti-D) in obstetrics in Australia. Australian and New Zealand Society of Blood Transfusion Ltd; Sidney: 2015. [Accessed on 06/02/2018]. Available at: http://www.ranzcog.edu.au/publications/statements/C-obs6.pdf. [Google Scholar]
- 10.Society of Obstetricians and Gynecologists of Canada. Prevention of Rh alloimmunization. J Obstet Gynecol Can. 2003;25:765–73. doi: 10.1016/s1701-2163(16)31006-4. [DOI] [PubMed] [Google Scholar]
- 11.Muñiz Diaz E, Oyonarte S, Rodriguez-Villanueva J, et al. Sociedad Espanola de Transfusion Sanguinea, Sociedad Espanola de Obstetricia y Ginecologia. Protocolo de Diagnostico y Prevencion de la Enfermedad Hemolitica del feto y del Recien Nacido. 2008. [Accessed on 06/02/2018]. Available at: http://www.sets.es/index.php/30-publicaciones/guias/208-protocolo-de-diagnostico-y-prevencion-de-la-enfermedad-hemolitica-del-feto-y-del-recien-nacido.
- 12.ACOG practice bulletin. Prevention of RhD alloimmunization. Number 4, May 1999 (replaces educational bulletin Number 147, October 1990). Clinical management guidelines for obstetrician-gynecologists. American College of Obstetrics and Gynecology. Int J Gynaecol Obstet. 1999;66:63–70. [PubMed] [Google Scholar]
- 13.The Irish Haematology Society, Institute of Obstetricians and Gynaecologists, Royal College of Physicians of Ireland and Obstetrics and Gynaecology Programme HSE Directorate for Quality and Clinical Strategy. Clinical Practice Guideline. The use of anti-D immunoglobulin for the prevention of RhD haemolytic disease of the newborn. [Accessed on 06/02/2018]. Guideline No 13. Version 2. 0 Date of publication: June 2012; revised June 2014. Available at: https://rcpi-live-cdn.s3.amazonaws.com/wp-content/uploads/2016/05/10.-Anti-D-Immunoglobin-for-prevention-of-RHD-Haemolytic-Disease-of-the-newborn.pdf.
- 14.Daniels G. Human Blood Groups. 3rd ed. London: Wiley-Blackwell; 2013. [Google Scholar]
- 15.Avent ND, Reid M. The Rh blood group system: a review. Blood. 2000;95:375–87. [PubMed] [Google Scholar]
- 16.Daniels G. Variants of RhD - current testing and clinical consequences. Br J Haematol. 2013;161:461–70. doi: 10.1111/bjh.12275. [DOI] [PubMed] [Google Scholar]
- 17.Teitelbaum L, Metcalfe A, Clarke G, et al. Cost and benefits of non-invasive fetal RhD determination in Alberta. Ultrasound Obstet Gynecol. 2015;45:84–8. doi: 10.1002/uog.14723. [DOI] [PubMed] [Google Scholar]
- 18.Kenny-Walsh E. Clinical outcomes after hepatitis C infection from contaminated anti-D immune globulin. New Engl J Med. 1999;340:1228–33. doi: 10.1056/NEJM199904223401602. [DOI] [PubMed] [Google Scholar]
- 19.Finning K, Martin P, Summers J, et al. Effect of high throughput RHD typing of fetal DNA in maternal plasma on use of anti-RhD immunoglobulin in RhD negative pregnant women: prospective feasibility study. BMJ. 2008;336:816–8. doi: 10.1136/bmj.39518.463206.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kent J, Farrell AM, Soothill P. Routine administration of anti-D: the ethical case for offering pregnant women fetal RHD genotyping and a review of policy and practice. BMC Pregnancy Childbirth. 2014;14:87–90. doi: 10.1186/1471-2393-14-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumpel BM. Lesson learnt from many years of experience using anti-D in humans for prevention of RhD immunization and haemolytic disease of the fetus and the newborn. Clin Exp Immunol. 2008;154:1–5. doi: 10.1111/j.1365-2249.2008.03735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolton-Maggs PHB, editor; Poles D, et al. on behalf of the Serious Hazards of Transfusion SHOT Steering Group. The 2016 Annual SHOT Report. 2017. [Accessed on 06/02/2018]. Available at: https://www.shotuk.org/wp-content/uploads/SHOT-Report-2016_web_11th-July.pdf.
- 23.Daniels G, Finning K, Martin O, Soothill P. Fetal blood group genotyping from DNA from maternal plasma: an important advance in the management and prevention of haemolytic disease of the fetus and the newborn. Vox Sang. 2004;87:225–32. doi: 10.1111/j.1423-0410.2004.00569.x. [DOI] [PubMed] [Google Scholar]
- 24.van der Schoot CE, Soussan AA, Koelewijn J, et al. Non-invasive antenatal RHD typing. Transfus Clin Biol. 2006;13:53–7. doi: 10.1016/j.tracli.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 25.Müller SP, Bartels I, Stein W, et al. The determination of the fetal D status from maternal plasma for decision taking on Rh prophylaxis is feasible. Transfusion. 2008;48:2292–301. doi: 10.1111/j.1537-2995.2008.01843.x. [DOI] [PubMed] [Google Scholar]
- 26.Daniels G, Finning K, Wade A, et al. Implementation of routine of fetal RH typing in all RHD-negative pregnant women: timing, costs, and efficiency. Vox Sang. 2012;103(Suppl 1):34. [Abstract] [Google Scholar]
- 27.Clausen FB, Steffensen R, Christiansen M, et al. Routine noninvasive prenatal screening for fetal RHD in plasma of RhD-negative pregnant women - 2 years of screening experience from Denmark. Prenat Diagn. 2014;34:1000–5. doi: 10.1002/pd.4419. [DOI] [PubMed] [Google Scholar]
- 28.Wikman AT, Tiblad E, Karlsson A, et al. Noninvasive single-exon fetal RHD determination in a routine screening program in early pregnancy. Obstet Gynecol. 2012;120:227–34. doi: 10.1097/AOG.0b013e31825d33d9. [DOI] [PubMed] [Google Scholar]
- 29.de Haas M, Thurik FF, van der Ploeg CP, et al. Sensitivity of fetal RHD screening for safe guidance of targeted anti-D immunoglobulin prophylaxis: prospective cohort study of a nationwide programme in the Netherlands. BMJ. 2016;355:i5789. doi: 10.1136/bmj.i5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clausen FB. Integration of noninvasive prenatal prediction of fetal blood group into clinical prenatal care. Prenat Diagn. 2014;34:409–15. doi: 10.1002/pd.4326. [DOI] [PubMed] [Google Scholar]
- 31.Oliveira J, Osòrio N, Rocha J, et al. Fetal RHD and RHCE genotyping in plasma of Rh negative pregnant women. Int J Biomed Lab Sci. 2012;1:50–8. [Google Scholar]
- 32.Legler TJ, Liu Z, Mavrou A, et al. Workshop report on the extraction of foetal DNA from maternal plasma. Prenat Diagn. 2007;27:824–9. doi: 10.1002/pd.1783. [DOI] [PubMed] [Google Scholar]
- 33.Lupi C, Perrone E, Basevi V, et al. La nascita in Emilia-Romagna. 13° Rapporto sui dati del Certificato di Assistenza al Parto (CedAP) - Anno 2015. Regione Emilia-Romagna. 2016. [Accessed on 06/02/2018]. Available at: http://salute.regione.emilia-romagna.it/siseps/sanita/cedap.
- 34.Scheffer PG, van der Schoot CE, Page-Christiaens GCML, de Haas M. Noninvasive fetal blood group genotyping of Rhesus D, c, E and of K in alloimmunised pregnant women: evaluation of a 7-year clinical experience. Br J Obstet Gynaecol. 2011;118:1340–8. doi: 10.1111/j.1471-0528.2011.03028.x. [DOI] [PubMed] [Google Scholar]
- 35.Ferrari M, Carrera P, Lampasona V, Galbiati S. New trend in non-invasive prenatal diagnosis. Clin Chim Acta. 2015;451:9–13. doi: 10.1016/j.cca.2014.12.026. [DOI] [PubMed] [Google Scholar]
- 36.Lo YM, Corbetta N, Chamberlain PF, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350:485–7. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- 37.Akolekar R, Finning K, Kappusamy R, et al. Fetal RHD genotyping in maternal plasma at 11–13 weeks of gestation. Fetal Diagn Ther. 2011;29:301–6. doi: 10.1159/000322959. [DOI] [PubMed] [Google Scholar]
- 38.High-throughput non-invasive prenatal testing for fetal RHD genotype. Diagnostics guidance National Institute for Health and Clinical Excellence. Nov 9, 2016. [Accessed on 06/02/2018]. Available at: https://www.nice.org.uk/guidance/dg25/resources/highthroughput-noninvasive-prenatal-testing-for-fetal-rhd-genotype-pdf-1053691935685.
- 39.Moise KJ, Gandhi M, Boring NH, et al. Circulating cell-free DNA to determine the fetal RHD status in all three trimesters of pregnancy. Obstet Gynecol. 2016;6:1340–6. doi: 10.1097/AOG.0000000000001741. [DOI] [PubMed] [Google Scholar]
- 40.Lun FM, Chiu RW, Chan KC, et al. Microfluidics digital PCR reveals a higher than expected fraction of fetal DNA in maternal plasma. Clin Chem. 2008;54:1664–72. doi: 10.1373/clinchem.2008.111385. [DOI] [PubMed] [Google Scholar]
- 41.Saito H, Sekizawa A, Morimoto T, et al. Prenatal DNA diagnosis of a single-gene disorder from maternal plasma. Lancet. 2000;356:1170. doi: 10.1016/S0140-6736(00)02767-7. [DOI] [PubMed] [Google Scholar]
- 42.Ding C, Chiu RW, Lau TK, et al. MS analysis of single-nucleotide differences in circulating nucleic acids: application to noninvasive prenatal diagnosis. Proc Natl Acad Sci USA. 2004;101:10762–7. doi: 10.1073/pnas.0403962101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bustamante-Aragones A, Gallego-Merlo J, Trujillo-Tiebas MJ, et al. New strategy for the prenatal detection/exclusion of paternal cystic fibrosis mutations in maternal plasma. J Cyst Fibros. 2008;7:505–10. doi: 10.1016/j.jcf.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 44.Hyett JA, Gardener G, Stojilkovic-Mikic T, et al. Reduction in diagnostic and therapeutic interventions by non-invasive determination of fetal sex in early pregnancy. Prenat Diagn. 2005;25:1111–6. doi: 10.1002/pd.1284. [DOI] [PubMed] [Google Scholar]
- 45.Costa JM, Benachi A, Gautier E, et al. First-trimester fetal sex determination in maternal serum using real-time PCR. Prenat Diagn. 2001;21:1070–4. doi: 10.1002/pd.219. [DOI] [PubMed] [Google Scholar]
- 46.Bustamante-Aragones A, Rodriguez de Alba M, Gonzalez-Gonzalez C, et al. Foetal sex determination in maternal blood from the seventh week of gestation and its role in diagnosing haemophilia in the foetuses of female carriers. Haemophilia. 2008;14:593–8. doi: 10.1111/j.1365-2516.2008.01670.x. [DOI] [PubMed] [Google Scholar]
- 47.Bartha JL, Finning K, Soothill PW. Fetal sex determination from maternal blood at 6 weeks of gestation when at risk for 21-hydroxylase deficiency. Obstet Gynecol. 2003;101:1135–6. doi: 10.1016/s0029-7844(03)00064-4. [DOI] [PubMed] [Google Scholar]
- 48.Chitty LS, Finning K, Wade A, et al. Diagnostic accuracy of routine antenatal determination of fetal RHD status across gestation: population based cohort study. BMJ. 2014;349:g5243. doi: 10.1136/bmj.g5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vivanti A, Benachi A, Huchet FX, et al. Diagnostic accuracy of fetal Rhesus D genotyping using cell-free fetal DNA during the first trimester of pregnancy. Am J Obstet Gynecol. 2016;215:606.e1–606.e5. doi: 10.1016/j.ajog.2016.06.054. [DOI] [PubMed] [Google Scholar]
- 50.Zhu Y, Zheng Y, Li L, et al. Diagnostic accuracy of non-invasive fetal RhD genotyping using cell-free fetal DNA: a meta analysis. J Matern Fetal Med. 2014;27:1839–44. doi: 10.3109/14767058.2014.882306. [DOI] [PubMed] [Google Scholar]
- 51.Finning KM, Martin PG, Soothill PW, Avent ND. Prediction of fetal D status from maternal plasma: introduction of a new noninvasive fetal RHD genotyping service. Transfusion. 2002;42:1079–85. doi: 10.1046/j.1537-2995.2002.00165.x. [DOI] [PubMed] [Google Scholar]
- 52.Finning K, Martin P, Daniels G. A clinical service in the UK to predict fetal Rh (Rhesus) D blood group using free fetal DNA in maternal plasma. Ann N Y Acad Sci. 2004;1022:119–23. doi: 10.1196/annals.1318.019. [DOI] [PubMed] [Google Scholar]
- 53.Rouillac-Le Sciellour C, Puillandre P, Gillot P, et al. Large scale prediagnosis study of fetal RHD genotyping by PCR on plasma DNA from RhD-negative pregnant women. Mol Diagn. 2004;8:23–31. doi: 10.1007/BF03260044. [DOI] [PubMed] [Google Scholar]
- 54.Rouillac-Le Sciellour C, Sèrazin V, Brossard Y, et al. Noninvasive fetal RHD genotyping from maternal plasma. Use of a new developed Free DNA Fetal Kit RhD. Transfus Clin Biol. 2007;14:572–7. doi: 10.1016/j.tracli.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 55.Minon JM, Gerard C, Senterre JM, et al. Routine fetal RHD genotyping with maternal plasma: a four year experience in Belgium. Transfusion. 2008;48:373–81. doi: 10.1111/j.1537-2995.2007.01533.x. [DOI] [PubMed] [Google Scholar]
- 56.Hyland CA, Gardener Gj, Davies H, et al. Evaluation of non-invasive prenatal RHD genotyping of the fetus. Med J Aust. 2009;191:21–5. doi: 10.5694/j.1326-5377.2009.tb02668.x. [DOI] [PubMed] [Google Scholar]
- 57.Wang XD, Wang BL, Ye SL, et al. Non-invasive foetal RHD genotyping via real-time PCR of foetal DNA from Chinese RhD-negative maternal plasma. Eur J Clin Invest. 2009;39:607–17. doi: 10.1111/j.1365-2362.2009.02148.x. [DOI] [PubMed] [Google Scholar]
- 58.Morano D, Girardi A, Carinci F, Patella A. [Prenatal non invasive determination of fetal Rh status on maternal plasma in first trimester of pregnancy by real-time polymerase chain reaction: an italian experience]. Riv It Ost Gin. 2010;27:242–8. [in Italian.] [Google Scholar]
- 59.Mohammed NB, Kakal F, Somani M, et al. Non-invasive prenatal determination of fetal RhD genotyping from maternal plasma: a preliminary study in Pakistan. JCPSP. 2010;20:246–9. [PubMed] [Google Scholar]
- 60.Chinen PA, Nardozza LMM, Martinhago CD, et al. Noninvasive determination of fetal Rh blood group, D antigen status by cell-free DNA analysis in maternal plasma: experience in a Brazilian population. Amer J Perinatol. 2010;27:759–62. doi: 10.1055/s-0030-1253560. [DOI] [PubMed] [Google Scholar]
- 61.Clausen FB, Christiansen M, Steffensen R, et al. Report of the first nationally implemented clinical routine screening for fetal RHD in D− pregnant women to ascertain the requirement for antenatal RhD prophylaxis. Transfusion. 2012;52:752–8. doi: 10.1111/j.1537-2995.2011.03362.x. [DOI] [PubMed] [Google Scholar]
- 62.De Haas M, van der Ploeg CPB, Scheffer PG, et al. A nation-wide fetal RHD screening program for targeted antenatal and postnatal anti-D. ISBT Scie Ser. 2012;7:164–7. [Google Scholar]
- 63.Grande M, Ordonez E, Cirigliano V, et al. Clinical application of midtrimester non-invasive fetal RHD genotyping and identification of RHD variants in a mixed-ethnic population. Prenat Diagn. 2013;33:173–8. doi: 10.1002/pd.4035. [DOI] [PubMed] [Google Scholar]
- 64.Macher HC, Noguerol P, Medrano-Campillo P, et al. Standardization non-invasive fetal RHD and SRY determination into clinical routine using a multiplex RT-PCR assay for fetal cell-free DNA in pregnant women plasma: results in clinical benefits and cost saving. Clin Chim Acta. 2012;413:490–4. doi: 10.1016/j.cca.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 65.Sbarsi I, Isernia P, Montanari L, et al. Implementing non-invasive RHD genotyping on cell-free foetal DNA from maternal plasma: the Pavia experience. Blood Transfus. 2012;10:34–8. doi: 10.2450/2011.0021-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dovč-Drnovšek T, Klemenc P, Toplak N, et al. Reliable determination of fetal RhD status by RHD genotyping from maternal plasma. Transfus Med Haemother. 2013;40:37–43. doi: 10.1159/000345682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tiblad E, Wikman AT, Ajne G, et al. Targeted routine antenatal anti-D prophylaxis in the prevention of RhD immunisation - outcome of a new antenatal screening and prevention program. PLoS ONE. 2013;8:e70984. doi: 10.1371/journal.pone.0070984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schmidt LC, Cabral ACV, Faria MA, et al. Noninvasive fetal RHD genotyping from maternal plasma in an admixed Brazilian population. Genet Mol Res. 2014;13:799–805. doi: 10.4238/2014.February.7.1. [DOI] [PubMed] [Google Scholar]
- 69.Trucco Boggione C, Lujàn Brajovich ME, Mattaloni SM, et al. Genotyping approach for non-invasive foetal RHD detection in an admixed population. Blood Transfus. 2017;15:66–73. doi: 10.2450/2016.0228-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Geifman-Holtzman O, Grotegut CA, Gaughan JP. Diagnostic accuracy of noninvasive fetal Rh genotyping from maternal blood - a meta-analysis. Am J Obstet Gynecol. 2006;195:1163–73. doi: 10.1016/j.ajog.2006.07.033. [DOI] [PubMed] [Google Scholar]
- 71.Scheffer PG, de Haas M, van der Schoot CE. The controversy about controls for fetal blood group genotyping by cell-free fetal DNA in maternal plasma. Curr Opin Hematol. 2011;18:467–73. doi: 10.1097/MOH.0b013e32834bab2d. [DOI] [PubMed] [Google Scholar]
- 72.Avent ND. RHD genotyping from maternal plasma: guidelines and technical challenges. Methods Mol Biol. 2008;444:185–201. doi: 10.1007/978-1-59745-066-9_14. [DOI] [PubMed] [Google Scholar]
- 73.Chitty LS, van der Schoot CE, Hahn S, Avent ND. SAFE - The Special Non-invasive Advances in Fetal and Neonatal Evaluation Network: aims and achievements. Prenat Diagn. 2008;28:83–8. doi: 10.1002/pd.1929. [DOI] [PubMed] [Google Scholar]
- 74.Daniels G, Finning K, Martin P, Massey E. Noninvasive prenatal diagnosis of fetal blood group phenotypes: current practice and future prospects. Prenat Diagn. 2009;29:101–7. doi: 10.1002/pd.2172. [DOI] [PubMed] [Google Scholar]
- 75.Van der Schoot CE, Thurik FF, Scheffer PG, et al. Non-invasive prenatal diagnosis - erythrocyte and platelet antigens. ISBT Science Series. 2015;10:192–6. [Google Scholar]