Abstract
Umbilical cord mesenchymal stem cells (UC-MSCs) exert strong immunomodulatory effects and can repair organs. However, their roles in radiation injury remain unclear. We show that in tree shrews with acute radiation injury, injected UC-MSCs significantly improved survival rates, reduced lung inflammation and apoptosis, prevented pulmonary fibrotic processes, recovered hematopoiesis, and increased blood counts. A protein microarray analysis showed that serum levels of the anti-inflammatory cytokines IL-10 and IL-13 and the growth factors BMP-5, BMP-7, HGF, insulin, NT-4, VEGFR3, and SCF were significantly higher, while those of the inflammatory cytokines IL-2, TIMP-2, TNF-α, IFN-γ, IL-1ra, and IL-8 and the fibrosis-related factors PDGF-BB, PDGF-AA, TGF-β1, IGFBP-2, and IGFBP-4 were significantly lower in UC-MSC-injected animals. A transcriptome analysis of PBMCs showed that the mRNA expression of C1q was upregulated, while that of HLA-DP was downregulated after UC-MSC injection. These results confirm the immunohistochemistry results. eGFP-labeled UC-MSCs were traced in vivo and found in the heart, liver, spleen, lungs, kidneys, thymus, small intestine and bone marrow. Our findings suggest that UC-MSC transplantation may be a novel therapeutic approach for treating acute radiation injury.
Keywords: Radio-pulmonary lesion, Umbilical cord mesenchymal stem cells, C1q, HLA-DP, Tissue repair
Introduction
As ionizing radiation has become widely used in industry and medical science, the incidence of radiation injury has significantly increased. Different radiation doses may cause different levels of damage at the gene, cell, and organ levels (Gorbunov and Kiang 2017). Hematopoietic cells and lung tissues are highly sensitive to radiation. Thus, restoring hematopoietic immune function and improving the structure and function of the lung are key to treating radiation injury (Majeed 2017). Bone marrow-derived mesenchymal stem cells (BMSCs) contribute to hematopoietic recovery and organ repair. However, the mechanisms underlying their effects remain unclear.
Mesenchymal stem cells (MSCs) are a group of adult cells with multidirectional differentiation potential that are derived from the mesenchyme. MSCs are widely distributed in the interstitium of connective tissues and in organs throughout the body. Among these tissues, MSCs are most abundant in bone marrow and cord blood but can be isolated from other organs and tissues, including the skin, blood vessels, adipose tissue and several populations of mesenchymal stem cells. MSCs therefore represent a heterogeneous group. Currently, research studies are mainly focused on BMSCs. However, the number of BMSCs and their potential to proliferate and differentiate decline with age. In addition, autologous transplantation of BMSCs depends on the patient’s state, limiting their application. UC-MSCs are a new representative population of MSCs that exhibit high plasticity, are relatively abundant, have stable biological properties, are easy to obtain, store, and transport, induce no allogeneic rejection, and are associated with no ethical controversy. Hence, they are a natural and ideal seed for cell therapies (Marupanthorn et al. 2017).
Few studies have explored the use of allograft transplantation of UC-MSCs in acute radiation injury, and the mechanism by which UC-MSCs exert their effects in the treatment of radiation-induced lung injury therefore remains unclear. In this study, a linear accelerator was used to irradiate tree shrews to establish an animal model. Tree shrews provide the following advantages: they are small in size and have a short breeding cycle, the cost of breeding them is low, they offer good repeatability, and their genetic background and biological characteristics are similar to those of humans. After allograft transplantation of UC-MSCs was performed in a tree shrew model of acute radiation injury, the post-radiation injury survival rate was calculated, the structure of the tissues was observed, and cytokine levels were detected using a protein chip. Differentially expressed genes were analyzed using transcriptomics. The aim of this study was to explore the protective mechanisms exerted by UC-MSCs in radiation injury.
Methods and materials
Experimental animals
In all, 150 healthy adult (6 months old, 108–118 g) female tree shrews and 10 pregnant tree shrews (38–41 days old, 152–172 g) were purchased from the Kunming Animal Institute (Kunming, Yunnan, China) and used for all experiments. The animal production license number was SCXK (Dian) K2012-0001. All experiments were performed in accordance with the guidelines and with the approval of the National Institute of Laboratory Resources Committee and the Kunming General Hospital Animal Management and Use Committee.
Isolation, culture, identification, and labeling of tree shrew UC-MSCs
Primary UC-MSCs were isolated and subcultured as follows (Pan et al. 2017). Umbilical cord tissues were collected from healthy tree shrews after caesarean birth. Small blood vessels and fascia were removed, and a 1 mm3 section of the umbilical cord was cut off, washed with PBS (Sigma, St. Louis, MO, USA), and cultured in DMEF/F12 medium with 1% penicillin–streptomycin (PSL01, Caisson Labs, SBA, Logan, UT, USA) and 20% fetal bovine serum (SV30087.02, HyClone, Thermo, Logan, UT, USA). The medium was changed every 3 days. The tissue mass was discarded after the cord had been cultured for 7 or 10 days. The remaining cells were then grown to 85% fusion, digested with 0.25% Trypsin-0.04% EDTA (Invitrogen, Carlsbad, CA, USA), and then passaged three times for the following experiments.
CCK-8 cell proliferation assays were performed as follows: logarithmic phase cells at a concentration of 5 × 104 cells/mL were inoculated in 96-well plates (Corning, Corning, NY, USA) (100 µL/well) and continuously analysed for 9 days (10 repeat wells per day; the medium was changed every 3 days). Beginning on the second day after inoculation, samples were tested at the same time every day, and 10 µL of CCK-8 solution (WH1199, Sigma) was then added to each well. After the cells were incubated at 37 °C for 4 h, absorbance was detected at 450 nm in each well with a microplate reader (RT-6000, Rayto, Shenzhen, China). Growth curves were drawn with time (t) as the abscissa and the mean absorbance value of all of the wells on the same day (A) placed on the ordinate axis.
Cell differentiation potential was analyzed as follows. Logarithmic phase cells were inoculated at a concentration of 1 × 105 cells/mL into 6-well plates (Corning) and induced according to the instructions included in adipogenesis differentiation kits (MKRMA-90031, OriCellTM, Cyagen, Santa Clara, CA, USA), osteogenic differentiation kits (HUXUC-90021, OriCellTM, Cyagen), and cartilage differentiation kits (MKRMA-90042, OriCellTM, Cyagen). The negative controls were set up separately. After the cells were induced for 21 days, the differentiated cells obtained from each of the above kits were stained with oil-red-O, alizarin red and alcian blue, respectively.
The eGFP + PURO lentivirus-transduced tree shrew UC-MSCs were obtained as follows. On the first day, UC-MSCs (2 × 104/well) were cultured at 37 °C overnight in 24-well plates. On the second day, viral vector particles (GM100102-2, Genomeditech, Shanghai, China) were thawed on ice before they were used for transduction. The viral vector stock was diluted to the desired concentration (cells:vector = 1:150) in DMEF/F12 medium containing 20% fetal bovine serum. Diluted viral vector was then added to each well. At 48 h after transduction was initiated, the lentiviral vector-containing medium was poured off and replaced with fresh medium. The cells were continuously cultured for an additional 24–48 h. The rate of transduction was determined by flow cytometry, and cell morphology was observed with an inverted fluorescence microscope.
Irradiation injury procedure and experimental groups
Unanesthetized tree shrews were placed in a homemade plastic box (25 cm × 6.6 cm × 4 cm) for whole-body single X-ray ionizing radiation (Elekta Synergy Platform, Linear Accelerator, Elekta Limited, Crawley, UK). All of these procedures were performed at the Radiation Center of Kunming General Hospital. The size of the irradiation field was 30 cm × 30 cm, the source wave distance was 100 cm, the dose rate was 480 cGy/min, and the total irradiation dose was 4.5 Gy.
The 150 tree shrews were randomly divided into two parts, the first part (n = 60) was used to observe weight changes and survival analysis, the second part (n = 90) was used for other experiments. The first part was randomly divided into three groups: (1) Treatment (n = 20): UC-MSCs injection (3 × 107 cells/kg) + radiation; (2) Model (n = 20): radiation + PBS injection; (3) Control (n = 20): PBS injection. The second part was randomly divided into four groups: (1) Treatment (n = 20): UC-MSCs injection (3 × 107 cells/kg) + radiation; (2) EGFP (n = 10): eGFP labeled UC-MSCs injection (3 × 107 cells/kg) + radiation; (3) Model (n = 40): radiation + PBS injection; (4) Control (n = 20): PBS injection. In all animals, the injection was via tail vein, the first injection was performed immediately after irradiation, and the injection day was defined as the first day. The tree shrews were treated once every 7 days for a total of 4 treatments.
Histology and lung inflammation score
Histology was evaluated in 4- to 6-µm-thick paraffin-embedded sections that were fixed in 4% paraformaldehyde (158127, Sigma). The sections were then stained with hematoxylin and eosin (HE) and Masson’s staining, submitted to TUNEL assays (11684817910, Roche, Shanghai, China), and observed under a light microscope. Nine fields were viewed in each lung section at 400 × magnification and scored for pneumonia as follows (Arrieta et al. 2009): 0 indicated a normal lung structure, 1–2 indicated a wider alveolar wall and the presence of < 10 inflammatory cells per high-power (HP) field, 3–4 indicated a significantly expanded alveolar wall and 10–20 inflammatory cells per HP field, 5–6 indicated a significantly wider alveolar wall and infiltration (> 20 per HP field) by inflammatory cells, and 7–8 indicated that the alveoli and alveolar walls were both infiltrated with inflammatory cells. Final calculations were performed to statistically analyze the average score in the nine fields.
Flow cytometry
Surface markers were detected on UC-MSCs as follows: suspended cells (1 × 106) were incubated with 5 µL of anti-human CD105-phycoerythrin (PE), CD90-PE, CD34-PE or CD31-PE antibodies (BD Biosciences, San Jose, CA, USA) at room temperature for 30 min in the dark. Isotype-controls were incubated at the same time in the same volume of buffer. After the cells were washed 3 times with PBS, they were centrifuged at a rate of 300–400 g for 5 min. The suspended cells were fixed with 400 µL of 4% paraformaldehyde and then analyzed. Apoptosis was detected in peripheral blood mononuclear cells (PBMCs) as follows. PBMCs were isolated from peripheral blood samples using lymphocyte separation medium (Solarbio, Beijing, China) according to the instructions included in the apoptosis kit (20170524, 4A Biotech, Beijing, China). Suspended PBMCs (1 × 106) were incubated with 5 µL of Annexin V/FITC at room temperature for 30 min in the dark. Then, 10 µL of PI (20 µg/mL) was immediately added, and the blank tubes and tubes containing cells labeled with a single antibody were set. Data were acquired with a FACSCalibur flow cytometer and analyzed with CELLQuest software (LSR Fortessa, Becton–Dickinson, San Jose, CA, USA).
Real-time PCR analysis
Total RNA was extracted from cultured cells or tissues using a kit (RP1202, BioTeKe, Beijing, China), and 10 µg of each sample was then amplified using a gene amplification instrument [TC-96/G/H(b)C, LifeECO, Hangzhou, China] and reverse-transcribed into cDNA. The primer sequences are shown in Tables 1, 2, and 3. DNA amplification was performed on a PCR cycler (C1000, Bio-Rad, Hercules, CA, USA) using the following conditions: an initial denaturation step for 1 min at 95 °C and 40 cycles of a denaturation step for 10 s at 95 °C, an annealing step for 15 s at the temperature indicated for each primer, and an extension step for 15 s at 72 °C. Relative mRNA expression levels were calculated using the 2−△△Ct relative quantification method (Fausto et al. 2017).
Table 1.
qRT-PCR primers used for validation of UC-MSCs multidirectional differentiation abilities analysis
| Gene symbol | Sequence (5′ → 3′) | Product length (bp) | Annealing temperature (°C) |
|---|---|---|---|
| Osteocytes | |||
| Runx2 | F: TCTGACTTCTGCTCTGGCCTTC | 152 | 60.7 |
| R: GTGGCTGGATAGTGCATTCGTGG | |||
| ALPI | F: GAGGAAGAGGACCCAGCCTTCTG | 164 | 60.7 |
| R: AGCTGTGCTTTCAGGATCCGAGT | |||
| OCN | F: CGGTGAGTGAGGACAAAGGTCAG | 167 | 60.7 |
| R: GGTGGAGACCCCTTGCCTAATTG | |||
| Adipocytes | |||
| PPARγ2 | F: CATTTTGGCCCACCAACTTCGGA | 158 | 60.7 |
| R: CATTGGATCGGCCCTTGGGAATG | |||
| LPL | F: TACTTCGGCTGGTCAGACTGGTG | 121 | 60.7 |
| R: GCAAACGGGACACTTTCTCCCTG | |||
| Chordrocytes | |||
| Sox6 | F: CCCTAAGTCAGGTGATGGAACAA | 137 | 57.0 |
| R: AGCAGACTGAAGTTACCCTTTCA | |||
| Col2a1 | F: ATCAGAGGCATGAGTTTGAGGG | 157 | 57.0 |
| R: CACATTGCGAGAACACTTCTACA | |||
Table 2.
qRT-PCR primers used for validation of pro-inflammatory cytokines analysis
| Gene symbol | Sequence (5′ → 3′) | Product length (bp) | Annealing temperature (°C) |
|---|---|---|---|
| IL-6 | F: ACCCCAGTTAATCTCATTCACCC | 159 | 62.3 |
| R:GTCAGGGGTGGTTATTGCATCT | |||
| TGF-β1 | F: ATTAAAGATGGAGAGAGGACTGCG | 180 | 55.0 |
| R: CTACAGTAGTGTTCCCCACTGGTC | |||
| TNF-α | F: ATCTGGAACCCGAGTGACAAG | 178 | 57.9 |
| R: CGTGAAGAGGACCTGGGAGTAG |
Table 3.
qRT-PCR primers used for validation of transcriptome analysis
| Gene ID Gene symbol |
Sequence (5′ → 3′) | Product length (bp) | Annealing temperature (°C) |
|---|---|---|---|
| TREES_T100020781 (CCL24) |
F: CTGGTGGTGAAGATGACCCC | 149 | 54.0 |
| R: CACTACCTTGCCCTCACAGG | |||
| TREES_T100019229 (JCHAIN) |
F: CAGGATAGCAGGAATCCGGG | 129 | 60.4 |
| R: AAGACAGTGCCACAGAGACC | |||
| TREES_T100008796 (C1Q) |
F: TTGTAGTCGTTGACGGCCAG | 147 | 62.3 |
| R: CAACCTCTGCGTGAACTTGC | |||
| TREES_T100012017 (SERHL2) |
F: CTCTCTGCAACAGGAGTCGC | 142 | 54.0 |
| R: TTCCCTGCTCCTTCCGACTG | |||
| TREES_T100016538 (HLA-DP) |
F: CTCTCAGCAACACCGTCAGT | 129 | 54.7 |
| R: TCCAAAGAAGTCCGTGGAGC | |||
| TREES_T100007200 (untitled) |
F: CAAGCTTGGCCGGTATGAAG | 197 | 54.7 |
| R: CACGGACTGTAGCTTTGGGTTC | |||
| TREES_T100021224 (untitled) |
F: GGGAGATCAAATGTAATGATGCCC | 150 | 62.3 |
| R: TAACACCATCCACCTTCCACG | |||
| TREES_T100001931 (FN1) |
F: TGCCTATCAGCAGGGTCTTTG | 143 | 60.4 |
| R: CGGTGTCCAAGTCACCATGA | |||
| GAPDH | F: GCTGGTGCTGAGTATGTTG | 129 | 57.0 |
| R: AGTCTTCTGGGTGGCAGTGATG |
Immunohistochemical staining of paraffin-embedded tissues
Di-methyl benzene-fixed paraffin sections of lung tissues were rehydrated, and endogenous peroxidase activity was quenched with 3% H2O2. The sections were incubated with primary antibodies overnight at 4 °C, rinsed, and then incubated with secondary antibodies at 37 °C for 1 h. Streptavidin Biotin Complex (SABC) was applied for 30 min at 37 °C, 3,3’-diaminobenzidine (DAB) was applied at room temperature for 20 min, and the tissues were then counter-stained with hematoxylin. The sections were dehydrated, mounted in resin, and analyzed using microscopy. After each of these steps, the sections were washed with PBS for 5 min (for a total of 4 washes). The following reagents were used for immunohistochemistry: mouse anti-GFP monoclonal antibodies (11814460001, Anti-GFP, Roche), Supervision TM anti-mouse reagent (HRP, D-3001,Changdao, Shanghai, China); mouse monoclonal antibodies for C1q [Anti-C1q antibodies (JL-1), ab71940, Abcam, Cambridge, MA, USA], Supervision TM anti-mouse reagent (HRP, D-3001, Changdao, Shanghai, China); rabbit polyclonal antibodies for HLA-DP (anti-HLA-DP antibodies, ab153886, Abcam), Supervision TM anti-rabbit reagent (HRP, D-3002, Changdao, Shanghai, China); a DAB colorimetric kit (DA1010, Solarbio, Beijing, China), and an SABC kit (SA1020, Diaclone, Besançon, France).
Blood tests
Blood samples were collected from tree shrews via the femoral vein on the 7th, 14th, 21st and 28th day after irradiation. EDTA-K2 anticoagulation was used to detect white blood cells (WBCs), red blood cells (RBCs), platelets (PLTs) and hemoglobin (HGB) in an automatic blood cell analyzer (Sysemex, XT2000i, kobe, Japan).
Protein chip detection of inflammatory cytokines and growth factors
On day 28 after cell transplantation, serum samples were collected from 1 mL of femoral vein peripheral blood using centrifugation at 340 g for 5 min. All procedures were performed in accordance with the instructions included in the protein chip for inflammatory cytokines (QAH-INF-3-1, Ray Biotech, Norcross, GA, USA) and growth factors (QAH-GF-1-1, Ray Biotech) kits. The chips were scanned with an infrared fluorescence scanner, and the signal intensity was converted into a picture and saved in a gray-scale TIFF format. Densitometry analysis was performed with AAH-INF-3 Software (Inga Jácome et al. 2016).
Transcriptome analysis of peripheral blood mononuclear cells
A total of six samples were used for RNA-Seq analysis. Samples were collected from the model group and treatment group on the 28th day after irradiation. RNA-Seq was performed on an Illumina HiSeq 2000 instrument. The raw reads were cleaned up by removing adaptor-polluted or low-quality reads to ensure the quality of the data that were used in further analyses. The reference genomes and annotation files were downloaded from the Ensembl database (http://www.ensembl.org/index.html). Bowtie/Bowtie2 was used to build the genome index, and clean data were mapped to the reference genome using TopHat v2.0.12. The fragment counts for each gene in each sample were counted with HTSeq v0.6.0, and the fragments per kilobase of transcript per million mapped reads (FPKM) was then calculated to estimate the expression level of the genes in each sample. DEGseq v1.18.0 was used to identify genes that were differentially expressed between the two samples in non-biological replicates. Genes with q ≤ 0.05 and a |log 2_ratio| ≥ 1 were identified as differentially expressed genes (DEGs). NCBI, Uniprot, GO, KEGG and other databases were used to annotate the DEGs (Li et al. 2016).
Statistical analysis
SPSS software (version 21.0, SPSS Inc., Chicago, IL, USA) was used to analyze all statistical data using one-way analysis of variance (ANOVA). Survival curves were analyzed using the Kaplan–Meier method, and values are shown as the mean ± SD. P < 0.05 was considered statistically significant.
Results
Isolation, culture, identification, and labeling of tree shrew umbilical cord mesenchymal stem cells
UC-MSCs were collected in the third passage and found to have similar sizes, fusiform shapes and parallel or spiral-shaped growth patterns. The cell growth curves had a typical “S” shape. The cells began the logarithmic growth phase on the 4th day and then entered plateau stage on the 6th day (Fig. 1a). The UC-MSCs were successfully transfected with eGFP, and the rate of positive labeling was as high as 100% (Fig. 1b). Flow cytometry analysis revealed that 98.7% of the UC-MSCs expressed CD105, 96.3% expressed CD90, 0.185% expressed CD34, and 0.043% expressed CD31 (Fig. 1c). Osteogenesis-induced UC-MSCs stained with alizarin red contained a large amount of orange-red calcium and calcium nodules, oil red O-stained adipose-induced UC-MSCs contained significantly higher numbers of red particles, and Alcian blue-stained chondrocyte-induced UC-MSCs were labeled with chondroitin proteoglycan (Fig. 1d). Real-time quantitative PCR indicated that significantly higher levels of osteocyte-specific genes (Runx2, ALPI, and OCN), adipocyte-specific genes (LPL and PPARγ2) and chondrocyte-specific genes (Sox6 and Col2a1) were expressed in the induced than in the non-induced UC-MSCs (Dominici et al. 2006) (P < 0.01) (Fig. 1e).
Fig. 1.
Umbilical cord collection and the isolation, culture, labeling and identification of UC-MSCs. a The isolation and culture of UC-MSCs and their growth curve. b UC-MSCs were labeled with eGFP, nuclei were stained blue with DAPI (4′,6-diamidino-2-phenylindole), and FACS analysis was used to determine the rate of eGFP transfection. c FACS analysis of surface markers (CD105, CD90, CD31 and CD34). d After 21 days of induction, osteogenic induction was analyzed using alizarin red staining, adipogenic induction was evaluated using Oil-Red-O staining, and chondrogenic differentiation was evaluated using Alcian blue staining. e The mRNA levels of osteocyte-specific genes (Runx2, ALPI, and OCN), adipocyte-specific genes (LPL and PPARγ2) and chondrocyte-specific genes (Sox6 and Col2a1) in control and separately induced cells. mRNA expression levels were analyzed by quantitative RT-PCR. Third-passage UC-MSCs were used as the controls, **P < 0.01, compared with control. The images in a and d are shown at × 100 magnification, and the images in b are shown at × 200 magnification, scale bars = 100µm. (Color figure online)
Pathological changes in vital tissues and organs after irradiation
Pathological changes were analyzed in vital tissues and organs on the 7th day after irradiation and compared to those observed in the normal control group. Here, we describe our post-irradiation findings. We found that the heart changed very little (Fig. 2a, b). The structure of the liver was complete, but necrotic, degenerative, and swollen cells were occasionally observed (Fig. 2c, d). The structure of the spleen was complete, and its acini lienalis and trabecula were clear, while the red pulp sinus of the spleen was slightly expanded and congested (Fig. 2e, f). The interstitial lung and pulmonary capillaries were congested and exhibited edema. Alveolar lumens and bronchioles had clearly been infiltrated by mononuclear, neutrophil, lymphocyte, and phagocytic hemosiderin cells, and a flaky hemorrhage was observed in addition to a slight thickening of the alveolar wall (Fig. 2g, h). The structure of the kidneys was generally complete, and renal tissues exhibited hyperemia and were slightly infiltrated by mononuclear and lymphocyte cells (Fig. 2i, j). After irradiation, the structure of the small intestinal villi and glands was intact but exhibited mild edema, the central lacteal was clear, and the tissue was infiltrated by very small numbers of inflammatory cells (Fig. 2k, l). The structure of the thymus was clear, there were thymic corpuscles in the medulla, and degenerative and necrotic lymphocyte cell debris was occasionally visible in the cortex (Fig. 2m, n). Finally, the bone marrow exhibited significant hyperemia, and there were fewer BMCs, especially granulocyte cells, but more osteoclasts (Fig. 2o, p). These findings indicate that whole-body radiation damage promoted inflammation in multiple organs and that the bone marrow and lung tissues were most sensitive to radiation.
Fig. 2.
Histopathological observation of vital organs on the 7th day after irradiation. a–p: Hematoxylin–eosin (H&E)-stained sections of tree shrew vital organs at 7 days after irradiation. A total of 6 tree shrews were used in 2 separate experiments. Normal tree shrews were used as controls. Representative images are shown at × 200 magnification, scale bars = 100µm. Nuclei were stained blue with hematoxylin
Effect of UC-MSCs on basic status and survival rates in tree shrews exposed to irradiation
During the 7 days after irradiation, the tree shrews in both the model and treatment groups had a poor general state. They were apathetic, exhibited poor activity, anorexia and sweating, and some of them had erected or lost hair and had difficulties in opening their eyes. The general state of the treatment group animals gradually improved through the 14th day after irradiation. They exhibited higher rates of water drinking, food intake, and activity and had less vertical hair and hair loss. The general state of the model group animals improved until the 21st day after irradiation, but their activity and dietary intake were slightly worse than those observed in the treatment group. The average body weights of the model and the treatment animals were significantly lower than those in the control group during the observation period (P < 0.01), but the weight in the treatment group was significantly higher than that in the model group on the 28th day (model, 114.50 ± 4.97 g; treatment, 121.40 ± 5.64 g; control, 130.51 ± 4.09 g) (P < 0.01). In addition, we calculated the weights of dead tree shrews and compared them with the surviving tree shrews during the same period. The results revealed that body weights was significantly lower in the dead tree shrews in the model group than in the control and treatment groups (P < 0.05), but there was no significant difference in body weight between the dead tree shrews and those that survived in the model group (control, 120.56 ± 4.42 g; treatment, 112.40 ± 7.63 g; model (survived), 108 ± 5.74 g; model (dead), 106.02 ± 3.73 g). These data indicate that UC-MSCs protected body weight (Fig. 3a). By the end of the 28-day observation period, the survival rate was 100% in both the control and treatment groups but 50% in the model group (median survival time, 21.5 days), indicating that tree shrew UC-MSCs improved post-irradiation survival in tree shrews (Fig. 3b).
Fig. 3.
Weight changes and survival analysis in irradiated tree shrews. a Weight changes in tree shrews after irradiation, **P < 0.01, compared with control; ΔP < 0.01, model compared with treatment. b Survival analysis of tree shrews after irradiation. Survival curves were analyzed using the Kaplan–Meier method. A total of 60 tree shrews were used for each control and experimental group in each separate experiment. Normal tree shrews were used as controls
Effect of UC-MSCs on post-irradiation peripheral blood cell counts in tree shrews
After irradiation, WBC, RBC, HGB and PLT counts gradually decreased in the tree shrews in the model and treatment groups. The WBC and PLT counts in the treatment and model groups decreased to their lowest levels on the 7th and 14th days, respectively, before gradually increasing. On the 28th day after UC-MSC transplantation, WBC and PLT counts (2.0 ± 0.4 × 109/L and 538.8 ± 183.3 × 109/L, respectively) were similar to those observed in the controls (2.1 ± 0.4 × 109/L and 598.6 ± 93.4 × 109/L, respectively) (P > 0.05) and significantly higher than those in the model group (1.6 ± 0.3 × 109/L and 394.3 ± 70.5 × 109/L, respectively) (P < 0.05). Both RBC and HGB counts were also lower on the 14th day after irradiation before gradually increasing, similar to the trend observed for WBC and PLT counts. RBC and HGB counts were significantly lower on the 14th and 21st days in the model and treatment groups than in the control group (P < 0.01), but there were no significant differences among the three groups at other time points (P > 0.05) (Fig. 4).
Fig. 4.
Changes in peripheral blood cell counts in irradiated tree shrews. WBC: White blood cell; PLT: Platelet; HGB: Hemoglobin; RBC: Red blood cell.**P < 0.01, compared with control group; ΔP < 0.05, model group vs treatment group. A total of 90 tree shrews were used for each control and experimental group in each separate experiment. Normal tree shrews were used as controls
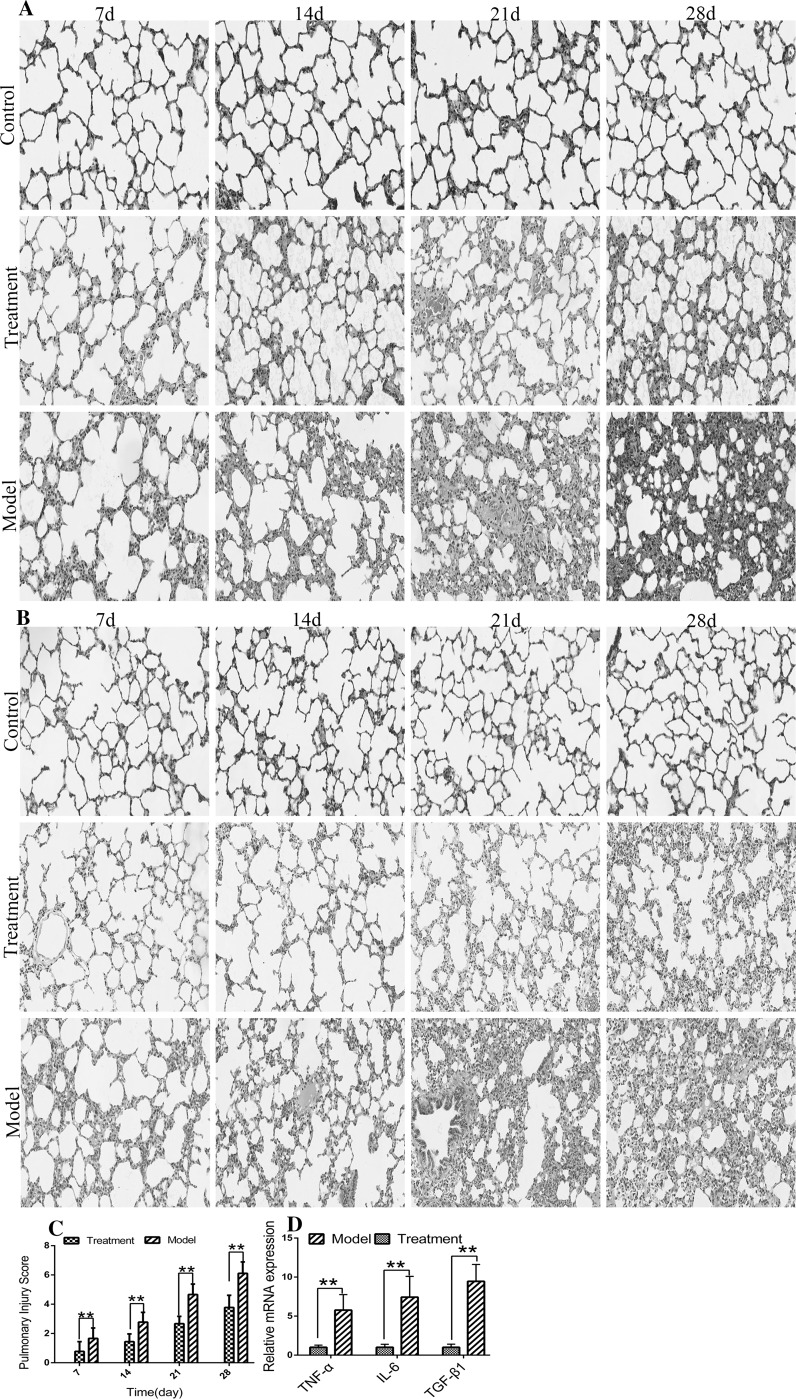
Effect of UC-MSCs on Pneumonia and Fibrosis in tree shrews after irradiation
HE performed in tree shrew lung tissues (Fig. 5a) showed that the irradiation group exhibited gradual congestion and edema of the pulmonary capillaries and that the alveoli and bronchiolar capillaries were infiltrated by mononuclear, neutrophilic and lymphocyte cells. These animals also had slightly thickened alveolar walls and exhibited alveolar fusion and collapse. More lung inflammation was observed in the treatment group than in the control group. However, the degree of inflammation was significantly lower than that observed in the model group. Masson’s staining of lung tissue (Fig. 5b) showed that over time, collagen fibers gradually proliferated and collagen deposition increased in the model group. A significantly lower degree of collagen deposition was observed in the treatment group than in the model group. Here, we used a quantitative score to estimate the degree of lung inflammation. The pneumonia score was higher in the model group than in the treatment group at each time point (P < 0.01) (Fig. 5c). To improve our understanding of why there was a difference in the degree of inflammation between the treatment and model groups, we detected the mRNA levels of TGF-β1, IFN-γ, and IL-6 in the lungs on the 28th day after irradiation. We found that the mRNA levels of TGF-β1, IFN-γ, and IL-6 were significantly higher in the model group than in the treatment group (P < 0.01) (Fig. 5d).
Fig. 5.
Lung injury assessment. a HE-stained sections of tree shrew lungs. b Masson-stained sections of tree shrew lungs. Collagen, mucus, and cartilage are shown in blue; plasma, cellulose, pulp and glial cells are shown in red; and nuclei are shown in blue or dark blue. c Injury scores in the lungs. d mRNA levels of TGF-β1, IFN-γ, and IL-6 in the lungs on the 28th day after irradiation. Images are shown at × 200 magnification, scale bars = 100um. Normal tree shrews were used as controls. Vertical bars indicate the mean ± SD, **P < 0.01, model compared with treatment. (Color figure online)
UC-MSC transplantation reduced cell apoptosis
On the 28th day after irradiation, TUNEL staining was performed in lung tissues (Fig. 6a). Among the three groups, the number of apoptotic cells was lowest in the control group, highest in the model group, and intermediate in the treatment group. The results of a flow cytometry analysis of PBMCs (Fig. 6b) suggested that UC-MSC transplantation significantly reduced radiation injury-induced apoptosis, and the differences observed among the three groups were significant (P < 0.01) (control, 7.2 ± 0.4; treatment, 14.0 ± 0.5; and model, 21.4 ± 0.3).
Fig. 6.
UC-MSC transplantation reduced cell apoptosis. a TUNEL-stained sections of tree shrew lungs on the 28th day after irradiation. Representative images are shown at × 100 magnification, scale bars = 100um. b The rate of cell apoptosis in PBMCs on the 28th day after irradiation. Apoptosis assays were performed using flow cytometry. Normal tree shrews were used as controls. Vertical bars indicate the mean ± SD,**P < 0.01, compared with treatment; ΔΔP < 0.01, model compared with control
Detection of peripheral blood growth factor and inflammatory levels with protein chips on the 28th day after irradiation
BMP-5, BMP-7, HGF, insulin, NT-4, VEGFR3 and SCF levels were significantly higher and the levels of IGFBP-2, IGFBP-4, PDGF-AA, and TGF-β1 were significantly lower in the treatment group than in the model group (P < 0.05) (Table 4). The levels of the anti-inflammatory cytokines IL-10 and IL-13 were significantly higher, while the levels of IL-2, TIMP-2, PDGF-BB, TNF-α, IFN-γ, IL-1ra and IL-8 were significantly lower, in the treatment group than in the model group (P < 0.05) (Table 5).
Table 4.
Comparison of peripheral blood growth factors in the treatment and the model on 28th day after irradiation (mean ± SD)
| Treatment (pg/mL) | Model (pg/mL) | |
|---|---|---|
| BMP-5 | 9349.0 (3688.4) | 1664.3 (800.9)* |
| BMP-7 | 3260.9 (1664.0) | 419.3 (149.2)* |
| IGFBP-2 | 3031.1 (491.8) | 4616.2 (1042.3)* |
| IGFBP-4 | 5383.4 (3821.2) | 26,113.5 (14,864.3)* |
| HGF | 559.2 (146.5) | 38.2 (12.6)** |
| Insulin | 687.8 (288.3) | 118.3 (28.8)* |
| NT-4 | 712.0 (179.7) | 118.7 (40.3)** |
| PDGF-AA | 817.1 (167.9) | 1691.2 (202.7)** |
| VEGF R3 | 1453.9 (493.8) | 240.4 (71.5)* |
| SCF | 824.9 (136.0) | 66.7 (36.5)** |
| TGFβ-1 | 461.1 (43.5) | 1957.6 (589.1)** |
*P < 0.05, **P < 0.01, compared with treatment group
Table 5.
Comparison of peripheral blood Inflammatory factors in the treatment and the model on 28th day after irradiation (mean ± SD)
| Treatment (pg/mL) | Model (pg/mL) | |
|---|---|---|
| IL-2 | 35.0 (25.4) | 220.8 (136.0)** |
| TIMP-2 | 9083.1 (445.8) | 10,613.1 (1115.5)* |
| PDGF-BB | 139.3 (61.4) | 477.4 (140.7)** |
| TNF-α | 124.1 (45.2) | 375.2 (63.1)** |
| IFN-γ | 79.5 (56.1) | 378.0 (76.6)** |
| IL-1ra | 76.7 (58.2) | 292.6 (120.0)* |
| IL-8 | 52.25 (30.8) | 158.5 (30.2)* |
| IL-10 | 157.7 (61.3) | 20.1 (12.3)* |
| Il-13 | 34.2 (2.3) | 12.9 (2.6)** |
*P < 0.05, **P < 0.01, compared with treatment group
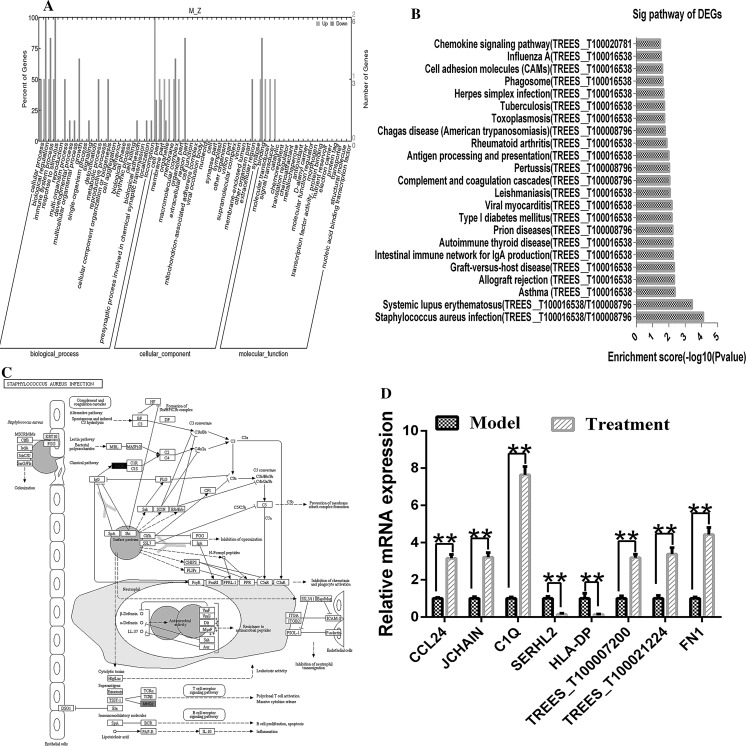
Transcriptome analysis of peripheral blood mononuclear cells
An Illumina HiSeq 2000 sequencing platform was used to perform transcriptome sequencing (RNA-Seq) on PBMCs obtained from three model group tree shrews and three treatment group tree shrews on the 28th day of the experiment. After contaminated reads, low-quality reads, and reads with more than 5% undermined bases were removed, the remaining clean data were compiled. These results are summarized in Table 6. We identified 8 genes that were differentially expressed (DEGs) between the model and the treatment groups (Table 7). These DEGs were annotated using the NCBI, Uniprot, GO and KEGG databases to obtain detailed descriptive information. The GO data were divided into three ontology groups, including biological processes, molecular functions and cellular components. Cellular processes, biological regulatory processes, immune system processes, and responses to stimuli were the most highly represented items among biological processes; binding factors, molecular transducers, and catalytic factors were the most highly represented items among molecular functions; and cell components, membrane components, organelles, and macromolecular complexes were the most highly represented items among cellular components (Fig. 7a). These DEGs were enriched in 23 metabolic pathways in a KEGG analysis (Fig. 7b), and infection with Staphylococcus aureus (map05150) (Fig. 7c) was the most highly represented pathway. TREES_T100016538 (HLA-DP) and TREES_T100008796 (C1Q) were involved in multiple pathways. The mRNA expression levels of the DEGs were consistent with the results of RT-PCR transcriptomic analysis when verified by RT-PCR (Fig. 7d).
Table 6.
Primary analysis of sequencing transcriptome data
| M1 | M2 | M3 | Z1 | Z2 | Z3 | |
|---|---|---|---|---|---|---|
| Raw Reads Number | 94,982,010 | 93,727,106 | 78,641,332 | 83,709,386 | 86,944,020 | 89,637,152 |
| Clean Reads Number | 69,455,356 | 68,067,494 | 58,641,546 | 60,329,252 | 64,380,534 | 64,493,658 |
| Clean Q30 Bases Rate (%) | 94.03 | 93.73 | 94.23 | 93.97 | 94.15 | 93.83 |
| Mapping Rate | 0.44 | 0.6 | 0.57 | 0.62 | 0.59 | 0.53 |
Table 7.
Differential genes of transcriptome analysis
| Gene ID Gene symbol |
Up/down | NT: description |
|---|---|---|
| TREES_T100020781 (CCL24) |
Down | Chemokine (C–C motif) ligand 24 (CCL24), mRNA |
| TREES_T100019229 (JCHAIN) |
Down | Immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides (IGJ), mRNA |
| TREES_T100008796 (C1Q) |
Down | Complement component 1, q subcomponent, C chain (C1QC), mRNA |
| TREES_T100012017 (SERHL2) |
Up | Serine hydrolase-like 2 (SERHL2), mRNA |
| TREES_T100016538 (HLA-DP) |
Up | HLA class II histocompatibility antigen, DP alpha 1 chain-like (LOC102498495), mRNA |
| TREES_T100007200 (untitled) |
Down | Felis catus immunoglobulin kappa light chain mRNA, complete cds |
| TREES_T100021224 (untitled) |
Down | Macaca mulatta clone 2.14 T cell receptor delta chain mRNA, complete cds |
| TREES_T100001931 (FN1) |
Down | Tupaia chinensis fibronectin 1 (FN1), mRNA |
Up, higher (upregulated) in the model than in the treatment group; Down, lower (downregulated) in the model than in the treatment group
Fig. 7.
Transcriptome analysis of peripheral blood mononuclear cells. a GO statistical histograms of differentially expressed genes. Upper: higher (upregulated) in the model group than in the treatment group; lower: lower (downregulated) in the model group than in the treatment group. b KEGG map of the assembled results. c The top enrichment score [− log 10 (P value)] of the significant enrichment pathway (red indicates nodes representing upregulated genes, and green indicates nodes representing downregulated genes). d A quantitative analysis of the expression of eight genes by RT-PCR in peripheral blood samples obtained from model and treatment group tree shrews. **P < 0.01, model compared with treatment. (Color figure online)
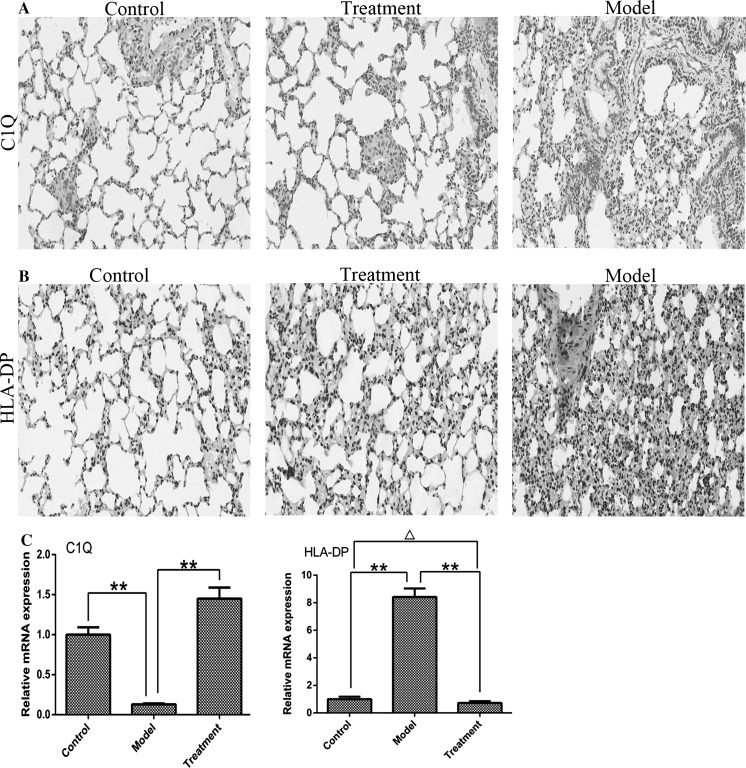
C1q deposition was increased and HLA-DP deposition was reduced in the lung tissues of irradiated tree shrews treated with UC-MSCs
To determine whether lung tissues exhibited changes in C1q and HLA-DP similar to those observed in the PBMC transcriptome results, we evaluated C1q and HLA-DP levels in the lungs with immunohistochemistry and RT-PCR. The results suggested that ionizing radiation led to excessive consumption of C1q, while cell transplantation slowed down this process (Fig. 8a). The relative mRNA expression level of C1q was significantly lower in the model group than in the control and treatment groups (P < 0.01) (Fig. 8c). In contrast, the protein expression level of HLA-DP was higher in irradiated lungs (Fig. 8b). Additionally, the relative mRNA expression level of HLA-DP was significantly higher in the model group than in the control and treatment groups (P < 0.01), and the difference between the control and treatment groups was significant (Fig. 8c).
Fig. 8.
After irradiated tree shrews were treated with UC-MSCs, C1q deposition was increased and HLA-DP deposition was reduced in lung tissues. a Immunohistochemistry (IHC) with anti-C1q antibodies (red). Representative images are shown at × 200 magnification, scale bars = 100um. b IHC with anti-HLA-DP antibodies (red). Representative images are shown at × 200 magnification, scale bars = 100um. Nuclei were stained blue with hematoxylin. c The mRNA expression levels of C1q and HLA-DP in the lungs after irradiation. Vertical bars indicate the mean ± SD, **P < 0.01, compared with model; ΔP < 0.05, treatment compared with control. Normal tree shrews were used as controls. (Color figure online)
Tracking UC-MSCs in vivo
EGFP-labeled UC-MSCs were used to track transplanted cells in vivo. On the 28th day after cell transplantation, important organs, such as the liver, heart, kidneys, spleen, lungs, small intestine, bone marrow and thymus, were removed. The EGFP protein was detected by immunohistochemistry in all of the above organs, with positive expression localized in the cytoplasm (red signal in Fig. 9a). In these experiments, we transplanted male tree shrew UC-MSCs into female tree shrews. This allowed us to use fluorescence in situ hybridization (FISH) and karyotype analysis to determine whether cells belonged to the recipient or donor animal. The result of FISH showed that only one x chromosome can be colored by a red probe, indicating male tree shrew's chromosome (Fig. 9b), and chromosome karyotype analysis of tree shrews showed that cells possessing XY chromosomes were present in the bone marrow of the recipient tree shrews (Fig. 9b, c), indicating that the transplanted cells survived in the recipient animals.
Fig. 9.
Distribution of UC-MSCs in major tree shrew organs. a Immunohistochemistry (IHC) with anti-eGFP antibodies (red). Images are shown at × 200 magnification, scale bars = 100um. Nuclei were stained blue with hematoxylin. Arrows indicate positively labeled cells. A total of 6 tree shrews were used for each control and EGFP group. Normal tree shrews were used as controls. b Fluorescence in situ hybridization (FISH) was used to detect tree shrew chromosomes, and red arrow indicates the X chromosome. c Chromosome karyotype analysis of tree shrews, with red arrow indicating the XY chromosomes. (Color figure online)
Discussion
In this study, we established a whole-body acute radiation injury model in tree shrews. We found that in tree shrews, the survival rate was significantly higher after UC-MSC transplantation (Fig. 3). Some studies (Sun et al. 2016) have suggested that ionizing radiation can inhibit division in various types of hematopoietic cells in the bone marrow, and this affects the release of mature cells from the peripheral blood. Therefore, blood counts can be used as an indicator of bone marrow suppression or injury. The precursor cells of WBCs, RBCs, and PLTs have the same sensitivity to radiation. However, when exposed to radiation, the blood counts of WBCs and PLTs decrease rapidly, resulting from their shorter life span, whereas the changes in RBC counts are delayed, resulting from its longer life span (Sun et al. 2016). Our results are similar to those presented in the literature (Sun et al. 2016). The recovery of hematopoiesis involves a variety of factors. Cell transplantation had little effect on early radioisotope blood levels but provided better protection to WBCs and PLTs than to RBCs and HGB. On the 28th day after irradiation, WBC and PLT counts were significantly higher in the treatment group than in the model group (P < 0.05). However, there was no significant difference in HGB and RBC counts between these groups (P > 0.05) (Fig. 4). In addition, ionizing radiation interferes with cell proliferation and differentiation by disrupting the hematopoietic microenvironment in bone marrow. The ability of UC-MSCs to home into the bone marrow may play a key role in the restoration of the bone marrow micro-environment (Fig. 9).
After cell transplantation, there were significantly fewer apoptotic cells in the peripheral blood and lung tissues. Complement proteins are key factors that regulate innate and adaptive immune responses by modulating enzymatic activity. They are also important participants in non-immune functions, such as subsidence in inflammation and apoptotic cell clearance, and they promote angiogenesis, accelerate wound healing, and activate and recruit stem cells. These conditions are necessary for the regulation of many human diseases, including infections and autoimmune diseases (Ricklin et al. 2016; Ho et al. 2016). Clq acts as a promoter of the classical complement activation pathway. It initiates and activates the complement cascade by recognizing the complement-binding site of the Fc region in IgG or IgM and then clearing antigen–antibody complexes (Cserhalmi et al. 2017). In addition, C1q recognizes apoptotic cells and rapidly initiates their phagocytosis via complement-dependent immunomodulatory pathways. This results in the clearance of apoptotic cells and avoids exposing intracellular autoantigens to the immune system (Kolev et al. 2014; Pulanco et al. 2016). C1q regulates phagocytes by regulating the expression of phagocytic genes (Spivia et al. 2014) and inhibits inflammatory responses by downregulating the expression of the inflammatory cytokines IL-1β, IL-6, TFN-α, and IFN-γ and upregulating the expression of IL-10 at both the mRNA and protein level (Kaur et al. 2016; Hulsebus et al. 2016). C1q is synthesized by monocytes and macrophages (Zhou et al. 1991), cultured PBMCs (Kaul and Loos 1995), spleen dendritic cells (Schwaeble et al. 1995), Kupffer cells (Armbrust et al. 1997), and microglial cells (Dietzschold et al. 1995; Singhrao et al. 1999). After exposure to ionizing radiation, C1q expression was decreased in the peripheral blood and lung tissues of the model group, potentially as a result of hypocomplementemia (over-activation of the classic complement pathway, leading to the overconsumption of C1q). In the absence of C1q, cells cannot activate the complement cascade to remove immune complexes, nor can macrophages promptly eliminate apoptotic cells. As a result, autoantigens are released by apoptotic bodies. This stimulates B cells to produce autoantibodies, which aggravates the inflammatory response. MSCs regulate the complement system (Stegert et al. 2015) to reduce the inflammatory response. Our results show that the level of C1q is closely related to the degree of disease development, but this correlation needs to be further studied.
The lungs contain tissues among the most sensitive to radiation. The most radiosensitive sub-unit of the lung is the alveolar-capillary complex. Type 1 endothelial cells cover 90% of the alveolar surface. Irradiation is thought to induce type 1 endothelial cells to shed (Ghafoori et al. 2008). Radiation also causes the loss of barrier functions by destroying epithelial and endothelial cells. These changes cause the accumulation of inflammatory cells and the release of a series of inflammatory factors, such as IL-6, IL-1ra, IFN-γ, and TGF-β. After MSC transplantation, MSCs secrete various cytokines into the blood circulation that then reach the injured tissue expressing inflammatory signals. Once there, they promote the proliferation of host cells at the damaged tissues. Following exposure to radiation, this initial inflammatory response can induce a vicious cycle of further inflammation and fibrosis (Giridhar et al. 2015).
After cell transplantation, inflammatory reactions and fibrosis were significantly improved in lung tissues. One study (Ao et al. 2008) has suggested that the mechanism underlying radiation-induced lung injury involves a variety of cell types, cytokines, proteases and signaling pathways. In terms of cytokines, MMP-2, MMP-9, TIMP, TGF-β1, IL-1, IL-2, ICAM-1, IL-6 and IL-8 levels are associated with radiation-induced lung injury (Chambers 2015), and some of these cytokines, including TGF-β1, IL-6, and TNF-α, have been clearly linked to the incidence of radiation-induced lung injury (Li et al. 2017). The mRNA levels of IL-6, TGF-β1, and TNF-α were significantly lower in the lungs after cell transplantation, similar to previous reports. UC-MSCs may interfere with fibrotic processes by regulating the expression of fibrotic and anti-fibrotic cytokines. PDGF promotes inflammatory infiltration by inducing chemotaxis in neutrophils and macrophages, thereby increasing the synthesis of type I and type III collagens and contributing to the proliferation of pulmonary interstitial cells and collagen deposition (Li et al. 2007). TGF-β1 inhibits proliferation in parenchymal cells, promotes proliferation in fibroblasts and osteoblasts, and increases matrix synthesis. BMP-7 reduces epithelial apoptosis, promotes cell proliferation, inhibits the release of proinflammatory cytokines, reduces inflammatory cell infiltration, reduces interstitial fibrosis and antagonizes TGF-β (Flechsig et al. 2012). HGF exerts a strong anti-fibrotic effect and plays an important role in repairing tissue damage and promoting the formation of normal structures (Willems et al. 2013). The results of our protein chip analysis verified that the levels of BMP-7 and HGF were increased, while the levels of PDGF-BB, PDGF-AA, and TGF-β1 were lower in peripheral blood following transplantation.
Irradiation upregulates IFN-γ, and this can induce a substantial increase in the expression of HLA-II (such as HLA-DP). HLA-DP molecules are mainly expressed on APC cells, such as B cells, monocytes, macrophages, dendritic cells, activated T cells, sperm and vascular endothelial cells. T lymphocytes are activated by changes in the relationships among antigen-presenting cells (APCs), lymphocytes and inflammatory cytokines, resulting in further increases in the expression of HLA-DP (Pasi et al. 2014). UC-MSCs downregulate the expression of HLA-DP (Fig. 8) while simultaneously inhibiting the expression of co-stimulatory molecules on the surfaces of APC cells to balance immune responses (Johnson and June 2017). One important anti-inflammatory role of UC-MSCs is to effectively remove bacteria (Giridhar et al. 2015). Our results show that infection with Staphylococcus aureus (map05150) (Fig. 7c) was the most highly represented pathway in a KEGG analysis. This may be a consequence of the regulation of C1q and HLA-DP genes by UC-MSCs.
There are three important theories concerning the therapeutic mechanisms by which UC-MSCs exert their effects (Li et al. 2015). First, UC-MSCs can replace damaged cells by relying on their potential for self-renewal and multi-lineage differentiation. Here, we transplanted eGFP-labeled UC-MSCs into tree shrews that were exposed to acute radiation injury and found that while the cells were distributed in the lungs and bone marrow in addition to other off-target tissues (such as heart, liver, spleen, kidneys, small intestine, and thymus), the number of cells that entered the damaged site was far lower than would be sufficient for tissue reengineering (Phinney and Prockop 2007; Zhang et al. 2011). Therefore, it is unlikely that replacement is the main mechanism by which UC-MSCs exert their protective role. Second, the paracrine activity of UC-MSCs may promote the proliferation and differentiation of endogenous stem cells, inhibit programmed cell death in host cells and regulate inflammation and immunity (Li et al. 2015). This theory is recognized as the most likely mechanism and is supported by a large number of scientists. Here, we found that UC-MSCs secrete an extensive array of cytokines, including SCF, HGF, insulin, and VEGFR3 (Table 4), that help to create a micro-environment that promotes intrinsic restorative processes (Wei et al. 2014; Arno et al. 2014). Third, UC-MSCs can interact with host cells through gap junctions, tight junctions, or desmosomes to affect host cell proliferation, migration and differentiation. The secretion of cytokines is probably the result of cell-to-cell contacts (Li et al. 2015). UC-MSCs may contact pulmonary endothelial cells to downregulate their expression of TGF-β1, IFN-γ, and IL-6 during injury repairs (Han et al. 2014).
In conclusion, the results of this study demonstrate that it is feasible to use UC-MSCs to treat acute radiation injury in tree shrews. UC-MSC transplantation improved the survival rate in tree shrews with acute radiation injury, reduced lung inflammation, prevented pulmonary fibrotic processes, recovered hematopoiesis and decreased cellular apoptosis. These changes may be associated with the modulation of immune cells (the downregulation of the expression of HLA-DP) and the levels of inflammatory cytokines (IL-10, IL-13, IL-2, TNF-α, IFN-γ, IL-1ra, and IL-8) and cytokines related to fibrosis (BMP-5, BMP-7, HGF, PDGF-BB, PDGF-AA, TGF-β1, IGFBP-2, and IGFBP-4). C1q may play an important role in the development and progression of acute radiation injury. In summary, UC-MSC transplantation may represent a novel and feasible therapeutic method for treating acute radiation injury.
Acknowledgements
This work was supported by funding from the Key Program of Kunming (No. 2015-1-S-00973), the National Natural Science Foundation of China (No. 31660655), and Applied Basic Research of Yunnan Province (Nos. 2016FB146, 2017FB042 and 2015FA039).
Author contributions
DBG, XQZ and XHP designed the study and wrote the paper; QQL and GMYL participated in the RT-PCR and flow cytometric analyses; GPR carried out the statistical analysis of the data; RQP, YHC and QW established the acute radiation injury model in the tree shrews and completed the UC-MSC transplantations; and JXW, JFL and QC cultured and labeled UC-MSCs.
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval and consent to participate
All experimental protocols were approved by the Experimental Animal Ethics Committee of Kunming General Hospital [Approval number (fast): 2,014,013].
References
- Ao X, Lubman DM, Davis MA, Xing X, Kong FM, Lawrence TS, Zhang M. Comparative proteomic analysis of radiation-induced changes in mouse lung: fibrosis-sensitive and -resistant strains. Radiat Res. 2008;169:417–425. doi: 10.1667/RR1173.1. [DOI] [PubMed] [Google Scholar]
- Armbrust T, Nordmann B, Kreissig M, Ramadori G. C1Q synthesis by tissue mononuclear phagocytes from normal and from damaged rat liver: up-regulation by dexamethasone, down-regulation by interferon gamma, and lipopolysaccharide. Hepatology. 1997;26:98–106. doi: 10.1053/jhep.1997.v26.pm0009214457. [DOI] [PubMed] [Google Scholar]
- Arno AI, Amini-Nik S, Blit PH, Al-Shehab M, Belo C, Herer E, Tien CH, Jeschke MG. Human Wharton’s jelly mesenchymal stem cells promote skin wound healing through paracrine signaling. Stem Cell Res Ther. 2014;5:28. doi: 10.1186/scrt417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrieta O, Gallardorincón D, Villarrealgarza C, Michel RM, Astorga-Ramos AM, Martínez-Barrera L, de la Garza J. High frequency of radiation pneumonitis in patients with locally advanced non-small cell lung cancer treated with concurrent radiotherapy and gemcitabine after induction with gemcitabine and carboplatin. J Thorac Oncol. 2009;4:845–852. doi: 10.1097/JTO.0b013e3181a97e17. [DOI] [PubMed] [Google Scholar]
- Chambers D (2015) Mesenchymal stromal cell-based therapies for lung disease. In: Bertoncello I, (ed) Stem cells in the lung. Development, repair and regeneration, Springer International Publishing, pp. 225–242
- Cserhalmi M, Csincsi ÁI, Mezei Z, Kopp A, Hebecker M, Uzonyi B, Józsi M. The murine factor H-related protein FHR-B promotes complement activation. Front Immunol. 2017;8:1145. doi: 10.3389/fimmu.2017.01145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzschold B, Schwaeble W, Schäfer MK, Hooper DC, Zehng YM, Petry F, Sheng H, Fink T, Loos M, Koprowski H, Weihe E. Expression of C1q, a subcomponent of the rat complement system, is dramatically enhanced in brains of rats with either Borna disease or experimental allergic encephalomyelitis. J Neurol Sci. 1995;130:11–16. doi: 10.1016/0022-510X(94)00269-T. [DOI] [PubMed] [Google Scholar]
- Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A, Prockop DJ, Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- Fausto AKS, Silva TDF, Romanel E, Vaslin MFS. microRNAs as reference genes for quantitative PCR in cotton. PLoS ONE. 2017;12:e0174722. doi: 10.1371/journal.pone.0174722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flechsig P, Dadrich M, Bickelhaupt S, Jenne J, Hauser K, Timke C, Peschke P, Hahn EW, Gröne HJ, Yingling J, Lahn M, Wirkner U, Huber PE. LY2109761 attenuates radiation-induced pulmonary murine fibrosis via reversal of TGF-beta and BMP-associated proinflammatory and proangiogenic signals. Clin Cancer Res. 2012;18:3616–3627. doi: 10.1158/1078-0432.CCR-11-2855. [DOI] [PubMed] [Google Scholar]
- Ghafoori P, Marks LB, Vujaskovic Z, Kelsey CR. Radiation-induced lung injury. Assessment, management, and prevention. Assessment Manage Prev Oncol. 2008;22:37–47. [PubMed] [Google Scholar]
- Giridhar P, Mallick S, Rath GK, Julka PK. Radiation induced lung injury: prediction, assessment and management. Asian Pac J Cancer Prev. 2015;16:2613–2617. doi: 10.7314/APJCP.2015.16.7.2613. [DOI] [PubMed] [Google Scholar]
- Gorbunov NV, Kiang JG. Ghrelin therapy decreases incidents of intracranial hemorrhage in mice after whole-body ionizing irradiation combined with burn trauma. Int J Mol Sci. 2017;18(pii):E1693. doi: 10.3390/ijms18081693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han DM, Wang ZD, Ding L, Zheng XL, Yan HM, Xue M, Zhu L, Liu J, Wang HX. Effect of umbilical cord MSC infusion on the pulmonary infection in haploidentical hematopoietic stem cell transplantation. J Exp Hematol. 2014;22:1084–1088. doi: 10.7534/j.issn.1009-2137.2014.04.037. [DOI] [PubMed] [Google Scholar]
- Ho MM, Manughian-Peter A, Spivia WR, Taylor A, Fraser DA. Macrophage molecular signaling and inflammatory responses during ingestion of atherogenic lipoproteins are modulated by complement protein C1q. Atherosclerosis. 2016;253:38–46. doi: 10.1016/j.atherosclerosis.2016.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsebus HJ, O’Conner SD, Smith EM, Jie C, Bohlson SS. Complement component C1q programs a pro-efferocytic phenotype while limiting TNFα production in primary mouse and human macrophages. Front Immunol. 2016;7:230. doi: 10.3389/fimmu.2016.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inga Jácome MC, Morales Chacòn LM, Vera Cuesta H, Maragoto Rizo C, Whilby Santiesteban M, Ramos Hernandez L, Noris García E, González Fraguela ME, Fernandez Verdecia CI, Vegas Hurtado Y, Siniscalco D, Gonçalves CA, Robinson-Agramonte ML. Peripheral inflammatory markers contributing to comorbidities in autism. Behav Sci. 2016;6(pii):E29. doi: 10.3390/bs6040029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LA, June CH. Driving gene-engineered T cell immunotherapy of cancer. Cell Res. 2017;27:38–58. doi: 10.1038/cr.2016.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul M, Loos M. Collagen-like complement component C1q is a membrane protein of human monocyte-derived macrophages that mediates endocytosis. J Immunol. 1995;155:5795–5802. [PubMed] [Google Scholar]
- Kaur A, Sultan SH, Murugaiah V, Pathan AA, Alhamlan FS, Karteris E, Kishore U. Human c1q induces apoptosis in an ovarian cancer cell lineviatumor necrosis factor pathway. Front Immunol. 2016;7:599. doi: 10.3389/fimmu.2016.00599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev M, Le FG, Kemper C. Complement–tapping into new sites and effector systems. Nat Rev Immunol. 2014;14:811–820. doi: 10.1038/nri3761. [DOI] [PubMed] [Google Scholar]
- Li M, Verena J, Claus B. The role of PDGF in radiation oncology. Radiat Onco. 2007;12:1–9. doi: 10.1186/1748-717X-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Xia M, Gao Y, Chen Y, Xu Y. Human umbilical cord mesenchymal stem cells: an overview of their potential in cell-based therapy. Expert Opin Biol Ther. 2015;15:1–14. doi: 10.1517/14712598.2015.1077303. [DOI] [PubMed] [Google Scholar]
- Li C, Xu D, Ye Q, Hong S, Jiang Y, Liu X, Zhang N, Shi L, Qin CF, Xu Z. Zika virus disrupts neural progenitor development and leads to microcephaly in mice. Cell Stem Cell. 2016;19:120–126. doi: 10.1016/j.stem.2016.04.017. [DOI] [PubMed] [Google Scholar]
- Li X, Ma D, Zha X, Quan D, Pan D, Sun M, Hu B, Zhao B. Ilomastat, a synthetic inhibitor of MMPs, prevents lung injury induced by γ-ray irradiation in mice. Oncotarget. 2017;8:60789. doi: 10.18632/oncotarget.18487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeed T. Mitigation of whole-body gamma radiation-induced damages by Clerodendron infortunatum in mammalian organisms. J Radiat Res. 2017;58:281–291. doi: 10.1093/jrr/rrw093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marupanthorn K, Tantrawatpan C, Kheolamai P, Tantikanlayaporn D, Manochantr S. Bone morphogenetic protein-2 enhances the osteogenic differentiation capacity of mesenchymal stromal cells derived from human bone marrow and umbilical cord. Int J Mol Med. 2017;39:654–662. doi: 10.3892/ijmm.2017.2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan XH, Huang X, Ruan GP, Pang RQ, Chen Q, Wang JX, He J, Zhao J, Cai XM, Zhao N, Chen Y, Zhu XQ. Umbilical cord mesenchymal stem cells are able to undergo differentiation into functional islet-like cells in type 2 diabetic tree shrews. Mol Cell Probes. 2017;34:1–12. doi: 10.1016/j.mcp.2017.04.002. [DOI] [PubMed] [Google Scholar]
- Pasi F, Paolini A, Nano R, Di Liberto R, Capelli E. Effects of single or combined treatments with radiation and chemotherapy on survival and danger signals expression in glioblastoma cell lines. Biomed Res Int. 2014;2014:453–497. doi: 10.1155/2014/453497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phinney DG, Prockop DJ. Concise review: mesenchymal stem/multipotent stromal cells: the state of transdifferentiation and modes of tissue repair-current views. Stem Cells. 2007;25:2896–2902. doi: 10.1634/stemcells.2007-0637. [DOI] [PubMed] [Google Scholar]
- Pulanco MC, Cosman J, Ho MM, Huynh J, Fing K, Turcu J, Fraser DA. Complement protein C1q enhances macrophage foam cell survival and efferocytosis. J Immunol. 2016;198:472–480. doi: 10.4049/jimmunol.1601445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklin D, Reis ES, Lambris JD. Complement in disease: a defence system turning offensive. Nat Rev Nephrol. 2016;12:383–401. doi: 10.1038/nrneph.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaeble W, Schäfer MK, Petry F, Fink T, Knebel D, Weihe E, Loos M. Follicular dendritic cells, interdigitating cells, and cells of the monocyte-macrophage lineage are the C1q-producing sources in the spleen. Identification of specific cell types by in situ hybridization and immunohistochemical analysis. J Immunol. 1995;155:4971–4978. [PubMed] [Google Scholar]
- Singhrao SK, Neal JW, Morgan BP, Gasque P. Increased complement biosynthesis by microglia and complement activation on neurons in Huntington’s disease. Exp Neurol. 1999;159:362–378. doi: 10.1006/exnr.1999.7170. [DOI] [PubMed] [Google Scholar]
- Spivia W, Magno PS, Le P, Fraser DA. Complement protein C1q promotes macrophage anti-inflammatory M2-like polarization during the clearance of atherogenic lipoproteins. Inflamm Res. 2014;63:885–893. doi: 10.1007/s00011-014-0762-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegert M, Bock M, Trendelenburg M. Clinical presentation of human C1q deficiency: How much of a lupus? Mol Immunol. 2015;67:3–11. doi: 10.1016/j.molimm.2015.03.007. [DOI] [PubMed] [Google Scholar]
- Sun L, Zhang H, Bi L, Shi YF, Xing C, Tang L, Jiang S, Yu K. Angiopoietin-1 facilitates recovery of hematopoiesis in radiated mice. Am J Transl Res. 2016;8:2011–2021. [PMC free article] [PubMed] [Google Scholar]
- Wei L, Zhang J, Xiao XB, Mai HX, Zheng K, Sun WL, Wang L, Liang F, Yang ZL, Liu Y, Wang YQ, Li ZF, Wang JN, Zhang WJ, You H. Multiple injections of human umbilical cord-derived mesenchymal stromal cells through the tail vein improve microcirculation and the microenvironment in a rat model of radiation myelopathy. J Transl Med. 2014;12:246. doi: 10.1186/s12967-014-0246-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems S, Verleden SE, Vanaudenaerde BM, Wynants M, Dooms C, Yserbyt J, Somers J, Verbeken EK, Verleden GM, Wuyts WA. Multiplex protein profiling of bronchoalveolar lavage in idiopathic pulmonary fibrosis and hypersensitivity pneumonitis. Ann Thorac Med. 2013;8:38–45. doi: 10.4103/1817-1737.105718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MJ, Sun JJ, Qian L, Liu Z, Zhang Z, Cao W, Li W, Xu Y. Human umbilical mesenchymal stem cells enhance the expression of neurotrophic factors and protect ataxic mice. Brain Research. 2011;1402:122–131. doi: 10.1016/j.brainres.2011.05.055. [DOI] [PubMed] [Google Scholar]
- Zhou AQ, Herriott MJ, Leu RW. Kinetics of the biosynthesis of complement subcomponent C1q by murine macrophages: LPS, immune complexes, and zymosan alone and in combination with interferon-gamma. J Leukoc Biol. 1991;50:453–463. doi: 10.1002/jlb.50.5.453. [DOI] [PubMed] [Google Scholar]