Abstract
Incomplete epigenetic reprogramming is one of the major factors affecting the development of embryos cloned by somatic cell nuclear transfer (SCNT). Histone 3 lysine 9 (H3K9) trimethylation has been identified as a key barrier to efficient reprogramming by SCNT. The aim of this study was to explore a method of downregulating H3K9me3 levels in donor cells by using histone lysine demethylase (KDM) protein. When sheep fetal fibroblast cells were treated with recombinant human KDM4D protein (rhKDM4D), the levels of H3K9 trimethylation and dimethylation were both significantly decreased. After SCNT, rhKDM4D-treated donor cells supported significantly higher percentage of cloned embryos developing into blastocysts as compared to non-treated control cells. Moreover, the blastocyst quality was also improved by rhKDM4D treatment of donor cells, as assessed by the total cell number in blastocysts and the expression of developmental genes including SOX2, NANOG and CDX2. These results indicate that treatment of donor cells with recombinant KDM4D protein can downregulate the levels of H3K9 trimethylation and dimethylation and improve the developmental competence of SCNT embryos. This strategy may be convenient to be used in KDM4-assisted SCNT procedure for improving the efficiency of cloning.
Electronic supplementary material
The online version of this article (10.1007/s10616-018-0224-6) contains supplementary material, which is available to authorized users.
Keywords: H3K9 methylation, KDM4D, Sheep, Somatic cell nuclear transfer
Introduction
The technology of somatic cell nuclear transfer (SCNT) in mammals has potential utility in agricultural and biomedical applications, but its efficiency is quite low due to poor embryo development. The developmental defects of SCNT embryos have been largely attributed to incomplete epigenetic reprogramming of donor cell genome. In particular, histone 3 lysine 9 (H3K9) trimethylation (me3) was recently identified as a key barrier to SCNT-mediated reprogramming (Matoba et al. 2014). In support of this notion, removal of the donor cell-characteristic H3K9me3 mark by overexpression of lysine-specific demethylase (KDM) has beneficial roles in the development of cloned mouse embryos from somatic cells (Matoba et al. 2014) as well as embryonic stem (ES) cells (Antony et al. 2013). These pioneer studies provide a promising approach for improving the SCNT efficiency.
The effectiveness of KDM-assisted SCNT has been confirmed recently in several species including mouse (Antony et al. 2013; Liu et al. 2016; Matoba et al. 2014; Wei et al. 2017), human (Chung et al. 2015), monkey (Liu et al. 2018) and porcine (Ruan et al. 2018). In most of these reports, the authors adopted a method of directly injecting KDM mRNAs into SCNT zygotes to downregulate the H3K9me3 levels (Chung et al. 2015; Liu et al. 2016, 2018; Matoba et al. 2014; Ruan et al. 2018). However, a major limitation of this approach is that the zygote injection is laborious and relatively difficult in technical manipulation. An alternative strategy is to reduce the H3K9me3 levels via overexpression of KDM4 or knockdown of H3K9 methyltransferases, Suv 39h1/2, in donor cells prior to nuclear transfer (Antony et al. 2013; Matoba et al. 2014; Wei et al. 2017). Treatment of donor cells would be simpler than zygote injection, however this procedure requires a DNA or siRNA transfection step and this step may be time-consuming and is usually inefficient for some cell types.
To overcome these limitations, this study explored the possibility to downregulate the H3K9me3 levels by treating the cultured donor cells with recombinant KDM4 protein. We showed that a simple incubation of sheep fibroblast cells with recombinant human KDM4D protein could induce a remarkable reduction of H3K9 methylation and improve the cloned embryo development after SCNT. Our study suggests that KDM treatment of donor cells would be an alternative method to KDM mRNA injection of zygotes for improving the SCNT procedure.
Materials and methods
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA) unless specifically stated.
Plasmid construction
Human cDNA encoding the catalytic domain of KDM4D (residues 12–342) was amplified by reverse-transcription (RT)-PCR from Hela cells using the following primers: 5′-GGATCCCAGAATCCAAATTGTAACATA-3′, and 5′-GTCGACCCCGGTCTTGCCCACGTTTC-3′. The cDNA fragment was cloned into a eukaryotic expression vector, pEASY-M2 (Transgen biotech, Beijing, China), which contains a sequence for Myc-tag. For manufacturing recombinant protein, the KDM4D cDNA fragment was constructed into a prokaryotic expression vector, pqe-80L (Qiagen, Dusseldorf, Germany), and a short sequence encoding a cell-penetrating peptide (CPP, containing 9 basic amino acid residues: RKKRRQRRR) was introduced to the 5′ upstream of KDM4D cDNA by PCR using the primer: 5′-GGATCCTACGGTCGTAAAAAACGTCGTCAGCGTCGTCGTCAGAATCCAAATTGTA-3′.
Protein expression and purification
KDM4D-pqe-80L vector was transformed into BL21 (DE3) E. coli and the cells were inoculated into 1 L LB medium. When cells reached an OD600 = 0.4–0.6, 0.5 mM isopropyl-β-d-thiogalactoside (IPTG) was added into the medium for inducing the KDM4D expression and cells were cultured overnight at room temperature. Cells were harvested by centrifugation at 17,000×g for 5 min at 4 °C, and the cell pellets were resuspended in native lysis buffer (50 mM NaH2PO4, 300 mM NaCl and 10 mM imidazol, pH 8.0) and disrupted by lysozyme on ice. After centrifugation at 17,000×g for 30 min at 4 °C, the supernatants were collected and protein was Ni2+ affinity purified by Ni-NTA agarose (Qiagen). The column was washed with washing solution and eluted with 250 mM imidazole. A centrifugal filter (Millipore, Bedford, MA, USA) was used for protein concentration and purification according to the standard protocol. Protein was dissolved in phosphate-buffered saline (PBS) and was filtered with a 0.22 μm-pore size millex. The protein concentration was determined by Bradford assay using an Eppendorf Biophotometer (Hamburg, Germany). Purified recombinant human KDM4D protein (rhKDM4D) was flash frozen in liquid nitrogen, and stored at − 80 °C.
Cell culture, transfection and treatment with KDM4D protein
A primary sheep fetal fibroblast (SFF) cell line was derived from a 70-day-old sheep fetus. The procedures of animal experiments were in accordance with the animal care policies of China Agricultural University and were approved by the Animal Ethics Committee at the university. Hela cells (ATCC, Manassas, VA, USA) or SFF cells were cultured in D-MEM/F-12 medium (Gibco, Invitrogen Corporation, Grand Island, NY, USA) supplemented with 10% (v/v) fetal bovine serum (FBS; HyClone, Logan, UT, USA) and 1% (v/v) penicillin/streptomycin. Cells were transfected with KDM4D-pEASY-M2 plasmid using Lipofectamine 3000 (Gibco, Invitrogen Corporation) according to the manufacturer’s protocol. For treatment with KDM4D protein, cells were cultured in medium containing rhKDM4D for 24 h and then used for nuclear transfer or detection.
Oocyte maturation and fertilization in vitro
The procedures of in vitro maturation (IVM) and fertilization (IVF) of ovine oocytes were described previously (Hou et al. 2008). Briefly, oocytes were aspirated from abattoir ovaries and cultured in TCM 199 supplemented with 20% (v/v) estrus sheep serum (eSS), at 38.5 °C, in 5% CO2 in saturated humide air for about 22 h. Frozen-thawed sperm were incubated in synthetic oviduct fluid (SOF) medium supplemented with 2% eSS for 30 min for swimming up. Highly motile spermatozoa were collected and co-incubated with matured oocytes in fertilization medium for 20–22 h. Fertilized ova were transferred into SOF supplemented with 8 mg/ml bovine serum albumin (BSA), 2% essential amino acids and 1% non-essential amino acids (SOFaa), and then cultured in this medium at 38.5 °C in 5% CO2, 7% O2 and 88% N2.
SCNT
SCNT was carried out as described previously (Fu et al. 2012). Briefly, matured oocytes were enucleated in manipulation medium, Hepes-buffered TCM199 supplemented with 10% (v/v) FBS and 5 μg/ml cytochalasin B. A donor cell was injected into the perivitelline space of the enucleated oocyte and the couplets were induced to fuse on a cell fusion apparatus (BLS CF-150/B, Budapest, Hungary). Fused embryos were activated by 5 μM ionomycin for 5 min, followed by incubation in 2 mM 6-dimethylaminopurine for 4 h. Finally, the cloned embryos were cultured in SOFaa, at 38.5 °C in 5% CO2, 7% O2 and 88% N2.
Immunofluorescence staining
Cells or embryos were fixed in 4% (v/v) paraformaldehyde at 4 °C overnight, permeated in 0.5% (v/v) Triton X-100 in PBS for 30 min at room temperature and blocked in 1% (m/v) BSA and 0.2% (v/v) Triton X-100 in PBS for 2 h at 37 °C. The samples were incubated with primary antibodies against Myc (Easybio, Beijing, China), dimethylated H3K9 (Cat. ab1220, Lot GR41249-1, Abcam, Cambridge, MA, USA) or trimethylated H3K9 (Cat. 07-442, Lot 33453, Upstate/Millipore, Lake Placid, NY, USA) at 37 °C for 1 h, followed by incubation with appropriate secondary antibodies for 1 h at 37 °C. The secondary antibodies included Delight 549 goat anti-mouse or anti-rabbit and Delight 488 goat anti-mouse IgG (Abbkine, Wuhan, Hubei, China). Cells were counterstained with 4,6-diamidino-2-phenylindole dihydrochloride (DAPI) and then mounted onto glass slides. Images were captured using Qlympus BX51 epifluorescence microscope (Olympus Corporation, Tokyo, Japan). The background of images was subtracted and fluorescence intensities were measured by manually outlining individual nuclei with ImageJ 1.37v software (National Institutes of Health, Bethesda, MD, USA).
Western blotting
Confluent cells cultured in 35-mm dishes were lysed with lysis buffer (Beyotime, Beijing, China). Cell lysates were separated by 12% SDS-PAGE gel electrophoresis and transferred onto nitrocellulose membranes. The membranes were blocked with 5% skim milk and incubated with anti-dimethylated or trimethylated H3K9 antibodies for 2 h at room temperature, followed by incubation with horseradish peroxidase (HRP)-conjugated secondary antibodies for another 2 h. House-keeping gene β-actin (Abcam) was used as an internal reference. The membranes were treated with super ECL Plus and the western blotting signals were detected by exposure on a film. The intensities of blotting signals were quantified by outlining the relevant bands on the film with Image J software.
Quantitative real-time PCR (qPCR)
Total RNA from 2 to 3 of pooled blastocysts for each sample was extracted using the Dynabeads mRNA DIRECT Micro Kit (Ambion, Austin, TX, USA) according to the standard protocol. RNA was reverse-transcribed to synthesize cDNA using FastQuant RT Kit (Tiangen Biotech, Beijing, China) according to the manufacturer’s protocol. The cDNA was used as template for qPCR. qPCR was carried out using SuperReal PreMix (Tiangen Biotech) on an ABI 7500 system (Applied Biosystems, Foster City, CA, USA). The primers used for analysis are listed in Supplementary Table 1.The thermal cycling conditions were 95 °C for 15 min, followed by 40 cycles at 95 °C for 10 s, 61.5 °C for 20 s and 72 °C for 32 s, and a melting cycle was 95 °C for 15 s, 60 °C for 1 min, 95 °C for 15 s and 60 °C for 15 s. Real-time fluorescence signals were collected during the conditions: 72 °C for 32 s. House-keeping gene (GAPDH) was used as an internal control. Relative expression levels of each gene analyzed were calculated relative to GAPDH and the control sample (IVF) by 2−ΔΔCT method. All samples were analyzed in triplicates and all experiments were repeated for three times.
Table 1.
Preimplantation development of cloned sheep embryos derived from KDM4D protein-treated cells
| Groupsa | No. of reconstructed embryosb | No. of cleaved (%, mean ± SEM) | No. of blastocysts (%, mean ± SEM) | Total cell number in blastocyst (mean ± SEM) |
|---|---|---|---|---|
| C-NT | 169 | 157 (92.6 ± 3.7) | 17 (11.3 ± 3.0)c | 83.0 ± 8.1c |
| T-NT | 170 | 162 (95.3 ± 2.0) | 31 (19.2 ± 1.2)d | 124.3 ± 17.0d |
Cleavage percentage: no. of cleaved/no. of reconstructed embryos. Blastocyst percentage: no. of blastocysts/no. of cleaved
aC-NT, embryos cloned from cells without KDM4D treatment; T-NT, embryos cloned from KDM4D-treated cells
bTotal numbers of embryos from 4 replicates
c,dWithin a row represent significant difference between groups (P < 0.05). The data were statistically analyzed by Student’s two-tailed t test
Statistical analysis
Data were statistically analyzed by Student’s two-tailed t test or One-way ANOVA using Graphpad Prime 5 Software. Results are presented as mean ± SEM. Differences were considered significant at three levels (*P < 0.05; **P < 0.01; ***P < 0.001).
Results and discussion
Human KDM4D has a H3K9 demethylase activity in sheep somatic cells
Human KDM4 (also known as JMJD2) family consists of four members (KDM4A–4D) that have an H3K9me3 demethylase activity. KDM4A, KDM4B and KDM4C also possess a function in demethylating trimethylated H3K36, while KDM4D is only specific for H3K9 demethylation (Krishnan and Trievel 2013; Whetstine et al. 2006). Moreover, KDM4D encodes a shorter protein than other KDM4 members (Whetstine et al. 2006), which would be more appropriate for production of recombinant protein. So far, three KDM4 members, including KDM4A, KDM4B and KDM4D, have been used to downregulate the H3K9me3 levels in SCNT embryos. However, the sequence for ovine KDM4 is unclear. Thus, we cloned the cDNA fragment encoding the catalytic domain of human KDM4D (residues 12–342).
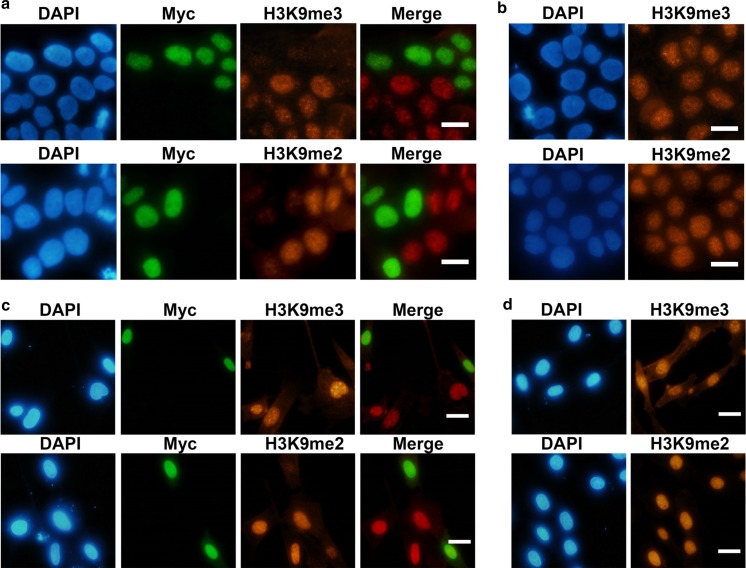
To determine the demethylase activity of the cloned KDM4D gene fragment, the plasmid vector for expressing the Myc-tagged KDM4D was transiently transfected into HeLa cells and SFF cells. Immunostaining showed that in both types of cells, the levels of H3K9me3 were remarkably reduced in Myc-positive cells, where KDM4D was overexpressed (Fig. 1a, c). In contrast, no demethylation was observed in non-transfected control cells (Fig. 1b, d). In addition, KDM4D was also able to demethylate dimethylated (me2) H3K9, consistent with a previous report (Whetstine et al. 2006). Thus, human KDM4D has a function of demethylating H3K9me3 as well as H3K9me2 in sheep cells, confirming that this enzyme functions across mammalian species.
Fig. 1.
Effects of human KDM4D on H3K9 methylation in cultured cells. Hela cells (a) and sheep fetal fibroblast (SFF) cells (c) were transfected with plasmid vector expressing Myc-tagged catalytic domain of human KDM4D. Forty-eight hours after transfection, the cells were co-immunostained for Myc and trimethylated (me3) or dimethylated (me2) H3K9. Non-transfected HeLa cells (b) and SFF cells (d) were immunostained as control. Nuclei were labeled with 4,6-diamidino-2-phenylindole dihydrochloride (DAPI). Scale bar = 20 μm
Treatment of cells with recombinant KDM4D protein reduces H3K9 methylation levels
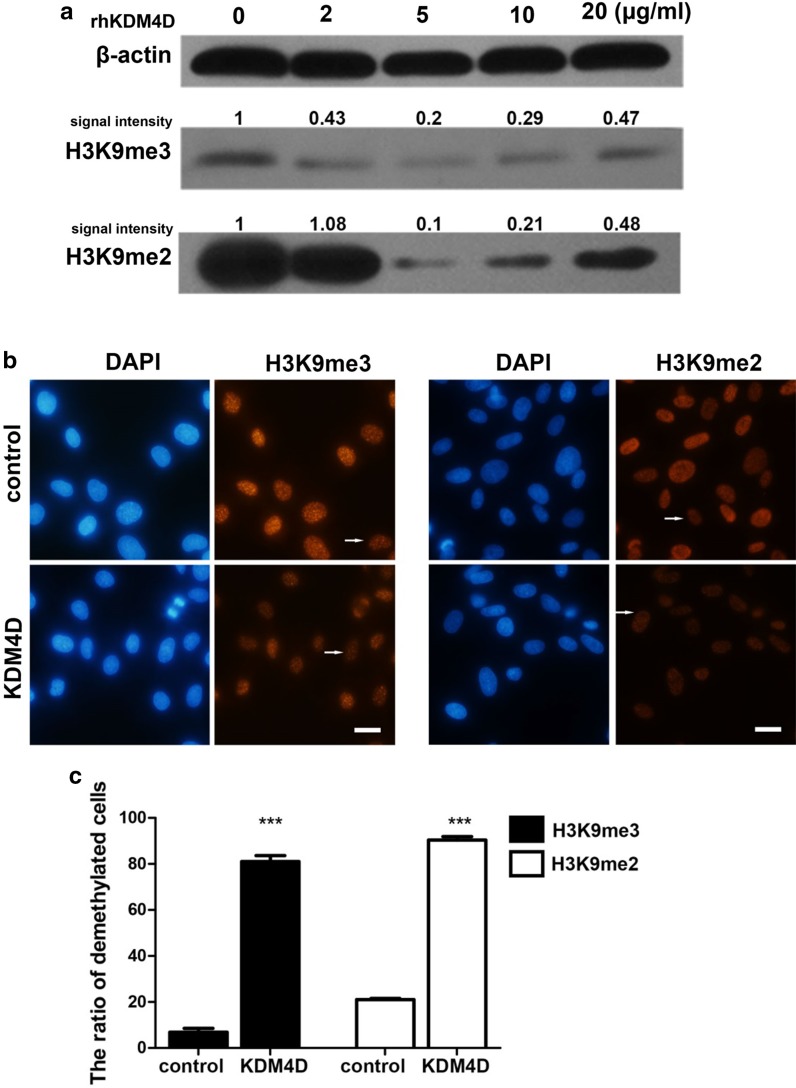
The human KDM4D gene was constructed into a prokaryotic expression vector for manufacturing rhKDM4D protein. To enhance the entry of protein into cells, a sequence encoding a cell-penetrating peptide was linked to the KDM4D gene. The recombinant protein was produced in E. coli and purified by affinity chromatography (Supplementary Figure 1). SFF cells were treated with different concentrations of purified rhKDM4D protein for 24 h and then sampled for detection. Western blotting analysis indicated that the concentration at 5 μg/ml resulted in a clear decline of H3K9me3 and H3K9me2 levels, while higher concentrations had less demethylating effects for unknown reasons (Fig. 2a). We therefore chose the concentration of 5 μg/ml for subsequent studies. Immunofluorescence staining confirmed that this concentration was able to induce a significant decrease of global H3K9me3 and H3K9me2 levels (Fig. 2b). More than 80% of rhKDM4D-treated cells showed remarkable H3K9me3/me2 demethylation, compared to 7% for H3K9me3 and 21% for H3K9me2, respectively, in control cells (Fig. 2c). These results suggest that rhKDM4D protein can efficiently enter the cells, probably with the aid of a cell-penetrating peptide attached to the protein, and thus exerting an enzymatic function in nucleus.
Fig. 2.
Effects of recombinant human KDM4D (rhKDM4D) protein on H3K9 methylation in SFF cells. a Western blotting analysis of H3K9me3 and H3K9me2 in cells treated with different concentrations of rhKDM4D protein for 24 h. β-actin served as loading controls. Signal intensity of bands was quantified using Image J and normalized to β-actin in control (0 μg/ml). The resulting values are shown. b Immunofluorescent detection of H3K9me3 and H3K9me2 levels in cells treated with rhKDM4D protein (5 μg/ml). Cells without KDM4D treatment were used as control. Nuclei were labeled with DAPI. Representative demethylated cells that were weakly immunostained for H3K9me3 or H3K9me2 are pointed by arrows. Scale bar = 20 μm. c Ratio of cells with demethylated H3K9 to total cell number. Numbers of analyzed nuclei from 4 replicated observations; H3K9me3: control (n = 114), KDM4D (n = 84); H3K9me2: control (n = 106), KDM4D (n = 123). Data are presented as mean ± SEM and statistically analyzed by Student’s two-tailed t test. ***P < 0.001
Downregulation of H3K9 methylation may also be achieved by using chemical inhibitors. In this regard, two small molecules, BIX-01294 and UNC0638, which specifically inhibit the activity of H3K9 dimethyltransferase G9A, have been used to reduce H3K9me2 levels in SCNT embryos (Fu et al. 2012, 2014; Huang et al. 2016, 2017; Wang et al. 2017). However, these inhibitors may have some non-specific cellular toxicity and their effectiveness in improving the cloning efficiency has not been fully demonstrated. Recent study on bovine suggested that serum starvation of donor cells could induce several epigenetic changes including reduction of H3K9me3, and these changes may improve the reprogramming of somatic cell into totipotency (Kallingappa et al. 2016). In contrast to the above mentioned approaches, KDM4D treatment is expected to be more specific to the H3K9 demethylation and has less non-specific adverse effects. Moreover, as KDM4D functions through an “active” demethylation pathway, it would induce a more rapid reduction of H3K9me3/me2 than methyltransferase inhibitors-mediated “passive” demethylation. More importantly, KDM4D can also demethylate H3K9me2 other than H3K9me3. Induction of H3K9me2 demethylation would also be beneficial to the development of SCNT embryos, as aberrant hypermethylation of H3K9me2 has been observed in cloned embryos in several species (Cao et al. 2015; Fu et al. 2012; Santos et al. 2003). Therefore, direct treatment of cultured cells with recombinant KDM4D protein could be an ideal approach of downregulating H3K9 methylation in cells for SCNT.
Treatment of donor cells with rhKDM4D protein improves the development of SCNT embryos
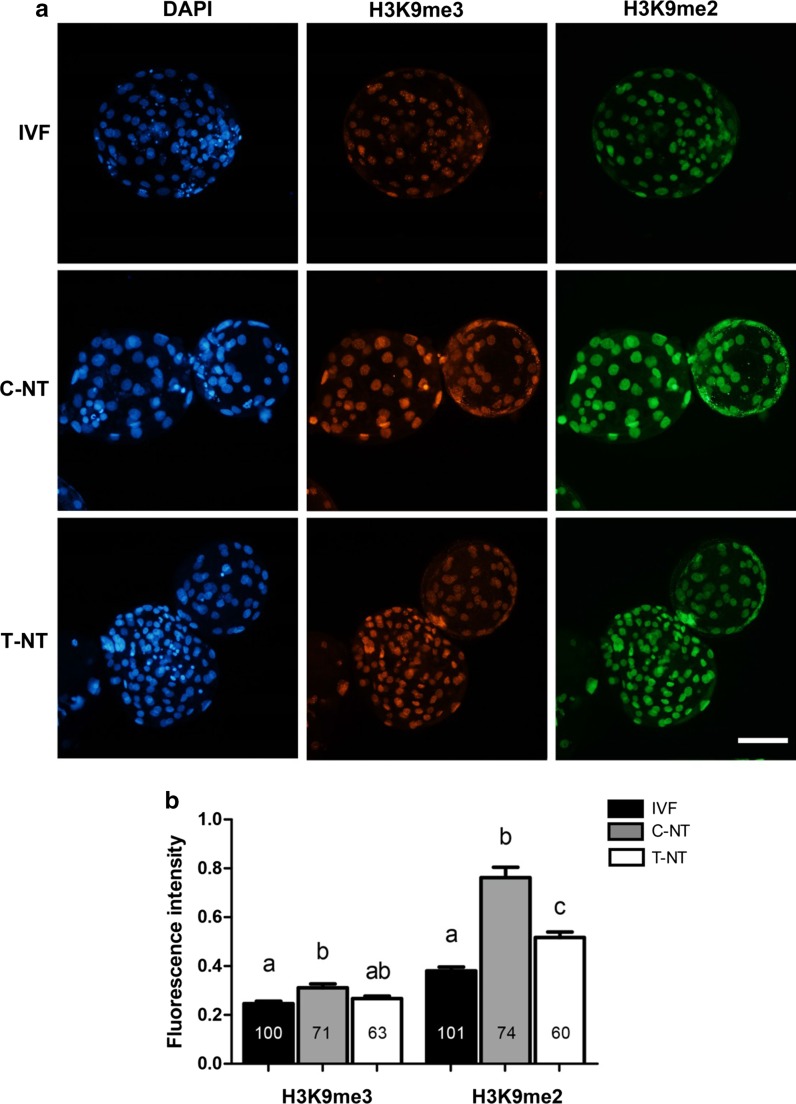
Having confirmed the H3K9 demethylating effects of rhKDM4D protein in cultured cells, we next evaluated the development of SCNT embryos produced from rhKDM4D-treated cells. As shown in Table 1, the rate of blastocyst development was significantly increased in rhKDM4D treatment group (T-NT) when compared to the non-treated control (C-NT) (19.2 vs 11.3%, P < 0.05). Moreover, the total cell number in blastocysts from T-NT group was significantly higher than that from C-NT group (124 vs 83, P < 0.05). Immunofluorescence staining showed that H3K9me3 and H3K9me2 were both hypermethylated in C-NT blastocysts as compared to IVF blastocysts, consistent with our previous observations of aberrant H3K9 hypermethylation in ovine SCNT embryos (Fu et al. 2012). While, in T-NT blastocysts, the H3K9me3 signals were decreased to a comparable level to that in IVF embryos (Fig. 3a, b). H3K9me2 levels also declined significantly in T-NT blastocyst compared with C-NT embryos, albeit still higher than that in the IVF counterparts (Fig. 3a, b). These results suggested that the defects of H3K9 methylation reprogramming in SCNT embryos were largely corrected by pretreatment of donor cells with rhKDM4D.
Fig. 3.
Levels of H3K9me3 and H3K9me2 in blastocysts. a Co-immunostaining of blastocysts with anti-H3K9me3 and anti-H3K9me2 antibodies. Nuclei were labeled with DAPI. Scale bar = 100 μm. b Quantification analysis of immunofluorescence intensities. Data are presented as mean ± SEM. A one-way ANOVA analysis was used to determine the differences between groups. Different superscripts above columns represent significant differences between groups (P < 0.05). Numbers within columns indicate the numbers of nuclei analyzed. IVF, in vitro fertilization embryos, n = 12; C-NT, cloned embryos from cells without KDM4D treatment, n = 12; T-NT, cloned embryos from KDM4D-treated cells, n = 9. The detection was replicated 3 times
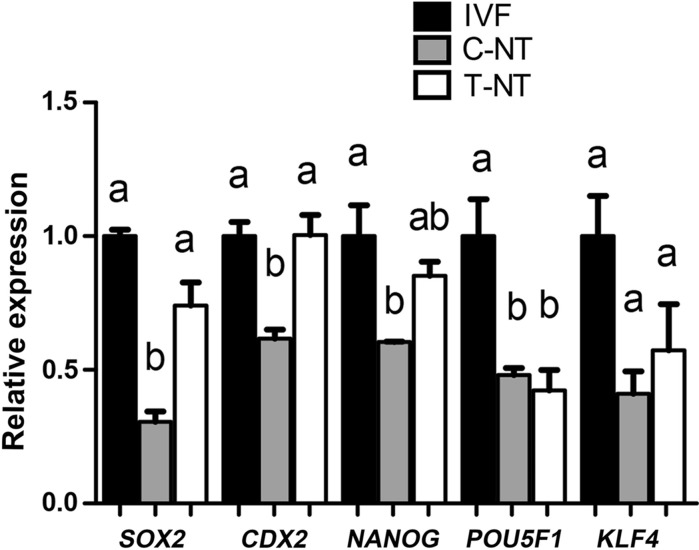
qPCR analysis indicated that the expression levels of several embryonic development-related genes, including POU5F1, NANOG, SOX2, and CDX2 were significantly lower in C-NT blastocysts than in their IVF counterparts, while in T-NT blastocysts, the expression of these genes except POU5F1, were upregulated and reached comparable levels to that of IVF blastocysts (Fig. 4). The regulating roles of KDM4 proteins on the expression of embryonic genes are not fully understood. In reprogramming of induced pluripotent stem cells, high levels of H3K9me3 are barrier to the activation of core pluripotent genes like Nanog and Sox2 (Chen et al. 2013; Sridharan et al. 2013). Kdm4c has been associated with the expression of Oct-4, Sox2 and Nanog in mouse ES cells (Loh et al. 2007). In addition to these pluripotent genes, Kdm4-induced H3K9me3 demethylation is involved in activation of complete gene networks including both protein-coding genes and nongenic repetitive elements, which are important for embryo development (Matoba et al. 2014; Wei et al. 2017). In our study, the expression of genes in both inner cell mass cells (NANOG and SOX2) and trophoblast cells (CDX2) of SCNT blastocysts was significantly improved. Collectively, these results demonstrated that rhKDM4D treated donor cells not only enhanced the development of SCNT embryos into blastocysts but also improved the quality of blastocysts. Future work is required to determine whether in vivo development ability of SCNT embryos is improved by this strategy.
Fig. 4.
Quantitative RT-PCR (qPCR) analysis for the expression of developmental genes in blastocysts. Data are presented as mean ± SEM. A one-way ANOVA analysis was used to determine the differences between groups. Different superscripts above columns represent significant differences between groups (P < 0.05). IVF, in vitro fertilization embryos; C-NT, cloned embryos from cells without KDM4D treatment; T-NT, cloned embryos from KDM4D treated cells
Conclusion
Our study has proposed a modified KDM4-assisted SCNT procedure and primarily demonstrated its feasibility in ovine SCNT. Treatment of donor cells with recombinant KDM4D protein can downregulate levels of H3K9me3 and H3K9me2 in cells and improve the developmental competence of SCNT embryos. This method may be convenient to be used for improving the cloning efficiency.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 31172208) and China Agriculture Research System (Grant No. CARS-39-04).
Compliance with ethical standards
Conflict of interest
The authors declare no financial or commercial conflict of interest.
References
- Antony J, Oback F, Chamley LW, Oback B, Laible G. Transient JMJD2B-mediated reduction of H3K9me3 levels improves reprogramming of embryonic stem cells into cloned embryos. Mol Cell Biol. 2013;33:974–983. doi: 10.1128/mcb.01014-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Li Y, Chen Z, Wang H, Zhang M, Zhou N, Wu R, Ling Y, Fang F, Li N, Zhang Y. Genome-wide dynamic profiling of histone methylation during nuclear transfer-mediated porcine somatic cell reprogramming. PLoS ONE. 2015;10:e0144897. doi: 10.1371/journal.pone.0144897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Liu H, Liu J, Qi J, Wei B, Yang J, Liang H, Chen Y, Chen J, Wu Y, Guo L, Zhu J, Zhao X, Peng T, Zhang Y, Chen S, Li X, Li D, Wang T, Pei D. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat Genet. 2013;45:34–42. doi: 10.1038/ng.2491. [DOI] [PubMed] [Google Scholar]
- Chung YG, Matoba S, Liu Y, Eum JH, Lu F, Jiang W, Lee JE, Sepilian V, Cha KY, Lee DR, Zhang Y. Histone demethylase expression enhances human somatic cell nuclear transfer efficiency and promotes derivation of pluripotent stem cells. Cell Stem Cell. 2015;17:758–766. doi: 10.1016/j.stem.2015.10.001. [DOI] [PubMed] [Google Scholar]
- Fu L, Zhang J, Yan FX, Guan H, An XR, Hou J. Abnormal histone H3K9 dimethylation but normal dimethyltransferase EHMT2 expression in cloned sheep embryos. Theriogenology. 2012;78:1929–1938. doi: 10.1016/j.theriogenology.2012.07.017. [DOI] [PubMed] [Google Scholar]
- Fu L, Yan FX, An XR, Hou J. Effects of the histone methyltransferase inhibitor UNC0638 on histone H3K9 dimethylation of cultured ovine somatic cells and development of resulting early cloned embryos. Reprod Domest Anim. 2014;49:e21–e25. doi: 10.1111/rda.12277. [DOI] [PubMed] [Google Scholar]
- Hou J, Liu L, Zhang J, Cui XH, Yan FX, Guan H, Chen YF, An XR. Epigenetic modification of histone 3 at lysine 9 in sheep zygotes and its relationship with DNA methylation. BMC Dev Biol. 2008;8:60. doi: 10.1186/1471-213x-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Zhang H, Yao J, Qin G, Wang F, Wang X, Luo A, Zheng Q, Cao C, Zhao J. BIX-01294 increases pig cloning efficiency by improving epigenetic reprogramming of somatic cell nuclei. Reproduction. 2016;151:39–49. doi: 10.1530/rep-15-0460. [DOI] [PubMed] [Google Scholar]
- Huang Y, Jiang X, Yu M, Huang R, Yao J, Li M, Zheng F, Yang X. Beneficial effects of diazepin–quinazolin–amine derivative (BIX-01294) on preimplantation development and molecular characteristics of cloned mouse embryos. Reprod Fertil Dev. 2017;29:1260–1269. doi: 10.1071/rd15463. [DOI] [PubMed] [Google Scholar]
- Kallingappa PK, Turner PM, Eichenlaub MP, Green AL, Oback FC, Chibnall AM, Wells DN, Oback B. Quiescence loosens epigenetic constraints in bovine somatic cells and improves their reprogramming into totipotency. Biol Reprod. 2016;95:16. doi: 10.1095/biolreprod.115.137109. [DOI] [PubMed] [Google Scholar]
- Krishnan S, Trievel RC. Structural and functional analysis of JMJD2D reveals molecular basis for site-specific demethylation among JMJD2 demethylases. Structure. 2013;21:98–108. doi: 10.1016/j.str.2012.10.018. [DOI] [PubMed] [Google Scholar]
- Liu W, Liu X, Wang C, Gao Y, Gao R, Kou X, Zhao Y, Li J, Wu Y, Xiu W, Wang S, Yin J, Liu W, Cai T, Wang H, Zhang Y, Gao S. Identification of key factors conquering developmental arrest of somatic cell cloned embryos by combining embryo biopsy and single-cell sequencing. Cell Discov. 2016;2:16010. doi: 10.1038/celldisc.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Cai Y, Wang Y, Nie Y, Zhang C, Xu Y, Zhang X, Lu Y, Wang Z, Poo M, Sun Q. Cloning of macaque monkeys by somatic cell nuclear transfer. Cell. 2018;172:1–7. doi: 10.1016/j.cell.2018.01.020. [DOI] [PubMed] [Google Scholar]
- Loh YH, Zhang W, Chen X, George J, Ng HH. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Gene Dev. 2007;21:2545–2557. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba S, Liu Y, Lu F, Iwabuchi KA, Shen L, Inoue A, Zhang Y. Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell. 2014;159:884–895. doi: 10.1016/j.cell.2014.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan D, Peng J, Wang X, Ouyang Z, Zou Q, Yang Y, Chen F, Ge W, Wu H, Liu Z, Zhao Y, Zhao B, Zhang Q, Lai C, Fan N, Zhou Z, Liu Q, Li N, Jin Q, Shi H, Xie J, Song H, Yang X, Chen J, Wang K, Li X, Lai L. XIST derepression in active X chromosome hinders pig somatic cell nuclear transfer. Stem Cell Rep. 2018;10:494–508. doi: 10.1016/j.stemcr.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F, Zakhartchenko V, Stojkovic M, Peters A, Jenuwein T, Wolf E, Reik W, Dean W. Epigenetic marking correlates with developmental potential in cloned bovine preimplantation embryos. Curr Biol. 2003;13:1116–1121. doi: 10.1016/S0960-9822(03)00419-6. [DOI] [PubMed] [Google Scholar]
- Sridharan R, Gonzales-Cope M, Chronis C, Bonora G, McKee R, Huang C, Patel S, Lopez D, Mishra N, Pellegrini M, Carey M, Garcia BA, Plath K. Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1gamma in reprogramming to pluripotency. Nat Cell Biol. 2013;15:872–882. doi: 10.1038/ncb2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang Y, Mao T, Yan B, Deng R, Wei B, Zhang Y, Liu J. Treatment donor cells with UNC0638 modify the abnormal histone H3K9 dimethylation and gene expression in cloned goat embryos. Small Rumin Res. 2017;156:27–32. doi: 10.1016/j.smallrumres.2017.08.011. [DOI] [Google Scholar]
- Wei J, Antony J, Meng F, MacLean P, Rhind R, Laible G, Oback B. KDM4B-mediated reduction of H3K9me3 and H3K36me3 levels improves somatic cell reprogramming into pluripotency. Sci Rep. 2017;7:7514. doi: 10.1038/s41598-017-06569-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, Shi Y. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.