Summary
The p53 tumor suppressor pathway is frequently inactivated in human cancers. However, there are some cancer types without commonly recognized alterations in p53 signaling. Here we report that histone demethylase KDM5A is involved in the regulation of p53 activity. KDM5A is significantly amplified in multiple types of cancers, an event that tends to be mutually exclusive to p53 mutation. We show that KDM5A acts as a negative regulator of p53 signaling through inhibition of p53 translation via suppression of a subgroup of eukaryotic translation initiation genes. Genetic deletion of KDM5A results in upregulation of p53 in multiple lineages of cancer cells and inhibits tumor growth in a p53-dependent manner. In addition, we have identified a regulatory loop between p53, miR-34, and KDM5A, whereby the induction of miR-34 leads to suppression of KDM5A. Thus, our findings reveal a mechanism by which KDM5A inhibits p53 translation to modulate cancer progression.
Subject Areas: Molecular Mechanism of Gene Regulation, Cancer
Graphical Abstract

Highlights
-
•
Genetic amplification of KDM5A tends to be negatively correlated with TP53 alterations
-
•
KDM5A inhibits p53 protein translation by suppressing a subset of translation genes
-
•
KDM5A regulates tumor growth in a p53-dependent manner
-
•
miR-34 targets KDM5A expression
Molecular Mechanism of Gene Regulation; Cancer
Introduction
The tumor suppressor p53 serves to maintain genomic integrity and regulates the expression of multiple genes that are engaged in cell cycle, apoptosis and metabolism (Lane and Levine, 2010, Vousden and Prives, 2009, Wang and Gu, 2014), as well as immunomodulation (Zitvogel and Kroemer, 2015). It is inactivated in over half of all human cancers via genetic mutations, or MDM2/MDMX- or human papilloma virus (HPV) E6-mediated degradation (Levine and Oren, 2009, Vogelstein and Kinzler, 2004, Vousden and Prives, 2009). Nevertheless, there are some tumor types where these mechanisms of p53 tumor suppressor pathway inactivation do not frequently occur at the time of diagnosis. One such tumor is neuroblastoma (Molenaar et al., 2012, Pugh et al., 2013), the most common pediatric extracranial solid malignancy, responsible for ∼15% of cancer deaths in children (Maris, 2010).
Accumulating evidence has implicated epigenetic changes as drivers of cancer (Chi et al., 2010, Dawson and Kouzarides, 2012, Greer and Shi, 2012, Kaelin and McKnight, 2013, Suva et al., 2013, Vogelstein et al., 2013), with many of these being due to alterations in the activity of histone lysine demethylases (KDM) (Dawson and Kouzarides, 2012, Greer and Shi, 2012, Kooistra and Helin, 2012). Several studies indicate that histone demethylases are involved in the regulation of p53 activity. KDM1A has been shown to remove the methyl groups from lysine 370 (K370me2) of the p53 protein directly, thereby suppressing its transcriptional activity (Huang et al., 2007). PHF2 facilitates p53-mediated cell death in response to chemotherapy (Lee et al., 2014), whereas KDM6B can be recruited to p53-bound promoters and enhancer elements with unknown function (Williams et al., 2014). Nevertheless, the roles of other histone demethylases in the regulation of p53 activity have not been well defined. Interestingly, a recent study has shown that methyltransferase SETD8 inhibits the p53 pathway by modulating p53K382me1 levels in high-risk neuroblastoma (Veschi et al., 2017), supporting the hypothesis that additional mechanisms may be involved in the regulation of p53 function in neuroblastoma.
KDM5A (also referred to as RBP2 or JARID1A), an H3K4me3/me2 histone demethylase (Christensen et al., 2007, Klose et al., 2007), is a potential oncogene that is highly expressed in many different cancers (Blair et al., 2011), supports a stem-like phenotype in cancer cells (Sharma et al., 2010), and promotes therapy resistance (Banelli et al., 2015, Hou et al., 2012, Sharma et al., 2010). Genetic ablation of KDM5A hinders tumorigenesis and metastasis (Cao et al., 2014, Lin et al., 2011, Teng et al., 2013). KDM5A is also involved in tumor angiogenesis (Li et al., 2014, Qi et al., 2014). Interestingly, KDM5A can be fused with NUP98 in over 10.5% of pediatric acute megakaryoblastic leukemia (de Rooij et al., 2013, van Zutven et al., 2006). A murine model demonstrated that NUP98/KDM5A is oncogenic in leukemogenesis (Wang et al., 2009). In this study, we examined the effect of all human histone demethylases on p53 activity and found that (1) several KDMs including KDM5A play a role in modulating p53 function; (2) KDM5A is amplified in many types of cancers, an event that tends to be mutually exclusive to p53 gene mutations; (3) KDM5A is overexpressed in high-stage and high-risk neuroblastomas, is associated with a poor outcome, and is negatively correlated with expression of p53 target genes; (4) loss of KDM5A significantly enhances p53 protein accumulation and inhibits tumor growth in wild-type p53, but not mutant p53-expressing colon cancer and neuroblastoma xenograft models; and (5) KDM5A inhibits p53 protein synthesis by suppressing the expression of a subgroup of protein translation genes. We also identified a signaling pathway between KDM5A and miR-34, a p53 target gene that is frequently deleted in high-risk neuroblastomas. Thus, our findings not only support a mechanism by which KDM5A regulates p53 function but also provide a rationale for targeting KDM5A in cancers with wild-type p53.
Results
Identification of Histone Demethylases that Modulate p53 Function
One of the most well-characterized p53 targets is CDKN1A (also known as p21), which bears two p53 binding sites (5′RE and 3′RE) in the promoter region (Zeng et al., 1997). We therefore used a luciferase reporter carrying the 5′RE and 3′RE of CDKN1A to monitor changes in p53 activity that might be affected by altering histone demethylase expression (Figure 1A). We used a focused pooled small interfering RNA (siRNA) library (Yang et al., 2017) (including 32 genes encoding JmjC domain proteins with 17 of them having histone lysine or arginine demethylase activity and 2 genes encoding FAD-dependent demethylases, KDM1A and KDM1B) to demonstrate histone demethylase activity in isogenic p53+/+ and p53−/− HCT116 colon cancer cell lines. These isogenic cell lines allowed us to assess the p53-dependent effects only since CDKN1A could be activated by other p53-independent epigenetic events such as histone deacetylase (HDAC) inhibition (Gui et al., 2004). We also transfected a Renilla luciferase reporter to serve as an internal control to normalize the CDKN1A firefly luciferase. We identified that depletion of KDM4D, KDM5A, KDM6B, PHF2, PHF8, C14orf169, and JMJD4 resulted in at least a 2-fold greater induction of CDKN1A luciferase activity in p53+/+ HCT116 cells compared with p53−/− HCT116 cells (Figure 1B). Here we focused on KDM5A for the reasons described below and therefore validated KDM5A-mediated p53 activity. We designed single siRNA oligos and performed RT-PCR. The results showed that depletion of KDM5A led to significant induction of CDKN1A in p53+/+ HCT116 cells (Figure 1C), consistent with the reporter assay screening.
Figure 1.
Identification of Histone Demethylases Engaged in Regulation of p53 Function
(A) Schematic showing the p21 luciferase reporter bearing two p53 binding sites that are subject to modulation by histone demethylases.
(B) Screening results showing the relative luciferase activity that is driven by p53 after siRNA knockdown of the indicated genes.
(C) RT-PCR for CDKN1A after 72-hr knockdown of KDM5A. Data are mean ± SEM.
(D) Genetic amplification and deletion of histone demethylases across 10,844 cancers. Circled red dots indicate that gene amplification is at its peak. Cutoff q value is 0.25.
(E) Contingency table for the analysis of the distribution frequency of genetic amplification/loss of KDM5A from TCGA PAN-CAN UCSC datasets.
(F) Amplification of KDM5A or MDM2 and TP53 mutations from TCGA PAN-CAN UCSC data. Chi-square test for statistical analyses, p < 0.001.
(G) The genetic alteration of TP53, MDM2, KDM5A, and CDKN2A from five cancer cohort data.
(H) The association of genetic alteration of TP53, MDM2, KDM5A, and CDKN2A from five cancer cohort datasets.
KDM5A Is Amplified in Several Different Cancers and Is Negatively Correlated with p53 Genetic Mutations
To assess whether any genomic alterations of the KDMs were associated with changes in p53 function, we first examined the genomic amplification or loss of KDMs, since these are important mechanisms by which cancer cells activate proto-oncogenes or inactivate tumor suppressors, using the Tumorscape program, which has high-resolution copy number data amassed from multiple cancer types (all generated through TCGA) (Beroukhim et al., 2010). We found that the KDM5A gene was significantly focally amplified across the entire dataset of 10,844 tumors and was located within a focal peak region of the amplicons (q value = 1.91 × 10−33) (Figure 1D and Table S1). KDM5A is significantly focally amplified in 12 of 33 independent cancer types (Table S2). Among these, it is located within a focal peak region of amplification in 11 cancer types (Table S2). Interestingly, after analysis of the tumors from TCGA-PAN cancer data that were well characterized for genetic alterations of KDM5A, MDM2, and TP53, we found that KDM5A is significantly associated with genetic mutations of TP53 (Figure 1E). Interestingly, tumors with KDM5A amplification tended to be enriched with wild-type p53 when compared with those with KDM5A loss (Figures 1E and 1F and Table S3), similar to MDM2 amplification/loss (Figure 1F). Furthermore, by using the cBioPortal program (Cerami et al., 2012, Gao et al., 2013) we combined five different types of cancer cohorts that had higher incidence of KDM5A amplification (>5%, Figure S1A) for analyses of genetic alteration data of TP53, KDM5A, MDM2, and CDKN2A (P14ARF) (Figure 1G). p14Arf is a classical tumor suppressor that antagonizes MDM2, thereby playing an important role in regulating p53 protein expression (Kamijo et al., 1998, Stott et al., 1998, Zhang et al., 1998). Among these genetic alterations, despite a tendency of co-occurrence genetic events between KDM5A and MDM2 (Figure 1H), KDM5A and MDM2 amplification tended to be mutually exclusive to TP53 mutations (Figure 1H). These genetic data suggest that, given the prevalence of its amplification in a variety of human cancers but the negative enrichment with p53 mutations, KDM5A might be a potential proto-oncogene in some types of cancers and may act on the p53 signaling pathway.
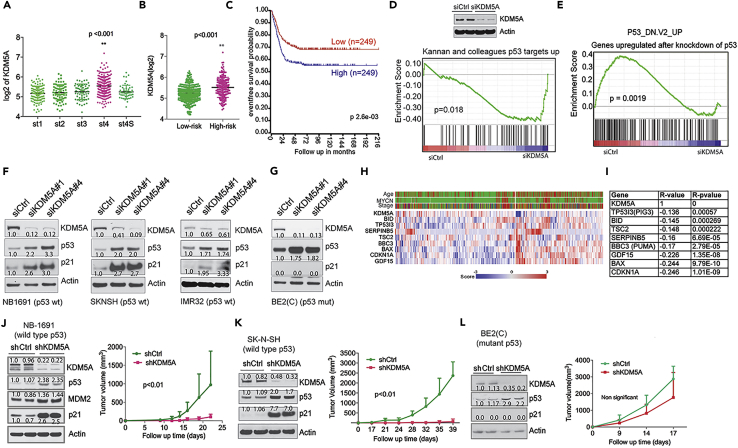
KDM5A Overexpression Occurs in High-Stage and High-Risk Neuroblastoma and Is Associated with a Poor Prognosis
The genetic amplification of oncogenes often leads to mRNA overexpression. We assumed that aberrant overexpression of KDM5A through mechanisms other than amplification may also be negatively correlated with p53 function. Neuroblastoma, the most common extracranial solid cancer in childhood, rarely bears genetic alterations of p53 (as well as p14Arf or MDM2) at diagnosis (Molenaar et al., 2012, Pugh et al., 2013). We, therefore, speculated that KDM5A might be important in neuroblastoma pathogenesis by suppressing p53 activity. Although amplification of KDM5A does not appear to be a common event in neuroblastoma, overexpression of the KDM5A gene was significantly higher in patients with stage 4 disease than in those with other stages, according to the RNA sequencing (RNA-seq) data from a cohort of 498 patients (Su et al., 2014) (Figure 2A). KDM5A was also significantly overexpressed in high-risk disease (Figure 2B). In addition, Kaplan-Meier analysis showed that high KDM5A expression (median value as a cutoff for KDM5A expression levels) was associated with poor outcome in children with neuroblastoma (Figure 2C).
Figure 2.
KDM5A Inhibits p53 Function in Neuroblastoma
(A) KDM5A expression in different International Neuroblastoma Staging System (INSS) stages of neuroblastoma (Cohort SEQC, GSE62564).
(B) KDM5A expression in low- and high-risk neuroblastomas (Cohort SEQC, GSE62564).
(C) Kaplan-Meier analysis of the association of event-free survival with KDM5A expression (Cohort SEQC, GSE62564).
(D) GSEA shows that genes upregulated by KDM5A knockdown in NB-1691 cells is associated with a gene set of p53 targets. Western blot analysis of indicated proteins after 72-hr knockdown of KDM5A.
(E) GSEA shows that genes downregulated by KDM5A knockdown in NB-1691 cells is associated with a gene set unregulated by p53 knockdown.
(F and G) Western blot analysis of indicated proteins after 72-hr knockdown of KDM5A in (F) NB1-691, SK-N-SH, and IMR-32 and (G) BE2(C) cells. Band intensity was quantified by ImageJ and normalized to loading control.
(H) Heatmap for gene expression of KDM5A and p53 targets from cohort GSE45547.
(I) Spearman correlation of KDM5A and p53 target genes from cohort GSE45547.
(J–L) Tumor volume curves of (J) NB-1691, (K) SK-N-SH, and (L) BE2(C) neuroblastoma xenografts with lentiviral-mediated shRNA knockdown of KDM5A or control (n = 5 per group). Data are mean ± SEM. Left panel shows western blot analysis of KDM5A knockdown by shRNA before tumor cell implantation. Band intensity was quantified by ImageJ and normalized to loading control. p value was calculated by ANOVA test.
KDM5A Inhibits p53 Signaling
Since KDM5A was highly expressed in high-risk neuroblastomas, we first characterized the function of KDM5A in neuroblastoma, by performing a global gene expression profile analysis after depletion of KDM5A in human neuroblastoma cells. Gene set enrichment analysis (GSEA) showed that depletion of KDM5A in NB-1691 cells (wild-type p53) resulted in induction of the p53 targets (Figure 2D) and negatively correlated with the gene signature upregulated after p53 knockdown (Figure 2E). However, there was no change in the p53 signaling pathways after KDM5A depletion in BE2(C) cells that have mutant p53 (Tweddle et al., 2001). Western blot analysis revealed that p53 and its target p21 were both induced by depletion of KDM5A in NB-1691, SK-N-SH, and IMR32 cells (Figure 2F), three cell lines with wild-type p53. Interestingly, we also observed induction of mutant p53, but not p21, in BE2(C) cells (Figure 2G), suggesting that KDM5A overexpression correlates with reduced p53 expression regardless of its mutation status. We further assessed the correlation of KDM5A and p53 targets. We chose several well-studied p53 target genes including CDKN1A, BBC3 (also named PUMA), BAX, and GDF15, which play important roles in cell-cycle arrest and apoptosis. The results showed that these genes were significantly negatively correlated with KDM5A overexpression (Figures 2H and 2I), supporting the hypothesis that KDM5A suppresses p53 function in neuroblastoma. To further assess the role of KDM5A in p53-mediated tumor suppression, we established stable KDM5A depletion using lentiviral small hairpin RNA (shRNA) in NB-1691, SK-N-SH, and BE2(C) cells for xenograft studies. Transduced cells containing the shRNAs were selected for in the presence of 1 μg/mL of puromycin to eliminate the non-transduced cells. KDM5A depletion led to significant tumor growth delay in p53 wild-type cancer cell lines NB-1691 and SK-N-SH (Figures 2J and 2K) but not the p53 mutant BE2(C) cell line (Figure 2L), in accordance with induction of p53 targets before the establishment of xenografts (Figures 2J–2L).
To address whether the above findings represented a general mechanism across different cancer lineages, we included several different types of cancer cell lines bearing wild-type p53 or no p53. First, we performed GSEA after depletion of KDM5A in p53+/+ and isogenic p53−/− HCT116 colon cancer cells. Again, this revealed that KDM5A loss resulted in high activity of p53 pathways in p53+/+ HCT116 cells (Figures 3A–3C) but not in p53−/− HCT116 cells (Figure S1B). Molecular concept analysis further demonstrated that loss of KDM5A function in p53+/+ HCT116 cells led to the expression of genes associated with various p53 signatures, as well as chemotherapy- and ionizing-radiation-induced signatures (Figure 3D). We then assessed protein expression of p53 targets and found that p21, PUMA, TIGAR, and MDM2 were upregulated after KDM5A depletion across several types of cancer cell lines including colon cancer, osteosarcoma, and lung cancer (Figures 3E–3G), in accordance with the induced p53 protein, demonstrating the ability of KDM5A to suppress p53 signaling in a wide variety of human cancer cell lines. The induction of p53 by loss of KDM5A was further validated by knocking out KDM5A using CRISPR/Cas9 at two different genomic loci of the KDM5A gene from five individual clones that were derived from single cells (Figure S1C). Conversely, overexpression of KDM5A reduced p53 expression, and its target p21 expression (Figure 3H). We further assessed the expression of KDM5A and p53 target genes in a variety of different types of cancer cohorts. The results showed that KDM5A overexpression was negatively correlated with the expression of p53 targets (Figures 3I–3L, and data not shown), suggesting that repression of p53 activity by KDM5A is common across many cancers. Consistent with the tumor suppressive functions of p53, depletion of KDM5A significantly reduced cell colony formation in p53+/+ but not p53−/− cancer cells (Figures S1D and S1E). We also generated lentiviral-mediated shRNA knockdown of KDM5A in isogenic p53+/+ and p53−/− HCT116 cells (Figure 3M), which allowed us to evaluate the p53 effect more specifically. Again, depletion of KDM5A resulted in significant reduction of tumor growth in the p53+/+ but not in the p53−/− xenografts (Figure 3M). Interestingly, after analysis of the expression of KDM5A and p53 in three HCT116 xenografts per group that were randomly chosen, we found that KDM5A re-expressed in all tumor samples of p53+/+ HCT116 but not in p53−/− HCT116 and p53 expression was restored back to the same levels as the control (Figure 3N), suggesting that shRNA knockdown of KDM5A confers a selective pressure on p53 wild-type cells, or that it was due to selection of rare clones that were not transduced by KDM5A shRNA. We therefore hypothesize that KDM5A may reflect the response of cancer cells to therapies that activate the p53 pathway. Indeed, the data from the Cancer Therapeutics Response Portal (Rees et al., 2016), which links genetic, lineage, and other cellular features of cancer cell lines to small-molecule sensitivity, showed that cancer cells expressing higher KDM5A were more sensitive to chemotherapeutic drugs such as etoposide, doxorubicin, and cytarabine (Figure S1F). However, KDM5B, a paralog of KDM5A, was correlated with drug sensitivity to tyrosine kinase inhibitors and mitogen-activated protein kinase inhibitors (Figure S1G). Taken together, these results demonstrate that KDM5A plays an important role in suppressing p53 function in neuroblastoma and other p53 wild-type-expressing tumors, further suggesting that KDM5A is a regulator of the p53 signaling pathway in most cancers if not all. Therefore, in the following studies, we mainly used the HCT116 colon cancer cell line as the model to dissect the molecular mechanism by which KDM5A regulates p53 expression because this cell line is more easily manipulated genetically and pharmacologically.
Figure 3.
KDM5A Broadly Suppresses p53 Signaling Pathway
(A) GSEA shows that genes upregulated after KDM5A depletion in p53+/+ HCT116 cells is associated with a gene set of Kannan TP53 targets up.
(B) GSEA shows that genes upregulated after KDM5A depletion in p53+/+ HCT116 cells is associated with a gene set of Inga p53 targets.
(C) GSEA shows that genes upregulated after KDM5A depletion in p53+/+ HCT116 cells is associated with a gene set of KEGG p53 signaling pathway.
(D) Molecular concept analysis of genes upregulated after KDM5A depletion in p53+/+ HCT116 cells.
(E–G) Western blot analysis with the indicated antibodies after 72-hr KDM5A depletion in (E) HCT116 (p53+/+ and p53−/−), (F) U2OS and SAOS2, and (G) A549 and H1299 cells. Band intensity was quantified by ImageJ and normalized to loading control.
(H) Effect of KDM5A overexpression in HCT116 cells. Cells transfected with either an empty vector or a plasmid overexpressing KDM5A were subject to western blot analysis. Band intensity was quantified by ImageJ and normalized to loading control.
(I–L) Spearman correlation of KDM5A and p53 target genes from cohort of renal cell carcinoma (I, TCGA ID: KIRC), medulloblastoma (J, GSE85217), uterine cancer (K, TCGA ID: UCEC), and prostate adenocarcinoma (L, TCGA ID: PRAD).
(M) Tumor volume curves of isogenic p53+/+ and p53−/− HCT116 xenografts with lentiviral-mediated shRNA knockdown of KDM5A or control (n = 5/group). Data are mean ± SEM. Left panel shows western blot analysis of KDM5A knockdown by shRNA before implantation. p value was calculated by ANOVA test.
(N) Western blot analysis of KDM5A and p53 expression in three terminal tumor samples/group in p53+/+ and p53−/− HCT116 xenografts. Band intensity was quantified by ImageJ and normalized to loading control.
KDM5A Inhibits p53 Protein Synthesis
We next sought to understand the mechanism by which KDM5A regulates the p53 signaling pathway in cancer cells. To define the mechanism by which KDM5A suppresses p53 expression, we first tested whether KDM5A directly regulates p53 transcription. Real-time PCR revealed that TP53 mRNA was modestly decreased, although CDKN1A was greatly upregulated by KDM5A depletion in p53 wild-type HCT116 cells (Figure 4A), suggesting that KDM5A regulates p53 expression at a post-transcriptional level. Normally, p53 is a very labile protein that is subject to proteasomal degradation (Kruse and Gu, 2009). In response to cellular stress, such as DNA damage, p53 undergoes post-translational modifications such as phosphorylation at serine 15 and serine 20, which result in the displacement of MDM2 from the N terminus of p53, with consequent stabilization of the p53 protein (Toledo and Wahl, 2006). We found that the DNA-damaging agent doxorubicin induced phosphorylation of p53 Serine 15 but KDM5A depletion did not (Figure 4B), nor did it reduce the protein-protein interaction between MDM2 and p53 (Figure 4C). KDM5A knockdown also did not significantly alter the half-life of p53 when protein synthesis was blocked by cycloheximide (Figure 4D, top panel), whereas treatment of cells with doxorubicin and Nutlin-3, a compound that disrupts the p53-MDM2 interaction, significantly stabilized p53 protein (Figure 4D, bottom panel). These data suggest that KDM5A does not regulate p53 expression through MDM2-mediated degradation. Conversely, when the proteasome inhibitor MG132 was used to block p53 degradation, the amount of newly synthesized p53 protein labeled with S35 was much greater after KDM5A depletion, although the total newly synthesized and total protein were not (Figures 4E and S2A), suggesting that KDM5A regulates p53 protein synthesis specifically.
Figure 4.
KDM5A Regulates p53 Translation
(A) TP53 and CDKN1A mRNA assessed by RT-PCR after 72-hr knockdown of KDM5A in p53+/+HCT116 cells. Data are mean ± SEM. Biological triplicates with technical triplicates for each were performed for Student's t test. **p < 0.01.
(B) Western blot after 48-hr knockdown of KDM5A in A549 cells, followed by 24-hr treatment with 1ug/mL of doxorubicin. Band intensity was quantified by ImageJ and normalized to loading control.
(C) Immunoprecipitation assay for assessment of p53-MDM2 interaction after 72-hr knockdown of KDM5A in p53+/+ HCT116 cells. Band intensity was quantified by ImageJ and normalized to loading control.
(D) Assessment of p53 half-life by western blot of protein from HCT116 cells treated with 50 μg/mL of cycloheximide after siRNA knockdown (48 hr) and 5 μM of Nutlin-3 (24 hr) or 0.5 μg/mL of doxorubicin (24hr). ImageJ was used to quantify the western blot bands for the graph on the right.
(E) Pulse-chase results with S35 labeling of HCT116 cells in the presence of 5 μM MG132 after 48-hr KDM5A knockdown. p53 band intensity was quantified by ImageJ and normalized to total proteins.
(F) A luciferase reporter assay demonstrating the effect of KDM5A on heterologous expression from a cassette bearing intact 5′UTR and -3′UTR of p53. After 24 hr siRNA knockdown of KDM5A in p53+/+ and p53−/− HCT116 cells by reverse transfection, 2.4 μg of p53-UTR firefly luciferase reporter plasmid and 0.1 μg pRL Renilla Luciferase Control Reporter Vector were transfected. After 48 hr, cells were harvested for the dual luciferase assay. Data are mean ± SEM. Biological triplicates with technical triplicates for each were performed for Student's t test. **p < 0.01.
(G) After 48-hr knockdown of KDM5A or Nutlin-3 treatment, HCT116 cells were treated with 100 nM of rocaglamide (Roc A) and 100 nM of cymarin for 24 hr. Western blot was used for assessment of p53 expression. Intensity of p53 bands with short exposure were quantified by ImageJ and normalized to loading control.
To further validate this, we used alternative approaches to assess the role of KDM5A in the regulation of p53 synthesis. First, we introduced into HCT116 cells a heterologous luciferase reporter in which the p53 coding sequence was replaced by the coding sequence for luciferase, whereas the 5′ and 3′ UTRs of p53 remained intact. The mRNA expression of this reporter was driven by a heterologous promoter, but its translation should be regulated by the same machinery as endogenous p53 because the 5′- and -3′-UTRs of p53 were intact. In these experiments, KDM5A knockdown significantly enhanced the luciferase activity in both p53+/+ and p53−/− HCT116 cells (Figure 4F), supporting the hypothesis that KDM5A regulates p53 translation. Next, we tested the effect of two compounds, rocaglamide (Roc A) and cymarin, which specifically inhibit protein translation (Didiot et al., 2013, Rodrigo et al., 2012). We found that when these two compounds were administered, the amount of p53 protein induced by KDM5A depletion was reduced to basal levels, without significantly downregulating p53 mRNA expression (Figures 4G and S2B); however, the p53 protein induced by Nutlin-3 compounds (Nutlin-3b is much less potent than Nutlin-3a), was only partially affected by protein translation inhibitors (Figure 4G). These data indicate that inhibition of protein translation reduced KDM5A-depletion- but not Nutlin-3-induced (via p53 stabilization) enhanced p53 expression. Finally, we found that KDM5A depletion resulted in increased p53 expression in RC10.1 cells (Figure S2C). Rapid degradation of p53 in this cell line normally occurs because these cells, derived from RKO cells, have been engineered to express the HPV E6 oncoprotein (Kessis et al., 1993). However, the increased p53 expression in these cells due to depletion of KDM5A partly overwhelmed the degradation by E6, allowing for the detection of p53. Taken together, these data further support our hypothesis that KDM5A inhibits p53 expression via a mechanism that is involved in p53 protein translation.
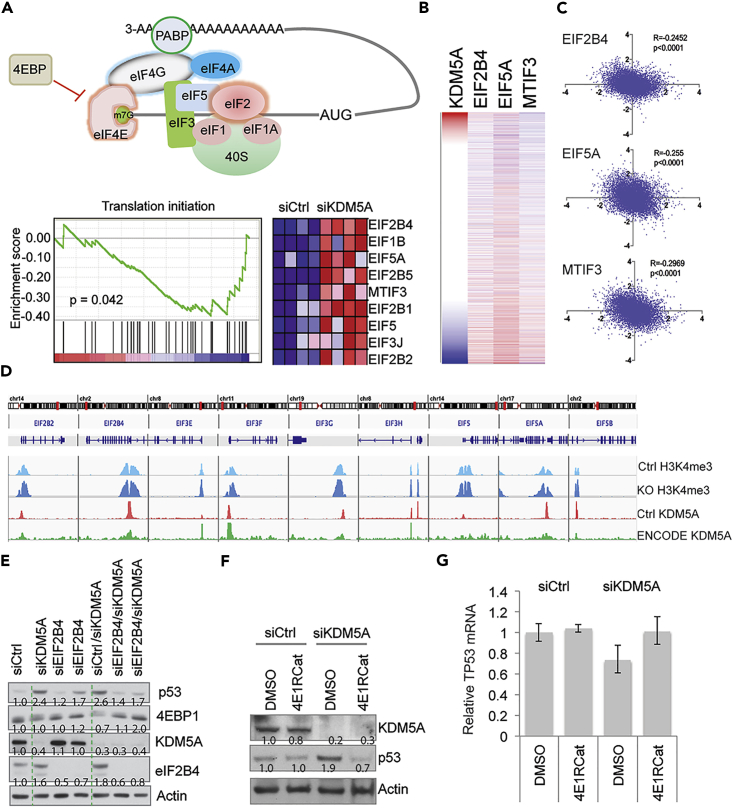
KDM5A Regulates the Expression of Genes Involved in Translation
Protein translation can be divided into four phases: initiation, elongation, termination, and ribosome recycling (Bhat et al., 2015). We sought to determine at what phase KDM5A was acting. Translation initiation is the most important and complicated step of the translation process and involves many factors and multiple complexes (Figure 5A). Our gene expression profiling analysis revealed that KDM5A depletion resulted in upregulation of a signature containing genes involved in translation initiation but not other steps of translation (Figure 5A), indicating that KDM5A inhibits a subgroup of translation initiation genes. We further analyzed the RNA-seq data from the PAN-CAN dataset and confirmed that expression of KDM5A and EIF2B4, EIF5A, and MTIF3 were inversely correlated (Figures 5B and 5C), supporting that KDM5A suppresses the expression of these genes. To explore whether KDM5A was directly involved in the expression of these translation initiation genes, we performed chromatin immunoprecipitation sequencing (ChIP-seq) analysis to determine whether KDM5A bound these genes. Our ChIP-seq data revealed that, among the approximately 5,000 genomic regions bound by KDM5A (Table S4), nearly 100 genes were involved in translation and many of these encoded ribosomal proteins, based on canonical signaling pathway analysis (Table S5). Many of these proteins engage in translation initiation (Figure 5D). We also analyzed the ENCODE ChIP-seq data in which data on KDM5A genomic binding in human embryonic stem cells are available. We found that the genomic loci of these target genes bound by KDM5A were significantly correlated (Figures 5D and S3A), which corroborated our ChIP-seq data.
Figure 5.
KDM5A Regulates p53 Expression by Modulating Translation Initiation
(A) Schematic showing the protein complexes involved in translation initiation. GSEA shows that the translation initiation pathway is upregulated by KDM5A depletion in isogenic p53+/+ and p53−/− HCT116 cells.
(B) Heatmap for gene expression of KDM5A, EIF2B4, EIF5, and MTIF3 from TCGA PAN-CAN data analyzed by UCSC Xena program.
(C) Spearman correlation of KDM5A and EIF2B4, EIF5A, and MTIF3 from PAN-CAN TCGA data.
(D) ChIP-seq data shows KDM5A binding at the genomic loci of translation initiation genes in HCT116 cells (pink) and human embryonic stem cells from ENCODE data (green), and H3K4me3 peaks at the genomic loci of translation initiation genes before (light blue) and after KDM5A disruption by CRISPR/Cas9 (dark blue). Peak scale of 100 for H3K4me3 and KDM5A in HCT116 cells, and 75 for KDM5A ENCODE data.
(E) Western blot assessment of p53 after 72-hr depletion of KDM5A and eIF2B4 in HCT116 cells.
(F) Western blot assessment of p53 expression in HCT116 cells treated with 4E1RCat for 24-hr after KDM5A depletion. Intensity of p53 bands were quantified by ImageJ and normalized to loading control.
(G) RT-PCR assessing the expression of TP53 mRNA from (E). Data are mean ± SEM. Technical triplicates for each were performed.
KDM5A is a histone demethylase that removes H3K4me3/me2. To explore the role of the histone demethylase activity of KDM5A in the regulation of translation initiation genes, we removed KDM5A expression in HCT116 cells using CRISPR/Cas9 and performed ChIP-seq to assess the H3K4me3 changes. The results showed that KDM5A genomic binding sites overlapped with H3K4me3 peaks (Figure 5D and Table S6), and deletion of KDM5A led to an increase in H3K4me3 levels at the promoter regions on these genes (Figure 5D), in line with its histone demethylase activity. Interestingly, we noticed that H3K4me3 was also upregulated at the locus of TP53 in KDM5A KO cells (Figure S3B), despite the mRNA levels of TP53 not being altered in these cells (Figure 4A).
To validate whether increased translation initiation was responsible for the upregulation of p53 protein, we studied eIF2B4, the component of the eIF2 complex that regulates the assembly of the 43S pre-initiation complex (PIC) during translation initiation (Bhat et al., 2015). Knockdown of eIF2B4 prevented the induction of p53 associated with depletion of KDM5A (Figure 5E), confirming that KDM5A regulates p53 expression by modulating translation initiation. Interestingly, we also found that downregulation of 4EBP1 due to KDM5A depletion was rescued by co-depletion of eIF2B4 (Figure 5E). 4EBP1 inhibits translation initiation by competing with eIF4E, which forms a complex with eIF4A and eIF4G, called eIF4F, leading to 48S PIC assembly (Bhat et al., 2015). We therefore tested eIF4A2 function and found that loss of eIF4A2 largely rescued the phenotype of p53 induction (Figure S3C). eIF4A2 was able to bind the p53 mRNA (Figure S3D). We further treated cells with 4E1RCat, a compound that interferes with protein translation initiation by preventing the assembly of the eIF4F complex (Cencic et al., 2011). The data showed that 4E1Rcat largely rescued the KDM5A effect on p53 protein accumulation (Figure 5F), but had no effect on TP53 mRNA expression (Figure 5G). To further analyze the correlation of KDM5A targets with signaling pathways, we used a connectivity map to compare the transcriptome of KDM5A with gene profiles resulting from gene knockdown, gene overexpression, or compound treatment. Despite the upregulation of a subgroup of translation genes (Figure 5A), the top hits highly correlated with KDM5A knockdown-induced transcriptomes were global protein synthesis inhibitors including emetine, verrucarin-a, and homoharringtonine (Table S7). Taken together, these data indicate that KDM5A regulates p53 translation by modulating the expression of protein translation genes.
Enhanced p53 Protein Expression after KDM5A Deletion Is Directly Regulated through Protein Synthesis of TP53
Although we had used pulse-chase with S35 labeling to show that KDM5A regulates p53 protein synthesis (Figure 4E), we further investigated the mechanism by using two approaches: (1) polysomal profiling to assess whether TP53 mRNA is more enriched in polysomes (high translation efficiency) than monosomes (low translation efficiency) and (2) Click-iT labeling to assess the newly synthesized p53 protein and total new proteins after KDM5A knockout (Figure 6). The results showed that p53 expression was greatly increased after KDM5A knockout (Figure 6A), consistent with results obtained by siRNA and shRNA of KDM5A. We then used a sucrose gradient (5%–50% of sucrose) method to perform the polysome fractionation (Figure 6B) and extracted total RNA from each fraction (Figure 6C), followed by quantification of the percentage of TP53 mRNA in each fraction (Figure 6D). Indeed, KDM5A deletion resulted in an enrichment of TP53 mRNA in the heavy polysome fractions (Figure 6D), suggesting a higher rate of p53 protein synthesis. Importantly, KDM5A knockout had no effect on the total steady-state p53 mRNA levels (Figure 6E).
Figure 6.
Enhanced p53 Protein Expression after KDM5A Deletion Is Directly Regulated by Protein Synthesis of TP53
(A) Western blot analysis of p53 and p21 proteins in KDM5A knockout HCT116 cells compared with HCT116 control cells.
(B) Schematic representation of sucrose separation of polysomes.
(C) Absorbance at 260 nm for all aligned polysome fractions (1–13). Values for HCT116 control cells or KDM5A knockout (KO) cells are shown as green or pink lines, respectively.
(D) Distribution of TP53 mRNA in each fraction of the polysome profile of either HCT116 control cells (green) or KDM5A-KO cells (pink). Values are represented as a percentage of TP53 mRNA for a single fraction and as a percentage of the total TP53 mRNA across all fractions. Data are mean ± SEM.
(E) The steady-state TP53 mRNA levels from control and KDM5A-KO input samples compared with 18S RNA. Technical triplicates for each were performed.
(F) Schematic representation for Click-iT metabolic labeling of newly synthesized proteins.
(G) AHA-labeled cells underwent the “Click-iT” either with or without biotin added. Increased biotinylated p53 is shown compared with total p53 protein immunoprecipitated (upper panel). Western blot analysis of input samples before immunoprecipitation (lower panel). Biotin or p53 band intensity in KDM5A knockout cells was quantified by ImageJ and normalized to controls.
(H) Total biotinylated protein comparing KDM5A-KO cells and control cells. Intensity of the biotin bands were quantified by ImageJ and normalized to loading control.
The Click-iT labeling, a similar approach to S35 pulse labeling, uses a methionine analog L-azidohomoalanine (AHA) to label newly synthesized proteins after methionine-free medium starvation (Figure 6F). Then a Click-iT chemistry-based reaction brings the alkyne-modified biotin to the AHA-incorporated polypeptides. The total p53 proteins, including old (non-labeled) and new (labeled) proteins, are immunoprecipitated, followed by immunoblotting for biotin and p53. The biotin antibody only detects newly synthesized p53 that was pulled down. Consistent with S35 radioactive labeling, the Click-iT labeling assay also showed that p53 protein synthesis was increased by KDM5A knockout (Figure 6G). However, the total new proteins were not affected by KDM5A deletion (Figure 6H), similar to what we have seen previously (Figure 4E). These data indicate that KDM5A specifically regulates p53 protein translation by modulating its mRNA protein synthesis.
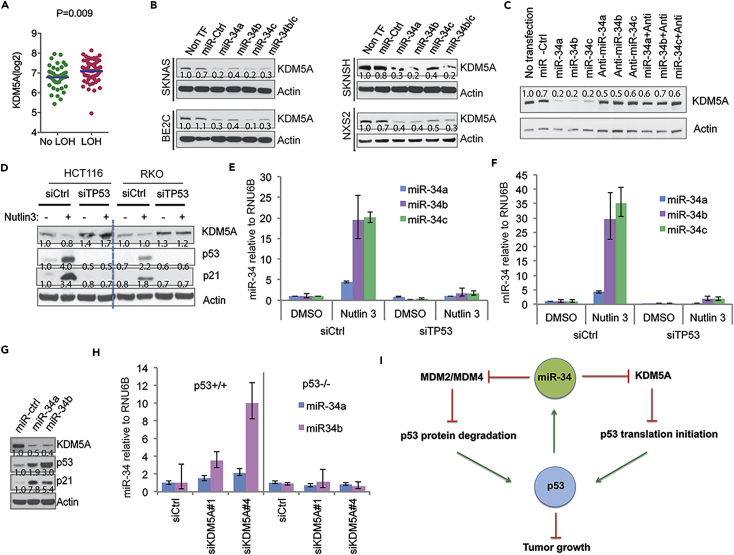
miR-34 Inhibits KDM5A Expression
Despite the importance of KDM5A, how it is regulated in cells is not clear. To determine what genetic factors might, in turn, regulate KDM5A expression, we analyzed published neuroblastoma microarray datasets with well-characterized genetic lesions and found that upregulation of KDM5A is correlated with loss of heterozygosity (LOH) of 1p36 and 11q23 in patients with neuroblastoma (Figure 7A). Interestingly, members of the miR-34 family of microRNAs (miRNAs), known to be direct targets of p53 (He et al., 2007) and mimics of which were being tested in a clinical trial for hepatocellular carcinoma, reside within these regions; miR-34a is located on chromosome 1p36, whereas its paralogs, the miR-34b/c cluster, is located on chromosome 11q23. LOH or silencing in either or both of these regions is common in a variety of human cancers (Bader, 2012), including neuroblastoma (Attiyeh et al., 2005), suggesting that these miRNAs may have a critical role in oncogenesis as tumor suppressors (Bader, 2012). To assess the functional link between miR-34 and KDM5A, we introduced miR-34 precursors into several neuroblastoma cell lines. Western blot analysis on protein from transfected cells showed that expression of KDM5A was downregulated by miR-34, suggesting that miR-34 targets KDM5A (Figure 7B). To further confirm the specificity of miR-34 in the regulation of KDM5A expression, we used an anti-miR to block miR-34 function, resulting in rescue of the expression of KDM5A (Figure 7C). The BE2(C) cell line has no detectable miR-34 expression (data not shown), and thus, introduction of anti-miR-34 into these cells has little effect on KDM5A expression.
Figure 7.
A Regulatory Feedback Loop between miR-34 and KDM5A
(A) The association of KDM5A gene expression in neuroblastomas (GSE3960 dataset) with LOH of 1p36 and/or 11q23 analyzed by Student's t test in the Prism program.
(B) Western blot assessing KDM5A expression in neuroblastoma cells transfected with miR-34 precursors for 72 hr. Band intensity was quantified by ImageJ and normalized to loading control.
(C) BE2(C) cells were transfected with miR-34 and anti-miRs for western blot analysis. Band intensity was quantified by ImageJ and normalized to loading control.
(D) HCT116 and RKO cells were transfected with p53 siRNA, followed by 48 hr of 5 μM Nutlin-3 treatment and analyzed by western blot. Band intensity was quantified by ImageJ and normalized to loading control.
(E and F) RT-PCR analysis of miR-34 expression in HCT116 (E) and RKO (F) from the same experiment as (D). Data are mean ± SEM.
(G) Western blot analysis of p53 and KDM5A expression after 72-hr transfection of miR-34. Band intensity was quantified by ImageJ and normalized to loading control.
(H) RT-PCR to assess miR-34 expression in isogenic p53+/+ and p53−/− HCT116 cells 48 hr after siRNA-mediated knockdown of KDM5A. Data are mean ± SEM.
(I) A working model of the interactions between the KDM5A and p53 signaling pathways.
To determine whether the induction of miR-34, through activation of p53, was able to recapitulate the signature of direct introduction of miR-34 precursors, HCT116 and RKO cells were treated with Nutlin-3 or doxorubicin (Figures 7D and S4A). Induction of p53 resulted in upregulation of miR-34 (Figures 7E and 7F) and concomitantly, downregulation of KDM5A (Figure 7D). Importantly, this was p53 dependent as depletion of p53 rescued the phenotype (Figures 7D–7F). We also found that activation of p53 in the neuroblastoma cell line NB-1691 caused upregulation of miR-34, but knockdown of p53 in these cells diminished these effects (Figure S4B), indicating a general mechanism. Importantly, introduction of miR-34 into HCT116 cells significantly upregulated p53 expression (Figure 7G). These data support our findings that miR-34 targets KDM5A through a p53-dependent process and suggest that a feedback loop exists. Indeed, KDM5A also regulated miR-34 in a p53-dependent manner with KDM5A knockdown resulting in miR-34 induction in p53+/+ but not in p53−/− cells (Figure 7H). Moreover, genes regulated by KDM5A were significantly enriched in the miR-34b/c signature revealed by GSEA analysis, further supporting the relationship between miR-34 and KDM5A (Figure S4C). In addition, we confirmed that KDM5B, another member of KDM5 family, was not affected by either doxorubicin or p53 knockdown (Figure S4D), indicating that KDM5A is specifically involved in the p53 pathway.
Discussion
p53 is commonly mutated in many different types of cancer. In addition, the classical p14Arf-MDM2 pathway plays an important role in regulating p53 protein expression (Kamijo et al., 1998, Stott et al., 1998, Zhang et al., 1998); this pathway is frequently deregulated in cancer. Nevertheless, some types of cancers such as neuroblastoma rarely bear p53 somatic mutations or alterations of the p14Arf-MDM2 pathway (Molenaar et al., 2012, Pugh et al., 2013), indicating that other mechanisms may exist that suppress p53 function in these cancers (Veschi et al., 2017). With the identification of the first true histone demethylase KDM1A/LSD1 (Shi et al., 2004), which is able to modulate p53 methylation, another class of JmjC histone demethylases has been shown to be deregulated in multiple cancers (Kandoth et al., 2013). However, the roles and working mechanisms of these histone demethylases in cancer have not been fully characterized. In this study, we identified several histone demethylases (KDM4D, KDM5A, KDM6B, PHF2, PHF8) and two additional JmjC-domain-containing proteins (C14orf169 and JMJD4) that are involved in regulating p53 activity. KDM4D is an H3K9me3/me2 demethylase and is able to induce p21 expression (Kim et al., 2012). However, overexpression of KDM4D causes global demethylation of H3K9me3, which may result in DNA damage and subsequent p53 activation. Similarly, loss of KDM4D may also result in a DNA damage response (Khoury-Haddad et al., 2015), and thus, activation of p53. KDM6B is an H3K27me3/me2 demethylase and can be recruited to p53-bound promoters and enhancer elements with unknown function (Williams et al., 2014). It is possible that KDM6B suppresses p53 function in this scenario. Although the histone demethylase activity of PHF2 is not definite, it has been shown to facilitate p53-mediated cell death in response to chemotherapy (Lee et al., 2014). PHF8 is an H3K9me2/me1 and H4K20me1 demethylase, but how it regulates p53 activity needs further studies. C14orf169 and JMJD4 may not have histone demethylase activity, and no previous studies show that they are involved in p53 function.
Although most genetic mutations in the coding sequences of TP53 disrupt the functions of p53, a rare polymorphism (rs78378222) in the 3′-UTR of TP53, which affects p53 protein translation, was found to confer susceptibility to multiple cancers, including neuroblastoma (Diskin et al., 2014, Stacey et al., 2011). These data suggest that repression of p53 protein translation contributes to oncogenesis. Here we found that the histone demethylase KDM5A was amplified in many types of cancers, and this was negatively correlated with TP53 mutation when compared with KDM5A loss. Most interestingly, we found that KDM5A regulates p53 function by inhibiting p53 protein translation, which may explain why KDM5A amplification is negatively correlated with TP53 mutation. A previous study had shown that ribosomal protein L26 (RPL26) and nucleolin could bind the 5′-UTR of p53 mRNA to control p53 translation and induction after DNA damage (Takagi et al., 2005). RNPC1 has been reported to repress p53 translation in lymphomas (Zhang et al., 2011). However, our data did not show a significant change in RNPC1, RPL26, or nucleolin expression after depletion of KDM5A, and loss of KDM5A did not induce a significant DNA damage response as assessed by measuring phosphorylated p53, suggesting that KDM5A works through a different mechanism. Indeed, KDM5A negatively regulated a subgroup of genes that are involved in translation initiation such as EIF5A and EIF2B4. Since depletion or knockout of KDM5A had no effect on global protein synthesis based on the assessment of newly synthesized proteins by using S35 labeling (Figure 4E) and Click-iT labeling technology (Figure 6), we speculate that two possible mechanisms (which are not mutually exclusive) may account for p53 synthesis regulated by KDM5A: (1) KDM5A regulates a specific program that alters the stoichiometry of translational complex components such as EIF2B4, leading to the activation of a specific translation machinery that promotes p53 synthesis. (2) Specific secondary structures of TP53 mRNA, as reported previously (Candeias et al., 2008, Fu et al., 1996, Ray et al., 2006), respond to the translation machinery induced by KDM5A. However, other additional mechanisms may exist; hence more studies are needed. In addition, whether there are other mRNAs whose translation is subject to KDM5A knockdown needs to be determined in the future.
Interestingly, by mining a database of a genome-scale genetic interaction map in yeast (Costanzo et al., 2010), we found that there was a positive genetic interaction between TIF5 and JHD2, the homologs of eIF5 and KDM5A in mammals; that is, the phenotype of TIF5 mutant was partially rescued by JHD2 mutation. Translation initiation factor eIF5 functions to regulate eIF2 activity. Although yeast does not have the p53 tumor suppressor gene, this may indicate that there is an evolutionarily conserved function for KDM5A in the regulation of protein translation. Consistent with the regulatory mechanism of KDM5A, expression of mutant p53 is also enhanced by loss of KDM5A. Mutation in p53 can be seen as a late event in multiple recurrent, chemotherapy-resistant cancers such as neuroblastoma (Keshelava et al., 2000), and mutant p53 may have additional, direct oncogenic actions that are different from the results of inhibiting p53 signaling early on in tumor progression (Muller and Vousden, 2014). Considering the context-dependent features of epigenetic modifiers in cancer, under some circumstances, loss of function of KDM5A might afford an additional growth advantage to affected cells with mutant p53. Despite the observation that KDM5A amplification tends to be mutually exclusive to TP53 mutation, there were a fraction of tumors showing co-occurrence of KDM5A amplification and TP53 mutation. This may suggest that either KDM5A has p53-independent function or that amplification of KDM5A confers selective pressure on p53, leading to p53 mutation during cancer progression. Nevertheless, genetic deletion of p14Arf and amplification of MDM2, two classical modulators of p53, also frequently overlap with TP53 mutations.
Although the mechanism by which KDM5A regulates the expression of its targets needs a deeper understanding, we found that KDM5A was located in the promoter regions of its downstream targets from both our own ChIP-seq data and ENCODE data and correlated with changes of the H3K4me3 mark, which is representative of active gene transcription. These data indicate that KDM5A directly regulates the expression of these genes. Previous studies also show that KDM5A can exert biological functions in a histone-demethylase-independent manner (Cao et al., 2014, DiTacchio et al., 2011, Secombe and Eisenman, 2007). Whether KDM5A regulates p53 translation through its histone demethylase activity needs to be further addressed in the future by using a potent and specific inhibitor.
Although p53 significantly induces p21 after KDM5A depletion, interestingly, we observed some induction of p21 expression in p53-null cells after the loss of KDM5A. The mechanism for this also needs to be further investigated. A recent study shows that KDM5A interacts with the nucleosome remodeling and deacetylase (NuRD) complex (Nishibuchi et al., 2014), which usually exerts gene repressive functions (Denslow and Wade, 2007), and HDAC is one of the core components of the NuRD complex (Denslow and Wade, 2007). It is well known that HDAC inhibitors induce p21 expression (Gui et al., 2004), so we speculate that KDM5A may modulate HDAC activity to suppress p21 induction, regardless of the p53 status.
We also found that miR-34s (transcriptional targets of p53) target KDM5A expression (Figure 7). A recent study showed that miR-34 targets MDM4 (Okada et al., 2014), which facilities MDM2-mediated function, resulting in p53 stabilization. Thus, miR-34 seems to bridge p53 protein stability and translation, thereby enhancing p53 activity (Figure 6I). Although this might suggest a feedback loop among these molecules, studies need to delve into more depth to consolidate this interesting observation.
In summary, we have identified a mechanism by which the histone demethylase KDM5A regulates p53 expression by modulating its translation initiation. Targeting KDM5A might be an option for treating p53 wild-type cancers.
Limitations of the Study
Our study has several limitations. First, the mechanism by which KDM5A affects the expression of a set of mRNA translation factors that controls p53 synthesis needs to be further investigated. Second, other messenger RNAs, in addition to p53, are likely to be regulated by KDM5A at the translational level; this needs to be investigated in future studies. In addition, the lack of a specific small-molecule KDM5A inhibitor prevents additional validation of the effect of KDM5A inhibition on tumor growth in the xenograft model. Last, the regulation of KDM5A by miR-34 also needs further in-depth studies to understand the upregulation of p53 by miR-34.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We thank William G. Kaelin, Jr., and Douglas R. Green for providing reagents and helpful discussion. We thank Bert Volgelstein and Michael Kastan for providing cell lines. We thank The Hartwell Center at St. Jude for ChIP-seq and Xenograft Core Facility for the pilot xenograft experiments. This work was supported by the Assisi Foundation of Memphis, the American Lebanese Syrian Associated Charities (ALSAC), the US Public Health Service Childhood Solid Tumor Program Project Grant No. CA23099, the Cancer Center Support Grant No. 21766 from the National Cancer Institute (A.M.D.), American Cancer Society-Research Scholar 130421-RSG-17-071-01-TBG (J.Y.), National Cancer Institute of the National Institutes of Health under Award Number R03CA212802 and R01CA229739 (J.Y.), and a Department of Defense Peer Reviewed Cancer Research Program Career Development Award W81XWH-13-1-0235 (Q.Y.). The K.M.M. laboratory is supported by the NIH National Cancer Institute (R01CA198279 and R01CA201268).
Author Contributions
Conceptualization: J.Y. and A.M.D.; Methodology: J.Y., P.-H.C., D.H., and C.J.; Investigation: J.Y., P.-H.C., H.D., C.J., A.A., N.G., A.A.Z., and D.Y.; Writing – Original Draft: J.Y.; Writing –Review & Editing: A.M.D., Q.Y., T.C., G.P.Z., Y.W, B.S., and C.L.; Formal Analysis: G.K., L.P., and Y.F.; Resources: R.C.K., K.M.M., Q.Y., Y.W., C.L., M.B., T.C.; Supervision: J.Y., A.M.D.; Funding Acquisition: J.Y., A.M.D., and K.M.
Declaration of Interests
The authors declare no competing interests.
Published: November 30, 2018
Footnotes
Supplemental Information includes Transparent Methods, four figures, and seven tables and can be found with this article online at https://doi.org/10.1016/j.isci.2018.10.012.
Data and Software Availability
All microarray and deep sequencing data have been deposited in GEO under accession number: GSE45967, GSE49854, GSE100511, GSE107221.
Supplemental Information
References
- Attiyeh E.F., London W.B., Mosse Y.P., Wang Q., Winter C., Khazi D., McGrady P.W., Seeger R.C., Look A.T., Shimada H. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N. Engl. J. Med. 2005;353:2243–2253. doi: 10.1056/NEJMoa052399. [DOI] [PubMed] [Google Scholar]
- Bader A.G. miR-34-a microRNA replacement therapy is headed to the clinic. Front Genet. 2012;3:120. doi: 10.3389/fgene.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banelli B., Carra E., Barbieri F., Wurth R., Parodi F., Pattarozzi A., Carosio R., Forlani A., Allemanni G., Marubbi D. The histone demethylase KDM5A is a key factor for the resistance to temozolomide in glioblastoma. Cell Cycle. 2015;14:3418–3429. doi: 10.1080/15384101.2015.1090063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R., Mermel C.H., Porter D., Wei G., Raychaudhuri S., Donovan J., Barretina J., Boehm J.S., Dobson J., Urashima M. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat M., Robichaud N., Hulea L., Sonenberg N., Pelletier J., Topisirovic I. Targeting the translation machinery in cancer. Nat. Rev. Drug Discov. 2015;14:261–278. doi: 10.1038/nrd4505. [DOI] [PubMed] [Google Scholar]
- Blair L.P., Cao J., Zou M.R., Sayegh J., Yan Q. Epigenetic regulation by lysine demethylase 5 (KDM5) enzymes in cancer. Cancers (Basel) 2011;3:1383–1404. doi: 10.3390/cancers3011383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candeias M.M., Malbert-Colas L., Powell D.J., Daskalogianni C., Maslon M.M., Naski N., Bourougaa K., Calvo F., Fahraeus R. P53 mRNA controls p53 activity by managing Mdm2 functions. Nat. Cell Biol. 2008;10:1098–1105. doi: 10.1038/ncb1770. [DOI] [PubMed] [Google Scholar]
- Cao J., Liu Z., Cheung W.K., Zhao M., Chen S.Y., Chan S.W., Booth C.J., Nguyen D.X., Yan Q. Histone demethylase RBP2 is critical for breast cancer progression and metastasis. Cell Rep. 2014;6:868–877. doi: 10.1016/j.celrep.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cencic R., Hall D.R., Robert F., Du Y., Min J., Li L., Qui M., Lewis I., Kurtkaya S., Dingledine R. Reversing chemoresistance by small molecule inhibition of the translation initiation complex eIF4F. Proc. Natl. Acad. Sci. U S A. 2011;108:1046–1051. doi: 10.1073/pnas.1011477108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P., Allis C.D., Wang G.G. Covalent histone modifications–miswritten, misinterpreted and mis-erased in human cancers. Nat. Rev. Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J., Agger K., Cloos P.A., Pasini D., Rose S., Sennels L., Rappsilber J., Hansen K.H., Salcini A.E., Helin K. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128:1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E.D., Sevier C.S., Ding H., Koh J.L., Toufighi K., Mostafavi S. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson M.A., Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- de Rooij J.D., Hollink I.H., Arentsen-Peters S.T., van Galen J.F., Berna Beverloo H., Baruchel A., Trka J., Reinhardt D., Sonneveld E., Zimmermann M. NUP98/JARID1A is a novel recurrent abnormality in pediatric acute megakaryoblastic leukemia with a distinct HOX gene expression pattern. Leukemia. 2013;27:2280–2288. doi: 10.1038/leu.2013.87. [DOI] [PubMed] [Google Scholar]
- Denslow S.A., Wade P.A. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26:5433–5438. doi: 10.1038/sj.onc.1210611. [DOI] [PubMed] [Google Scholar]
- Didiot M.C., Hewett J., Varin T., Freuler F., Selinger D., Nick H., Reinhardt J., Buckler A., Myer V., Schuffenhauer A. Identification of cardiac glycoside molecules as inhibitors of c-Myc IRES-mediated translation. J. Biomol. Screen. 2013;18:407–419. doi: 10.1177/1087057112466698. [DOI] [PubMed] [Google Scholar]
- Diskin S.J., Capasso M., Diamond M., Oldridge D.A., Conkrite K., Bosse K.R., Russell M.R., Iolascon A., Hakonarson H., Devoto M. Rare variants in TP53 and susceptibility to neuroblastoma. J. Natl. Cancer Inst. 2014;106:dju047. doi: 10.1093/jnci/dju047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiTacchio L., Le H.D., Vollmers C., Hatori M., Witcher M., Secombe J., Panda S. Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and influences the circadian clock. Science. 2011;333:1881–1885. doi: 10.1126/science.1206022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L., Minden M.D., Benchimol S. Translational regulation of human p53 gene expression. EMBO J. 1996;15:4392–4401. [PMC free article] [PubMed] [Google Scholar]
- Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer E.L., Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui C.Y., Ngo L., Xu W.S., Richon V.M., Marks P.A. Histone deacetylase (HDAC) inhibitor activation of p21WAF1 involves changes in promoter-associated proteins, including HDAC1. Proc. Natl. Acad. Sci. U S A. 2004;101:1241–1246. doi: 10.1073/pnas.0307708100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., He X., Lim L.P., de Stanchina E., Xuan Z., Liang Y., Xue W., Zender L., Magnus J., Ridzon D. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J., Wu J., Dombkowski A., Zhang K., Holowatyj A., Boerner J.L., Yang Z.Q. Genomic amplification and a role in drug-resistance for the KDM5A histone demethylase in breast cancer. Am. J. Transl. Res. 2012;4:247–256. [PMC free article] [PubMed] [Google Scholar]
- Huang J., Sengupta R., Espejo A.B., Lee M.G., Dorsey J.A., Richter M., Opravil S., Shiekhattar R., Bedford M.T., Jenuwein T. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449:105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- Kaelin W.G., Jr., McKnight S.L. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamijo T., Weber J.D., Zambetti G., Zindy F., Roussel M.F., Sherr C.J. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc. Natl. Acad. Sci. U S A. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth C., McLellan M.D., Vandin F., Ye K., Niu B., Lu C., Xie M., Zhang Q., McMichael J.F., Wyczalkowski M.A. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshelava N., Zuo J.J., Waidyaratne N.S., Triche T.J., Reynolds C.P. p53 mutations and loss of p53 function confer multidrug resistance in neuroblastoma. Med. Pediatr. Oncol. 2000;35:563–568. doi: 10.1002/1096-911x(20001201)35:6<563::aid-mpo15>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Kessis T.D., Slebos R.J., Nelson W.G., Kastan M.B., Plunkett B.S., Han S.M., Lorincz A.T., Hedrick L., Cho K.R. Human papillomavirus 16 E6 expression disrupts the p53-mediated cellular response to DNA damage. Proc. Natl. Acad. Sci. U S A. 1993;90:3988–3992. doi: 10.1073/pnas.90.9.3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury-Haddad H., Nadar-Ponniah P.T., Awwad S., Ayoub N. The emerging role of lysine demethylases in DNA damage response: dissecting the recruitment mode of KDM4D/JMJD2D to DNA damage sites. Cell Cycle. 2015;14:950–958. doi: 10.1080/15384101.2015.1014147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.D., Oh S., Shin S., Janknecht R. Regulation of tumor suppressor p53 and HCT116 cell physiology by histone demethylase JMJD2D/KDM4D. PLoS One. 2012;7:e34618. doi: 10.1371/journal.pone.0034618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose R.J., Yan Q., Tothova Z., Yamane K., Erdjument-Bromage H., Tempst P., Gilliland D.G., Zhang Y., Kaelin W.G., Jr. The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell. 2007;128:889–900. doi: 10.1016/j.cell.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Kooistra S.M., Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat. Rev. Mol. Cell Biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- Kruse J.P., Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D., Levine A. p53 Research: the past thirty years and the next thirty years. Cold Spring Harb. Perspect. Biol. 2010;2:a000893. doi: 10.1101/cshperspect.a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.H., Park J.W., Sung H.S., Choi Y.J., Kim W.H., Lee H.S., Chung H.J., Shin H.W., Cho C.H., Kim T.Y. PHF2 histone demethylase acts as a tumor suppressor in association with p53 in cancer. Oncogene. 2014;34:2897–2909. doi: 10.1038/onc.2014.219. [DOI] [PubMed] [Google Scholar]
- Levine A.J., Oren M. The first 30 years of p53: growing ever more complex. Nat. Rev. Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Wang L., Song P., Geng X., Liang X., Zhou M., Wang Y., Chen C., Jia J., Zeng J. Critical role of histone demethylase RBP2 in human gastric cancer angiogenesis. Mol. Cancer. 2014;13:81. doi: 10.1186/1476-4598-13-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., Cao J., Liu J., Beshiri M.L., Fujiwara Y., Francis J., Cherniack A.D., Geisen C., Blair L.P., Zou M.R. Loss of the retinoblastoma binding protein 2 (RBP2) histone demethylase suppresses tumorigenesis in mice lacking Rb1 or Men1. Proc. Natl. Acad. Sci. U S A. 2011;108:13379–13386. doi: 10.1073/pnas.1110104108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris J.M. Recent advances in neuroblastoma. N. Engl. J. Med. 2010;362:2202–2211. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar J.J., Koster J., Zwijnenburg D.A., van Sluis P., Valentijn L.J., van der Ploeg I., Hamdi M., van Nes J., Westerman B.A., van Arkel J. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- Muller P.A., Vousden K.H. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell. 2014;25:304–317. doi: 10.1016/j.ccr.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishibuchi G., Shibata Y., Hayakawa T., Hayakawa N., Ohtani Y., Sinmyozu K., Tagami H., Nakayama J. Physical and functional interactions between the histone H3K4 demethylase KDM5A and the nucleosome remodeling and deacetylase (NuRD) complex. J. Biol. Chem. 2014;289:28956–28970. doi: 10.1074/jbc.M114.573725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada N., Lin C.P., Ribeiro M.C., Biton A., Lai G., He X., Bu P., Vogel H., Jablons D.M., Keller A.C. A positive feedback between p53 and miR-34 miRNAs mediates tumor suppression. Genes Dev. 2014;28:438–450. doi: 10.1101/gad.233585.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh T.J., Morozova O., Attiyeh E.F., Asgharzadeh S., Wei J.S., Auclair D., Carter S.L., Cibulskis K., Hanna M., Kiezun A. The genetic landscape of high-risk neuroblastoma. Nat. Genet. 2013;45:279–284. doi: 10.1038/ng.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L., Zhu F., Li S.H., Si L.B., Hu L.K., Tian H. Retinoblastoma binding protein 2 (RBP2) promotes HIF-1alpha-VEGF-induced angiogenesis of non-small cell lung cancer via the Akt pathway. PLoS One. 2014;9:e106032. doi: 10.1371/journal.pone.0106032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray P.S., Grover R., Das S. Two internal ribosome entry sites mediate the translation of p53 isoforms. EMBO Rep. 2006;7:404–410. doi: 10.1038/sj.embor.7400623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees M.G., Seashore-Ludlow B., Cheah J.H., Adams D.J., Price E.V., Gill S., Javaid S., Coletti M.E., Jones V.L., Bodycombe N.E. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat. Chem. Biol. 2016;12:109–116. doi: 10.1038/nchembio.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo C.M., Cencic R., Roche S.P., Pelletier J., Porco J.A. Synthesis of rocaglamide hydroxamates and related compounds as eukaryotic translation inhibitors: synthetic and biological studies. J. Med. Chem. 2012;55:558–562. doi: 10.1021/jm201263k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secombe J., Eisenman R.N. The function and regulation of the JARID1 family of histone H3 lysine 4 demethylases: the Myc connection. Cell Cycle. 2007;6:1324–1328. doi: 10.4161/cc.6.11.4269. [DOI] [PubMed] [Google Scholar]
- Sharma S.V., Lee D.Y., Li B., Quinlan M.P., Takahashi F., Maheswaran S., McDermott U., Azizian N., Zou L., Fischbach M.A. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Lan F., Matson C., Mulligan P., Whetstine J.R., Cole P.A., Casero R.A., Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Stacey S.N., Sulem P., Jonasdottir A., Masson G., Gudmundsson J., Gudbjartsson D.F., Magnusson O.T., Gudjonsson S.A., Sigurgeirsson B., Thorisdottir K. A germline variant in the TP53 polyadenylation signal confers cancer susceptibility. Nat. Genet. 2011;43:1098–1103. doi: 10.1038/ng.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stott F.J., Bates S., James M.C., McConnell B.B., Starborg M., Brookes S., Palmero I., Ryan K., Hara E., Vousden K.H. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z., Fang H., Hong H., Shi L., Zhang W., Zhang W., Zhang Y., Dong Z., Lancashire L.J., Bessarabova M. An investigation of biomarkers derived from legacy microarray data for their utility in the RNA-seq era. Genome Biol. 2014;15:523. doi: 10.1186/s13059-014-0523-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suva M.L., Riggi N., Bernstein B.E. Epigenetic reprogramming in cancer. Science. 2013;339:1567–1570. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi M., Absalon M.J., McLure K.G., Kastan M.B. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- Teng Y.C., Lee C.F., Li Y.S., Chen Y.R., Hsiao P.W., Chan M.Y., Lin F.M., Huang H.D., Chen Y.T., Jeng Y.M. Histone demethylase RBP2 promotes lung tumorigenesis and cancer metastasis. Cancer Res. 2013;73:4711–4721. doi: 10.1158/0008-5472.CAN-12-3165. [DOI] [PubMed] [Google Scholar]
- Toledo F., Wahl G.M. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat. Rev. Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- Tweddle D.A., Malcolm A.J., Bown N., Pearson A.D., Lunec J. Evidence for the development of p53 mutations after cytotoxic therapy in a neuroblastoma cell line. Cancer Res. 2001;61:8–13. [PubMed] [Google Scholar]
- van Zutven L.J., Onen E., Velthuizen S.C., van Drunen E., von Bergh A.R., van den Heuvel-Eibrink M.M., Veronese A., Mecucci C., Negrini M., de Greef G.E. Identification of NUP98 abnormalities in acute leukemia: JARID1A (12p13) as a new partner gene. Genes Chromosomes Cancer. 2006;45:437–446. doi: 10.1002/gcc.20308. [DOI] [PubMed] [Google Scholar]
- Veschi V., Liu Z., Voss T.C., Ozbun L., Gryder B., Yan C., Hu Y., Ma A., Jin J., Mazur S.J. Epigenetic siRNA and chemical screens identify SETD8 inhibition as a therapeutic strategy for p53 activation in high-risk neuroblastoma. Cancer Cell. 2017;31:50–63. doi: 10.1016/j.ccell.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B., Kinzler K.W. Cancer genes and the pathways they control. Nat. Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- Vogelstein B., Papadopoulos N., Velculescu V.E., Zhou S., Diaz L.A., Jr., Kinzler K.W. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden K.H., Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Wang G.G., Song J., Wang Z., Dormann H.L., Casadio F., Li H., Luo J.L., Patel D.J., Allis C.D. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 2009;459:847–851. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.J., Gu W. To be, or not to be: functional dilemma of p53 metabolic regulation. Curr. Opin. Oncol. 2014;26:78–85. doi: 10.1097/CCO.0000000000000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K., Christensen J., Rappsilber J., Nielsen A.L., Johansen J.V., Helin K. The histone lysine demethylase JMJD3/KDM6B is recruited to p53 bound promoters and enhancer elements in a p53 dependent manner. PLoS One. 2014;9:e96545. doi: 10.1371/journal.pone.0096545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Milasta S., Hu D., AlTahan A.M., Interiano R.B., Zhou J., Davidson J., Low J., Lin W., Bao J. Targeting histone demethylases in MYC-driven neuroblastomas with ciclopirox. Cancer Res. 2017;77:4626–4638. doi: 10.1158/0008-5472.CAN-16-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y.X., Somasundaram K., el-Deiry W.S. AP2 inhibits cancer cell growth and activates p21WAF1/CIP1 expression. Nat. Genet. 1997;15:78–82. doi: 10.1038/ng0197-78. [DOI] [PubMed] [Google Scholar]
- Zhang J., Cho S.J., Shu L., Yan W., Guerrero T., Kent M., Skorupski K., Chen H., Chen X. Translational repression of p53 by RNPC1, a p53 target overexpressed in lymphomas. Genes Dev. 2011;25:1528–1543. doi: 10.1101/gad.2069311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Xiong Y., Yarbrough W.G. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- Zitvogel L., Kroemer G. CANCER. A p53-regulated immune checkpoint relevant to cancer. Science. 2015;349:476–477. doi: 10.1126/science.aac8475. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.