Abstract
DNA N6-methyladenine (6mA) modifications expand the information capacity of DNA and have long been known to exist in bacterial genomes. Xanthomonas oryzae pv. Oryzicola (Xoc) is the causative agent of bacterial leaf streak, an emerging and destructive disease in rice worldwide. However, the genome-wide distribution patterns and potential functions of 6mA in Xoc are largely unknown. In this study, we analyzed the levels and global distribution patterns of 6mA modification in genomic DNA of seven Xoc strains (BLS256, BLS279, CFBP2286, CFBP7331, CFBP7341, L8 and RS105). The 6mA modification was found to be widely distributed across the seven Xoc genomes, accounting for percent of 3.80, 3.10, 3.70, 4.20, 3.40, 2.10, and 3.10 of the total adenines in BLS256, BLS279, CFBP2286, CFBP7331, CFBP7341, L8, and RS105, respectively. Notably, more than 82% of 6mA sites were located within gene bodies in all seven strains. Two specific motifs for 6 mA modification, ARGT and AVCG, were prevalent in all seven strains. Comparison of putative DNA methylation motifs from the seven strains reveals that Xoc have a specific DNA methylation system. Furthermore, the 6 mA modification of rpfC dramatically decreased during Xoc infection indicates the important role for Xoc adaption to environment.
Introduction
DNA methylation, a base modification, does not alter the underlying DNA sequence, but adds additional information to bases through the addition of a methyl group. A group of enzymes, called DNA methyltransferases, catalyze the methylation process, and the methylated bases are assigned a name that reflects the atom harboring the methyl group1,2. Methylation on the fifth position of the pyrimidine ring of cytosine (5-methylcytosine, 5mC) is the predominant DNA methylation modification in eukaryotes3,4. The existence and abundance of a methyl group at the sixth position of the purine ring of adenine (N6-methyldeoxyadenosine, 6mA) was firstly reported in eukaryotes3,5. But several studies showed that the 6 mA base was present at extremely low levels in genomic DNA of higher eukaryotes6. The development of high throughput sequencing has greatly promoted the research and identification of 6 mA in fungi, plants, animals and humans4,7–12, which can further reveal the genome-wide distribution patterns of 6 mA modifications as well as different functions in biological processes among organisms. 6 mA modification was found as a prevalent DNA methylation in prokaryotes, which is used as a signal for epigenetic regulation13.
DNA 6 mA modification is ubiquitous in microbial genomes and plays an important role in regulating the biological processes in bacteria. It discriminates the host DNA from foreign pathogenic DNA and protects the host genome via the restriction-modification system, associated with defense against bacteriophages14,15. In addition, 6 mA is also involved in bacterial DNA replication and repair16,17, cell-cycle progression, and gene regulation15,18,19. Recently, the genome-wide distribution patterns of 6 mA have been investigated extensively using single molecule real-time (SMRT) sequencing20, which allows genome-wide mapping of m6A in bacteria at single-nucleotide resolution and at single-molecule level.
Single molecule real-time (SMRT) sequencing has been used for identification of 6 mA modifications in several bacterial genera, including Helicobacter pylori, Lactobacillus spp., Mycobacterium tuberculosis, Escherichia coli, Campylobacter coli and Xanthomonas spp.21–25. Among them, Xanthomonas spp. is the only agricultural genus for methylation detection. Xanthomonas causes serious and devastating diseases in more than 400 plants, including important economic and agricultural crops such as tomato, pepper, soybean and rice26. Bacterial leaf streak in rice, a devastating diseases in the world, is caused by Xanthomonas oryzae pv. oryzicola (Xoc)27. However, the genome-wide distribution patterns and potential functions of 6 mA in Xoc are still unknown. In this study, we measured the levels and global distribution patterns of 6 mA in genomic DNA of seven Xoc strains (BLS256, BLS279, CFBP2286, CFBP7331, CFBP7341, L8 and RS105), and compared DNA methylation motifs among seven strains.
Materials and Methods
Identification of 6 mA in Xoc genome
The raw data files of SMRT sequencing reads in h5 format were downloaded from the NCBI SRA database (Table S1)28. For each strain, 4–7 SMRT cells were used to achieve ~200 × coverage (Table S1). All cells used the P4C2 chemistry. Then, PacBio SMRT analysis platform (version 2.3.0) was used to detect DNA 6 mA modification of each strain (http://www.pacb.com/products-and-services/analytical-software/smrt-analysis/analysis-applications/epigenetics/). The detailed analysis workflow is as follows: Firstly, the raw reads were aligned to the corresponding reference genome of each strain by pbalign with the parameters ‘–seed = 1 –minAccuracy = 0.75 –minLength = 50 –concordant –algorithmOptions = “-useQuality” –algorithmOptions = ‘ -minMatch 12 -bestn 10 -minPctIdentity 70.0” (the reference resources are listed in Table S1). Furthermore, the polymerase kinetics information was loaded after alignment by loadChemistry.py and loadPulses scripts of raw h5 format files with‘-metrics DeletionQV, IPD, InsertionQV, PulseWidth, QualityValue, MergeQV, SubstitutionQV, DeletionTag’. The post-aligned datasets were sorted by using cmph5tools. The m6A was identified by using ipdSummary.py script with‘–methylFraction –identify m6A, m4C –numWorkers 4′. 6 mA sites with less than 25-fold coverage per chromosome of each strain were excluded for further analysis.
Bioinformatics analysis
The genome-wide 6 mA profiles across all chromosomes of seven Xoc strains were generated using Circos29. The gene bodies, intergenic regions and translation stop codons were defined by using an annotated file (gff format) of each strain by using in-house shell scripts. For each 6 mA modification site, we extracted 20 bp from the upstream and downstream sequences of the 6 mA site. The MEME was then used to predict conserved motifs in the flanking regions30.
6 mA levels of Xoc virulence-related genes during infection
Twenty-five-day-old leaves from the rice cultivar TP309 were inoculated with Xoc RS105 using the injection method. Bacteria for inoculation were taken from PSA plates and re-suspended in water at an OD600 of 0.3. 10 μl bacteria were injected into one rice leaf and 15 replicates were performed. Then the leaves were cut and DNA was isolated. The 6 mA levels of some virulence-related genes were analyzed according to previously described methods31.
Results
6 mA overview in Xoc genome
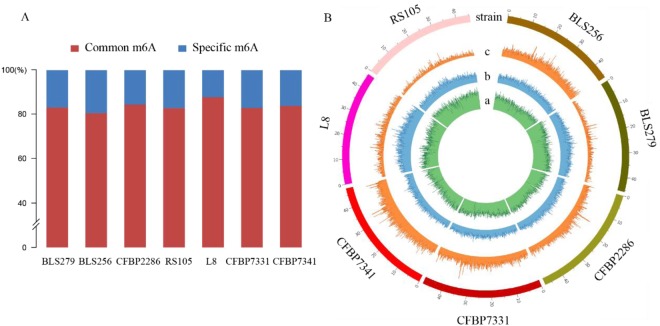
Analyzing SMRT sequencing datasets from Xoc genomic DNA, we detected 32751, 26395, 32779, 38009, 30694, 17900 and 26460 DNA 6 mA methylation sites in seven strains BLS256, BLS279, CFBP2286, CFBP7331, CFBP7341, L8 and RS105, respectively (Table 1 and Supplementary 1.xlsx). The density (6 mA/A) is about 2.1% to 4.2% of the total adenines in the Xoc genomic DNA (Table 1), which was close to the strain Xcv 85–10 of Xanthomonas campestris pv. vesicatoria (3.84%)25 and the Hesseltinella vesiculosa (2.8%)12,but higher than those in Caenorhabditis elegans (~0.7%)8, Chlamydomonas (~0.4%)7, Drosophila (0.07%)9, Human (0.051%) and Arabidopsis thaliana (0.048%)4. Among all of the 6 mA modification sites in the seven strains, nearly one-fifth was unique in each strain, and a large number of 6 mA sites (>80%) were identified in at least two strains (Fig. 1A and Supplementary 1.xlsx).
Table 1.
Statistical overview of 6 mA modification in genomic DNA of seven Xanthomonas oryzae pv. oryzicolastrains.
| Strain | Genome size (Mb) | Total A number | m6A number | m6A ratio | Specific m6A | Common m6A |
|---|---|---|---|---|---|---|
| BLS256 | 4.608 | 868176 | 32751 | 3.80% | 6354 | 26397 |
| BLS279 | 4.569 | 864106 | 26395 | 3.10% | 4459 | 21936 |
| CFBP2286 | 4.774 | 894661 | 32779 | 3.70% | 5093 | 27686 |
| CFBP7331 | 4.776 | 905931 | 38009 | 4.20% | 6496 | 31513 |
| CFBP7341 | 4.785 | 905944 | 30694 | 3.40% | 4954 | 25740 |
| L8 | 4.574 | 865646 | 17900 | 2.10% | 2196 | 15704 |
| RS105 | 4.558 | 862859 | 26460 | 3.10% | 4546 | 21914 |
Figure 1.
Distribution of 6 mA in seven Xoc strains. (A) Common and specific m6A sites; (B) circus plot of 6 mA in Xoc genome. a, density of 6 mA with fraction 0–0.3; b, density of 6 mA with fraction 0.3–0.7; c, density of 6 mA with fraction 0.7–1).
The distribution and modification level of 6 mA was presented in circos plot format in which concentric rings represent the density distribution of 6 mA across all seven strains in the given category. Densities were divided into three categories, namely, low (0–30%, green circle), middle (30–70%, blue circle) and high (70–100%, orange circle). The 6 mA density in low modification level group was dominant in all strains (Fig. 1B).
Analysis of 6 mA-methylated genes
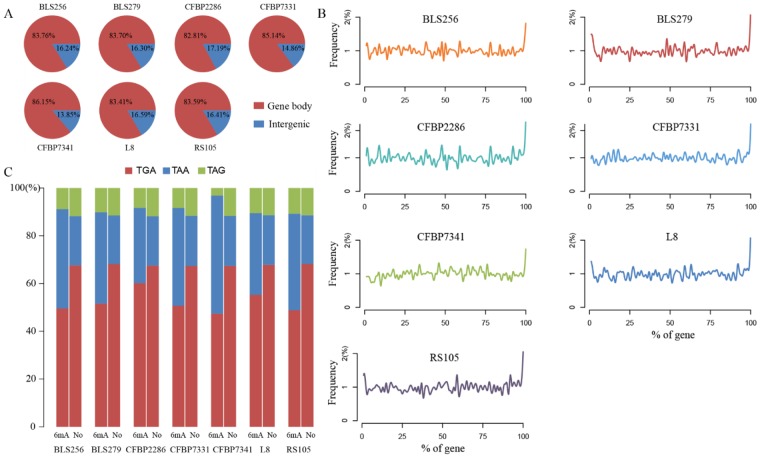
We further analyzed the gene bodies and intergenic regions to examine 6 mA distribution around functional elements according to the annotated genome, and found that more than 82% of 6 mA sites were located in the gene bodies (Fig. 2A). Furthermore, while 82–93% of genes were methylated, the 6 mA methylation ratios of protein coding genes were higher than all methylated genes (Table 2). In all gene bodies, more than one 6 mA site was identified throughout the majority methylated genes (Fig. S1), and the number of 6 mA sites was associated with the gene length (Fig. S2). As the gene length increases, there is a tendency for increased number of methylation sites.
Figure 2.
6 mA enrichment analysis in methylated genes. (A) Distribution of 6 mA sites in gene bodies and intergenic regions in seven strains; (B) frequency of 6 mA at relative position of protein coding genes; (C) stop codon usage in 6mA-methylated genes and no methylation genes)
Table 2.
The m6A methylation ratio in all and protein coding genes.
| Strains | All | Protein coding | ||||
|---|---|---|---|---|---|---|
| Total no. | m6A no. | Ratio | Total no. | m6A no. | Ratio | |
| BLS256 | 4493 | 4162 | 92.63% | 4267 | 4024 | 94.31% |
| BLS279 | 4485 | 3944 | 87.94% | 4261 | 3886 | 91.20% |
| CFBP2286 | 4709 | 4253 | 90.32% | 4482 | 4180 | 93.26% |
| CFBP7331 | 4711 | 4343 | 92.19% | 4488 | 4276 | 95.28% |
| CFBP7341 | 4716 | 4227 | 89.63% | 4493 | 4174 | 92.90% |
| L8 | 4479 | 3677 | 82.09% | 4254 | 3631 | 85.35% |
| RS105 | 4480 | 3977 | 88.77% | 4255 | 3916 | 92.03% |
To further investigate the 6 mA locations in protein coding genes, the 6 mA relative distance in these genes were analyzed. The 6 mA sites were enriched at 3′ end of coding regions (Fig. 2B) which was different from those in human. Based on the annotated genome, these enriched locations were the terminal of coding regions where the stop codon usage were TAG, TAA and TGA. TGA was the stop codon detected in highest proportion in all of the analyzed sites, both modified and non-modified, detected at 50% and 68%, respectively. (Fig. 2C and Table S2). Notably, the proportion of TAA in 6 mA predicted sites were higher than that in normal codons, whereas TAG and TGA levels were reduced (Fig. 2C).
Identification of consensus motifs for 6 mA in Xoc
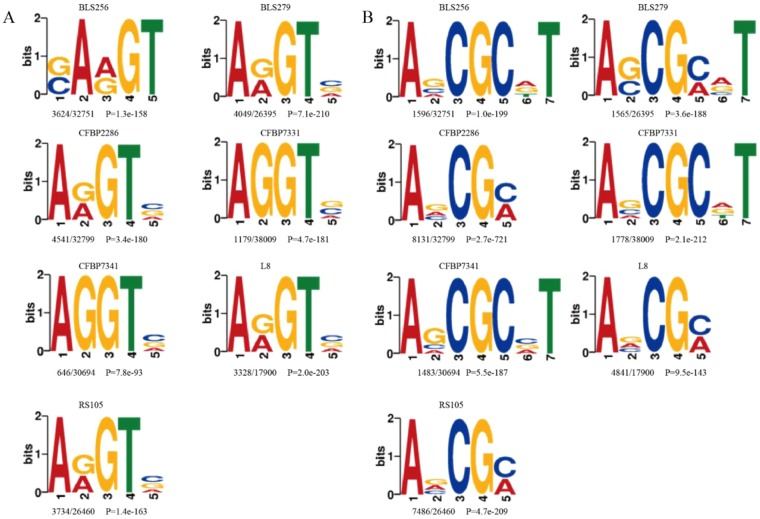
To determine whether the identified 6 mA sites share consensus sequence element(s) in seven strains, we extracted the upstream and downstream 20 bp sequences from the 6 mA sites, and performed a default search for significant consensus motifs enriched in these regions using MEME. There were three significantly enriched motifs sequences AGG, ARGT and AVCG in all seven strains (Figs 3 and S3). The sequences AGG detected in Xoc was consistent with the motif sequences in C. elegans, A. thaliana and human. The most significant strain containing AGG sequences was CFBP7341 (p = 2.8e-1884), which was present in approximately one-fourth of the methylated sites (Fig. S3). Interestingly, besides AGG, two novel enriched DNA methylation motifs (ARGT and AVCG) were identified in all seven Xoc strains (Fig. 3). The p-values of motifs in all strains were less than 7.8e-93, which indicated the identified motifs are indeed prevalent in 6 mA sites.
Figure 3.
The identified consensus motifs containing 6 mA sites in seven strains. The number of occurrences of each motif relative to the total number of 6mA-containing motifs and the corresponding p-value generated by MEME are shown under the sequence logo.
6mA might be important for Xoc infection
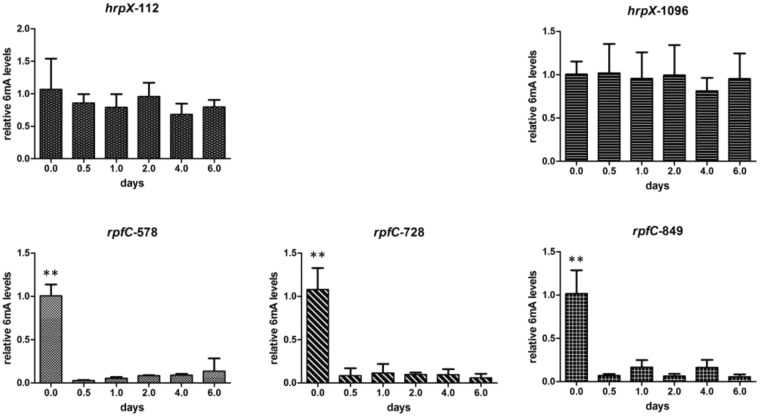
Xoc enters the host through the stomata or physical wounds and multiplies in the vascular regions. Its virulence is regulated by different mechanisms, but the pathways regulated by RpfC and HrpX are essential to successful infection32,33. Moreover, rpfC and hrpX have 6 mA sites (Table 3). Therefore, we chose these two genes to study the 6 mA levels during infection. As shown in Fig. 4, the methylation levels of three 6 mA sites in rpfC decreased significantly when growth in host (p < 0.01), but those in hrpX were not significantly changed, indicating that the 6 mA modification of rpfC is involved in Xoc adaptation to host environment. As RpfC is involved in quorum sensing and regulates the extracellular polysaccharide (EPS) levels, extracellular enzyme activity and motility in Xanthomonas oryzae32,34; However, HrpX regulates the expression of genes encoding the type-III secretion system which directly transports bacterial effectors into rice cells and regulates their functions33,35. These results imply that 6 mA might be important for Xoc adaptation to environment not for Xoc-rice direct interaction.
Table 3.
The m6A methylation sites in rpfC and hrpX.
| genes | 6mA site | position | strand |
|---|---|---|---|
| hrpX | CGCAGAGATCGCTGCAAAGTAGGTCGAAAGGATCATGCCGG | 112 | − |
| hrpX | ACAAGCCTTGTTGCTCTACAACCGCTATGCGCTGGACGCGG | 1096 | + |
| rpfC | ATTCGTGACTCATATTGGCCAGGAAACGGCTCTTGGCCTGG | 578 | − |
| rpfC | TCTGCTGTCGCTGGTGGAAGAGGTGCTGGATATTTCCGCGA | 728 | + |
| rpfC | CCGCAGGCCAGGACGCGCGGACTGGATTACGGCACCGAGGT | 849 | + |
Figure 4.
6 mA levels of special methylation sites in rpfC and hrpX during Xoc infection. The methylation levels at 0 days (DNA isolation immediately injection) were set as 1, and others were compared.
Discussion
N6-methyladenine (6 mA) is mainly found in prokaryotic genomes. The atlas of m6A modification sites have been elucidated in several bacteria strains by using SMRT sequencing, which opened up a new direction for epigenetics research22–25. In this paper, we studied the genome-wide distribution of DNA 6 mA modification among in seven Xoc strains, the pathogen responsible for bacterial leaf streak disease in rice. We found that that 6 mA sites are widely distributed in the Xoc genome and are enriched in gene bodies. Two previously unreported motifs, ARGT and AVCG, are involved in 6 mA modification among seven strains, which helps to understand 6 mA distribution patterns in Xoc.
SMRT sequencing datasets of seven Xoc strains were obtained from the NCBI SRA database28. We observed that 6 mA methylation density was similar in each strain (2.1–4.2%), and similar to the previously reported in bacterial genomes (1.9% in Escherichia coli, 2.7% in Campylobacter coli and 0.17–3.8% in Xanthomonas campestris)22,24,25. But our results were relatively higher than the density reported in C. elegans (~0.7%)8, Chlamydomonas (~0.4%)7, Drosophila (0.07%)9, Human (0.051%) and A. thaliana (0.048%). Interestingly, prokaryotic genomes have generally higher 6 mA levels than eukaryotes, which may be related to the defense mechanism of bacteriophage infection36,37. Adenine and cytosine methylation of bacterial DNA protects it from the action of the corresponding restriction endonuclease, whereas unmethylated sites of foreign nucleic acids, such as bacteriophage DNA are cleaved.
Sequence motifs are short recurring nucleotide sequences present throughout the genome. One 6 mA genomic distribution with the prevalent motif sequence AGG, similar to that in C. elegans, A. thaliana and human4,8, was detected in seven Xoc strains. Previous studies showed that the AGG motif sequence was widespread in eukaryotic species, and the motif GATC was likely the most ancient 6 mA motif exists in bacteria22,24,25. In this study, we find that the motif AGG, that was previously unreported in bacteria, was consistently present in all seven Xoc strains analyzed here (Fig. S3). Moreover, the motif GATC that consistent with previous reports were detected in the partial Xoc strains (Fig. S4). Importantly and interestingly, two specific motifs ARGT and AVCG were identified in all seven Xoc strains, which have not been reported in other organisms. These results indicate that the 6 mA modification pattern was not conserved across the species genome and is not species-specific, likely reflecting the potential diverse biological functions.
Xoc is an extracellular pathogen and infects rice through the stomata or physical wounds and grows in the vascular regions. The type II, III, VI secretion systems, quorum sensing and motility are all important for Xoc-rice interaction. RpfC and HrpX are two key regulators of Xoc virulence32,33. RpfC is involved in the regulation of quorum sensing, motility and Type II secretion system32, but HrpX regulates Type III secretion system that directly determines the Xoc-host interaction33. Therefore, the identified rpfC and hrpX 6 mA modification might affect Xoc adaption to environment and direct interaction with rice. The different changes of 6 mA modification levels in rpfC and hrpX indicate that the 6 mA modification in Xoc is associated with environmental adaptation.
In summary, we found that 6 mA was widely present in Xoc genomic DNA and detected two specific epidemic motifs, ARGT and AVCG, which promoted 6 mA modification. This study suggests that the 6 mA modification in Xoc is associated with environmental adaptation.
Electronic supplementary material
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31871326, 31760316, 31600667), Guangdong Natural Science Foundation (2015A030313127), China Postdoctoral Science Foundation (2017M612798). Priming Scientific Research Foundation of Hainan University (KYQD(ZR)1721) and Science Foundation for The Youth Teachers of Hainan University in 2017 (hdkyxj201702).
Author Contributions
C.L.X. and S.Q.X. conceived the project and designed the experiments; Z.Y.L. J.F.X. and K.K.J. collected and preprocess SMRT sequencing data; S.Q.X. detected 6 mA in samples and performed the informatics analysis; J.T. and Q.B.X. performed the experiments for investigating the function of 6 mA in vivo; S.Q.X. and C.L.X. wrote the paper; F.L. and L.Y.D. revised the paper.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Chuan-Le Xiao and Shang-Qian Xie contributed equally.
Contributor Information
Jun Tao, Email: taoj@hainu.edu.cn.
Liang-Ying Dai, Email: daily@hunau.net.
Feng Luo, Email: luofeng@clemson.edu.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-34559-5.
References
- 1.Bart A, van Passel MW, van Amsterdam K, van der Ende A. Direct detection of methylation in genomic DNA. Nucleic acids research. 2005;33:e124. doi: 10.1093/nar/gni121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nature reviews. Genetics. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 3.Vanyushin BF, Tkacheva SG, Belozersky AN. Rare bases in animalDNA. Nature. 1970;225:948–949. doi: 10.1038/225948a0. [DOI] [PubMed] [Google Scholar]
- 4.Liang Z, et al. DNA N(6)-Adenine Methylation in Arabidopsis thaliana. Developmental cell. 2018;45:406–416 e403. doi: 10.1016/j.devcel.2018.03.012. [DOI] [PubMed] [Google Scholar]
- 5.Unger G, Venner H. Remarks on minor bases in spermatic desoxyribonucleic acid. Hoppe-Seyler’s Zeitschrift fur physiologische Chemie. 1966;344:280–283. doi: 10.1515/bchm2.1966.344.1-3.280. [DOI] [PubMed] [Google Scholar]
- 6.Ratel D, Ravanat JL, Berger F, Wion D. N6-methyladenine: the other methylated base of DNA. BioEssays: news and reviews in molecular, cellular and developmental biology. 2006;28:309–315. doi: 10.1002/bies.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu Y, et al. N6-methyldeoxyadenosine marks active transcription start sites in Chlamydomonas. Cell. 2015;161:879–892. doi: 10.1016/j.cell.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greer EL, et al. DNA Methylation on N6-Adenine in C. elegans. Cell. 2015;161:868–878. doi: 10.1016/j.cell.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang G, et al. N6-methyladenine DNA modification in Drosophila. Cell. 2015;161:893–906. doi: 10.1016/j.cell.2015.04.018. [DOI] [PubMed] [Google Scholar]
- 10.Liu J, et al. Abundant DNA 6mA methylation during early embryogenesis of zebrafish and pig. Nature communications. 2016;7:13052. doi: 10.1038/ncomms13052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu TP, et al. DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature. 2016;532:329–333. doi: 10.1038/nature17640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mondo SJ, et al. Widespread adenine N6-methylation of active genes in fungi. Nature genetics. 2017;49:964–968. doi: 10.1038/ng.3859. [DOI] [PubMed] [Google Scholar]
- 13.Vanyushin BF, Belozersky AN, Kokurina NA, Kadirova DX. 5-methylcytosine and 6-methylamino-purine in bacterialDNA. Nature. 1968;218:1066–1067. doi: 10.1038/2181066a0. [DOI] [PubMed] [Google Scholar]
- 14.Arber W, Linn S. DNA modification and restriction. Annual review of biochemistry. 1969;38:467–500. doi: 10.1146/annurev.bi.38.070169.002343. [DOI] [PubMed] [Google Scholar]
- 15.Collier J, McAdams HH, Shapiro LA. DNA methylation ratchet governs progression through a bacterial cell cycle. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:17111–17116. doi: 10.1073/pnas.0708112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Messer W, Noyer-Weidner M. Timing and targeting: the biological functions of Dam methylation in E. coli. Cell. 1988;54:735–737. doi: 10.1016/S0092-8674(88)90911-7. [DOI] [PubMed] [Google Scholar]
- 17.Lu M, Campbell JL, Boye E, Kleckner N. SeqA: a negative modulator of replication initiation in E. coli. Cell. 1994;77:413–426. doi: 10.1016/0092-8674(94)90156-2. [DOI] [PubMed] [Google Scholar]
- 18.Wion D, Casadesus J. N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nature reviews. Microbiology. 2006;4:183–192. doi: 10.1038/nrmicro1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo GZ, Blanco MA, Greer EL, He C, Shi Y. DNA N(6)-methyladenine: a new epigenetic mark in eukaryotes? Nature reviews. Molecular cell biology. 2015;16:705–710. doi: 10.1038/nrm4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 21.Lee WC, et al. The complete methylome of Helicobacter pylori UM032. BMC genomics. 2015;16:424. doi: 10.1186/s12864-015-1585-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang G, et al. Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nature biotechnology. 2012;30:1232–1239. doi: 10.1038/nbt.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang W, Sun Z, Menghe B, Zhang H. Short communication: Single molecule, real-time sequencing technology revealed species- and strain-specific methylation patterns of 2 Lactobacillus strains. Journal of dairy science. 2015;98:3020–3024. doi: 10.3168/jds.2014-9272. [DOI] [PubMed] [Google Scholar]
- 24.Zautner AE, et al. SMRT sequencing of the Campylobacter coli BfR-CA-9557 genome sequence reveals unique methylation motifs. BMC genomics. 2015;16:1088. doi: 10.1186/s12864-015-2317-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seong HJ, et al. Methylome Analysis of Two Xanthomonas spp. Using Single-Molecule Real-Time Sequencing. The plant pathologyjournal. 2016;32:500–507. doi: 10.5423/PPJ.FT.10.2016.0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ryan RP, et al. Pathogenomics of Xanthomonas: understanding bacterium-plant interactions. Nature reviews. Microbiology. 2011;9:344–355. doi: 10.1038/nrmicro2558. [DOI] [PubMed] [Google Scholar]
- 27.Zou LF, et al. Elucidation of the hrp clusters of Xanthomonas oryzae pv. oryzicola that control the hypersensitive response in nonhost tobacco and pathogenicity in susceptible host rice. Applied and environmental microbiology. 2006;72:6212–6224. doi: 10.1128/AEM.00511-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilkins KE, Booher NJ, Wang L, Bogdanove AJ. TAL effectors and activation of predicted host targets distinguish Asian from African strains of the rice pathogen Xanthomonas oryzae pv. oryzicola while strict conservation suggests universal importance of five TAL effectors. Frontiers in plant science. 2015;6:536. doi: 10.3389/fpls.2015.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome research. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic acids research. 2009;37:W202–208,. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong T, et al. Selective detection of N6-methyladenine in DNA via metal ion-mediated replication and rolling circle amplification. Chemical science. 2017;8:200–205. doi: 10.1039/c6sc02271e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, et al. The HD-GYP domain protein RpfG of Xanthomonas oryzae pv. oryzicola regulates synthesis of extracellular polysaccharides that contribute to biofilm formation and virulence on rice. PloS one. 2013;8:e59428. doi: 10.1371/journal.pone.0059428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Xiao Y, Zou L, Zou H, Chen G. Identification of HrpX regulon genes in Xanthomonas oryzae pv. oryzicola using a GFP visualization technique. Archives of microbiology. 2012;194:281–291. doi: 10.1007/s00203-011-0758-x. [DOI] [PubMed] [Google Scholar]
- 34.He YW, Wu J, Cha JS, Zhang LH. Rice bacterial blight pathogen Xanthomonas oryzae pv. oryzae produces multiple DSF-family signals in regulation of virulence factor production. BMC microbiology. 2010;10:187. doi: 10.1186/1471-2180-10-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ikawa Yumi, Tsuge Seiji. The quantitative regulation of thehrpregulator HrpX is involved in sugar-source-dependenthrpgene expression inXanthomonas oryzaepv.oryzae. FEMS Microbiology Letters. 2016;363(10):fnw071. doi: 10.1093/femsle/fnw071. [DOI] [PubMed] [Google Scholar]
- 36.Bertani G, Weigle JJ. Host controlled variation in bacterial viruses. Journal of bacteriology. 1953;65:113–121. doi: 10.1128/jb.65.2.113-121.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luria SE, Human ML. A nonhereditary, host-induced variation of bacterial viruses. Journal of bacteriology. 1952;64:557–569. doi: 10.1128/jb.64.4.557-569.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.