Abstract
The S100 protein family contains 20 functionally expressed members, which are commonly dysregulated in cancer. Their wide range of functions includes cell proliferation, cell differentiation, regulation of transcription factors, inflammation, chemotaxis, and angiogenesis. S100 proteins have in several types of cancer proven to be biomarkers for disease progression and prognosis. Acute myeloid leukemia (AML) is a highly heterogeneous and aggressive disease in which immature myeloblasts replace normal hematopoietic cells in the bone marrow. This review focuses on the S100 protein family members, which commonly are dysregulated in AML, and on the consequences of their dysregulation in the disorder. Like in other cancers, it appears as if S100 proteins are potential biomarkers for leukemogenesis. Furthermore, several S100 members seem to be involved in maintaining the leukemic phenotype. For these reasons, specific S100 proteins might serve as prognostic biomarkers, especially in the patient subset with intermediate/undetermined risk, and as potential targets for patient-adjusted therapy. Because the question of the most suitable candidate S100 biomarkers in AML still is under discussion, because particular AML subgroups lead to specific S100 signatures, and because downstream effects and the significance of co-expression of potential S100 binding partners in AML are not fully elucidated yet, we conclude that a panel of S100 proteins will probably be best suited for prognostic purposes.
Introduction
Acute myeloid leukemia (AML) comprises a biologically and genetically heterogeneous group of disorders characterized by an accumulation of immature myeloblasts in the bone marrow [1], [2]. These blasts proliferate rapidly and have a block in differentiation and increased resistance towards apoptosis; as a result, they outgrow normal hematopoietic cells. The median age at diagnosis is about 65 years, and in most cases, AML occurs de novo; but AML can also be treatment-related or be secondary to myelodysplastic syndromes (MDSs) or myeloproliferative neoplasia [3]. The backbone of AML treatment for younger/fit patients is induction therapy with cytarabine in combination with an anthracyclin, followed by consolidation therapy [4]. Due to a high rate of relapse, the overall 5-year survival is below 50% also for those patients who can receive the most intensive treatment; for the remaining patients, the prognosis is even worse [4], [5].
AML is highly heterogeneous with regard to cell morphology, cytogenetics, and gene mutations. More mature blasts are characterized by their loss of the stem cell marker CD34 [6]. Morphologically, AML has been divided into eight distinct groups in the FAB system (FAB M0-M7), where the cells are categorized as showing no/minimal signs of differentiation (FAB M0/M1) or presenting a more mature phenotype (FAB M5-7) [7]. Out of these groups, only FAB M3 — acute promyelocytic leukemia (APL) — entails its own treatment regimen [8] and is now considered by hematologists as a distinct disorder rather than an AML subtype. Furthermore, 50%-60% of AML patients carry chromosomal alterations, which are divided into low/favorable, intermediate, and high/adverse risk [2], [8]. Finally, more than half of the patients show gene mutations in their leukemic cells, with FMS-related tyrosine kinase 3-internal tandem duplications (Flt3-ITD, negative prognosis) and nucleophosmin insertions (NPM1-ins, positive prognosis in absence of Flt3-ITD) being the most prominent [4], [9]. In recent years, mutations in isocitrate dehydrogenases 1 and 2 (IDH1/2), which are present in about 20% of AML patients, have been particularly in the focus of research (e.g., [10], [11]).
Due to its aggressiveness and still abysmal outcome as compared to, for instance, childhood acute lymphoblastic leukemia (ALL), there is a need for more patient-adjusted therapy than standard induction and consolidation treatment in AML. Furthermore, because AML is highly heterogeneous, identifying predictive biomarkers and/or targets that are common across AML subgroups would be of great value. S100 proteins might have the potential to predict prognosis in, for instance, the patients who fall in the relatively large AML category with intermediate risk or to aid as suitable targets for tailored antileukemic treatment.
The S100 Protein Family
Structure and Function
The S100 protein family is restricted to vertebrates and comprises 20 functionally expressed proteins. Additionally, the related S100 fused type protein (SFTP) family contains seven members. The S100A subgroup (S100A1-S100A16, where S100A15 has been renamed into S100A7A as it proved to be a paralog of S100A7 [12]), together with the SFTP family, is encoded on chromosome 1q21 in the so-called epidermal differentiation complex, a hotspot for genomic rearrangements [13], [14]. The remaining four members are spread throughout the genome: S100B is encoded on chromosome 21; S100G is encoded on the X chromosome; S100P is encoded on chromosome 4; and, finally, S100Z is located to chromosome 5 [15].
S100 proteins exist as monomers, homo- and heterodimers, and multimers; the various forms employ distinct functions, e.g., extracellular functions appear to be conducted by oligomers [16]. The protein family, with exception of S100A10 [15], binds Ca2+ — and some members additionally can bind Zn2+, Cu2+, and Mn2+ [17] — when high levels of calcium are available, which is in contrast to constitutively expressed Ca2+-binding proteins like calmodulin [18]. Each S100 protein contains two Ca2+-binding helix-loop-helix motifs, so-called EF-hands [19]. The C-terminal, canonical EF-hand contains 12 amino acids and has a 10-50 times higher Ca2+-binding affinity than the 14–amino acid containing N-terminal, S100-specific pseudo–EF-hand [20]. Between these two EF-hands is a hinge spanning 10-12 amino acids [20], which is replaced by multiple tandem repeats in the SFTP family [21]. Upon binding of Ca2+, the proteins undergo conformational changes, which expose hydrophobic amino acids in the first helix of the C-terminal EF-hand and in the hinge region, which in succession interact with hydrophobic patches of the target protein [13], [22]. The affinity towards calcium is increased up to 300 times during this process [23], [24]. Since the hydrophobic interaction is essential for ligand recognition, the hydrophobic patches of S100 proteins are the regions with the lowest sequence similarities among its members [20].
Most S100 proteins are Ca2+ sensors and will bind targets, and thus translate the changes in intracellular calcium concentrations into response, after influx of Ca2+ through voltage-gated or receptor-mediated channels [22]. Others, like S100G [15], are Ca2+ buffers and control calcium homeostasis, but the boundaries between these two groups appear to be more blurred than initially assumed [22].
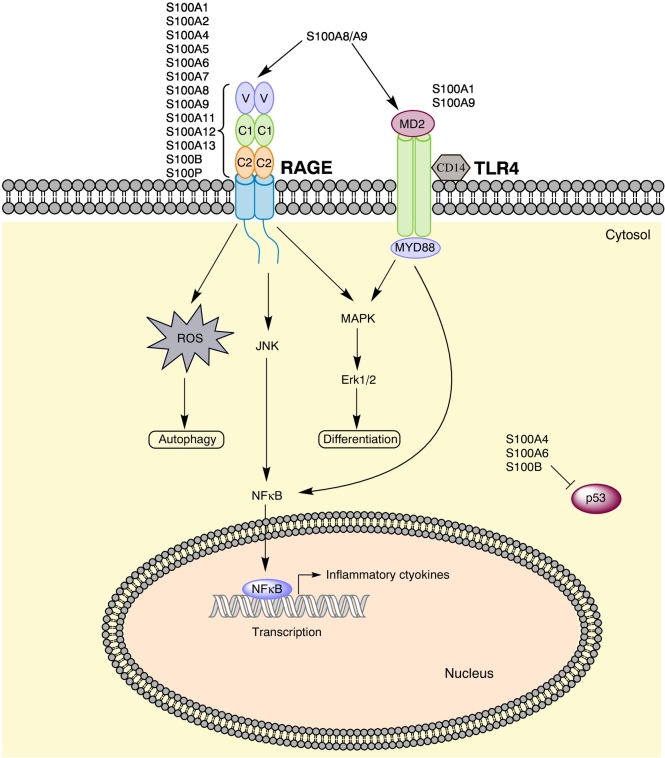
S100 proteins have a wide variation of functions; the reasons for that are: 1) tissue-, cell-, and time-dependent expression [13], [22]; 2) dimerization and oligomerization with different partners [16]; 3) binding of different metal ions and with varying affinities [16]; 4) posttranslational modifications [13]; and 5) employment of intracellular functions, extracellular functions, or both [22]. Among important intracellular functions are the regulation of cell proliferation, cell differentiation, and apoptosis [15], in addition to targeting enzymes, subunits of the cytoskeleton like F-actin and myosin, nucleic acids, and transcription factors [16], [25]. Outside the cells, the S100 family interacts with a variety of receptors, with the most important being G-protein–coupled receptors, scavenger receptors such as CD36 [15], toll-like receptor 4 (TLR4) [26], and the S100 general receptor for advanced glycation end products (RAGE) [16]. The latter, in turn, induces the formation of reactive oxygen species (ROS) and activates MAPK pathways, NFκB and Stat3, the PI3K-Akt-mTOR pathway, and small GTPases [16], [27]. S100 proteins also act as cytokines or damage-associated molecular patterns (DAMPs or alarmins) themselves [28] as they are involved in processes such as induction of cytokines and growth factors, activation of both the innate and adaptive immune system, chemotaxis and cell migration, and tissue development and repair [15], [25]. Figure 1 highlights the S100 downstream signaling pathways with most impact on tumorigenesis but also anticancerous effects such as cell differentiation.
Figure 1.
Important S100 signaling pathways. Most S100 proteins signal via the RAGE receptor, whereas a small selection also signals via TLR4. The pathways sum up the most important downstream signaling effects of S100A8 and S100A9, the most thoroughly studied S100 proteins. S100A8 can induce autophagy via RAGE, while S100A9 may induce cell differentiation via TLR4/Erk-signaling. Additionally, many S100 members can bind p53; the figure highlights the three proteins that are recognized inhibitors of the tumor suppressor.
With regard to all these diverse functions, of which many are essential in cancer development and progression, and the fact that S100 proteins are prone to genetic rearrangements at 1q21, it may not come as a surprise that S100 proteins frequently are dysregulated in cancer [13].
S100 Proteins in Tumorigenesis
There is now consensus that cancer results from the interaction between tumor cells (the seed) and the microenvironment (the soil). This interaction drives processes necessary for cancer progression, such as tumor cell growth, angiogenesis, and tumor invasion. Cross talk between tumor cells and stromal elements, e.g., through cytokine secretion, may also stimulate cancer cell growth but in addition remodels the stromal niche towards a tumor-promoting environment [29]. Furthermore, tumor cells can attract immune cells, e.g., leukocytes, which secrete growth factors and cytokines that either might support the tumor niche or inhibit functionally competent immune cells [29]. The processes involved in metastasis are similar to those of primary tumors, and both are often preceded by chronic inflammation [29]. Regarding AML, one may use the term “niche-driven oncogenesis” as remodeling of the mesenchymal niche can induce cytogenetic alterations in hematopoietic stem cells (HSCs), eventually resulting in AML [30].
S100 proteins contribute to tumorigenesis and metastasis in various ways. Members of the S100 family can provide a local inflammatory environment for cancer development and progression [20]. S100B upregulation in malignant gliomas, for instance, induces CCL2 expression and subsequently enhances infiltration of tumor-associated macrophages, which are important components in inflammation [31]. As soluble mediators, S100 proteins themselves are involved in the cross talk between tumor cells and the stroma. They can also function as chemoattractants. An example for this behavior is S100A4: tumor cells induce the secretion of this mediator from stromal fibroblasts [28]. S100A4 then modifies the tumor microenvironment through its function as chemoattractant which leads to macrophage and T-cell infiltration into the tumor niche [29]. Furthermore, S100 proteins can exhibit their tumorigenic effect through receptor binding and downstream signaling [29]. The RAGE receptor for instance is expressed on many cells in the tumor environment, e.g., endothelial cells and fibroblasts [27].
Dysregulation of S100 proteins is a common feature in cancer, and in most cancers, the protein expression is upregulated [13]. It appears as if every type of cancer has its specific S100 profile, and the latter may also vary among different stages and subtypes within a malignancy [13]. S100 profiling for cancer is further complicated by the fact that S100 protein overexpression also is associated with various noncancerous conditions, such as cardiovascular, neurological, and inflammatory diseases [13]. In some cancers, individual S100 proteins even appear to act as tumor suppressors [32], [33].
In recent years, reviews on the role of the S100 protein family in various types of cancers, e.g., breast [34], [35], pancreatic [27], colorectal [22], and lung cancer [36], have been published. This review aims to address the role of S100 proteins in hematological malignancies, with a special focus on AML.
S100 Proteins in AML
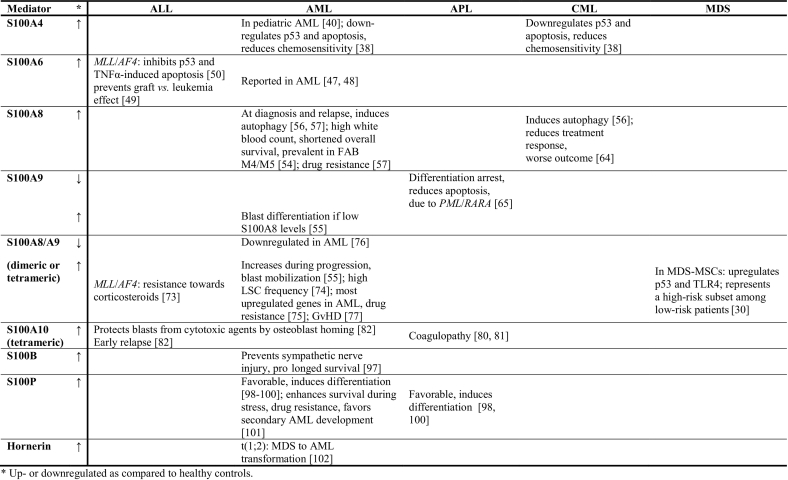
This chapter focuses on those members of the S100 and SFTP families which have been studied in AML and in three related malignancies: 1) chronic myeloid leukemia (CML) which in blast crisis resembles AML; 2) ALL, the other common form of acute leukemia; and 3) MDS, which resembles a preleukemic state where approximately 30% of the patients progress to AML. It is likely that S100 protein features in these malignancies will be comparable to the effects in AML. We will also include the effect of S100 protein expression on stromal cells, which interact with leukemic cells in the bone marrow niche. The most prominent effects on different types of leukemia are summarized in Table 1.
Table 1.
Observed Effects of S100 Protein Family Member Up- or Downregulation in Leukemias
S100A2
The effect of this mediator has, to our knowledge, not been studied in leukemia. However, it is an intriguing protein as it has been described both as tumor suppressor and as initiator of tumorigenesis and metastasis dependent on the cancer type. Its function as chemoattractant and its contradictory role as either inhibitor or inducer of p53 depending on the studied system [20], [37] might also be of importance in hematological malignancies.
S100A4
The protein was originally named “metastatin,” highlighting its importance in metastasis. S100A4 is infamous in cancer, as its expression is widely accepted to be a marker for poor prognosis [15], [20], [29] and resistance towards treatment [27], [38]. It exerts its prometastatic function by being a negative regulator of p53, positive regulator of cell migration, and stimulator of angiogenesis [20], [29]. However, S100A4 appears to only have minor effect on tumor initiation and progression [39].
S100A4 has been studied in both AML and CML. An early study showed that S100A4 mRNA expression in 52 pediatric AML patients was three-fold higher than in healthy controls [40]. Another group proposed a possible mechanism for why the expression of the repressor of retinoic acid signaling (PRAME) is a positive prognostic factor in leukemias as opposed to solid tumors [41]. PRAME is overexpressed in, among others, AML [42], [43], while it is very low in normal CD34+ cells [40], and high expression is correlated with both increased overall and event-free survival [44], [45] and reduced risk for relapse [41]. High PRAME expression downregulates S100A4 and subsequently increases p53 activity and apoptosis in the AML cell line KG-1. Knockdown of PRAME, on the other hand, rescued S100A4, thus sequestering and disabling of p53 in K562 (CML cell line in blast crisis). It was also shown that S100A4 expression has influence on chemosensitivity in CML [38]: K562, which showed low constitutive expression of S100A4, was more chemosensitive towards 4-hydroperoxy-cyclophosphamide than KU812, which expresses higher S100A4 levels. Furthermore, the treatment itself increased S100A4 levels in the cell lines [38].
S100A4 might also exert an effect on leukemic cells through the bone marrow niche because the protein is expressed by the tumor stroma [13], [36]. Among the expressing cells are also mesenchymal stem cells (MSCs), and S100A4 might be involved in MSC differentiation and proliferation [39].
S100A6
This is another member of the S100 family whose upregulation is linked with cell proliferation and tumorigenesis [15], [20] in many cancers, but also low expression is occasionally observed [46]. One interesting mechanism is the relationship between S100A6 and the tumor suppressor p53. The latter indirectly regulates S100A6 expression, and it is therefore speculated that insufficient suppression by mutated p53 is a reason for S100A6 overexpression and increased proliferation in cancer. On the other hand, S100A6 appears to protect wild-type p53 from degradation and thus contribute to apoptosis [46].
In hematological malignancies, increased levels of the protein have been reported in AML [47], [48]. The effect of S100A6 expression has thoroughly been studied in acute leukemias with rearrangements in the mixed-lineage leukemia (MLL) gene [49], [50], [51], [52]. Chromosomal aberrations in 11q23 are common in acute leukemias and especially in infants and leukemia secondary to treatment with DNA topoisomerase II inhibitors [49], [51]. The prognosis of MLL rearrangements is highly dependent on the chromosomal partner, and more than 70 have been identified so far [49]. In their studies, Tamai and coworkers have concentrated on the MLL/AF4 fusion gene, t(4;11)(q21;q23), which is the most prevalent translocation in ALL, accounting for more than 50% of infant and about 7% of adult ALL. The prognosis for t(4;11) is poor, even following allogeneic stem cell transplantation (allo-SCT), and thus represents the largest challenge in childhood ALL [49], [50], [51]. In their studies, the group identified the various roles of S100A6 in the malignancy. First, t(4;11) requires a “second hit” to initiate leukemia; especially mutations in the Flt3 tyrosine kinase domain (Flt3-TKD) are potent second hits. Compared to only t(4;11), S100A6 expression is increased 13-fold when t(4;11) is combined with Flt3-KTD. Knockdown of S100A6 on the other hand inhibited leukemic proliferation [52]. Second, high S100A6 expression can inhibit TNFα-induced apoptosis through its inhibition of p53 acetylation and subsequent repressed upregulation of the caspase 8-caspase 3 apoptotic pathway. This effect was exclusively observed in cell lines with t(4;11). The results were confirmed in mouse models of t(4;11) lymphoma [50]. Third, the S100A6-targeting drug Amlexanox rescued p53-caspase 3 initiated apoptosis upon TNFα treatment in t(4;11) cell lines. In mice, the inhibitor could counteract leukemic infiltration in t(4;11) B-cell leukemia and inhibited the upregulation of S100A6 [51]. Finally, knockdown of S100A6 in murine models showed to induce graft versus leukemia (GvL) effect after allo-SCT which is mainly mediated by TNFα; thus, the absence of the GvL effect might explain why prognosis is poor in t(4;11) ALL even following allo-SCT [49]. Inhibition of S100A6 might therefore be a promising strategy for patients with MLL genetic rearrangements or high constitutive S100A6 levels. However, S100A6 is almost exclusively expressed in proliferating cells [22], [36]. Therefore, quiescent cells in the bone marrow might escape S100A6 inhibition and ultimately lead to relapse.
S100A7/S100A7A
S100A7 is among the proteins that in some cancers are reported to be tumor suppressors, while they appear to be involved in metastasis in other cancers [20]. These two proteins may play a role in AML as they are chemotactic for granulocytes, monocytes, macrophages, and lymphocytes [15], [20] and induce the secretion of proinflammatory cytokines [28].
S100A8 and S100A9
Of the S100 protein family, these two members have been most thoroughly studied in AML. The homodimers are less abundant than the heterodimer S100A8/A9 due to lower stability of the former. However, the two individual proteins appear to play quite contradictory roles in AML; thus, this chapter will discuss S100A8, S100A9, and the heterodimer separately.
S100A8
S100A8 is a marker for myeloid cell differentiation as it is expressed by normal CD34− bone marrow cells but not by immature myelocytes/blasts [53], [54]. At proinflammatory conditions or during oxidative stress, S100A8 can be induced in macrophages, dendritic cells (DCs), microvascular endothelial cells, and fibroblasts [15]. In general, S100A8 appears to play a role in leukemogenesis as a regulator of myelopoiesis, where the protein contributes to maintaining the undifferentiated phenotype [55].
In both childhood and adult AML studies (31 and 189 patients, respectively), S100A8 bone marrow mRNA levels were significantly higher at diagnosis and relapse as compared to healthy controls or patients in complete remission and, thus, indicated the clinical status of the disease [56], [57].
The role of S100A8 has been studied by proteomics in childhood AML with a normal or rare/noninformative karyotype, defined as intermediate risk. High S100A8 levels correlated with white blood count and shorter overall survival [54]. In a larger patient cohort, S100A8 levels were also linked with cells with morphological signs of differentiation as patients classified as FAB M4/M5 (monocytic differentiation) had significantly higher levels than FAB M0/M1 (no/minimal differentiation) [57]. Further, overall survival in patients with a normal karyotype resembled that of adverse or favorable cytogenetics depending on high or low S100A8 mRNA expression levels, respectively. Finally, HL-60 cells with low initial S100A8 levels showed increased expression after treatment with etoposide in a dose-dependent matter, which interfered with the effect of the drug [57].
This drug-resistant effect of S100A8 appears to be caused by autophagy — the metabolic process in which intracellular aggregates, misfolded proteins, or damaged organelles are degraded by lysosomes in response to cellular stress [58]. Autophagy is recognized as important for therapy-related resistance in hematological malignancies [59], [60], [61], [62] as it is induced by DNA-damaging agents, radiotherapy, and molecular targeting [56]. In supernatants from HL-60 and K562, S100A8 was increased after treatment with vincristine, adrenomedullin, and As2O3. When S100A8 expression was inhibited, chemosensitivity and apoptosis were increased, while autophagy was reduced. The authors showed that S100A8-induced ROS production is essential for autophagy in AML (Figure 1) through disruption of the interaction between Bcl-2 and Beclin1 [56]. In a follow-up study, S100A8 mRNA overexpression in leukemic cell lines indicated to be linked with drug resistance and increased basal autophagy [63].
Finally, S100A8 appears to be negatively correlated with both treatment and sustained long-term major molecular response also in CML in a study with 37 patients and might therefore be a marker for aggressiveness, i.e., progression towards blast crisis [64].
S100A9
Like S100A8, also S100A9 is a marker of myeloid differentiation and is increased in granulocytes and monocytes [65], [66]. But unlike S100A8 which signals via RAGE, S100A9 mainly signals via TLR4 [15] and can also associate with CD147/basigin [67], [68]. The receptors lead to the induction of proinflammatory cytokines such as IL-6, CXCL8, and TNFα, and matrix metalloproteinases [13], [15], [25]. S100A9 is widely acknowledged to be involved in tumorigenesis, as it is chemotoactic for leukocytes and inhibits differentiation of macrophages and DCs [28], [69]. However, recent studies show divergent effects for S100A9 in AML.
In APL, the FAB M3 AML subtype, S100A9 shows the lowest gene transcribes among all FAB classes [65]. S100A9 levels are inversely correlated with expression levels of PML/RARA, the fusion gene, which defines APL. Upon treatment with all-trans retinoic acid (ATRA) and, to a lesser degree, with As2O3, S100A9 expression levels increase. Furthermore, higher S100A9 mRNA levels correlated with higher expression of PU.1, an important transcription factor for myeloid differentiation [65]. The results were confirmed in the APL cell line NB4, where higher levels of S100A9 induced apoptosis through reduced Bcl-2 levels and cleavage of caspase 3, and leukemic cell growth suppression. Thus, at least in APL, higher S100A9 levels are linked with myeloid differentiation, leukemia growth suppression, and increased treatment response [65].
Also in non-APL AML, high S100A9 levels were correlated with myeloid differentiation, but this ability was dependent on S100A8 expression levels [55]. S100A9 had to exceed S100A8 concentrations by at least 10-fold; otherwise, S100A8 blocked differentiation. The authors argue that the differentiation potential is mediated by TLR4 because the downstream kinases Erk1/2 and JNK are known to be important for granulocyte and monocyte differentiation [55]. However, treatment with exogenous S100A9 induced leukemia cell differentiation only in FAB M4/M5 patients, which according to mRNA data also showed the highest basal levels of S100A8, S100A9, and TLR4 [55]. Thus, the S100A9-TLR4-MAPK/Erk axis might be a potential target for overcoming differentiation arrest in cells presenting a monocytic phenotype (Figure 1).
S100A8/A9
Since the mRNA levels of the individual partners of the heterodimer calprotectin appear to have opposing effects in AML, it might not come as a surprise that the studies of the heterodimer show conflicting results. First, S100A8/A9 is overexpressed in many cancers [55] and associated with inflammation as it is an activator of monocytes and macrophages, and leads to neutrophil infiltration [20]. Through the characteristics of its individual partners, S100A8/A9 can signal via both TLR4 and RAGE and promotes tumor development and invasiveness both by increasing cell growth, including leukemic cell proliferation, via RAGE and by excretion of proinflammatory cytokines via TLR4-NFκB [15]. Interestingly, S100A8/A9 is also associated with an anti-inflammatory effect, reduced cancer growth, and apoptosis. The tetradimer can induce the latter by sequestering Zn2+, which is antiapoptotic [70]. The tumor-promoting effects appear in most cancers to be valid only for low S100A8/A9 levels [71]; at high concentrations, several cells (macrophages, bone marrow cells, lymphocytes, and fibroblasts) are inhibited, and apoptosis is induced [22], [25], [72].
In the already mentioned MLL/AF4 ALL, S100A8/A9 overexpression was associated with failure to induce free cytostatic Ca2+ and with corticosteroid resistance [73]. In AML, S100A8/A9 mRNA levels increase during AML progression and show correlation with mobilization of leukemia cells from the bone marrow into peripheral blood [55]. Furthermore, S100A8 and S100A9 were the most upregulated genes in patients with the highest frequencies of leukemic stem cells [74], indicative of an adverse prognosis. A recent study showed that the mRNA levels of S100A8/A9 are the most overexpressed as compared to healthy controls [75]. High levels were linked with resistance towards the novel drugs quizartinib (Flt3 inhibitor) and especially venetoclax (Bcl-2 inhibitor), which resulted in the absence of free cytosolic Ca2+ and inhibition of apoptosis in cell line studies [75]. Another study came to opposite conclusions: S100A8 and especially S100A9 were among the most downregulated genes as compared with healthy controls [76]. However, the latter study focused on FAB M1/M2 (little or granulocytic differentiation) patients who present with lower S100A8/A9 basal levels than blasts with monocytic differentiation [55], and S100A9 could then be used to distinctively differentiate FAB M1 from FAB M2 [76]. Nevertheless, the authors were able to reproduce their results, i.e., consequently downregulated S100A8/A9 in AML, analyzing microarrays (GDS acquisitions: 1059, 2251, and 3057) deposited online [76].
S100A8/A9 and, to some extent, also S100A7 and S100A12 are furthermore among the DAMPs which are associated with development of graft versus host disease (GvHD) after allo-SCT, a potentially lethal complication in which donor T-cells attack host tissues. But it remains unclear whether elevated S100 protein levels in monocytes after allo-SCT initiate GvHD or whether their increase rather results from release from necrotic/apoptotic cells after establishment of GvHD [77], [78]. As a remark, these elevated S100A8/A9 levels were not detected in blood samples but in saliva and stool [77], [79].
The role of the DAMPs S100A8 and S100A9 has also been studied in the tumor environment of the preleukemic conditions Schwachman-Diamond syndrome (SDS) and MDS [30]. In SDS, downregulation of the affected Sbds gene in MSCs leads to upregulation of both p53 and S100A8/A9, with the subsequent activation of TLR4-MAPK/NKκB. S100A8/A9 upregulation was sufficient to induce genotoxic stress in HSCs [30], increasing the propensity for AML progression. Also in MDS, S100A8/A9 MSC expression was correlated with p53 and TLR4 upregulation. Furthermore, in a cohort consisting of low-risk MDS patients, high S100A8/A9 levels resulted in approximately doubled (corresponding to almost 30%) risk for AML transformation, as well as the time interval for AML development was significantly reduced [30]. It appeared as if the MSC expression level of S100A8/A9 in these syndromes was more important than that of myeloid cells. In addition, the protein expression was observed in a specific subset of CD271+ MSCs, which in the bone marrow are in direct contact with CD34+ HSCs; thus, spatial proximity to the source of S100A8/A9 seems to be of importance [30].
S100A10
This particular S100 family member has mutations in its EF-hands rendering it Ca2+-independent and thus permanently activated [15], [17]. It exerts its main biological function in a heterotetrameric complex together with annexin 2 [17], [20], [80].
In AML, S100A10 is associated with coagulopathy in FAB M3 patients [80], [81]. Approximately 5% of APL patients die early due to severe bleedings, and up to 90% of patients suffer from coagulopathy at diagnosis [81]. The S100A10/annexin 2 heterotetramer is elevated in APL cells; at the cell surface, the tetramer increases the affinities of urokinase and especially tissue plasminogen activators (uPa and tPa) for their substrate plasminogen, which thereafter is converted to active plasmin. The latter leads to hyperfibrinolysis, i.e., plasmin-mediated degradation of the extracellular matrix, which in turn can cause extensive bleeding [80], [81]. As much as 90% of the observed plasmin concentration seems to be due to S100A10 [80]. The fusion gene PML/RARA induces increased levels of annexin 2 but not of S100A10. However, the concentrations of both proteins increase, indicating posttranslational regulation of S100A10 by elevated annexin 2 levels [81]. Depletion of S100A10 or downregulation of PML/RARA by ATRA in NB4 cells had approximately the same effect and reduced both plasmin formation and fibrinolysis [80]. In addition, in nine patients with de novo APL, either treatment with ATRA or with an S100A10 antibody before APL therapy reduced blast invasiveness [81]. Thus, downregulation of S100A10 might be the reason why the risk of death from severe bleeding immediately declines after the first dose of ATRA in APL patients.
S100A10 further seems to play a role in protecting acute leukemia cells from cytotoxic agents. Annexin 2 leads to homing to the bone marrow and adhesion of HSCs to osteoblasts [82]. The latter are known to protect primary ALL and AML blasts from chemotherapy [83]. In this respect, elevated S100A10 mRNA levels were identified in childhood B-cell ALL, and increased levels also correlated with early relapse [82]. Thus, inhibition of annexin 2, S100A10, or the formation of the heterotetramer might be a means of overcoming treatment resistance and early relapse in a patient subset.
S100A12
S100A12 is expressed by a variety of myeloid cells and induces chemotaxis of leukocytes, but not lymphocytes, after binding to RAGE [20], [84]. It is associated with chronic inflammation through sustained recruitment of monocytes and neutrophils and might therefore also play a role in hematological malignancies. S100A12 expression can be induced by lipopolysaccharide (LPS) and TNFα, and — when it acts as a cytokine — its effects are comparable to those of CCL2, CCL3, CCL4, and CCL5 [84], i.e., cytokines that commonly are increased in AML and of which CCL5 has been linked with cancer progression in, e.g., lymphoma [85].
S100A13
This protein has not been extensively studied in leukemia. However, the S100A6 inhibitor Amlexanox additionally targets S100A13 [51]. During stress, S100A13 induces the excretion of, among others, acidic fibroblast growth factor (aFGF) and contributes to cancer hallmarks as angiogenesis, cell differentiation, and tumor growth [15], [51]. Since Amlexanox inhibits the heat-shock protein (HSP)–induced release of S100A13 [86] and HSPs are commonly expressed in AML and their expression additionally increased when the blasts undergo apoptosis [87], [88], the downstream effects of S100A13 might contribute to leukemogenesis or to maintaining the disease.
S100A16
S100A16 might exert an indirect effect on AML. As with many other S100 proteins, also S100A16 is upregulated in several types of cancer [15]. It might play a role in the bone marrow niche because S100A16 inhibits osteogenic differentiation of MSCs in expense of adipogenesis [89]. Both MSCs and osteoblasts have shown to protect AML blasts from chemotherapy [83], [90], but also bone marrow adipocytes appear to exert effects on leukemic cells: adipocytes provide energy and have shown to protect both ALL and CML blasts from chemotherapy [91], [92], [93]. However, AML itself causes MSCs to differentiate towards osteoblasts rather than to adipocytes [94]. Thus, high S100A16 levels in the bone marrow niche might alter the cross talk between the MSC progeny and AML cells, but more research is necessary in order to uncover if this alteration will result in improved or worsened prognosis.
S100B
This S100 protein is mainly linked with neurological disorders and is for instance upregulated in brain metastasis [36]. However, it is also a metastatic marker in other cancers and is then one of the S100 proteins with contradictory prognostic impact depending on the cancer type. In breast cancer, high S100B levels are correlated with better outcome in the endocrine-resistant negative subtype [95], whereas increased posttreatment levels in the endocrine-resistant positive subtype are linked with reduced survival [96].
S100B is further expressed by DCs and lymphocytes and leads to increase in cell proliferation and migration, while inhibiting apoptosis and differentiation [15]. In AML, S100B expression in bone marrow neural tissues seems to be of importance because sympathetic nerve injury is closely related to hematological diseases [97]. S100B is a marker for nonmyelated Schwann cells, which are part of the healthy bone marrow niche, where they facilitate HSC hibernation. Damage of these cells can lead to AML progression. In concurrence with that, elevated S100B mRNA levels in AML showed a correlation trend towards prolonged patient survival [97].
S100P
As many other S100 proteins, also high S100P levels are associated with cell growth, carcinogenesis, and metastasis [20], [22], [27]. However, like for S100B, elevated S100P levels appear to be favorable in AML.
S100P is transiently expressed during early leukocyte differentiation and is probably essential in this process [98]. In agreement with that, S100P shows low expression in K562 [38] and in AML blasts, whereas high levels are connected with a favorable prognosis [99]. S100P has mainly been studied in association with cytokinins in the treatment of AML. Cytokinins are purine derivatives that act as hormones in plants but which also exert antiproliferative effects in cancer cells and induce granulocyte differentiation in myeloid leukemias through activation of MAPK [98]. Different kinds of cytokinins and their derivatives (isopentenyladenosine, cotylenin A, methyljasmonate, and 4,5-didehydrojasmonate) had in cell line studies (AML: HL-60, THP-1; APL: NB4; lymphoma: U937) approximately the same effects with regard to granulocytic/monocytic differentiation as ATRA and vitamin D3 [98], [100]. But the similar effects were mediated through different mechanisms, as S100P was the most upregulated gene only after treatment with cytokinins [98], [100]. Thus, induction of S100P expression might be a possible means to counteract the differentiation arrest in AML.
On the other hand, S100P has been proposed to play a role in the cellular defense mechanism and to enhance survival under stress [101]. Treatment-related MDS — and further transformation to AML — is seen after cytotoxic therapy with bifunctional alkylating agents that form DNA interstrand cross-links. When HL-60 was exposed to this type of alkylating agents at a low dosage, thus facilitating recovery of cell growth, S100P showed the highest increase in mRNA levels [101]. The elevated levels led to cell cycle arrest at the G2/M transition point, and it took about 10 days to resume cell growth. Furthermore, the cells were then resistant to reexposure of the drug [101]. Thus, S100P might be involved in processes leading to survival of cells with DNA interstrand cross-links, which later on might initiate hematological malignancies. However, it must be pointed out that not all tested cell lines responded in the same way as HL-60 [101]. Hence, dosage of the alkylating agents appears to be of major importance.
S100Z
S100Z is downregulated in several cancers [15]. It interacts with S100P and is, together with its partner, highly upregulated by alkylating agents, with the highest levels reported in leukocytes and spleen [101]. S100Z might therefore play a role in the S100P-mediated effects on cell survival during stress.
Hornerin
Of the seven SFTPs, hornerin is the one which to the highest degree appears to be involved in other processes than epidermal maturation and skin/mucosa cancers.
The relation between hornerin and AML was demonstrated in a case study. This particular patient presented with the rare chromosomal alteration t(1;2)(q21;q37) and had recently progressed from MDS [102]. Thus, the translocation was situated at 1q21 where most S100 proteins and the SFTP family are encoded. Chromosomal alterations, including duplications and translocations in 1q21-25, which result in gene degeneration or fusion genes that interfere with normal proliferation and differentiation, are frequently detected in hematological malignancies. In this case, the translocation did not interrupt the hornerin open reading frame but resulted in its aberrant expression in bone marrow cells. Additional 5 out of 90 patients suffering from hematological diseases showed elevated hornerin mRNA levels [102]. The authors speculate 1) that S100 fusion genes may be involved in neoplasia, 2) that translocations have an effect similar to activation of an oncogene, and 3) that hornerin activation was responsible for disease progression for the t(1;2) patient [102]. The latter notion is supported by studies on breast and liver cancer, where hornerin expression levels increased from the preinvasive phase to the invasive carcinoma [21] or were correlated with poor outcome in hepatocellular carcinoma [103], respectively.
S100 Proteins as Biomarkers in AML: Potential and Pitfalls
Like in many other cancers as well as noncancerous conditions, the expression of S100 proteins appears to be altered in AML. Overexpression of S100A4, A6, A8, A9, A10, and A12 and downregulation of S100P seem to maintain the leukemic phenotype. More extensive studies will have to be conducted in order to determine whether one single member of this family or rather a panel of them will be best to predict factors such as prognosis, disease progression, and treatment response.
At least two caveats arise from this review. First, S100 protein expression levels have been determined with different techniques, and the latter were conducted on samples obtained from various sources. Gene expression, both by microarray and PCR techniques, has been measured in both peripheral blood blasts and bone marrow cells. Furthermore, protein expression was detected using Western blotting, immunostaining, ELISA, flow cytometry, or proteomic techniques, and samples comprised peripheral blood cells, bone marrow cells, primary cell culture supernatants, serum, plasma, saliva, and stool. Protein expression is regulated both posttranscriptionally and posttranslationally, and proteins are further prone to modifications which alter their function. Even though there is a correlation between mRNA and protein levels, this connection is not strong, and about 60% of protein concentrations are not reflected by the mRNA levels [104], [105]. An example is the deviation between S100A10 gene versus protein expression levels [81]; thus, one should be careful when comparing protein with gene expression levels. Also, sample source can influence expression levels. A recent example is osteopontin: high serum/plasma osteopontin levels are a recognized negative prognostic marker in AML [106], [107]; however, osteopontin seems to be upregulated in cell culture supernatants of patients with prolonged survival [108]. Therefore, when implementing S100 proteins as biological markers in AML, sample source and selected method will have to be standardized. Second, the expression levels of several of the S100 proteins are correlated with cell maturation/differentiation. Particularly, S100A8 and A9 are associated with FAB M4/M5. Thus, at some occasions, upregulated S100 proteins will not be correlated with pathology but simply reflect the AML phenotype.
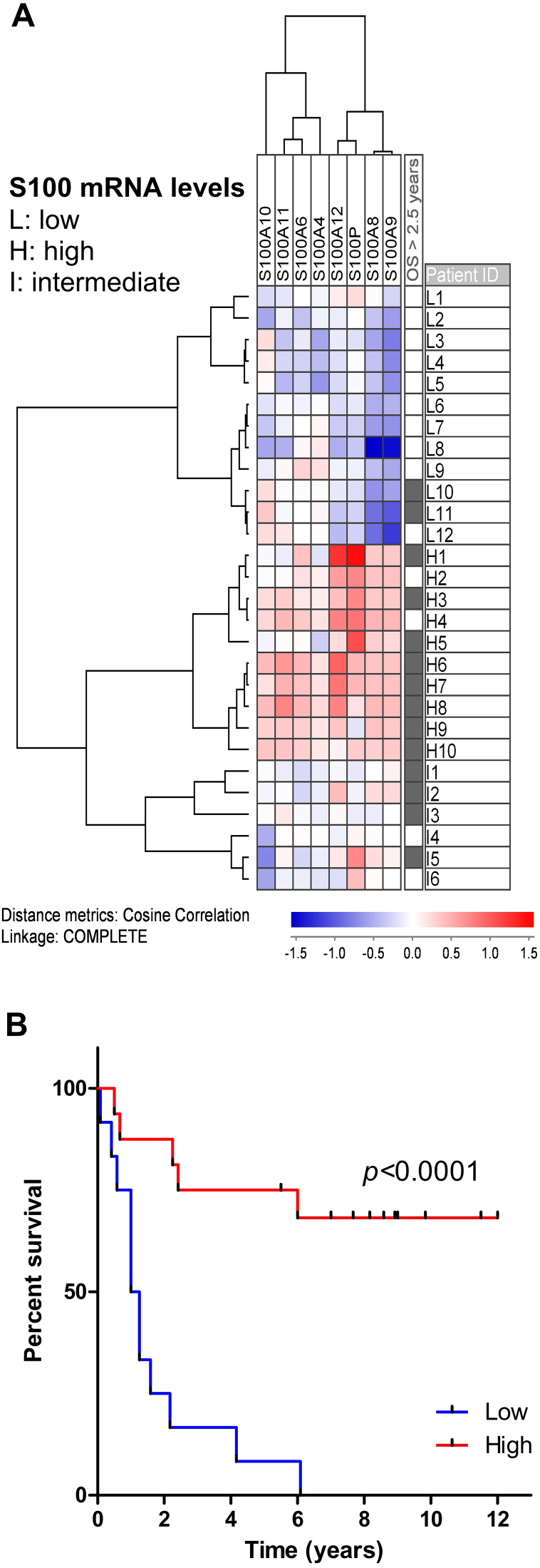
In a recent study, we observed that mRNA levels for S100A6, A8, A9, and A11 were upregulated in AML patients with high cell culture supernatant concentrations of specific proteases and protease inhibitors, including CD147/basigin — a S100A9 receptor — and the S100A10-related uPa [109]. Like S100A9, basigin was found to be overexpressed in FAB M4/M5 patients and the more mature CD34− cell fraction and was additionally correlated with an upregulation of S100A4 and the S100A9 receptor TLR4 [109]. Interestingly, there was a large overlap between the patients with high cellular expression of proteases and those with a high cytokine excretion profile, where the latter had been linked with improved survival [110]. Therefore, we analyzed whether S100 protein mRNA expression also was correlated with survival. Even though only 28 of the patients from the microarray study qualified for inclusion in the analysis, the 12 patients with low S100 protein mRNA levels (Figure 2A; previously unpublished data) had a significantly (P < .0001, log-rank test) shorter survival compared to the patients with higher levels (Figure 2B). Eight of the S100 proteins were differentially expressed among patients; of these, low levels of S100A8 and A9 seem to correlate with an adverse phenotype, whereas high levels of S100A12 and S100P appear to be linked with a more favorable phenotype (Figure 2A).
Figure 2.
High S100 protein expression is associated with increased patient survival. (A) Only patients who received intensive induction therapy were included in the cluster; the mRNA levels were median normalized and log(10) transformed prior to unsupervised clustering using the program JExpress. Patients could be divided into two main subgroups according to low (patients L1-L12) and high (patients H1-H10)/intermediate (patients I1-I6) mRNA expression levels. The upper/low expression subgroup showed especially low (blue color) S100A8/A9 expression, whereas the group below, characterized by high expression, showed especially elevated (red color) levels for S100A12 and S100P. Patients with long-term survival (>2.5 years) are indicated to the right. The S100A4 and A10 values represent the mean value of two probes for these genes. B) Kaplan-Meier (calculated by SPSS version 25) comparison of patients with low S100 protein expression (L1-L12) versus patients with median or high levels, i.e., the two subgroups obtained in the cluster. In this patient cohort, elevated levels of S100 proteins are correlated with prolonged patient survival (log-rank test). Because the plot takes into account all eight differently expressed S100 proteins, the improved patient survival might be due to the impact of a single S100 member, to the whole panel of these eight proteins, or to a group of co-expressed genes, which remain to be identified yet.
To summarize, it might be advisable to use a panel of S100 proteins as biomarkers instead of single members for several reasons. First, a distinctive S100 protein profile is, in many cancers, stage and subtype specific. Furthermore, the profile at diagnosis at its best can provide information about prognosis and/or treatment options, whereas monitoring of the protein profile can yield insight into treatment response [13]. Second, even though there exist strong candidates for single biomarkers, such as S100A4, A8, A9, and S100P, the expression of single proteins is prone to bias due to specific AML subgroups or the simultaneous presence of an AML-unrelated malignancy, which both can lead to dysregulation of specific S100 members. Third, sampling of a protein profile also takes into account that S100 proteins may be co-expressed. Remarkably, even though most of the S100 family members cluster together and have a high degree of sequence similarity, the expression of S100 proteins is commonly not synchronized [111]. On the contrary, there is evidence that S100 protein dysregulation during cancer is regulated by promoter hypo- or hypermethylation [111]. In this respect, co-expression of S100A14 and A16 has been observed in breast cancer and resulted in increased tumor invasion and thus worsened prognosis [112]. Furthermore, nuclear co-expression of S100A4 and p53 was seen in colorectal cancer. In combination with TP53 wt, the cells underwent apoptosis. The authors therefore speculated that S100A4 might select for cancer-promoting mutations in TP53 [113]. Finally, as long as the role of particular S100 members in AML remains unclear and not all aspects of their impact on other molecules or pathways, such as the above-mentioned p53 which frequently is dysregulated in AML [114], are resolved, a panel of biomarkers offers the best possibility to capture those candidate proteins, which define AML.
Conclusion
S100 proteins are frequently dysregulated in malignancies, including AML. More studies on the effect of S100 proteins in AML have to be undertaken in order to determine which members of the protein family have the highest impact on leukemogenesis and from which sample source gene and/or protein expression levels should be obtained. Furthermore, since disease heterogeneity apparently blurs some of the effects, which single mediators exert, it might be necessary to concentrate the studies, at least at first, on patient subsets sharing the same morphology (e.g., FAB classes), cytogenetic aberrations, or other common features.
Ethical Approval
The study was approved by the local Ethics Committee (Regional Ethics Committee III, University of Bergen), and peripheral blood samples were collected after written informed consent.
Authors’ Contributions
Annette K. Brenner wrote the manuscript and designed the figures. Øystein Bruserud designed the study and critically reviewed and edited the manuscript.
Footnotes
Funding: The study received financial support from the Norwegian Cancer Society (Grant Numbers 15752, 17813, 62370, 145007, and 145008), Helse-Vest (Grant Number 911788), and the University of Bergen.
Conflict of Interest: The authors declare no conflict of interests.
References
- 1.Diaz de la Guardia R, Lopez-Millan B, Lavoie JR, Bueno C, Castano J, Gomez-Casares M, Vives S, Palomo L, Juan M, Delgado J. Detailed characterization of mesenchymal stem/stromal cells from a large cohort of AML patients demonstrates a definitive link to treatment outcomes. Stem Cell Reports. 2017;8:1573–1586. doi: 10.1016/j.stemcr.2017.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Estey E, Döhner H. Acute myeloid leukaemia. Lancet. 2006;368:1894–1907. doi: 10.1016/S0140-6736(06)69780-8. [DOI] [PubMed] [Google Scholar]
- 3.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, Bloomfield CD, Cazzola M, Vardiman JW. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 4.Burnett A, Wetzler M, Loewenberg B. Therapeutic advances in acute myeloid leukemia. J Clin Oncol. 2011;29:487–494. doi: 10.1200/JCO.2010.30.1820. [DOI] [PubMed] [Google Scholar]
- 5.Stone RM, O'Donnell MR, Sekeres MA. Acute myeloid leukemia. Hematology Am Soc Hematol Educ Program. 2004:98–117. doi: 10.1182/asheducation-2004.1.98. [DOI] [PubMed] [Google Scholar]
- 6.Sidney LE, Branch MJ, Dunphy SE, Dua HS, Hopkinson A. Concise review: evidence for CD34 as a common marker for diverse progenitors. Stem Cells. 2014;32:1380–1389. doi: 10.1002/stem.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW. IARC Press; Lyon: 2008. WHO classification of tumours of haemotopoietic and lymphoid tissues. [Google Scholar]
- 8.Hasserjian RP. Acute myeloid leukemia: advances in diagnosis and classification. Int J Lab Hematol. 2013;35:358–366. doi: 10.1111/ijlh.12081. [DOI] [PubMed] [Google Scholar]
- 9.Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Buchner T, Dombret H, Ebert BL, Fenaux P, Larson RA. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DiNardo CD, Stein EM, de Botton S, Roboz GJ, Altman JK, Mims AS, Swords R, Collins RH, Mannis GN, Pollyea DA. Durable remissions with ivosidenib in IDH1-mutated relapsed or refractory AML. N Engl J Med. 2018;378:2386–2398. doi: 10.1056/NEJMoa1716984. [DOI] [PubMed] [Google Scholar]
- 11.Levis M. Targeting IDH: the next big thing in AML. Blood. 2013;122:2770–2771. doi: 10.1182/blood-2013-09-522441. [DOI] [PubMed] [Google Scholar]
- 12.Marenholz I, Lovering RC, Heizmann CW. An update of the S100 nomenclature. Biochim Biophys Acta. 2006;1763:1282–1283. doi: 10.1016/j.bbamcr.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 13.Bresnick AR, Weber DJ, Zimmer DB. S100 proteins in cancer. Nat Rev Cancer. 2015;15:96–109. doi: 10.1038/nrc3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenfeld JA, Traylor RN, Schaefer GB, McPherson EW, Ballif BC, Klopocki E, Mundlos S, Shaffer LG, Aylsworth AS. Proximal microdeletions and microduplications of 1q21.1 contribute to variable abnormal phenotypes. Eur J Human Genet. 2012;20:754–761. doi: 10.1038/ejhg.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donato R, Cannon BR, Sorci G, Riuzzi F, Hsu K, Weber DJ, Geczy CL. Functions of S100 proteins. Curr Mol Med. 2013;13:24–57. [PMC free article] [PubMed] [Google Scholar]
- 16.Kuberappa PH, Bagalad BS, Ananthaneni A, Kiresur MA, Srinivas GV. Certainty of S100 from physiology to pathology. J Clin Diagn Res. 2016;10:Ze10–15. doi: 10.7860/JCDR/2016/17949.8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hermann A, Donato R, Weiger TM, Chazin WJ. S100 calcium binding proteins and ion channels. Front Pharmacol. 2012;3:67. doi: 10.3389/fphar.2012.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saimi Y, Kung C. Calmodulin as an ion channel subunit. Annu Rev Physiol. 2002;64:289–311. doi: 10.1146/annurev.physiol.64.100301.111649. [DOI] [PubMed] [Google Scholar]
- 19.Heizmann CW. Intracellular calcium-binding proteins: structure and possible functions. J Cardiovasc Pharmacol. 1986;8(Suppl. 8):S7–12. [PubMed] [Google Scholar]
- 20.Gross SR, Sin CG, Barraclough R, Rudland PS. Joining S100 proteins and migration: for better or for worse, in sickness and in health. Cell Mol Life Sci. 2014;71:1551–1579. doi: 10.1007/s00018-013-1400-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi J, Kim DI, Kim J, Kim BH, Kim A. Hornerin is involved in breast cancer progression. J Breast Cancer. 2016;19:142–147. doi: 10.4048/jbc.2016.19.2.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moravkova P, Kohoutova D, Rejchrt S, Cyrany J, Bures J. Role of S100 proteins in colorectal carcinogenesis. Gastroenterol Res Pract. 2016;2016:2632703. doi: 10.1155/2016/2632703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malashkevich VN, Varney KM, Garrett SC, Wilder PT, Knight D, Charpentier TH, Ramagopal UA, Almo SC, Weber DJ, Bresnick AR. Structure of Ca2+−bound S100A4 and its interaction with peptides derived from nonmuscle myosin-IIA. Biochemistry. 2008;47:5111–5126. doi: 10.1021/bi702537s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Markowitz J, Rustandi RR, Varney KM, Wilder PT, Udan R, Wu SL, Horrocks WD, Weber DJ. Calcium-binding properties of wild-type and EF-hand mutants of S100B in the presence and absence of a peptide derived from the C-terminal negative regulatory domain of p53. Biochemistry. 2005;44:7305–7314. doi: 10.1021/bi050321t. [DOI] [PubMed] [Google Scholar]
- 25.Tesarova P, Kalousova M, Zima T, Tesar V. HMGB1, S100 proteins and other RAGE ligands in cancer - markers, mediators and putative therapeutic targets. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2016;160:1–10. doi: 10.5507/bp.2016.003. [DOI] [PubMed] [Google Scholar]
- 26.Ehrchen JM, Sunderkotter C, Foell D, Vogl T, Roth J. The endogenous Toll-like receptor 4 agonist S100A8/S100A9 (calprotectin) as innate amplifier of infection, autoimmunity, and cancer. J Leukoc Biol. 2009;86:557–566. doi: 10.1189/jlb.1008647. [DOI] [PubMed] [Google Scholar]
- 27.Leclerc E, Vetter SW. The role of S100 proteins and their receptor RAGE in pancreatic cancer. Biochim Biophys Acta. 2015;1852:2706–2711. doi: 10.1016/j.bbadis.2015.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H, Xu C, Jin Q, Liu Z. S100 protein family in human cancer. Am J Cancer Res. 2014;4:89–115. [PMC free article] [PubMed] [Google Scholar]
- 29.Lukanidin E, Sleeman JP. Building the niche: the role of the S100 proteins in metastatic growth. Semin Cancer Biol. 2012;22:216–225. doi: 10.1016/j.semcancer.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 30.Zambetti NA, Ping Z, Chen S, Kenswil KJG, Mylona MA, Sanders MA, Hoogenboezem RM, Bindels EMJ, Adisty MN, Van Strien PMH. Mesenchymal inflammation drives genotoxic stress in hematopoietic stem cells and predicts disease evolution in human pre-leukemia. Cell Stem Cell. 2016;19:613–627. doi: 10.1016/j.stem.2016.08.021. [DOI] [PubMed] [Google Scholar]
- 31.Wang H, Zhang L, Zhang IY, Chen X, Da Fonseca A, Wu S, Ren H, Badie S, Sadeghi S, Ouyang M. S100B promotes glioma growth through chemoattraction of myeloid-derived macrophages. Clin Cancer Res. 2013;19:3764–3775. doi: 10.1158/1078-0432.CCR-12-3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagy N, Brenner C, Markadieu N, Chaboteaux C, Camby I, Schafer BW, Pochet R, Heizmann CW, Salmon I, Kiss R. S100A2, a putative tumor suppressor gene, regulates in vitro squamous cell carcinoma migration. Lab Invest. 2001;81:599–612. doi: 10.1038/labinvest.3780269. [DOI] [PubMed] [Google Scholar]
- 33.Ohuchida K, Mizumoto K, Ohhashi S, Yamaguchi H, Konomi H, Nagai E, Yamaguchi K, Tsuneyoshi M, Tanaka M. S100A11, a putative tumor suppressor gene, is overexpressed in pancreatic carcinogenesis. Clin Cancer Res. 2006;12:5417–5422. doi: 10.1158/1078-0432.CCR-06-0222. [DOI] [PubMed] [Google Scholar]
- 34.Li F, Men X, Zhang W. S100 protein in breast tumor. Indian J Cancer. 2014;51(Suppl. 3):e67–e71. doi: 10.4103/0019-509X.154046. [DOI] [PubMed] [Google Scholar]
- 35.Zhang S, Wang Z, Liu W, Lei R, Shan J, Li L, Wang X. Distinct prognostic values of S100 mRNA expression in breast cancer. Sci Rep. 2017;7 doi: 10.1038/srep39786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang T, Huo X, Chong Z, Khan H, Liu R, Wang T. A review of S100 protein family in lung cancer. Clin Chim Acta. 2018;476:54–59. doi: 10.1016/j.cca.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 37.Pan SC, Li CY, Kuo CY, Kuo YZ, Fang WY, Huang YH, Hsieh TC, Kao HY, Kuo Y, Kang YR. The p53-S100A2 positive feedback loop negatively regulates epithelialization in cutaneous wound healing. Sci Rep. 2018;8:5458. doi: 10.1038/s41598-018-23697-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He X, Deng Y, Yue W. Investigating critical genes and gene interaction networks that mediate cyclophosphamide sensitivity in chronic myelogenous leukemia. Mol Med Rep. 2017;16:523–532. doi: 10.3892/mmr.2017.6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Atlasi Y, Noori R, Marolin I, Franken P, Brandao J, Biermann K, Collini P, Grigorian M, Lukanidin E, Ambartsumian N. The role of S100a4 (Mts1) in Apc- and Smad4-driven tumour onset and progression. Eur J Cancer. 2016;68:114–124. doi: 10.1016/j.ejca.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 40.Steinbach D, Pfaffendorf N, Wittig S, Gruhn B. PRAME expression is not associated with down-regulation of retinoic acid signaling in primary acute myeloid leukemia. Cancer Genet Cytogenet. 2007;177:51–54. doi: 10.1016/j.cancergencyto.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 41.Xu Y, Rong LJ, Meng SL, Hou FL, Zhang JH, Pan G. PRAME promotes in vitro leukemia cells death by regulating S100A4/p53 signaling. Eur Rev Med Pharmacol Sci. 2016;20:1057–1063. [PubMed] [Google Scholar]
- 42.Ding K, Wang XM, Fu R, Ruan EB, Liu H, Shao ZH. PRAME gene expression in acute leukemia and its clinical significance. Cancer Biol Med. 2012;9:73–76. doi: 10.3969/j.issn.2095-3941.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steinbach D, Bader P, Willasch A, Bartholomae S, Debatin KM, Zimmermann M, Creutzig U, Reinhardt D, Gruhn B. Prospective validation of a new method of monitoring minimal residual disease in childhood acute myelogenous leukemia. Clin Cancer Res. 2015;21:1353–1359. doi: 10.1158/1078-0432.CCR-14-1999. [DOI] [PubMed] [Google Scholar]
- 44.Tajeddine N, Gala JL, Louis M, Van Schoor M, Tombal B, Gailly P. Tumor-associated antigen preferentially expressed antigen of melanoma (PRAME) induces caspase-independent cell death in vitro and reduces tumorigenicity in vivo. Cancer Res. 2005;65:7348–7355. doi: 10.1158/0008-5472.CAN-04-4011. [DOI] [PubMed] [Google Scholar]
- 45.Tajeddine N, Louis M, Vermylen C, Gala JL, Tombal B, Gailly P. Tumor associated antigen PRAME is a marker of favorable prognosis in childhood acute myeloid leukemia patients and modifies the expression of S100A4, Hsp 27, p21, IL-8 and IGFBP-2 in vitro and in vivo. Leuk Lymphoma. 2008;49:1123–1131. doi: 10.1080/10428190802035933. [DOI] [PubMed] [Google Scholar]
- 46.Donato R, Sorci G, Giambanco I. S100A6 protein: functional roles. Cell Mol Life Sci. 2017;74:2749–2760. doi: 10.1007/s00018-017-2526-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Calabretta B, Kaczmarek L, Mars W, Ochoa D, Gibson CW, Hirschhorn RR, Baserga R. Cell-cycle-specific genes differentially expressed in human leukemias. Proc Natl Acad Sci U S A. 1985;82:4463–4467. doi: 10.1073/pnas.82.13.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferrari S, Calabretta B, deRiel JK, Battini R, Ghezzo F, Lauret E, Griffin C, Emanuel BS, Gurrieri F, Baserga R. Structural and functional analysis of a growth-regulated gene, the human calcyclin. J Biol Chem. 1987;262:8325–8332. [PubMed] [Google Scholar]
- 49.Tamai H, Miyake K, Yamaguchi H, Shimada T, Dan K, Inokuchi K. Inhibition of S100A6 induces GVL effects in MLL/AF4-positive ALL in human PBMC-SCID mice. Bone Marrow Transplant. 2014;49:699–703. doi: 10.1038/bmt.2014.18. [DOI] [PubMed] [Google Scholar]
- 50.Tamai H, Miyake K, Yamaguchi H, Takatori M, Dan K, Inokuchi K, Shimada T. Resistance of MLL-AFF1-positive acute lymphoblastic leukemia to tumor necrosis factor-alpha is mediated by S100A6 upregulation. Blood Cancer J. 2011;1:e38. doi: 10.1038/bcj.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tamai H, Yamaguchi H, Miyake K, Takatori M, Kitano T, Yamanaka S, Yui S, Fukunaga K, Nakayama K, Inokuchi K. Amlexanox downregulates S100A6 to sensitize KMT2A/AFF1-positive acute lymphoblastic leukemia to TNFalpha treatment. Cancer Res. 2017;77:4426–4433. doi: 10.1158/0008-5472.CAN-16-2974. [DOI] [PubMed] [Google Scholar]
- 52.Yamaguchi H, Hanawa H, Uchida N, Inamai M, Sawaguchi K, Mitamura Y, Shimada T, Dan K, Inokuchi K. Multistep pathogenesis of leukemia via the MLL-AF4 chimeric gene/Flt3 gene tyrosine kinase domain (TKD) mutation-related enhancement of S100A6 expression. Exp Hematol. 2009;37:701–714. doi: 10.1016/j.exphem.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 53.Edgeworth J, Gorman M, Bennett R, Freemont P, Hogg N. Identification of p8,14 as a highly abundant heterodimeric calcium binding protein complex of myeloid cells. J Biol Chem. 1991;266:7706–7713. [PubMed] [Google Scholar]
- 54.Nicolas E, Ramus C, Berthier S, Arlotto M, Bouamrani A, Lefebvre C, Morel F, Garin J, Ifrah N, Berger F. Expression of S100A8 in leukemic cells predicts poor survival in de novo AML patients. Leukemia. 2011;25:57–65. doi: 10.1038/leu.2010.251. [DOI] [PubMed] [Google Scholar]
- 55.Laouedj M, Tardif MR, Gil L, Raquil MA, Lachhab A, Pelletier M, Tessier PA, Barabe F. S100A9 induces differentiation of acute myeloid leukemia cells through TLR4. Blood. 2017;129:1980–1990. doi: 10.1182/blood-2016-09-738005. [DOI] [PubMed] [Google Scholar]
- 56.Yang L, Yang M, Zhang H, Wang Z, Yu Y, Xie M, Zhao M, Liu L, Cao L. S100A8-targeting siRNA enhances arsenic trioxide-induced myeloid leukemia cell death by down-regulating autophagy. Int J Mol Med. 2012;29:65–72. doi: 10.3892/ijmm.2011.806. [DOI] [PubMed] [Google Scholar]
- 57.Yang XY, Zhang MY, Zhou Q, Wu SY, Zhao Y, Gu WY, Pan J, Cen JN, Chen ZX, Guo WG. High expression of S100A8 gene is associated with drug resistance to etoposide and poor prognosis in acute myeloid leukemia through influencing the apoptosis pathway. OncoTargets Ther. 2016;9:4887–4899. doi: 10.2147/OTT.S101594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bellodi C, Lidonnici MR, Hamilton A, Helgason GV, Soliera AR, Ronchetti M, Galavotti S, Young KW, Selmi T, Yacobi R. Targeting autophagy potentiates tyrosine kinase inhibitor-induced cell death in Philadelphia chromosome-positive cells, including primary CML stem cells. J Clin Invest. 2009;119:1109–1123. doi: 10.1172/JCI35660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carew JS, Nawrocki ST, Kahue CN, Zhang H, Yang C, Chung L, Houghton JA, Huang P, Giles FJ, Cleveland JL. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood. 2007;110:313–322. doi: 10.1182/blood-2006-10-050260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Livesey KM, Tang D, Zeh HJ, Lotze MT. Autophagy inhibition in combination cancer treatment. Curr Opin Investig Drugs. 2009;(10):1269–1279. [PubMed] [Google Scholar]
- 62.Maclean KH, Dorsey FC, Cleveland JL, Kastan MB. Targeting lysosomal degradation induces p53-dependent cell death and prevents cancer in mouse models of lymphomagenesis. J Clin Invest. 2008;118:79–88. doi: 10.1172/JCI33700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang M, Zeng P, Kang R, Yu Y, Yang L, Tang D, Cao L. S100A8 contributes to drug resistance by promoting autophagy in leukemia cells. PLoS One. 2014;9 doi: 10.1371/journal.pone.0097242. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 64.Alaiya AA, Aljurf M, Shinwari Z, Almohareb F, Malhan H, Alzahrani H, Owaidah T, Fox J, Alsharif F, Mohamed SY. Protein signatures as potential surrogate biomarkers for stratification and prediction of treatment response in chronic myeloid leukemia patients. Int J Oncol. 2016;49:913–933. doi: 10.3892/ijo.2016.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu Y, Zhang F, Zhang S, Deng W, Fan H, Wang H, Zhang J. Regulatory mechanism and functional analysis of S100A9 in acute promyelocytic leukemia cells. Front Med. 2017;11:87–96. doi: 10.1007/s11684-016-0469-4. [DOI] [PubMed] [Google Scholar]
- 66.Lagasse E, Weissman IL. Mouse MRP8 and MRP14, two intracellular calcium-binding proteins associated with the development of the myeloid lineage. Blood. 1992;79:1907–1915. [PubMed] [Google Scholar]
- 67.Hibino T, Sakaguchi M, Miyamoto S, Yamamoto M, Motoyama A, Hosoi J, Shimokata T, Ito T, Tsuboi R, Huh NH. S100A9 is a novel ligand of EMMPRIN that promotes melanoma metastasis. Cancer Res. 2013;73:172–183. doi: 10.1158/0008-5472.CAN-11-3843. [DOI] [PubMed] [Google Scholar]
- 68.Muramatsu T. Basigin (CD147), a multifunctional transmembrane glycoprotein with various binding partners. J Biochem. 2016;159:481–490. doi: 10.1093/jb/mvv127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cheng P, Corzo CA, Luetteke N, Yu B, Nagaraj S, Bui MM, Ortiz M, Nacken W, Sorg C, Vogl T. Inhibition of dendritic cell differentiation and accumulation of myeloid-derived suppressor cells in cancer is regulated by S100A9 protein. J Exp Med. 2008;205:2235–2249. doi: 10.1084/jem.20080132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang S, Song R, Wang Z, Jing Z, Wang S, Ma J. S100A8/A9 in inflammation. Front Immunol. 2018;9:1298. doi: 10.3389/fimmu.2018.01298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ghavami S, Rashedi I, Dattilo BM, Eshraghi M, Chazin WJ, Hashemi M, Wesselborg S, Kerkhoff C, Los M. S100A8/A9 at low concentration promotes tumor cell growth via RAGE ligation and MAP kinase-dependent pathway. J Leukoc Biol. 2008;83:1484–1492. doi: 10.1189/jlb.0607397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yui S, Nakatani Y, Mikami M. Calprotectin (S100A8/S100A9), an inflammatory protein complex from neutrophils with a broad apoptosis-inducing activity. Biol Pharm Bull. 2003;26:753–760. doi: 10.1248/bpb.26.753. [DOI] [PubMed] [Google Scholar]
- 73.Spijkers-Hagelstein JA, Schneider P, Hulleman E, de Boer J, Williams O, Pieters R, Stam RW. Elevated S100A8/S100A9 expression causes glucocorticoid resistance in MLL-rearranged infant acute lymphoblastic leukemia. Leukemia. 2012;26:1255–1265. doi: 10.1038/leu.2011.388. [DOI] [PubMed] [Google Scholar]
- 74.Wilhelm BT, Briau M, Austin P, Faubert A, Boucher G, Chagnon P, Hope K, Girard S, Mayotte N, Landry JR. RNA-seq analysis of 2 closely related leukemia clones that differ in their self-renewal capacity. Blood. 2011;117:e27–e38. doi: 10.1182/blood-2010-07-293332. [DOI] [PubMed] [Google Scholar]
- 75.Liu M., Karjalainen R., Kumar A., Parsons A., He L., Malani D., Porkka K., Heckman C.A. PS955. European Hematology Association (EHA) congress in Stockholm; Sweden: 2018. The calcium binding protein S100A8/S100A9 complex is associated with drug resistance in acute myeloid leukemia. [Google Scholar]
- 76.Handschuh L, Kazmierczak M, Milewski MC, Goralski M, Luczak M, Wojtaszewska M, Uszczynska-Ratajczak B, Lewandowski K, Komarnicki M, Figlerowicz M. Gene expression profiling of acute myeloid leukemia samples from adult patients with AML-M1 and -M2 through boutique microarrays, real-time PCR and droplet digital PCR. Int J Oncol. 2018;52:656–678. doi: 10.3892/ijo.2017.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Apostolova P, Zeiser R. The role of danger signals and ectonucleotidases in acute graft-versus-host disease. Hum Immunol. 2016;77:1037–1047. doi: 10.1016/j.humimm.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 78.Chiusolo P, Giammarco S, Fanali C, Bellesi S, Metafuni E, Sica S, Iavarone F, Cabras T, Messana I, Leone G. Salivary proteomic analysis and acute graft-versus-host disease after allogeneic hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2013;19:888–892. doi: 10.1016/j.bbmt.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 79.Reinhardt K, Foell D, Vogl T, Mezger M, Wittkowski H, Fend F, Federmann B, Gille C, Feuchtinger T, Lang P. Monocyte-induced development of Th17 cells and the release of S100 proteins are involved in the pathogenesis of graft-versus-host disease. J Immunol. 2014;193:3355–3365. doi: 10.4049/jimmunol.1400983. [DOI] [PubMed] [Google Scholar]
- 80.O'Connell PA, Madureira PA, Berman JN, Liwski RS, Waisman DM. Regulation of S100A10 by the PML-RAR-alpha oncoprotein. Blood. 2011;117:4095–4105. doi: 10.1182/blood-2010-07-298851. [DOI] [PubMed] [Google Scholar]
- 81.Huang D, Yang Y, Sun J, Dong X, Wang J, Liu H, Lu C, Chen X, Shao J, Yan J. Annexin A2-S100A10 heterotetramer is upregulated by PML/RARalpha fusion protein and promotes plasminogen-dependent fibrinolysis and matrix invasion in acute promyelocytic leukemia. Front Med. 2017;11:410–422. doi: 10.1007/s11684-017-0527-6. [DOI] [PubMed] [Google Scholar]
- 82.Gopalakrishnapillai A, Kolb EA, Dhanan P, Mason RW, Napper A, Barwe SP. Disruption of annexin II /p11 interaction suppresses leukemia cell binding, homing and engraftment, and sensitizes the leukemia cells to chemotherapy. PLoS One. 2015;10 doi: 10.1371/journal.pone.0140564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kremer KN, Dudakovic A, McGee-Lawrence ME, Philips RL, Hess AD, Smith BD, van Wijnen AJ, Karp JE, Kaufmann SH, Westendorf JJ. Osteoblasts protect AML cells from SDF-1-induced apoptosis. J Cell Biochem. 2014;115:1128–1137. doi: 10.1002/jcb.24755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang Z, Tao T, Raftery MJ, Youssef P, Di Girolamo N, Geczy CL. Proinflammatory properties of the human S100 protein S100A12. J Leukoc Biol. 2001;69:986–994. [PubMed] [Google Scholar]
- 85.Aldinucci D, Colombatti A. The inflammatory chemokine CCL5 and cancer progression. Mediators Inflamm. 2014;2014:292376. doi: 10.1155/2014/292376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rani SG, Mohan SK, Yu C. Molecular level interactions of S100A13 with amlexanox: inhibitor for formation of the multiprotein complex in the nonclassical pathway of acidic fibroblast growth factor. Biochemistry. 2010;49:2585–2592. doi: 10.1021/bi9019077. [DOI] [PubMed] [Google Scholar]
- 87.Fredly H, Ersvær E, Gjertsen BT, Bruserud Ø. Immunogenic apoptosis in human acute myeloid leukemia (AML): primary human AML cells expose calreticulin and release heat shock protein (HSP) 70 and HSP90 during apoptosis. Oncol Rep. 2011;25:1549–1556. doi: 10.3892/or.2011.1229. [DOI] [PubMed] [Google Scholar]
- 88.Reikvam H, Hatfield KJ, Ersvær E, Hovland R, Skavland J, Gjertsen BT, Petersen K, Bruserud Ø. Expression profile of heat shock proteins in acute myeloid leukaemia patients reveals a distinct signature strongly associated with FLT3 mutation status-consequences and potentials for pharmacological intervention. Br J Haematol. 2012;156:468–480. doi: 10.1111/j.1365-2141.2011.08960.x. [DOI] [PubMed] [Google Scholar]
- 89.Li D, Zhang R, Zhu W, Xue Y, Zhang Y, Huang Q, Liu M, Liu Y. S100A16 inhibits osteogenesis but stimulates adipogenesis. Mol Biol Rep. 2013;40:3465–3473. doi: 10.1007/s11033-012-2413-2. [DOI] [PubMed] [Google Scholar]
- 90.Brenner AK, Nepstad I, Bruserud Ø. Mesenchymal stem cells support survival and proliferation of primary human acute myeloid leukemia cells through heterogeneous molecular mechanisms. Front Immunol. 2017;8:106. doi: 10.3389/fimmu.2017.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu H, Zhai Y, Zhao W, Wan Y, Lu W, Yang S, Yu Y, Wei Y, Li Z, Shi J. Consolidation chemotherapy prevents relapse by indirectly regulating bone marrow adipogenesis in patients with acute myeloid leukemia. Cell Physiol Biochem. 2018;45:2389–2400. doi: 10.1159/000488225. [DOI] [PubMed] [Google Scholar]
- 92.Sheng X, Tucci J, Parmentier JH, Ji L, Behan JW, Heisterkamp N, Mittelman SD. Adipocytes cause leukemia cell resistance to daunorubicin via oxidative stress response. Oncotarget. 2016;7:73147–73159. doi: 10.18632/oncotarget.12246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ye H, Adane B, Khan N, Sullivan T, Minhajuddin M, Gasparetto M, Stevens B, Pei S, Balys M, Ashton JM. Leukemic stem cells evade chemotherapy by metabolic adaptation to an adipose tissue niche. Cell Stem Cell. 2016;19:23–37. doi: 10.1016/j.stem.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Boyd AL, Reid JC, Salci KR, Aslostovar L, Benoit YD, Shapovalova Z, Nakanishi M, Porras DP, Almakadi M, Campbell CJV. Acute myeloid leukaemia disrupts endogenous myelo-erythropoiesis by compromising the adipocyte bone marrow niche. Nat Cell Biol. 2017;19:1336–1347. doi: 10.1038/ncb3625. [DOI] [PubMed] [Google Scholar]
- 95.Yen MC, Huang YC, Kan JY, Kuo PL, Hou MF, Hsu YL. S100B expression in breast cancer as a predictive marker for cancer metastasis. Int J Oncol. 2018;52:433–440. doi: 10.3892/ijo.2017.4226. [DOI] [PubMed] [Google Scholar]
- 96.Charmsaz S, Hughes E, Bane FT, Tibbitts P, McIlroy M, Byrne C, Cocchiglia S, McBryan J, Hennessy BT, Dwyer RM. S100beta as a serum marker in endocrine resistant breast cancer. BMC Med. 2017;15:79. doi: 10.1186/s12916-017-0836-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen C, Liu Y, Hua M, Li X, Ji C, Ma D. Neuropathy correlated with imbalanced Foxp3/IL-17 in bone marrow microenvironment of patients with acute myeloid leukemia. Oncotarget. 2016;7:24455–24465. doi: 10.18632/oncotarget.8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ishii Y, Kasukabe T, Honma Y. Immediate up-regulation of the calcium-binding protein S100P and its involvement in the cytokinin-induced differentiation of human myeloid leukemia cells. Biochim Biophys Acta. 2005;1745:156–165. doi: 10.1016/j.bbamcr.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 99.Bullinger L, Dohner K, Bair E, Frohling S, Schlenk RF, Tibshirani R, Dohner H, Pollack JR. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med. 2004;350:1605–1616. doi: 10.1056/NEJMoa031046. [DOI] [PubMed] [Google Scholar]
- 100.Tsumura H, Akimoto M, Kiyota H, Ishii Y, Ishikura H, Honma Y. Gene expression profiles in differentiating leukemia cells induced by methyl jasmonate are similar to those of cytokinins and methyl jasmonate analogs induce the differentiation of human leukemia cells in primary culture. Leukemia. 2009;23:753–760. doi: 10.1038/leu.2008.347. [DOI] [PubMed] [Google Scholar]
- 101.Jiang F, Shults K, Flye L, Hashimoto Y, Van Der Meer R, Xie J, Kravtsov V, Price J, Head DR, Briggs RC. S100P is selectively upregulated in tumor cell lines challenged with DNA cross-linking agents. Leuk Res. 2005;29:1181–1190. doi: 10.1016/j.leukres.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 102.Wang L, Wang YY, Cao Q, Chen Z, Chen SJ. Hornerin gene was involved in a case of acute myeloid leukemia transformed from myelodysplastic syndrome with t(1;2)(q21;q37) Leukemia. 2006;20:2184–2187. doi: 10.1038/sj.leu.2404436. [DOI] [PubMed] [Google Scholar]
- 103.Fu SJ, Shen SL, Li SQ, Hua YP, Hu WJ, Guo B, Peng BG. Hornerin promotes tumor progression and is associated with poor prognosis in hepatocellular carcinoma. BMC Cancer. 2018;18:815. doi: 10.1186/s12885-018-4719-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Maier T, Guell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583:3966–3973. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 105.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liersch R, Gerss J, Schliemann C, Bayer M, Schwoppe C, Biermann C, Appelmann I, Kessler T, Lowenberg B, Buchner T. Osteopontin is a prognostic factor for survival of acute myeloid leukemia patients. Blood. 2012;119:5215–5220. doi: 10.1182/blood-2011-11-389692. [DOI] [PubMed] [Google Scholar]
- 107.Powell JA, Thomas D, Barry EF, Kok CH, McClure BJ, Tsykin A, To LB, Brown A, Lewis ID, Herbert K. Expression profiling of a hemopoietic cell survival transcriptome implicates osteopontin as a functional prognostic factor in AML. Blood. 2009;114:4859–4870. doi: 10.1182/blood-2009-02-204818. [DOI] [PubMed] [Google Scholar]
- 108.Brenner AK, Aasebø E, Hernandez-Valladares M, Selheim F, Berven F, Bruserud Ø. Rethinking the role of osteopontin in human acute myeloid leukemia. Leuk Lymphoma. 2017;58:1494–1497. doi: 10.1080/10428194.2016.1243682. [DOI] [PubMed] [Google Scholar]
- 109.Honnemyr M, Bruserud Ø, Brenner AK. The constitutive protease release by primary human acute myeloid leukemia cells. J Cancer Res Clin Oncol. 2017;143:1985–1998. doi: 10.1007/s00432-017-2458-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Brenner AK, Tvedt TH, Nepstad I, Rye KP, Hagen KM, Reikvam H, Bruserud Ø. Patients with acute myeloid leukemia can be subclassified based on the constitutive cytokine release of the leukemic cells; the possible clinical relevance and the importance of cellular iron metabolism. Expert Opin Ther Targets. 2017;21:357–369. doi: 10.1080/14728222.2017.1300255. [DOI] [PubMed] [Google Scholar]
- 111.Lesniak W. Epigenetic regulation of S100 protein expression. Clin Epigenetics. 2011;2:77–83. doi: 10.1007/s13148-011-0023-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tanaka M, Ichikawa-Tomikawa N, Shishito N, Nishiura K, Miura T, Hozumi A, Chiba H, Yoshida S, Ohtake T, Sugino T. Co-expression of S100A14 and S100A16 correlates with a poor prognosis in human breast cancer and promotes cancer cell invasion. BMC Cancer. 2015;15:53. doi: 10.1186/s12885-015-1059-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Berge G, Costea DE, Berg M, Rasmussen H, Grotterod I, Lothe RA, Maelandsmo GM, Flatmark K. Coexpression and nuclear colocalization of metastasis-promoting protein S100A4 and p53 without mutual regulation in colorectal carcinoma. Amino Acids. 2011;41:875–884. doi: 10.1007/s00726-010-0514-6. [DOI] [PubMed] [Google Scholar]
- 114.Quintás-Cardama A, Hu C, Qutub A, Qiu YH, Zhang X, Post SM, Zhang N, Coombes K, Kornblau SM. p53 pathway dysfunction is highly prevalent in acute myeloid leukemia independent of TP53 mutational status. Leukemia. 2016;31:1296. doi: 10.1038/leu.2016.350. [DOI] [PubMed] [Google Scholar]