Abstract
Introduction
Less than 10% of early-onset Alzheimer's disease (EOAD) is explained by known mutations.
Methods
We conducted genetic linkage analysis of 68 well-phenotyped Caribbean Hispanic families without clear inheritance patterns or mutations in APP, PSEN1, and PSEN2 and with two or more individuals with EOAD.
Results
We identified 16 (logarithm of odds > 3.6) linked regions, including eight novel loci for EOAD (2p15, 5q14.1, 11p15.1, 13q21.22, 13q33.1, 16p12.1, 20p12.1, and 20q11.21) and eight regions previously associated with late-onset Alzheimer's disease. The strongest signal was observed at 16p12.1 (25 cM, 33 Mb; heterogeneity logarithm of odds = 5.3), ∼3 Mb upstream of the ceroid lipofuscinosis 3 (CLN3) gene associated with juvenile neuronal ceroid lipofuscinosis (JNCL), which functions in retromer trafficking and has been reported to alter intracellular processing of the amyloid precursor protein.
Discussion
This study supports the notion that the genetic architectures of unexplained EOAD and late-onset AD overlap partially, but not fully.
Keywords: Early-onset Alzheimer's disease, Non-Mendelian, Genetics, Linkage analysis, Linkage loci
1. Introduction
To clarify the molecular mechanisms underlying Alzheimer's disease (AD; OMIM # 104300), several large-scale genomic studies have been conducted over the past decade, and additional studies are ongoing [1], [2], [3], [4], [5], [6], [7], [8], [9], [10]. However, most of these studies of AD have focused on non-Hispanic white participants affected by the late-onset form of the disease (late-onset Alzheimer's disease [LOAD]; age at onset > 65 years), or the study of early-onset Alzheimer's disease (EOAD) (EOAD; age at onset ≤ 65 years) cases in families with clear autosomal dominant inheritance patterns, typical of pathogenic mutations in APP, PSEN1, or PSEN2. Mutations in these three genes, however, explain less than 10% of EOAD [11], [12] and less than 1% of all AD. Few studies have been performed in families with early-onset disease lacking known mutations with Mendelian inheritance, which can have a mix of early- and late-onset disease. The few studies that have assessed this EOAD subgroup have suggested that the genetic architectures partially overlap with LOAD, but not completely [13], [14], [15], [16], [17], [18]. Thus, studying EOAD in patients without known mutations (i.e., unexplained EOAD) is a critical gap that provides a unique opportunity to clarify disease mechanisms and discover novel targets for prevention or treatment. To begin addressing this issue, we conducted a genetic linkage analysis of 68 well-phenotyped Caribbean Hispanic families without clear inheritance patterns, with two or more early-onset cases but lacking mutations in APP, PSEN1, and PSEN2. The frequency of AD among multiplex families from the Dominican Republic was found to be approximately five-fold higher than in a similarly aged non-Hispanic white population from the United States [19].
2. Materials and methods
2.1. Ethics statement
Study participants were recruited as part of the EFIGA study (Estudio Familiar de Influencia Genetica de Alzheimer).Written informed consent for the study was obtained from all subjects and/or authorized representatives and study partners. The EFIGA study was approved by the institutional review board of the New York State Psychiatric Institute.
2.2. Description of study sample
The 68 Caribbean Hispanic families included in the linkage analyses are part of the EFIGA cohort, which has been previously described in detail [20]. In brief, EFIGA participants have been recruited since January 1998 from clinics in the Dominican Republic and Puerto Rico, as well as the Alzheimer Disease Research Center Memory Disorders Clinic at Columbia University in New York City. Participants are followed up every 18 months; at each visit, participants completed a standardized assessment that included ascertainment of medical history, physical and neurological examination, and an extensive neuropsychological battery [21] for evaluation of cognitive impairment, which measures cognitive function in key domains affected by aging and dementia, including memory, visuospatial ability, psychomotor speed, and executive function. The battery includes the Selective Reminding Test [22], the Benton Visual Retention Test recognition and matching trials [23], the Rosen Drawing Test [7], the Boston Naming Test [8], the Controlled Oral Word Association Test [9], the Category Fluency Test [10], the Color Trails Test [11], the Similarities subtest from the Wechsler Adult Intelligence Scale [12], and the orientation items from the Mini-Mental State Examination [24]. Brief tests of writing and reading comprehension and formal measures of reading recognition were also administered [14], [15], [25]. Functional status was assessed using the Disability and Functional Limitation Instrument [26], which contains self- and observer ratings in the following areas: instrumental activities, such as using the telephone, handling money, and completing chores; personal self-maintenance activities, such as bathing, dressing, using the toilet; perceived difficulty with memory, language, and visuospatial function, mobility, activities, and social participation. The Clinical Dementia Rating Scale [27] was completed. The diagnosis of AD was made at a consensus conference of physicians and neuropsychologists based on guidelines from the National Institute of Neurological and Communicative Disorders and Stroke–the Alzheimer Disease and Related Disorders Association [28].To increase the likelihood of detecting novel rare variants increasing risk of EOAD, we restricted the analyses to Caribbean Hispanic families free of known mutations at established AD Mendelian loci (APP, PSEN1, PSEN2, MAPT, or GRN) and at least two family members with EOAD (i.e., age at onset < 65 years). Six hundred thirty-six individuals in the resulting 68 Caribbean Hispanic families had genome-wide genotyping data available and were included in the final analyses.
2.3. Genotyping and data quality control
Genome-wide genotyping was performed using the Illumina Human Hap 650k and Illumina 1M arrays. After excluding single-nucleotide polymorphisms (SNPs) with a call rate less than 98%, the data derived from the various platforms were merged into a single data set for analysis. SNPs with minor allele frequencies less than 0.01, as well as variants not in Hardy-Weinberg equilibrium (P < 10−6) in controls, were subsequently excluded yielding a final set of 1,420,917 variants for analysis. Employing PLINK1.9 (https://www.cog-genomics.org/plink2/data), x-chromosome SNPs were used to determine and exclude participants whose reported sex differed from the genomic sex assignment.
2.4. Statistical analyses
Because 23 families had the number of nonfounders exceeding the computation limit for MERLIN, we trimmed uninformative family members (based on an individual's position in the pedigree and/or absence of genotyping) using PowerTrim [29] to reduce bit size to 24 before performing MERLIN analyses. To examine and correct the relationships among family members before the linkage scan, we employed the program MAKEPED to detect errors in the family structure, followed by PREST-PLUS [30] to confirm the accuracy of family member relationships using a set of 50,000 independent SNP markers (correlation coefficient R2 = 1) with a minor allele frequencies ≥1%. Based on the resulting information, we excluded individuals who were found to be biologically unrelated and corrected relationships where necessary.
We then performed parametric two-point affecteds-only and two-point age-penetrance models for AD using MERLIN (http://www.sph.umich.edu/csg/abecasis/Merlin/), applying heterogeneity logarithm of odds (HLOD) models to allow for detection of linkage in the presence of locus heterogeneity [31], and including both early- and late-onset cases in the analyses. Parameters for the parametric two-point models assumed dominant inheritance, a disease allele frequency of 0.001, and penetrance measures of 0.01, 0.90, and 0.90 (representing NN, NA, and AA genotypes, respectively). Age-dependent penetrance employed in the analyses is listed in Supplementary Table 1. Two-point parametric analysis utilized all SNPs for each of the analyses. According to Lander and Kruglyak [32], the significance threshold for the parametric two-point linkage scans was set at HLOD ≥ 3.6 (P = 2 × 10−5). Linkage regions were considered independent if the locations of their peak HLOD or LOD scores were separated by >20 cM. Linkage peaks were considered concordant with previous linkage peaks if they were ≤10 cM apart. We subsequently followed up the identified linkage peaks meeting this threshold by joint linkage and association analyses using PSEUDOMARKER (http://www.helsinki.fi/∼tsjuntun/pseudomarker/) applying a disease allele frequency of 0.001 and penetrance measures of 0.01, 0.90, and 0.90 (representing AA, AB, and BB genotypes, respectively). Adjustment for multiple testing in the joint linkage and association analysis was performed using Bonferroni correction, establishing the threshold for significance at P = .004. Parametric multipoint analysis was performed on regions previously reported in the Alzheimer's Disease Sequencing Project [33], [34] but not identified in the parametric two-point models in this sample (2q22, 3q13, 4q34, 5p13, 6q25, 7p14, 7p21, 8q22, 9p22, 9q33, 10p13, 11q12, 13q14, 14q13, 19q13).
3. Results
Characteristics of the study sample are shown in Table 1. In the 68 families included in the analyses, there were in total 304 affected individuals (on average 4.4 per family), 135 (44%) of these had EOAD and 169 (55.6%) individuals had LOAD. Three hundred twenty-three persons were unaffected. Mean age of the EOAD cases was 59.0 ± 5.7 years. A total of 41.4% of subjects were carriers of one apolipoprotein E ε4 (APOEe4) allele, 13.1% were homozygous carriers. The average number of patients with EOAD per family was 2.3.
Table 1.
Characteristics of the data set
| Families, n | 68 |
| Sample included in linkage analyses | |
| Participants, n | 636 |
| Women, n (%) | 403 (63.4) |
| Unaffected, n | 323 |
| Age at last examination of unaffected individuals, mean (SD) | 62.0 (10.4) |
| EOAD, n | 135 |
| Mean AAO EOAD in family | 59.0 (5.7) |
| LOAD, n | 169 |
| Mean AAO LOAD in family | 76.0 (6.7) |
| Ambiguous, n | 9 |
| APOE ε4 allele frequency, n (%)∗ | |
| -/- | 255 (40.1) |
| -/ε4 | 263 (41.4) |
| ε4/ε4 | 83 (13.1) |
Abbreviations: AAO, age at onset; EOAD, early-onset Alzheimer's disease; LOAD, late-onset Alzheimer's disease; APOE, apolipoprotein E; SD, standard deviation.
All small subsets (n = 35) were missing APOE genotyping.
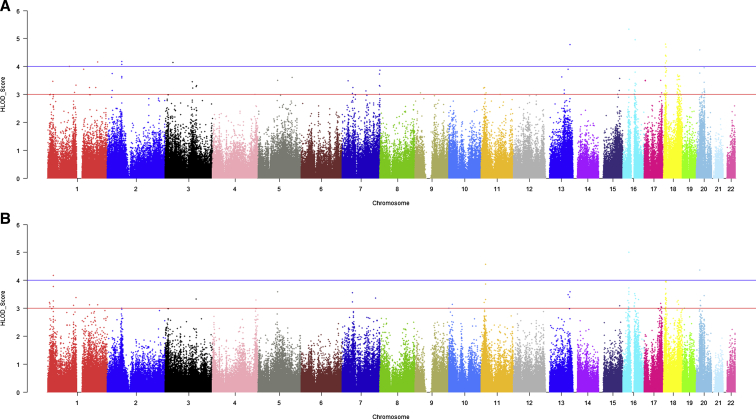
We first conducted two-point affecteds-only and two-point age-dependent penetrance models. In these analyses, we identified 16 linkage regions with HLOD scores equal to or exceeding 3.6 in either model (Fig. 1, Table 2). Although eight of these peaks were previously reported (1p36.1, 1q32.2, 2p24.1, 5q31.3, 7q36.3, 16q12.1, 18p11.3, and 18q22), eight additional peaks were novel (2p15, 5q14.1, 11p15.1, 13q21.22, 13q33.1, 16p12.1, 20p12.1, and 20q11.21). Four loci (13q33.1, 16p12.1, 18p11.23, and 20p12.1) had significant linkage signals under both models. In addition, two previously reported regions on chromosomes 3q13.3 and 3q23 [38] yielded HLOD scores suggestive of linkage in the affecteds-only model (HLOD 3.5 and 3.3, respectively). Although the APOE region on chromosome 19q13.2 did not reach the threshold of HLOD ≥ 3.6, there was evidence of suggestive linkage within a 5 cM range in the age-dependent penetrance model (HLOD = 2.4). Biologically plausible candidate genes under each peak are summarized in Table 2 and discussed in detail below.
Fig. 1.
(A) Two-point genome-wide linkage analysis results from the single-marker affecteds-only model. (B) Two-point genome-wide linkage analysis results from the age-dependent penetrance model. Abbreviation: HLOD, heterogeneity logarithm of odds.
Table 2.
Two-point genome-wide linkage analysis results from single-marker affecteds-only and age-dependent penetrance models (HLOD ≥ 3.6)
| CHR | Cytogenic band | Marker Name | BP (hg19) | cM | HLOD (affecteds-only model) | HLOD (age-dependent penetrance model) | Candidate genes under the linkage peak | Previous evidence for region |
|---|---|---|---|---|---|---|---|---|
| 1a | p36.12 | rs4654814 | 23,094,421 | 49.08 | 2.5 | 4.2 | EPHB2 | [35] |
| 1a | p36.12 | rs4655107 | 23,094,454 | 49.08 | 1.9 | 3.8 | ||
| 1b | q32.2 | rs10779486 | 208,739,309 | 217.43 | 4.2 | 3.1 | CR1 | [34] |
| 2a | p24.1 | rs10191266 | 20,824,708 | 41.9 | 3.8 | 2 | RAB10 | [33] |
| 2b | p15 | kgp1860064 | 61,891,504 | 83.95 | 4.1 | 3 | CDH8 | |
| 2b | p15 | kgp9340583 | 61,897,742 | 83.96 | 4.1 | 3 | ||
| 2b | p15 | kgp14358944 | 61,912,011 | 83.96 | 4.2 | 3 | ||
| 2b | p15 | kgp5707669 | 61,945,708 | 83.98 | 3.6 | 2.5 | ||
| 5a | q14.2 | rs13180356 | 82,395,874 | 99.66 | 3.5 | 3.6 | XRCC4, MEF2C | |
| 5b | q31.3 | rs249725 | 141875313 | 148.75 | 3.6 | 1.4 | APBB3 | [34] |
| 7a | q36.3 | rs2365514 | 156,468,759 | 182.93 | 3.7 | 1.9 | EPHA1 | [33] |
| 7a | q36.3 | rs13229349 | 158,524,530 | 273.16 | 3.9 | 1.3 | ||
| 11a | p15.1 | rs2278732 | 18,764,113 | 32.62 | 2 | 3.9 | PTPN5 | |
| 11a | p15.1 | rs1106865 | 18,782,131 | 32.65 | 3 | 4.6 | ||
| 13a | q21.33 | rs4597193 | 69,343,101 | 61.67 | 3.6 | 2.6 | FBXL3 | |
| 13b | q32.1 | rs9556428 | 95,548,547 | 88.89 | 3.9 | 3.5 | DNAJC3, VPS36 | |
| 13b | q33.1 | rs4772445 | 102,803,631 | 100.23 | 4.8 | 3.6 | ||
| 16a | p12.1 | rs1013534 | 25,426,202 | 51.61 | 5.3 | 5 | CLN3, APOBR, IL4R | |
| 16a | p12.1 | rs11646441 | 25,849,559 | 52.43 | 2.2 | 3.7 | ||
| 16a | p12.1 | rs4578651 | 25,870,810 | 52.47 | 2.3 | 3.6 | ||
| 16a | p12.1 | rs9922199 | 27,179,523 | 55.63 | 3.6 | 1.8 | ||
| 16b | q12.1 | rs8053972 | 51,565,812 | 65.16 | 3.8 | 3.5 | ADCY7, FTO | [34] |
| 18a | p11.31 | rs571298 | 5,983,611 | 20 | 4 | 2.6 | LAMA1, PTPRM, ANKRD12, RAB12, NDUFV2 | [36] |
| 18a | p11.31 | rs6506440 | 6,781,016 | 23.44 | 4.1 | 1.8 | ||
| 18a | p11.31 | rs665265 | 7,017,599 | 24.66 | 2.2 | 3.7 | ||
| 18a | p11.23 | rs679561 | 8,303,370 | 30.73 | 3.7 | 3.4 | ||
| 18a | p11.23 | rs685144 | 8,346,342 | 30.9 | 3.8 | 2.8 | ||
| 18a | p11.23 | rs656568 | 8,351,986 | 30.93 | 4.4 | 4 | ||
| 18a | p11.23 | rs9950784 | 8,432,545 | 31.24 | 4.8 | 3.5 | ||
| 18a | p11.22 | rs11081390 | 8,507,522 | 31.54 | 3.9 | 1.8 | ||
| 18a | p11.23 | rs7233676 | 8,511,586 | 31.56 | 2.2 | 3.9 | ||
| 18a | p11.22 | rs1442685 | 8,605,665 | 31.9 | 4.4 | 3.4 | ||
| 18a | p11.22 | rs4797331 | 8,799,709 | 32.61 | 4.7 | 2.9 | ||
| 18a | p11.22 | rs3810053 | 8,820,886 | 32.69 | 2.5 | 4 | ||
| 18a | p11.22 | rs7506330 | 9,177,894 | 33.73 | 3.8 | 2.5 | ||
| 18a | p11.21 | rs12455464 | 10,901,807 | 39.62 | 3.8 | 3.3 | ||
| 18a | p11.21 | rs8088825 | 11,512,551 | 41.29 | 4.2 | 2.4 | ||
| 18b | q21.32 | rs1942863 | 57,745,744 | 87.67 | 3.7 | 1.6 | BCL2 | [36], [37] |
| 18b | q21.33 | rs7236310 | 59,098,446 | 89.92 | 3.7 | 1.5 | ||
| 18b | q22.1 | rs176139 | 62,781,618 | 96.45 | 3.6 | 1.7 | ||
| 18b | q22.1 | rs9319758 | 65,852,874 | 100.889 | 3.7 | 2.5 | ||
| 20a | p12.1 | rs1431441 | 13,026,863 | 36.05 | 3.8 | 2.4 | SPTLC3 | |
| 20a | p12.1 | rs6041821 | 13,036,800 | 36.07 | 4.6 | 4.4 | ||
| 20b | q11.21 | rs293554 | 31,085,857 | 53.84 | 3.9 | 3.5 | NOL4L |
Abbreviations: HLOD, heterogeneity logarithm of odds; BP, base pairs in GCHr17/hg19; cM, centimorgan (Kosambi).
The highest linkage peak was observed at SNP marker rs1013534 on chromosome 16p12.1 (25 cM, 33 Mb) under both the affecteds-only (HLOD = 5.3) and age-dependent penetrance models (HLOD = 5.0). SNP rs1013534 is intergenically located between ZKSCAN2 and CYCSP39, both of which have relatively unknown function. Joint linkage and association analysis of the novel identified linkage peaks confirmed significant association and linkage at loci 2p15, 5q14.1, 11p15.1, 16p12.1, and 20p12.1, after correction for multiple testing. Parametric multipoint analysis on regions previously reported in the Alzheimer's Disease Sequencing Project [33], [34] but not identified in the parametric two-point models in this sample (2q22, 3q13, 4q34, 5p13, 6q25, 7p14, 7p21, 8q22, 9p22, 9q33, 10p13, 11q12, 13q14, 14q13, 19q13) did not show evidence for linkage in this sample.
4. Discussion
In 68 Caribbean Hispanic families with multiple members with EOAD but free of known mutations, we identified 16 regions with HLOD scores equal to or above 3.6, five of these loci were supported by evidence for significant joint linkage and association (Pjoint = 0.004). Eight loci were previously reported (1p36, 1q32, 2p24, 5q31, 7q36, 16q12, 18p11, and 18q21). The 1p36.1 locus was reported by a linkage study on Finnish LOAD families [35]. Notably, rs4654814 and rs4655107, constituting the markers with strongest linkage signals at this locus, are located in the EPHB2 gene encoding another member of the Eph receptor family of receptor tyrosine kinase transmembrane glycoproteins. In addition to the implication of EPHA1 in AD etiology as described previously [2], [6], [10], [39], there is mounting evidence from cell biological experiments and animal studies for the involvement of this protein family, and EPHB2 in particular. It has been shown that amyloid β (Aβ)–derived diffusible ligands interact with EphB2 and trigger its degradation [40]. EphB2 is a key regulator of synaptic localization of N-methyl-D-aspartate (NMDA) receptors, and its depletion in normal mice reduces NMDAR currents and impairs long-term potentiation, both of which are critical for memory formation [40]. Increasing EphB2 levels in a mouse model of AD improves memory deficits, phosphorylation, and surface expression of GluN2B-containing NMDA receptors [40], [41], [42]. Overexpression of EphB2 also rescues the Aβ-derived diffusible ligands–induced depletion of the expression of EphB2 and GluN2B-containing NMDA receptors trafficking in cultured hippocampal neurons [41]. These results suggest that improving the decreased expression of EphB2 and subsequent GluN2B-containing NMDA receptors trafficking in the hippocampus may be a promising strategy for AD treatment.
Locus 1q32.2 harbors the AD candidate gene CR1 [6]. The linkage peak on 2p24.1 includes RAB10 (a member of the RAS superfamily of small GTPases that are key regulators of membrane trafficking and critical for neuronal development), and 5q31.3 includes the APBB3 gene encoding a member of the Aβ (A4) precursor protein–binding family B binding to the intracellular domain of the amyloid precursor protein potentially modulating its internalization. 7q36.3 contains the AD candidate gene EPHA1, a member of the Eph receptor family of receptor tyrosine kinase transmembrane glycoproteins identified by genome-wide association studies [2], [6], [39]. A biologically plausible candidate gene under the linkage peak at chromosome 16q12.1 includes ADCY7 encoding a membrane-bound adenylate cyclase that catalyzes the formation of cyclic adenosine monophosphate from adenosine triphosphate, and the fat and obesity–associated gene (FTO) involved in obesity-related traits and insulin resistance, which has previously been associated with AD in genetic association studies [43]. The 18p11 and 18q22.1 loci have been previously observed in a linkage study derived from an isolated population of Amish families [44]. Biologically plausible genes at the 18p11 locus include LAMA1, PTPRM, ANKRD12, RAB12, and NDUFV2. LAMA1 encodes one of the α1 subunits of laminin. Laminins, a family of extracellular matrix glycoproteins, make up a major component of the basement membrane of many tissues including the endothelium of blood vessel walls and might contribute to vascular homeostasis [45]. The α1 subunit of laminin is expressed in the basal lamina of blood vessels in the central nervous system, mostly confined to capillary walls [46]. There is strong evidence that cerebrovascular dysregulation plays a role in neurodegeneration and AD [47]. A recent whole-exome sequencing study in the Amish population identified a synonymous variant in LAMA1, rs73938538 [48]. PTPRM encodes protein tyrosine phosphatase, receptor type M, a member of the protein tyrosine phosphatase (PTP) family. PTPs are signaling molecules regulating a variety of cellular processes including cell growth, differentiation, mitotic cycle, and oncogenic transformation. ANKRD12 encodes a member of the ankyrin repeats–containing cofactor family, which inhibit the transcriptional activity of nuclear receptors through the recruitment of histone deacetylases. RAB12 encodes a member of the family of the small GTPases Rab, which are, as described previously, key regulators of membrane trafficking and critical for neuronal development. NDUFV2 encodes a subunit of the NADH-ubiquinone oxidoreductase complex (complex I) of the mitochondrial respiratory chain, which catalyzes the transfer of electrons from NADH to ubiquinone. Mutations in this gene have been implicated in Parkinson's disease, bipolar disorder, and schizophrenia [49], [50]. The 18q21 linkage region harbors BCL2. The BCL2 proteins family are key regulators of evolutionally conserved pathways of apoptosis and involved in regulation of neuronal survival [51]. There is evidence that reduction of BCL2 results in Aβ-induced neuronal cell death [52].
Novel linkage regions were observed at chromosomes 2p15, 5q14, 11p15, 13q21, 13q33, 16p12, 20p12, and 20q11. The strongest of these novel signals was detected at marker rs1013534 on chromosome 16p12.1 (25 Mb, 51.6 cM) under both the affecteds-only (HLOD = 5.3) and age-dependent penetrance models (HLOD = 5.0) (Table 2, Fig. 1). This linkage region is located ∼3 Mb upstream of the ceroid lipofuscinosis 3 (CLN3) gene associated with juvenile neuronal ceroid lipofuscinosis (JNCL). CLN3 protein functions in trafficking of the mannose-6-phosphate receptor (M6PR), a key cargo of retromer [53]. Retromer is a multimodular protein assembly that has been implicated in the pathogenesis of LOAD [54], [55] and is considered the “master conductor” of endosomal sorting and trafficking [56]. There is evidence that CLN3 alters intracellular processing of the amyloid precursor protein [57]. Additional plausible candidate genes at this locus include APOBR (encoding apolipoprotein B receptor involved in endothelial dysfunction and atherothrombogenesis), and IL4R encoding interleukin receptor 4 involved in immune response. Both vascular disease and immune response are molecular mechanisms involved in AD etiology [58], [59], [60], [61], [62]. The markers with strongest linkage signals at the 2p15 locus are located in CDH8. CDH8 codes for a calcium-dependent cell adhesion protein implicated in synaptic adhesion and axonal growth and guidance. Neuronal cadherin interacts with presenilin-1 [63], and cell adhesion molecules may be decreased in mild cognitive impairment and AD [64], suggesting the possibility of a mechanistic relationship to AD that warrants investigation. A SNP adjacent to this gene was associated with rate of longitudinal hippocampal structural change over 12 months in the ADNI cohort [65]. In addition, there is evidence that reduced expression of CDH8 results in abnormal activation of RE-1 silencing transcription factor (REST), which represses genes that promote cell death and AD pathology, protects neurons from oxidative stress and Aβ-protein toxicity, and is lost in mild cognitive impairment and AD [66]. CDH23, another member of the cadherin superfamily, has been previously associated with AD in an epigenetic association study [67]. Although the locus at 5q14.2 does not contain any previously reported genes, it is located ∼5 Mb upstream of the AD candidate gene MEF2C identified in the IGAP meta-analysis [6]. The marker exhibiting the strongest HLOD score (rs13180356) is located in the XRCC4 gene functioning in the repair of DNA double-strand breaks and associated with SSMED syndrome characterized by short stature, microcephaly, and endocrine dysfunction [68].
The locus at 11p15.1 does not include any known candidate genes from previous genetic studies. However, the two SNPs (rs2278732 and rs1106865) exerting the strongest LOD score are located in the PTPN5 gene encoding striatal-enriched protein tyrosine phosphatase (STEP). STEP is a central nervous system–enriched protein implicated in multiple neurologic and neuropsychiatric disorders, which regulates key signaling proteins required for synaptic strengthening and NMDA and α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor trafficking and has been implicated in multiple neurologic and neuropsychiatric disorders. Both high and low levels of STEP disrupt synaptic function and contribute to learning and behavioral deficits. High levels of STEP are present in human postmortem samples and animal models of AD, Parkinson's disease, and schizophrenia and in animal models of fragile X syndrome [69], [70], [71]. Low levels of STEP activity are present in additional disorders that include ischemia, Huntington's chorea, alcohol abuse, and stress disorders. STEP acts by dephosphorylating regulatory tyrosine residues in substrates that include subunits of both NMDA and α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid glutamate receptors, thereby leading to internalization of these receptor complexes [72]. Additional targets of STEP include the kinases ERK1/2, Fyn, and Pyk2 that are inactivated by dephosphorylation of regulatory tyrosines within their activation loop [73], [74], [75], [76], thus modulating downstream signaling pathways. When STEP activity is elevated, as occurs in AD, the increased internalization of glutamate receptors disrupts synaptic function and contributes to the cognitive deficits that are present. Importantly, the STEP inhibitor TC-2153 significantly improves cognitive function in 3 × Tg-AD mice [69].
A biologically plausible candidate gene within a 10 Mb range of the 13q21.33 peak is FBXL3 encoding a member of the F-box protein family, which functions in phosphorylation-dependent ubiquitination. A family member of this protein, FBXL7, has been recently reported in a GWAS of AD in Caribbean Hispanics [77]. Two plausible genes at the 13q33.1 locus are DNAJC3 and VPS36, both of which are involved in intracellular sorting of proteins. VPS36 is a component of the ESCRT-II (endosomal sorting complex required for transport II) complex. The ESCRT complexes regulate the biogenesis of multivesicular bodies and the sorting of ubiquitinated cargos onto intraluminal vesicles within these multivesicular bodies [78]. DNAJC3 is involved in the unfolded protein response during endoplasmic reticulum stress. As a co-chaperone of HSPA8/HSC70 promotes normal protein folding, it stimulates its ATPase activity. Loss-of-function mutations in DNAJC3 result in multisystemic neurodegeneration [79]. The linkage signal at chromosome 20p12.1 is located in the SPTLC3 gene encoding a subunit of serine palmitoyltransferase catalyzing the rate-limiting step of the de novo synthesis of sphingolipids that are critical regulators of membrane dynamics in the nervous system [80]. The marker with strongest signal at the 20q11 locus is located in the NOL4L gene whose function is largely unknown.
Three of the novel linkage peaks did not meet the threshold for significance in joint linkage and association analyses. Discordance between linkage and joint linkage and association analysis is to some extent expected given the different statistical algorithms underlying both approaches. Genetic linkage analysis identifies genomic loci that are shared between affected individuals within the same family by testing for co-segregation of chromosomal segments from a common ancestor with affection status. In contrast, association analysis examines differences in allele frequencies between affected and unaffected subjects taking the pedigree relationships into account. Although association analysis is more powerful in detecting smaller effects in the population, linkage analysis is more powerful for finding large effects in a small number of related individuals and is more robust to genetic heterogeneity.
As described previously, the frequency of AD among multiplex families from the Dominican Republic was found to be approximately five-fold higher than in a similarly aged non-Hispanic white population from the United States [19]. In addition, this population shows a moderate degree of inbreeding [81]. Inbreeding can modify disease risk due to excess homozygosity of recessive alleles [82]. A recent study examining the concordance for AD among parent-offspring pairs suggested that as much as 90% of EOAD cases with AD might be the result of autosomal recessive inheritance [12]. In line with this notion, a previous study identified a higher presence of long runs of homozygosity in Caribbean Hispanic AD cases compared with healthy controls [83]. The present linkage analyses of multiplex Caribbean Hispanic families with two or more EOAD cases unexplained by known early-onset mutations are in line with the notion of a strong heritable component. It identified both loci previously reported in linkage analyses of LOAD families harboring several known AD candidate genes including CR1 and EPHA1, as well as novel loci on chromosomes 2p15, 5q14, 11p15, 13q21, 13q33, 16p12, 20p12, and 20q11 most of which also harbor plausible candidate genes. As described previously, several of the genes under the previously identified and novel peaks cluster in established AD pathways identified in genomic studies of family-based or case-control data sets on LOAD, including amyloid precursor protein/Aβ processing, endosomal sorting, inflammation and immune response, and synaptic transmission. Acknowledging that linkage analyses do not identify specific genes or variants but rather genomic regions potentially harboring causative variants, this observation—together with the finding that we identified both regions overlapping with linkage analyses from late-onset data sets as well as novel regions—is in line with the notion that the mechanisms underlying unexplained EOAD might partially, but not fully, overlap with the late-onset form. The finding of numerous linkage regions instead of a few shared loci further suggests that there is substantial locus heterogeneity within this AD subtype. Both the known and novel linkage regions need to be more closely examined by next-generation sequencing analyses to identify the underlying responsible variants and their functional consequences. In addition, studies in other ethnic groups are needed to determine generalizability of these loci across ethnic groups and potentially identify additional regions.
Research in Context.
-
1.
Systematic review: The authors reviewed the literature using PubMed. Few studies have studied families with EOAD without clear Mendelian inheritance that can have a mix of early- and late-onset cases and account for over 90% of EOAD.
-
2.
Interpretation: Our findings support the notion that the genetic architectures of unexplained EOAD and late-onset AD overlap partially, but not fully.
-
3.
Future directions: Sequencing efforts are needed that focus on individuals with unexplained EOAD and screen these regions likely to harbor rare variants contributing to disease.
Acknowledgments
This work was supported by the National Institutes of Health [R37AG015473, RF1AG015473, RF1AG054080].
Footnotes
The authors have declared that no conflict of interest exists.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.dadm.2018.07.007.
Supplementary data
References
- 1.Guerreiro R., Wojtas A., Bras J., Carrasquillo M., Rogaeva E., Majounie E. TREM2 variants in Alzheimer's disease. N Engl J Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hollingworth P., Harold D., Sims R., Gerrish A., Lambert J.C., Carrasquillo M.M. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jonsson T., Stefansson H., Steinberg S., Jonsdottir I., Jonsson P.V., Snaedal J. Variant of TREM2 associated with the risk of Alzheimer's disease. N Engl J Med. 2013;368:107–116. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jonsson T., Stefansson K. TREM2 and neurodegenerative disease. N Engl J Med. 2013;369:1568–1569. doi: 10.1056/NEJMc1306509. [DOI] [PubMed] [Google Scholar]
- 5.Lambert J.C., Grenier-Boley B., Harold D., Zelenika D., Chouraki V., Kamatani Y. Genome-wide haplotype association study identifies the FRMD4A gene as a risk locus for Alzheimer's disease. Mol Psychiatry. 2013;18:461–470. doi: 10.1038/mp.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lambert J.C., Ibrahim-Verbaas C.A., Harold D., Naj A.C., Sims R., Bellenguez C. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reitz C., Jun G., Naj A., Rajbhandary R., Vardarajan B.N., Wang L.S. Variants in the ATP-binding cassette transporter (ABCA7), apolipoprotein E 4,and the risk of late-onset Alzheimer disease in African Americans. JAMA. 2013;309:1483–1492. doi: 10.1001/jama.2013.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reitz C., Mayeux R. TREM2 and neurodegenerative disease. N Engl J Med. 2013;369:1564–1565. doi: 10.1056/NEJMc1306509#SA1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seshadri S., Fitzpatrick A.L., Ikram M.A., DeStefano A.L., Gudnason V., Boada M. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vardarajan B.N., Ghani M., Kahn A., Sheikh S., Sato C., Barral S. Rare coding mutations identified by sequencing of Alzheimer disease genome-wide association studies loci. Ann Neurol. 2015;78:487–498. doi: 10.1002/ana.24466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cacace R., Sleegers K., Van Broeckhoven C. Molecular genetics of early-onset Alzheimer's disease revisited. Alzheimers Dement. 2016;12:733–748. doi: 10.1016/j.jalz.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 12.Wingo T.S., Lah J.J., Levey A.I., Cutler D.J. Autosomal recessive causes likely in early-onset Alzheimer disease. Arch Neurol. 2012;69:59–64. doi: 10.1001/archneurol.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Duijn C.M., de Knijff P., Cruts M., Wehnert A., Havekes L.M., Hofman A. Apolipoprotein E4 allele in a population-based study of early-onset Alzheimer's disease. Nat Genet. 1994;7:74–78. doi: 10.1038/ng0594-74. [DOI] [PubMed] [Google Scholar]
- 14.Pottier C., Hannequin D., Coutant S., Rovelet-Lecrux A., Wallon D., Rousseau S. High frequency of potentially pathogenic SORL1 mutations in autosomal dominant early-onset Alzheimer disease. Mol Psychiatry. 2012;17:875–879. doi: 10.1038/mp.2012.15. [DOI] [PubMed] [Google Scholar]
- 15.Pastor P., Roe C.M., Villegas A., Bedoya G., Chakraverty S., Garcia G. Apolipoprotein Eepsilon4 modifies Alzheimer's disease onset in an E280A PS1 kindred. Ann Neurol. 2003;54:163–169. doi: 10.1002/ana.10636. [DOI] [PubMed] [Google Scholar]
- 16.Dermaut B., Croes E.A., Rademakers R., Van den Broeck M., Cruts M., Hofman A. PRNP Val129 homozygosity increases risk for early-onset Alzheimer's disease. Ann Neurol. 2003;53:409–412. doi: 10.1002/ana.10507. [DOI] [PubMed] [Google Scholar]
- 17.Jonsson T., Atwal J.K., Steinberg S., Snaedal J., Jonsson P.V., Bjornsson S. A mutation in APP protects against Alzheimer's disease and age-related cognitive decline. Nature. 2012;488:96–99. doi: 10.1038/nature11283. [DOI] [PubMed] [Google Scholar]
- 18.Brouwers N., Sleegers K., Van Broeckhoven C. Molecular genetics of Alzheimer's disease: An update. Ann Med. 2008;40:562–583. doi: 10.1080/07853890802186905. [DOI] [PubMed] [Google Scholar]
- 19.Vardarajan B.N., Faber K.M., Bird T.D., Bennett D.A., Rosenberg R., Boeve B.F. Age-specific incidence rates for dementia and Alzheimer disease in NIA-LOAD/NCRAD and EFIGA families: National Institute on Aging Genetics Initiative for Late-Onset Alzheimer Disease/National Cell Repository for Alzheimer Disease (NIA-LOAD/NCRAD) and Estudio Familiar de Influencia Genetica en Alzheimer (EFIGA) JAMA Neurol. 2014;71:315–323. doi: 10.1001/jamaneurol.2013.5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romas S.N., Santana V., Williamson J., Ciappa A., Lee J.H., Rondon H.Z. Familial Alzheimer disease among Caribbean Hispanics: A reexamination of its association with APOE. Arch Neurol. 2002;59:87–91. doi: 10.1001/archneur.59.1.87. [DOI] [PubMed] [Google Scholar]
- 21.Stern Y., Andrews H., Pittman J., Sano M., Tatemichi T., Lantigua R. Diagnosis of dementia in a heterogeneous population. Development of a neuropsychological paradigm-based diagnosis of dementia and quantified correction for the effects of education. Arch Neurol. 1992;49:453–460. doi: 10.1001/archneur.1992.00530290035009. [DOI] [PubMed] [Google Scholar]
- 22.Buschke H., Fuld P.A. Evaluating storage, retention, and retrieval in disordered memory and learning. Neurology. 1974;24:1019–1025. doi: 10.1212/wnl.24.11.1019. [DOI] [PubMed] [Google Scholar]
- 23.Benton A.L. Development of finger-localization capacity in school children. Child Dev. 1955;26:225–230. [PubMed] [Google Scholar]
- 24.Folstein M.F., Folstein S.E., McHugh P.R. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 25.Del Ser T., Gonzalez-Montalvo J.I., Martinez-Espinosa S., Delgado-Villapalos C., Bermejo F. Estimation of premorbid intelligence in Spanish people with the Word Accentuation Test and its application to the diagnosis of dementia. Brain Cogn. 1997;33:343–356. doi: 10.1006/brcg.1997.0877. [DOI] [PubMed] [Google Scholar]
- 26.Golden R.R., Teresi J.A., Gurland B.J. Development of indicator scales for the Comprehensive Assessment and Referral Evaluation (CARE) interview schedule. J Gerontol. 1984;39:138–146. doi: 10.1093/geronj/39.2.138. [DOI] [PubMed] [Google Scholar]
- 27.Morris J.C., Edland S., Clark C., Galasko D., Koss E., Mohs R. The consortium to establish a registry for Alzheimer's disease (CERAD). Part IV. Rates of cognitive change in the longitudinal assessment of probable Alzheimer's disease. Neurology. 1993;43:2457–2465. doi: 10.1212/wnl.43.12.2457. [DOI] [PubMed] [Google Scholar]
- 28.McKhann G., Drachman D., Folstein M., Katzman R., Price D., Stadlan E.M. Clinical diagnosis of Alzheimer's disease: Report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 29.Thornton T.A., Haines J.L. PowerTrim: An automated decision support algorithm for preprocessing family-based genetic data. Am J Hum Genet. 2003;72:1280–1281. doi: 10.1086/374823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun L., Dimitromanolakis A. PREST-plus identifies pedigree errors and cryptic relatedness in the GAW18 sample using genome-wide SNP data. BMC Proc. 2014;8:S23. doi: 10.1186/1753-6561-8-S1-S23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ott J. Linkage probability and its approximate confidence interval under possible heterogeneity. Genet Epidemiol Suppl. 1986;1:251–257. doi: 10.1002/gepi.1370030739. [DOI] [PubMed] [Google Scholar]
- 32.Lander E., Kruglyak L. Genetic dissection of complex traits: Guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 33.Barral S., Cheng R., Reitz C., Vardarajan B., Lee J., Kunkle B. Linkage analyses in Caribbean Hispanic families identify novel loci associated with familial late-onset Alzheimer's disease. Alzheimers Dement. 2015;11:1397–1406. doi: 10.1016/j.jalz.2015.07.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kunkle B.W., Jaworski J., Barral S., Vardarajan B., Beecham G.W., Martin E.R. Genome-wide linkage analyses of non-Hispanic white families identify novel loci for familial late-onset Alzheimer's disease. Alzheimers Dement. 2016;12:2–10. doi: 10.1016/j.jalz.2015.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hiltunen M., Mannermaa A., Thompson D., Easton D., Pirskanen M., Helisalmi S. Genome-wide linkage disequilibrium mapping of late-onset Alzheimer's disease in Finland. Neurology. 2001;57:1663–1668. doi: 10.1212/wnl.57.9.1663. [DOI] [PubMed] [Google Scholar]
- 36.Ashley-Koch A.E., Shao Y., Rimmler J.B., Gaskell P.C., Welsh-Bohmer K.A., Jackson C.E. An autosomal genomic screen for dementia in an extended Amish family. Neurosci Lett. 2005;379:199–204. doi: 10.1016/j.neulet.2004.12.065. [DOI] [PubMed] [Google Scholar]
- 37.Pericak-Vance M.A., Grubber J., Bailey L.R., Hedges D., West S., Santoro L. Identification of novel genes in late-onset Alzheimer's disease. Exp Gerontol. 2000;35:1343–1352. doi: 10.1016/s0531-5565(00)00196-0. [DOI] [PubMed] [Google Scholar]
- 38.Lee J.H., Cheng R., Santana V., Williamson J., Lantigua R., Medrano M. Expanded genomewide scan implicates a novel locus at 3q28 among Caribbean hispanics with familial Alzheimer disease. Arch Neurol. 2006;63:1591–1598. doi: 10.1001/archneur.63.11.1591. [DOI] [PubMed] [Google Scholar]
- 39.Naj A.C., Jun G., Beecham G.W., Wang L.S., Vardarajan B.N., Buros J. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cisse M., Halabisky B., Harris J., Devidze N., Dubal D.B., Sun B. Reversing EphB2 depletion rescues cognitive functions in Alzheimer model. Nature. 2011;469:47–52. doi: 10.1038/nature09635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu R., Wei P., Jin L., Zheng T., Chen W.Y., Liu X.Y. Overexpression of EphB2 in hippocampus rescues impaired NMDA receptors trafficking and cognitive dysfunction in Alzheimer model. Cell Death Dis. 2017;8:e2717. doi: 10.1038/cddis.2017.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi X.D., Sun K., Hu R., Liu X.Y., Hu Q.M., Sun X.Y. Blocking the Interaction between EphB2 and ADDLs by a Small Peptide Rescues Impaired Synaptic Plasticity and Memory Deficits in a Mouse Model of Alzheimer's Disease. J Neurosci. 2016;36:11959–11973. doi: 10.1523/JNEUROSCI.1327-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reitz C., Tosto G., Mayeux R., Luchsinger J.A. Genetic variants in the Fat and Obesity Associated (FTO) gene and risk of Alzheimer's disease. PLoS One. 2012;7:e50354. doi: 10.1371/journal.pone.0050354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cummings A.C., Jiang L., Velez Edwards D.R., McCauley J.L., Laux R., McFarland L.L. Genome-wide association and linkage study in the Amish detects a novel candidate late-onset Alzheimer disease gene. Ann Hum Genet. 2012;76:342–351. doi: 10.1111/j.1469-1809.2012.00721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yousif L.F., Di Russo J., Sorokin L. Laminin isoforms in endothelial and perivascular basement membranes. Cell Adh Migr. 2013;7:101–110. doi: 10.4161/cam.22680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Virtanen I., Gullberg D., Rissanen J., Kivilaakso E., Kiviluoto T., Laitinen L.A. Laminin alpha1-chain shows a restricted distribution in epithelial basement membranes of fetal and adult human tissues. Exp Cell Res. 2000;257:298–309. doi: 10.1006/excr.2000.4883. [DOI] [PubMed] [Google Scholar]
- 47.Cullen K.M., Kocsi Z., Stone J. Microvascular pathology in the aging human brain: Evidence that senile plaques are sites of microhaemorrhages. Neurobiol Aging. 2006;27:1786–1796. doi: 10.1016/j.neurobiolaging.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 48.D'Aoust L.N., Cummings A.C., Laux R., Fuzzell D., Caywood L., Reinhart-Mercer L. Examination of candidate exonic variants for association to Alzheimer disease in the Amish. PLoS One. 2015;10:e0118043. doi: 10.1371/journal.pone.0118043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nishioka K., Vilarino-Guell C., Cobb S.A., Kachergus J.M., Ross O.A., Hentati E. Genetic variation of the mitochondrial complex I subunit NDUFV2 and Parkinson's disease. Parkinsonism Relat Disord. 2010;16:686–687. doi: 10.1016/j.parkreldis.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu C., Li P.P., Kennedy J.L., Green M., Hughes B., Cooke R.G. Further support for association of the mitochondrial complex I subunit gene NDUFV2 with bipolar disorder. Bipolar Disord. 2008;10:105–110. doi: 10.1111/j.1399-5618.2008.00535.x. [DOI] [PubMed] [Google Scholar]
- 51.Davies A.M. The Bcl-2 family of proteins, and the regulation of neuronal survival. Trends Neurosci. 1995;18:355–358. doi: 10.1016/0166-2236(95)93928-q. [DOI] [PubMed] [Google Scholar]
- 52.Kudo W., Lee H.P., Smith M.A., Zhu X., Matsuyama S., Lee H.G. Inhibition of Bax protects neuronal cells from oligomeric Abeta neurotoxicity. Cell Death Dis. 2012;3:e309. doi: 10.1038/cddis.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Metcalf D.J., Calvi A.A., Seaman M., Mitchison H.M., Cutler D.F. Loss of the Batten disease gene CLN3 prevents exit from the TGN of the mannose 6-phosphate receptor. Traffic. 2008;9:1905–1914. doi: 10.1111/j.1600-0854.2008.00807.x. [DOI] [PubMed] [Google Scholar]
- 54.Small S.A., Petsko G.A. Retromer in Alzheimer disease, Parkinson disease and other neurological disorders. Nat Rev Neurosci. 2015;16:126–132. doi: 10.1038/nrn3896. [DOI] [PubMed] [Google Scholar]
- 55.Muhammad A., Flores I., Zhang H., Yu R., Staniszewski A., Planel E. Retromer deficiency observed in Alzheimer's disease causes hippocampal dysfunction, neurodegeneration, and Abeta accumulation. Proc Natl Acad Sci U S A. 2008;105:7327–7332. doi: 10.1073/pnas.0802545105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burd C., Cullen P.J. Retromer: A master conductor of endosome sorting. Cold Spring Harb Perspect Biol. 2014;6 doi: 10.1101/cshperspect.a016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Golabek A.A., Kida E., Walus M., Kaczmarski W., Michalewski M., Wisniewski K.E. CLN3 protein regulates lysosomal pH and alters intracellular processing of Alzheimer's amyloid-beta protein precursor and cathepsin D in human cells. Mol Genet Metab. 2000;70:203–213. doi: 10.1006/mgme.2000.3006. [DOI] [PubMed] [Google Scholar]
- 58.Dichgans M., Leys D. Vascular Cognitive Impairment. Circ Res. 2017;120:573–591. doi: 10.1161/CIRCRESAHA.116.308426. [DOI] [PubMed] [Google Scholar]
- 59.Gorelick P.B., Scuteri A., Black S.E., Decarli C., Greenberg S.M., Iadecola C. Vascular contributions to cognitive impairment and dementia: a statement for healthcare professionals from the american heart association/american stroke association. Stroke. 2011;42:2672–2713. doi: 10.1161/STR.0b013e3182299496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vijayan M., Reddy P.H. Stroke, Vascular Dementia, and Alzheimer's Disease: Molecular Links. J Alzheimers Dis. 2016;54:427–443. doi: 10.3233/JAD-160527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Efthymiou A.G., Goate A.M. Late onset Alzheimer's disease genetics implicates microglial pathways in disease risk. Mol Neurodegener. 2017;12:43. doi: 10.1186/s13024-017-0184-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ulrich J.D., Ulland T.K., Colonna M., Holtzman D.M. Elucidating the Role of TREM2 in Alzheimer's Disease. Neuron. 2017;94:237–248. doi: 10.1016/j.neuron.2017.02.042. [DOI] [PubMed] [Google Scholar]
- 63.Uemura K., Kuzuya A., Aoyagi N., Ando K., Shimozono Y., Ninomiya H. Amyloid beta inhibits ectodomain shedding of N-cadherin via down-regulation of cell-surface NMDA receptor. Neuroscience. 2007;145:5–10. doi: 10.1016/j.neuroscience.2006.12.022. [DOI] [PubMed] [Google Scholar]
- 64.Hochstrasser T., Weiss E., Marksteiner J., Humpel C. Soluble cell adhesion molecules in monocytes of Alzheimer's disease and mild cognitive impairment. Exp Gerontol. 2010;45:70–74. doi: 10.1016/j.exger.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saykin A.J., Shen L., Foroud T.M., Potkin S.G., Swaminathan S., Kim S. Alzheimer's Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: Genetics core aims, progress, and plans. Alzheimers Dement. 2010;6:265–273. doi: 10.1016/j.jalz.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lu T., Aron L., Zullo J., Pan Y., Kim H., Chen Y. REST and stress resistance in ageing and Alzheimer's disease. Nature. 2014;507:448–454. doi: 10.1038/nature13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.De Jager P.L., Srivastava G., Lunnon K., Burgess J., Schalkwyk L.C., Yu L. Alzheimer's disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat Neurosci. 2014;17:1156–1163. doi: 10.1038/nn.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.de Bruin C., Mericq V., Andrew S.F., van Duyvenvoorde H.A., Verkaik N.S., Losekoot M. An XRCC4 splice mutation associated with severe short stature, gonadal failure, and early-onset metabolic syndrome. J Clin Endocrinol Metab. 2015;100:E789–E798. doi: 10.1210/jc.2015-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu J., Chatterjee M., Baguley T.D., Brouillette J., Kurup P., Ghosh D. Inhibitor of the tyrosine phosphatase STEP reverses cognitive deficits in a mouse model of Alzheimer's disease. PLoS Biol. 2014;12:e1001923. doi: 10.1371/journal.pbio.1001923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chin J., Palop J.J., Puolivali J., Massaro C., Bien-Ly N., Gerstein H. Fyn kinase induces synaptic and cognitive impairments in a transgenic mouse model of Alzheimer's disease. J Neurosci. 2005;25:9694–9703. doi: 10.1523/JNEUROSCI.2980-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Snyder E.M., Nong Y., Almeida C.G., Paul S., Moran T., Choi E.Y. Regulation of NMDA receptor trafficking by amyloid-beta. Nat Neurosci. 2005;8:1051–1058. doi: 10.1038/nn1503. [DOI] [PubMed] [Google Scholar]
- 72.Kamceva M., Benedict J., Nairn A.C., Lombroso P.J. Role of Striatal-Enriched Tyrosine Phosphatase in Neuronal Function. Neural Plast. 2016;2016:8136925. doi: 10.1155/2016/8136925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Munoz J.J., Tarrega C., Blanco-Aparicio C., Pulido R. Differential interaction of the tyrosine phosphatases PTP-SL, STEP and HePTP with the mitogen-activated protein kinases ERK1/2 and p38alpha is determined by a kinase specificity sequence and influenced by reducing agents. Biochem J. 2003;372:193–201. doi: 10.1042/BJ20021941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Paul S., Nairn A.C., Wang P., Lombroso P.J. NMDA-mediated activation of the tyrosine phosphatase STEP regulates the duration of ERK signaling. Nat Neurosci. 2003;6:34–42. doi: 10.1038/nn989. [DOI] [PubMed] [Google Scholar]
- 75.Nguyen T.H., Liu J., Lombroso P.J. Striatal enriched phosphatase 61 dephosphorylates Fyn at phosphotyrosine 420. J Biol Chem. 2002;277:24274–24279. doi: 10.1074/jbc.M111683200. [DOI] [PubMed] [Google Scholar]
- 76.Xu J., Kurup P., Bartos J.A., Patriarchi T., Hell J.W., Lombroso P.J. Striatal-enriched protein-tyrosine phosphatase (STEP) regulates Pyk2 kinase activity. J Biol Chem. 2012;287:20942–20956. doi: 10.1074/jbc.M112.368654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tosto G., Fu H., Vardarajan B.N., Lee J.H., Cheng R., Reyes-Dumeyer D. F-box/LRR-repeat protein 7 is genetically associated with Alzheimer's disease. Ann Clin Transl Neurol. 2015;2:810–820. doi: 10.1002/acn3.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Carlton J. The ESCRT machinery: A cellular apparatus for sorting and scission. Biochem Soc Trans. 2010;38:1397–1412. doi: 10.1042/BST0381397. [DOI] [PubMed] [Google Scholar]
- 79.Yan W., Frank C.L., Korth M.J., Sopher B.L., Novoa I., Ron D. Control of PERK eIF2alpha kinase activity by the endoplasmic reticulum stress-induced molecular chaperone P58IPK. Proc Natl Acad Sci U S A. 2002;99:15920–15925. doi: 10.1073/pnas.252341799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Posse de Chaves E., Sipione S. Sphingolipids and gangliosides of the nervous system in membrane function and dysfunction. FEBS Lett. 2010;584:1748–1759. doi: 10.1016/j.febslet.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 81.Vardarajan B.N., Schaid D.J., Reitz C., Lantigua R., Medrano M., Jimenez-Velazquez I.Z. Inbreeding among Caribbean Hispanics from the Dominican Republic and its effects on risk of Alzheimer disease. Genet Med. 2015;17:639–643. doi: 10.1038/gim.2014.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rudan I., Rudan D., Campbell H., Carothers A., Wright A., Smolej-Narancic N. Inbreeding and risk of late onset complex disease. J Med Genet. 2003;40:925–932. doi: 10.1136/jmg.40.12.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ghani M., Sato C., Lee J.H., Reitz C., Moreno D., Mayeux R. Evidence of recessive Alzheimer disease loci in a Caribbean Hispanic data set: genome-wide survey of runs of homozygosity. JAMA Neurol. 2013;70:1261–1267. doi: 10.1001/jamaneurol.2013.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.