Abstract
The lysosomal degradation of G protein-coupled receptors (GPCRs) is essential for receptor signaling and down regulation. Once internalized, GPCRs are sorted within the endocytic pathway and packaged into intraluminal vesicles (ILVs) that bud inward to form the multivesicular endosome (MVE). The mechanisms that control GPCR sorting and ILV formation are poorly understood. Quantitative strategies are important for evaluating the function of adaptor and scaffold proteins that regulate sorting of GPCRs at MVEs. In this chapter, we outline two strategies for the quantification and visualization of GPCR sorting into the lumen of MVEs. The first protocol utilizes a biochemical approach to assay the sorting of GPCRs in a population of cells, whereas the second strategy examines GPCR sorting in individual cells using immunofluorescence confocal microscopy. Combined, these assays can be used to establish the kinetics of activated GPCR lysosomal trafficking in response to specific ligands, as well as evaluate the contribution of endosomal adaptors to GPCR sorting at MVEs. The protocols presented in this chapter can be adapted to analyze GPCR sorting in a myriad of cell types and tissues, and expanded to analyze the mechanisms that regulate MVE sorting of other cargoes.
INTRODUCTION
G protein-coupled receptors (GPCRs) comprise the largest family of mammalian transmembrane signaling receptors and are important drug targets for the treatment of multiple diseases, including cancer and cardiovascular disease. Endosomal sorting and lysosomal degradation are critical for regulating GPCR signaling, and defects in receptor degradation impact many pathophysiological conditions. At late endosomes, activated GPCRs are packaged into membrane invaginations that bud inward to form intraluminal vesicles (ILVs) (Marchese, Paing, Temple, & Trejo, 2008). Once formed, multivesicular endosomes (MVEs) fuse with lysosomes, facilitating receptor degradation. A diverse array of adaptor and scaffold proteins mediate GPCR sorting into ILVs. GPCRs targeted for degradation by ubiquitination are bound by ubiquitin-binding subunits of the ESCRT (endosomal sorting complexes required for transport) complexes (Hislop & von Zastrow, 2011; Marchese et al., 2003; Shenoy et al., 2008). The ESCRT complexes are evolutionary conserved (Hurley & Emr, 2006) and facilitate the biogenesis of MVEs containing GPCRs and other transmembrane proteins (Babst, 2005) through a process that involves cargo binding at early endosomes by the ESCRT-0/I complexes. The ESCRT-I complex recruits members of the ESCRT-II complex, which mediates ILV formation (Im, Wollert, Boura, & Hurley, 2009) and encaptures ubiquitinated cargo (Wollert & Hurley, 2010). Receptors are then deubiquitinated and the ESCRT-III complex mediates the scission of ILVs (Hurley & Hanson, 2010). However, some GPCRs are sorted into ILVs without being ubiquitinated. The adaptor proteins GASP1 and ALIX facilitate the interaction between specific GPCRs and the ESCRT complexes. GASP1 binds to the C-terminal tail of multiple GPCRs and facilitates interaction with all four ESCRT complexes (Cho et al., 2013; Henry, White, Marsh, von Zastrow, & Hislop, 2011). In addition, the adaptor protein ALIX directs the sorting of protease-activated receptor 1 (PAR1), a GPCR for the coagulant protease thrombin. ALIX binds to a YPXnL motif within the second intracellular loop of PAR1, and facilitates interaction between PAR1 and the ESCRT-III complex, bypassing the ubiquitin-binding ESCRT complexes (Dores, Chen, et al., 2012). However, the molecular mechanisms that regulate these pathways are not well understood. In addition, these pathways are defined by studies of a small number of human GPCRs, suggesting that many more multivesicular endosomal sorting pathways could exist.
Robust and quantitative strategies to assay GPCR sorting into the lumen of MVEs are important tools for investigating the regulation and dynamics of receptor sorting at the late endosome. This chapter will outline two experimental strategies for quantifying and visualizing GPCR sorting into the lumen of MVEs. The first strategy surveys GPCR sorting in a population of cells by analyzing the amount of GPCRs that are incorporated into MVEs and protected from protease cleavage. The second strategy employs confocal immunofluorescence microscopy to visualize and quantify the sorting of GPCRs into the lumen of expanded endosomes in individual cells. Together, these strategies can be used to quantify defects in GPCR endosomal sorting following the genomic or posttranscriptional manipulation of target endocytic adaptor and scaffold proteins, and define the dynamics of GPCR sorting into MVEs in a variety of cell types.
1. OBJECTIVES AND RATIONALE
There are multiple approaches to studying the sorting of GPCRs into MVEs. Traditionally, immune-electron microscopy (IEM) has served as the benchmark for imaging GPCRs in ILVs and at the limiting membrane of MVEs. IEM allows for high resolution at high magnification and reveals valuable qualitative information regarding the subcellular and subendosomal localization of GPCRs in relation to other endosomal markers. However, electron micrographs are difficult to quantify, and time-course experiments are resource intensive. In addition, antibodies for many GPCRs are unavailable or not sensitive enough for IEM. In the following sections, we will outline two strategies that complement IEM studies, providing quantitative biochemical and visual evidence of GPCR sorting into the lumen of MVEs.
The proteinase-protection assay is a robust method for studying the trafficking of GPCRs at MVEs. The assay requires gentle permeabilization of the plasma membrane that leaves internal endosomal membranes intact. This process has been described for studying endosome acidification (Diaz & Stahl, 1989), and is a powerful tool for quantifying GPCR levels within protective endosomal compartments. Following permeabilization of the plasma membrane, GPCRs that are presented at the plasma membrane and at the limiting membranes of endosomes are susceptible to proteinase degradation, whereas receptors that have been internalized into MVEs remain protected from proteinase cleavage. The levels of GPCRs within protective endosomes are measured by immunoblot analysis and quantified using densitometry or other methods such as the Odyssey system (LI-COR). This strategy allows for the survey of GPCR sorting in an entire population of cells, and can be expanded to encompass multiple time points in a single experiment.
Immunofluorescence confocal microscopy is a robust method for assaying the localization of GPCRs within the cell and is widely used to investigate the role of adaptor and scaffold proteins in GPCR endosomal trafficking. However, the relatively small size of endosomes prevents quantitative analysis of receptor sorting into ILVs of MVEs using fluorescence microscopy techniques. Research focused on the biogenesis and regulation of endosomes has produced a number of useful tools for labeling specific endosomal populations and manipulating endosome morphology. Rab GTPases regulate fusion of vesicles and endosomes (Rink, Ghigo, Kalaidzidis, & Zerial, 2005) and define discrete endosomal compartments, including early, late, and recycling endosomes (Simons & Zerial, 1993). Specifically, Rab5 mediates homotypic fusion of endocytic vesicles that form early endosomes (Stenmark et al., 1994), and a constitutive active form of Rab5 (Rab5-Q79L) induces the formation of enlarged endosomes through increased endosomal membrane fusion (Stenmark et al., 1994) that retains the ability to sort endocytic cargo. The expression of Rab5-Q79L fused to green fluorescence protein (GFP) labels expanded endosome limiting membrane with green fluorescence, facilitating the visualization of GPCR sorting into the endosomal lumen (Dores, Paing, et al., 2012; Hislop, Henry, & von Zastrow, 2011). This technique complements the proteinase-protection assay and provides direct visual evidence of GPCR sorting into endosomes.
2. MATERIALS AND INSTRUMENTS
2.1. REAGENTS
2.1.1. Cell culture, media, and general reagents
HeLa cells
Plasmid constructs encoding: FLAG-PAR1 in pBJ vector (Trejo, Altschuler, Fu, Mostov, & Coughlin, 2000), Rab5-Q79L-GFP in pcDNA (Gullapalli et al., 2004)
Dulbecco’s Modification of Eagles Medium (DMEM) + glucose, l-glutamine,
pyruvate
Fetal bovine serum
Penicillin Streptomycin Fibronectin
Polyethylenimine (PEI)
HEPES buffer
Bovine serum albumen (BSA)
Phosphate-buffered salt (PBS) without calcium or magnesium
2.1.2. Antibodies
Immunoblotting Antibodies: Rabbit polyclonal anti-FLAG (Rockland Immunochemicals, Cat. No. 600-401-383), mouse monoclonal anti-early endosomal antigen-1 (EEA1) (BD Biosciences, Cat. No. 610457).
Immunofluorescence Antibodies: Rabbit polyclonal anti-FLAG (Rockland Immunochemicals, Cat. No. 600-401-383), Alexa Fluor® 594-conjugated goat anti-rabbit (Life Technologies, Cat. No. A-11012).
2.1.3. Proteinase K protection assay reagents
Dipotassium phosphate (K2HPO4)
Monopotassium phosphate (KH2PO4)
1 M magnesium chloride (MgCl2)
Sucrose
Digitonin
Proteinase K
Phenylmethylsulfonyl fluoride (PMSF)
Triton X-100
2.1.4. Confocal immunofluorescence microscopy assay reagents
Paraformaldehyde (PFA)
Saponin
Goat serum
Methanol
Nonfat dry milk
3 M sodium acetate
FluorSave™ Reagent (Millipore)
2.2. INSTRUMENTATION
Microscope: Olympus IX81, spinning-disk confocal imaging system (DSU), PlanApo 60x oil objective (1.4 NA), ORCA-ER digital camera (Hamamatsu Photonics).
Software: SlideBook 5.0 software (Intelligent Imaging Innovations).
3. METHODS
3.1. CELL PLATING AND TRANSFECTION
HeLa cells are cultured in DMEM supplemented with 10% (v/v) fetal bovine serum, penicillin, and streptomycin at 37 °C in a 5% CO2 incubator. For proteinase-protection assays, cells are split into fibronectin-coated 6-well polystyrene tissue culture plates at 5.0 × 105 cells/well and grown for 48 h until 80–90% confluent. For immunofluorescence confocal microscopy experiments, circular glass coverslips (Fisherbrand Cat. No. 12-454-100, 18CIR-1) that have been ethanol-dipped and dried are placed in 12-well polystyrene tissue culture plates. Coverslips are then precoated with 300 μL fibronectin for 15 min at room temperature. Cells are diluted to 0.75 × 105 cells/well and grown 24 h until 30–40% confluent. Cells are then transfected with plasmid expressing Rab5-Q79L-GFP using PEI. Transfected cells are grown 24–48 h until cells are 70–80% confluent prior to processing.
3.2. PROTEINASE-PROTECTION ASSAY
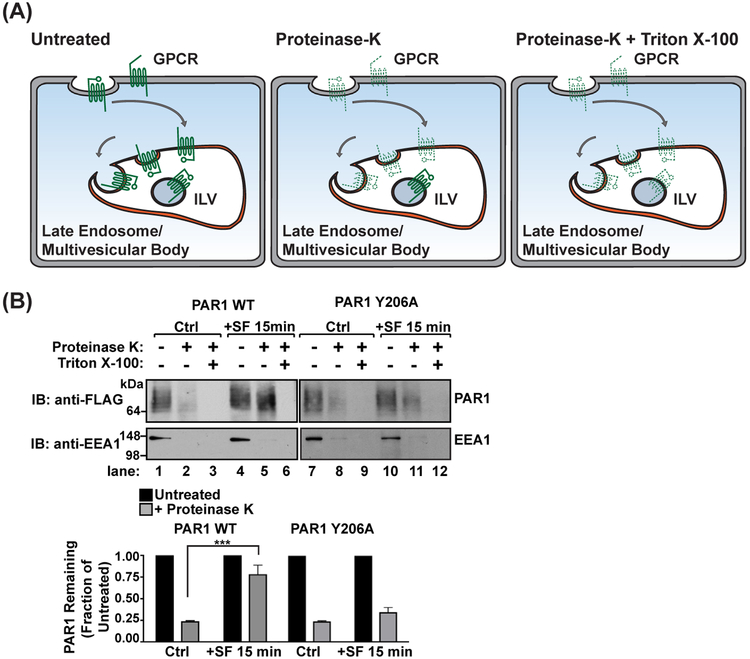
Cells are first incubated with DMEM supplemented with 20 mM HEPES, 1 mg/mL BSA, and 2 mM leupeptin to inhibit lysosomal degradation at 37 °C. Prior to stimulation, cells are incubated in a water-bath incubator at 37 °C for 1 h, then stimulated for the appropriate times with agonist. The duration of stimulation will vary depending upon the GPCR, but will coincide with wild-type (WT) receptor degradation. Following stimulation, cells are placed on ice to halt membrane trafficking and washed with ice-cold PBS without Mg2+ or Ca2+. Cells are then gently lifted from the plates using a cell scraper and pelleted by centrifugation. Cells are resuspended in DMEM media supplemented with 6.5 μg/mL digitonin to gently permeabilize the plasma membrane, leaving endosomal membranes intact. Following digitonin treatment, membranes are pelleted by high-speed centrifugation, then resuspended in homogenization buffer which is split into three equal fractions. The first fraction is left untreated and represents the total amount of receptor in limiting membranes as well as inside protective endosomal compartments (Figure 1(A)). The second fraction is treated with 2.5 ng/mL proteinase K, which will degrade any receptors on limiting membranes of endosomes or the plasma membrane (Figure 1(A)). The third fraction is treated with 2.5 ng/mL proteinase K and 0.1% Triton X-100 detergent. This treatment will degrade all membranes and serve as a control for proteinase K activity (Figure 1(A)). Samples are then treated with 2X Laemmli sample buffer supplemented with 20 mM PMSF, then analyzed by immunoblotting (Figure 1(B)). As an additional control for digitonin treatment, samples are probed with antibodies targeted to early endosome antigen 1 (EEA1), a cytosolic protein that associates strongly with the cytoplasmic surface of endosomes (Figure 1(B)). Immunoblots are quantified by densitometry using ImageJ or appropriate software. Statistical analysis of GPCR levels by 2-way ANOVA requires a minimum of three replicate experiments.
FIGURE 1.
Proteinase-protection endosomal sorting assay. (A) A schematic of proteinase treatment assay with predicted results. Untreated samples represent the total amount of GPCR. Treatment with proteinase K degrades all GPCRs exposed on the plasma membrane or limiting membranes of internal endosomes (dashed lines), whereas GPCRs sorted into intraluminal vesicles (ILVs) are protected from proteinase activity (solid lines). Membranes treated with Triton X-100 detergent acts as a control for proteinase K activity, exposing all receptors to proteinase K degradation. (B) Agonist-stimulated PAR1 sorting into protective endosomal compartments is mediated by the YPXnL motif localized in the second intracellular loop. HeLa cells expressing FLAG-PAR1 WT or FLAG-PAR1 Y206A were stimulated with 100 μM SFLLRN for 15 min. Cells were harvested and treated with digitonin to permeabilize the plasma membrane, then treated with either H2O (untreated), proteinase K, or proteinase K with Triton X-100 detergent. The amount of PAR1 was analyzed by immunoblot and quantified using densitometry (***, p < 0.001, n = 3, 2-way ANOVA)
3.2.1. Workflow of experiment
Incubate cells for 1 h in 1 mL 37 °C DMEM supplemented with 2 mM HEPES, 1 mg/mL BSA, and 2 mM leupeptin
Stimulate cells with agonist diluted in 1 mL DMEM supplemented with 2 mM HEPES and 1 mg/mL BSA preheated to 37 °C
Remove cells from 37 °C water bath and place on ice
Wash 1× with 1 mL ice-cold PBS (Ca2+/Mg2+-free)
Incubate cells in 1 mL ice-cold PBS for 5 min
Scrape cells and collect into prelabeled 1.5 mL tubes
Pellet cells at 1500 rpm for 5 min at 4 °C
- Resuspend cell pellets in 1 mL DMEM supplemented with 20 mM HEPES, 1 mg/mL BSA, 6.5 μg/mL digitonin
- Incubate for 5 min at room temperature
- Incubate for 30 min on ice
Centrifuge at 14,000 rpm for 5 min at 4 °C to pellet membranes
Resuspend cell pellets in 300 μL homogenization buffer (See Table 1)
- Combine 100 μL of sample with either of the following:
- 1 μL H2O treat (no enzyme)
- 2.5 ng/mL proteinase K (add 1 μL of 250 ng/mL stock proteinase K)
- 2.5 ng/mL proteinase K + 0.1% Triton X-100 (add 1 μL of 250 ng/mL stock proteinase K and 1 μL 10% Triton X-100 stock)
Incubate at room temperature for 10 min
Stop reaction by adding 100 μL 2X Laemmli sample buffer containing 20 mM PMSF
Boil at 100 °C for 3 min prior to loading on to an SDS-PAGE gel
Table 1.
Homogenization Buffer (pH 6.7)
| 100 mM K2HPO4/KH2PO4 | 0.87 g K2HPO4/0.68 g KH2PO4 |
| 5 mM MgCl2 | 500 μL (1 M stock) |
| 250 mM sucrose | 8.6 g |
| H2O | 100 mL |
3.2.2. Considerations
We have observed that digitonin and proteinase K have a limited shelf life typically between 2 and 3 weeks following dilution when stored at 4 °C. Enzymes in solution can be frozen and stored for 2–3 months at −20 °C. Samples may require vortexing immediately following boiling due to the presence of genomic DNA.
3.2.3. Results
We found that stimulation of WT PAR1 with a synthetic peptide agonist SFLLRN (SF) induces translocation into protective endosomal compartments. In the absence of agonist, stimulation of PAR1 is sensitive to proteinase K treatment (Figure 1(B), lane 2), indicating that the majority of PAR1 is localized to limiting membranes at steady state. In contrast, PAR1 stimulated with SFLLRN for 15 min is protected from proteinase K degradation (Figure 1(B), lane 5). PAR1 is degraded in samples treated with proteinase K and Triton X-100 detergent, confirming proteinase K activity (Figure 1(B), lanes 3 and 6). In comparison, PAR1 Y206A, a mutant that cannot bind to the adaptor protein ALIX and is not degraded (Dores, Chen, et al., 2012), fails to be protected from proteinase K degradation following agonist stimulation (Figure 1(B), lane 11). The degradation of EEA1 indicates that digitonin treatment permeabilized the plasma membrane, allowing proteinase K access to proteins anchored to the cytosolic side of endosomal limiting membranes. These data provide quantitative evidence that activated PAR1 is sorted into the lumen of endosomes and protected from protease cleavage, and suggest that mutation of the ALIX binding site on PAR1 blocks receptor sorting into the endosomal lumen.
3.3. QUANTIFICATION OF GPCR SORTING AT MVEs EXPANDED BY Rab5-Q79L EXPRESSION
Cells should be covered or protected from direct light during incubations to prevent photo bleaching of the Rab5-Q79L-GFP. Cells transfected with Rab5-Q79L-GFP were seeded onto 18 mm glass coverslips (Fisherbrand Cat. No. 12-454-100, 18CIR-1) and preincubated in DMEM supplemented with 20 mM HEPES, 1 mg/mL BSA, and 2 mM leupeptin to inhibit lysosomal degradation. Following pretreatment, cells are shifted to ice and chilled to halt membrane trafficking. Primary antibody against the GPCR is then added to each well to label the surface cohort of receptor while maintaining leupeptin treatment. Following primary antibody labeling for 1 h, cells are washed two times with ice-cold DMEM. Cells are then stimulated with prewarmed 37 °C media containing agonist and shifted to a 37 °C water bath. Unstimulated control plates are maintained on ice. Following stimulation, cells are shifted back to ice and washed once with ice-cold PBS. Cells are fixed with 4% PFA, washed with ice-cold PBS, then permeabilized with a short methanol treatment. Permeabilized cells are gently washed twice with ice-cold PBS. The coverslips are removed from the wells onto a flat surface coated with parafilm at room temperature. Cells are incubated in quench buffer that is pipetted directly on top of the coverslips. Cells are then incubated with wash buffer, followed by incubation with secondary antibody diluted in wash buffer for 1 h at room temperature. Following incubation with secondary antibody, coverslips are washed with room temperature PBS and then mounted onto glass slides using FluorSave™ reagent.
3.3.1. Workflow of experiment
Incubate cells for 1 h in 0.4 mL 37 °C DMEM supplemented with 2 mM HEPES, 1 mg/mL BSA, and 2 mM leupeptin
Place cells on ice, incubate 10 min to chill cells
- Dilute primary antibody in ice-cold DMEM supplemented with 2 mM HEPES and 1 mg/mL BSA and add to wells
- For a 1:1000 antibody dilution: 0.5 μL primary antibody (1.0 μg/μL)/well diluted in 100 μL DMEM (500 μL total volume/well)
Incubate on ice for 1 h to surface-label receptor expressed in cells
Wash 2× with 500 μL ice-cold DMEM supplemented with 2 mM HEPES and 1 mg/mL BSA
Stimulate cells with agonist diluted in 500 μL DMEM supplemented with 2 mM HEPES and 1 mg/mL BSA, preheated to 37 °C
Remove cells from 37 °C water bath and place on ice
Wash 1× with 500 μL ice-cold PBS (Ca2+/Mg2+-free)
Incubate on ice with 500 μL ice-cold 4% PFA (pH 7.2) for 5 min to fix cells
Wash 1× with 500 μL ice-cold PBS (Ca2+/Mg2+-free)
Add 500 μL ice-cold 100% methanol for 30 s to permeabilize cells
Wash 2× with 500 μL ice-cold PBS (Ca2+/Mg2+-free)
Remove coverslips from wells and place onto a flat surface covered with parafilm at room temperature
Incubate 3× with 300 μL quench buffer (See Table 2) for 5 min at room temperature, bubbled on top of coverslips
Incubate 3× with 300 μL wash buffer (See Table 2) for 5 min
Incubate with 300 μL secondary antibody diluted in wash buffer for 1 h at room temperature
Wash 4× with 300 μL PBS (Ca2+/Mg2+-free) for 5 min at room temperature
Place 25 μL FluorSave™ reagent (Millipore) onto glass slide, then invert coverslip onto drop
Aspirate excess FluorSave™ reagent from edges of coverslip
Dry slides 1–24 h prior to collecting images
Table 2.
Quench and Wash Buffers
| Quench Buffer | Wash Buffer | |
|---|---|---|
| 1% Nonfat dry milk | 0.15 g | 0.3g |
| 150 mM sodium acetate | 750 μL (3 M stock) | |
| 1 × PBS | 14.25 mL | 30 mL |
| Total volume | 15 mL | 30 mL |
3.3.2. Considerations and alternative approaches
Due to the short duration of methanol treatment, we recommend treating four or fewer samples at a time. Methanol permeabilization may not be optimal for labeling certain GPCRs and cytosolic proteins. We have also used saponin to permeabilize cells prior to antibody labeling with minimal effect on the morphology of Rab5-Q79L-expanded endosomes. Following fixing, coverslips are removed from wells and placed on a flat surface covered with parafilm at room temperature. Samples are incubated for 30 min in blocking buffer (PBS supplemented with 1% goat serum and 0.03% saponin) to facilitate permeabilization. Samples are washed with PBS and then treated for 1 h with secondary antibody diluted in blocking buffer. Unbound antibody is washed away with PBS and coverslips are mounted to glass slides using FluorSave™ reagent.
3.3.2.1. Workflow of saponin permeabilization
Complete steps 1–10 above
Remove coverslips and place onto a flat surface covered with parafilm at room temperature, cell-side up
Incubate for 30 min at room temperature with 300 μL blocking buffer (PBS supplemented with 1% goat serum and 0.03% saponin)
Wash 2× with 300 μL PBS, no incubation
Label for 1 h at room temperature with 300 μL secondary antibody diluted in blocking buffer
Wash sequentially 3× with 300 μL PBS, no incubation
Place 25 μL FluorSave™ reagent onto glass slide, then invert coverslip onto drop
Aspirate excess FluorSave™ reagent from edges of coverslip
Dry slides 1–24 h prior to collecting images
3.3.3. Results and fluorescence intensity analysis
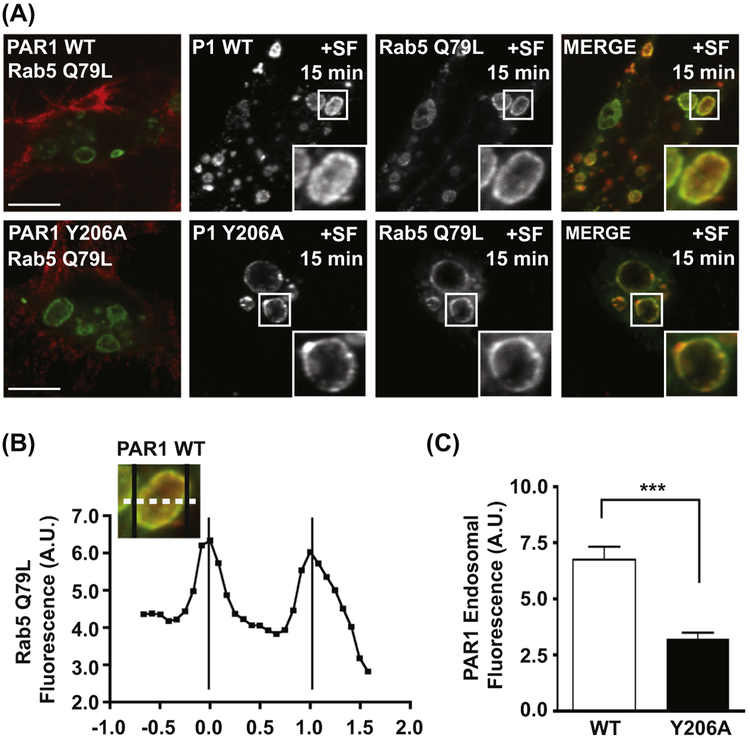
Activated PAR1 sorts into ILVs of MVEs enlarged by Rab5-Q79L expression. The expression of Rab5-Q79L-GFP induces the expansion of endosomal compartments, but does not affect the surface expression of unstimulated PAR1 (Figure 2(A)). Treatment of cells with the PAR1 peptide agonist SFLLRN for 15 min induces WT receptor internalization and sorting to the limiting membrane and the lumen of Rab5-positive endosomes (Figure 2(A)). In comparison, agonist stimulation of the PAR1 Y206A mutant induces trafficking to the limiting membrane of Rab5-positive endosomes, however, the Y206A mutant receptor is not sorted into the lumen of expanded endosomes (Figure 2(A)). To quantify the amount of PAR1 sorted into the lumen of endosomes expanded by Rab5-Q79L expression, we used a line scan tool available in the SlideBook 5.0 microscope software to measure fluorescence intensity. Alternatively, line scans can be performed using ImageJ. The fluorescence intensity of Rab5-Q79L-GFP defines the limiting membrane of the endosome (Figure 2(B)), and fluorescence intensity of PAR1 is represented as an average of intensity values measured between the Rab5-maxima. Consistent with our proteinase-protection assay, PAR1 WT fluorescence intensity is significantly higher in the lumen of expanded MVEs compared to PAR1 Y206A (Figure 2(C)). These results provide visual confirmation that activated PAR1 is sorted into the lumen of endosomes, and that ALIX binding to PAR1 is required for receptor sorting into ILVs.
FIGURE 2.
Visualization and quantification of GPCR sorting into expanded endosomes. (A) Agonist-stimulated PAR1 sorting into the lumen of expanded endosomes is mediated by the YPXnL motif. HeLa cells expressing FLAG-PAR1 WT or FLAG-PAR1 Y206A were transfected with Rab5-Q79L-GFP, then surface-labeled with anti-FLAG antibody. Cells were stimulated for 15 min with 100 μM SFLLRN, then fixed and analyzed by confocal microscopy. (B) Line scan analysis of Rab5-Q79L-GFP endosomes. GFP-fluorescence intensity was quantified by line scan (dashed line) and plotted to determine the limiting edges of the endosome (black vertical lines). (C) The PAR1 Y206A mutant is defective for sorting into the lumen of endosomes. PAR1 fluorescence intensity was measured within the limits defined by Rab5-Q79L-GFP (B) and averaged from multiple expanded endosomes (***, p < 0.001, n = 6, Student’s t-test). (See color plate)
SUMMARY
In this chapter, we have described two techniques for the quantification and visualization of GPCR sorting into MVEs. We have outlined a biochemical strategy for the quantification of GPCR sorting into protective endosomal compartments, and we have also provided a protocol for visualizing GPCR sorting into MVEs using confocal immunofluorescence microscopy. These methods can be used to investigate the function of endocytic adaptor and scaffold proteins in the endolysosomal sorting of specific GPCRs. These strategies also provide temporal and spatial information about the sorting of GPCRs in cell populations and within individual cells. Our results demonstrate the strength of these techniques in defining the components and regulatory mechanisms of novel endosomal sorting pathways for GPCRs.
REFERENCES
- Babst M (2005). A protein’s final ESCRT. Traffic, 6(1), 2–9. [DOI] [PubMed] [Google Scholar]
- Cho DI, Zheng M, Min C, Kwon KJ, Shin CY, Choi HK, et al. (2013). ARF6 and GASP-1 are post-endocytic sorting proteins selectively involved in the intracellular trafficking of dopamine D(2) receptors mediated by GRK and PKC in transfected cells. British Journal of Pharmacology, 168(6), 1355–1374. 10.1111/bph.12025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz R, & Stahl PD (1989). Chapter 2 Digitonin permeabilization procedures for the study of endosome acidification and function In Alan MT (Ed.), Methods in cell biology (Vol. 31, pp. 25–43). Academic Press. [DOI] [PubMed] [Google Scholar]
- Dores MR, Chen B, Lin H, Soh UJK, Paing MM, Montagne WA, et al. (2012). ALIX binds a YPX3L motif of the GPCR PAR1 and mediates ubiquitin-independent ESCRT-III/MVB sorting. The Journal of Cell Biology, 197(3), 407–419. 10.1083/jcb.201110031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dores MR, Paing MM, Lin H, Montagne WA, Marchese A, & Trejo J (2012). AP-3 regulates PAR1 ubiquitin-independent MVB/lysosomal sorting via an ALIX-mediated pathway. Molecular Biology of the Cell, 23(18), 3612–3623. 10.1091/mbc.E12-03-0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullapalli A, Garrett TA, Paing MM, Griffin CT, Yang Y, & Trejo J (2004). A role for sorting nexin 2 in epidermal growth factor receptor down-regulation: evidence for distinct functions of sorting nexin 1 and 2 in protein trafficking. Molecular Biology of the Cell, 15(5), 2143–2155. 10.1091/mbc.E03-09-0711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry AG, White IJ, Marsh M, von Zastrow M, & Hislop JN (2011). The role of ubiquitination in lysosomal trafficking of delta-opioid receptors. Traffic, 12(2), 170–184. 10.1111/j.1600-0854.2010.01145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hislop JN, Henry AG, & von Zastrow M (2011). Ubiquitination in the first cytoplasmic loop of μ-opioid receptors reveals a hierarchical mechanism of lysosomal down-regulation. The Journal of Biological Chemistry, 286(46), 40193–40204. 10.1074/jbc.M111.288555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hislop JN, & von Zastrow M (2011). Role of ubiquitination in endocytic trafficking of G-protein-coupled receptors. Traffic, 12(2), 137–148. 10.1111/j.1600-0854.2010.01121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, & Emr SD (2006). The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annual Review of Biophysics and Biomolecular Structure, 35, 277–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, & Hanson PI (2010). Membrane budding and scission by the ESCRT machinery: it’s all in the neck. Nature Reviews Molecular Cell Biology, 11(8), 556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im YJ, Wollert T, Boura E, & Hurley JH (2009). Structure and function of the ESCRT-II-III interface in multivesicular body biogenesis. Developmental Cell, 17(2), 234–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchese A, Paing MM, Temple BRS, & Trejo J (2008). G protein-coupled receptor sorting to endosomes and lysosomes. Annual Review of Pharmacology and Toxicology, 48(1), 601–629. 10.1146/annurev.pharmtox.48.113006.094646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchese A, Raiborg C, Santini F, Keen JH, Stenmark H, & Benovic JL (2003). The E3 ubiquitin ligase AIP4 mediates ubiquitination and sorting of the G protein-coupled receptor CXCR4. Developmental Cell, 5(5), 709–722. [DOI] [PubMed] [Google Scholar]
- Rink J, Ghigo E, Kalaidzidis Y, & Zerial M (2005). Rab conversion as a mechanism of progression from early to late endosomes. Cell, 122(5), 735–749. [DOI] [PubMed] [Google Scholar]
- Shenoy SK, Xiao K, Venkataramanan V, Snyder PM, Freedman NJ, & Weissman AM (2008). Nedd4 mediates agonist-dependent ubiquitination, lysosomal targeting, and degradation of the beta2-adrenergic receptor. The Journal of Biological Chemistry, 283(32), 22166–22176. 10.1074/jbc.M709668200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K, & Zerial M (1993). Rab proteins and the road maps for intracellular transport. Neuron, 11, 789–799. [DOI] [PubMed] [Google Scholar]
- Stenmark H, Parton RG, Steele-Mortimer O, Lutcke A, Gruenberg J, & Zerial M (1994). Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. The EMBO Journal, 13(6), 1287–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trejo J, Altschuler Y, Fu H-W, Mostov KE, & Coughlin SR (2000). Protease-activated Receptor-1 down-regulation. The Journal of Biological Chemistry, 275(40), 31255–31265. 10.1074/jbc.M003770200. [DOI] [PubMed] [Google Scholar]
- Wollert T, & Hurley JH (2010). Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature, 464(7290), 864–869. [DOI] [PMC free article] [PubMed] [Google Scholar]