Abstract
Background
MYO18B has been identified as a novel tumor suppressor gene in several cancers. However, its specific roles in the progression of hepatocellular carcinoma (HCC) has not been well defined.
Methods
We firstly identified the expression and prognostic values of MYO18B in HCC using TCGA cohort and our clinical data. Then, MYO18B knockdown by RNA inference was implemented to investigate the effects of MYO18B on HCC cells. Quantitative RT-PCR and Western blot were used to determine gene and protein expression levels. CCK-8 and colony formation assays were performed to examine cell proliferation capacity. Wound healing and transwell assays were used to evaluate the migration and invasion of HepG2 cells.
Results
MYO18B was overexpressed and correlated with poor prognosis in HCC. MYO18B expression was an independent risk factor for overall survival. Knockdown of MYO18B significantly inhibited the proliferation, migration and invasion of HepG2 cells. Meanwhile, MYO18B knockdown could effectively suppress the phosphorylation of PI3K, AKT, mTOR and P70S6K, suggesting that MYO18B might promote HCC progression by targeting PI3K/AKT/mTOR signaling pathway.
Conclusions
MYO18B promoted tumor growth and migration via the activation of PI3K/AKT/mTOR signaling pathway. MYO18B might be a promising target for clinical intervention of HCC.
Keywords: Hepatocellular carcinoma, MYO18B, PI3K/AKT/mTOR signaling pathway, Prognosis
Background
Hepatocellular carcinoma (HCC) is the most common primary liver cancer, representing the fifth most common cancer and the third leading cause of cancer death worldwide [1]. It is well known that persistent infections by HBV or HCV are the primary inducers of chronic liver disease, thereby resulting in liver cirrhosis and HCC [2, 3]. Aflatoxin exposure and alcohol abuse are also important risk factors for developing HCC [2]. Surveillance by biannual ultrasonography is recommended for such patients [4, 5]. Current treatment options mainly include resection, liver transplantation and interventional radiology [5]. Although recent advances in therapies have achieved improved prognoses for patients with HCC, it is still involved in a poor survival rate due to late diagnosis [6]. Currently, patients with advanced HCC lack effective therapies, representing a unique clinical challenge. Given the current burden of HCC, identifying biomarkers associated with the progression and prognosis of HCC is a promising approach to make early diagnosis, predict prognosis and develop novel therapeutic strategies.
Over the past decade, significant effort has focused on the detection of molecular alterations in HCC [7–10]. Various kinds of biomarkers (such as AFP, VEGF, hepatocyte growth factor) have been proven to harbor prognostic implications, but only serum AFP has been widely used to complement HCC surveillance and guide treatment decisions [11–13]. However, AFP levels display limitations in sensitivity and specificity [14, 15]. It is imperative to screen adequate predictive or prognostic biomarkers. MYO18B is a myosin family gene located at chromosome 22q12.1. Genetic instability of chromosome arm 22q has been detected in patients with HCC [16, 17], suggesting the presence of a tumor-related gene on this chromosome arm that is involved in HCC carcinogenesis. Zhu et al. [18] indicated that tumor suppressor genes on chromosome 22q11.2-22q12.1 may contribute to the pathogenesis and development of HCC. Moreover, MYO18B has been identified as a tumor suppressor gene whose inactivation is associated with the progression of lung cancer [19, 20], colorectal cancer [21] and ovarian cancer [22]. These observations raise the possibility that MYO18B is a potential cancer marker. However, the specific role of MYO18B in HCC progression is still unclear.
In this study, we firstly identified the expression differences of MYO18B between HCC tissues and healthy tissues and its prognostic value using the public data from TCGA database, and validated the results using an independent clinical cohort. Then, we investigated the specific role of MYO18B in HCC by experimental technique. This work purposed to reveal the significance of MYO18B and its underlying mechanism in the pathogenesis of HCC. The result would be of great emphasis for future control strategy of patients with HCC.
Methods
Patients
In this study, we identified the expression and prognostic value of MYO18B in HCC using two independent cohorts. The RNA-seq data of patients with HCC were obtained from TCGA data portal (https://cancergenome.nih.gov/), which contains 374 HCC samples and 50 normal samples. The other cohort contained a total of 80 patients with HCC who had undergone a resection of primary tumors at The Second Affiliated Hospital of Shantou University Medical College between 2007 and 2009. All patients were histologically confirmed as HCC. TNM classification of hepatocellular carcinoma follows 8th edition AJCC Cancer Staging system. The patients were followed up for 80 months after surgery. None of these patients received radiotherapy or chemotherapy before surgery. The clinical information of these patients was listed in Table 1. The adjacent liver tissue was obtained for control.
Table 1.
Correlation between MYO18B expression and clinical characteristics in patients with hepatocellular carcinoma
| Characteristics | Expression of MYO18B | P value | |
|---|---|---|---|
| Low | High | ||
| Age | 0.361 | ||
| < 60 | 18 | 14 | |
| ≥ 60 | 22 | 26 | |
| Gender | 0.260 | ||
| Female | 20 | 15 | |
| male | 20 | 25 | |
| Grade | 1.000 | ||
| G1 + G2 | 26 | 26 | |
| G3 + G4 | 14 | 14 | |
| Pathologic-Stage | 0.025 | ||
| I + II | 24 | 14 | |
| III + IV | 16 | 26 | |
| Pathologic-T | 0.014 | ||
| T1 + T2 | 26 | 15 | |
| T3 + T4 | 14 | 25 | |
| Pathologic-N | 1.000 | ||
| N0 | 37 | 36 | |
| N1 | 3 | 4 | |
| Pathologic-M | 1.000 | ||
| M0 | 39 | 38 | |
| M1 | 1 | 2 | |
T tumor status, N regional lymph node status, M metastasis status
Cell culture
Human HCC cell line HepG2 was obtained from Cell Bank of the Type Culture Collection, Chinese Academy of Sciences (Shanghai, China), and was cultured in RPMI-1640 supplemented with 10% serum, 100 U/ml penicillin, and 0.1 mg/ml streptomycin at 37 °C in a humidified incubator with 5% CO2.
Transient transfection
Cells were seeded in 6-well plates at a concentration of 1 × 105 per well. The next day, cells were transfected with siRNA (experimental group, si-MYO18B, Cat no. sc-61,119, Santa Cluz Biotechnology, Shanghai, China) or non-specific control siRNA (negative control group, si-con) using Lipofectamine 2000 (Invitrogen Life Technologies, Karlsruhe, Germany) according to the manufacture’s protocol. The transfected cells were harvested after 48-h incubation for subsequent experiments.
Quantitative real-time PCR (qRT-PCR) assay
Total RNA was extracted from cultured cells or frozen human tissues using TRIzol Reagent (Invitrogen, Grand Island, NY, USA), and was then converted into cDNA using a PrimeScript 1st Strand cDNA Synthesis Kit (Takara, Dalian, China) according to the manufacture’s protocol. qRT-PCR assay was carried out using SYBR Green (Takara, Dalian, China). The samples were amplified as the following protocol: 95 °C for 5 min, 40 cycles of 95 °C for 30 s, 60 °C for 40 s, and 72 °C for 1 min. Primer sequences for MYO18B and GAPDH were as follows: MYO18B forward, 5’-GGAAGCAGTTAGCTGTCGC-3′ and reverse, 5’-TTGACTGGTCGTCCTGAGAGA-3; GAPDH forward, 5’-GGAGCGAGATCCCTCCAAAAT-3′, and reverse, 5’-GGCTGTTGTCATACTTCTCATGG-3′. The relative quantification was determined by 2−ΔΔCt method and normalized to GAPDH. All experiments were repeated at least three times.
Western blot
After 48-h transfection, cells were washed with PBS twice, harvested and lysed in RIPA buffer (Termo Scientifc, Rockford, IL, USA) containing protease inhibitors on ice for 30 min. The protein concentration was determined by the BCA method. Next, protein samples (20 μg) were equally loaded onto SDS-PAGE and electrotransferred to PVDF membranes. Subsequently, the membranes were blocked with 5% non-fat milk for 1 h and incubated overnight with primary antibodies (dilution 1:1000) against: MYO18B (Cat no. CBS-PA278900, Wuhan Huamei Biological Engineering Co., Ltd., Hubei, China), phosphoinositide-3 kinase (PI3K, Cat no. 4255, Cell Signaling Technology, Inc., Danvers, MA, USA), phosphorylated- (p-) PI3K (Cat no. 13857, Cell Signaling Technology, Inc.), AKT (Cat no. 9272, Cell Signaling Technology, Inc.), p-AKT (Cat no. 13038, Cell Signaling Technology, Inc.), mTOR (Cat no. 2972, Cell Signaling Technology, Inc.), p-mTOR (Cat no. 2971, Cell Signaling Technology, Inc.), p-P70S6K (Cat no. 9204, Cell Signaling Technology, Inc.), and GAPDH (Cat no. 8884, Cell Signaling Technology, Inc.). The membranes were washed with TBST buffer three times and then incubated with secondary antibodies (Anti-rabbit IgG, HRP-conjugated goat anti-rabbit, dilution 1:3000, Cat no. 7074, Cell Signaling Technology, Inc., Danvers, MA, USA) for 1 h at 25 °C. The membranes were then rinsed three times with blocking solution and visualized by the ECL detection system. The experiments were repeated at least three times for analysis. The intensity of signal on each membrane was determined using Quantity One software. GAPDH was used as a loading control for normalization of protein quantity.
Cell proliferation assay
After transfection, cells were seeded at a density of 1000 per well in 96-well plates. Then cell viability was detected at 24 h, 48 h, 72 h and 96 h using a CCK-8 kit (Sigma-Aldrich, Merck KGaA, Darmstadt, Germany) following the manufacture’s instruction. The absorbance at 450 nm was measured using a microplate reader (Thermo Scientific Microplate Reader, Waltham, Massachusetts, USA). All assays were repeated at least three times.
Colony formation assay
After transfection, cells were seeded at a density of 500 per well in 6-well plates and incubated in culture medium with 10% FBS for 2 weeks. During this period, the medium was replaced every 3 or 4 days. After 2 weeks, when cell colonies were formed, cells were fixed with 4% paraformaldehyde for 30 min and stained with 0.1% crystal violet (Sigma, St. Louis, MO, USA) for 30 min. Visible colonies were imaged and counted using ChemiDoc XRS image screening system (Bio-Rad). Each sample was repeated at least three times.
Wound healing assay
Wound healing assay was used to measure cell migration capacity. Cells suspended in RPMI 1640 medium with 10% serum were seeded in 6-well plates at a density of 5 × 105 cells/ml and incubated for 24 h to 100% confluence. A cell-free wound was made by scratching plates with a 100 μl plastic pipette tip. Afterwards, the cells were cultured with RPMI 1640 medium for another 24 h. The wound widths were recorded after 0 h and 24 h under an Olympus BX51 microscope (Olympus Corporation, Tokyo, Japan).
Transwell migration and invasion assays
Transwell migration and invasion assays were performed in 24-well plates using an 8 μm pore transwell chamber (BD Biosciences, Lake Franklin, New Jersey, USA) with/without Matrigel.
In the migration assay, cells (5 × 103) were seeded in the upper chamber in 100 μl serum-free RPMI 1640 medium. In the invasion assay, the transwell chambers were first coated with Matrigel, then cells (1 × 105) suspended in 100 μl serum-free RPMI 1640 medium were seeded in the matrigel-coated upper chamber. The lower chambers were filled with 500 μl culture medium containing 10% serum. After incubation at 37 °C in 5% CO2 overnight, the wells were washed with PBS and the cells attached to the low surface were fixed with paraformaldehyde for 30 min, and stained with 0.1% crystal violet for 20 min. Cells were imaged and counted under an Olympus BX51 microscope (Olympus Corp., Shinjuku, Tokyo, Japan) over five random fields in each well. Each assay was performed in triplicate.
Statistical analysis
Experimental values were expressed as the mean ± standard deviation, and were analyzed by SPSS Statistics software (version 22.0, Chicago, IL, USA). The data preprocessing and differential expression analysis in TCGA were performed using edgeR package [23] (version 3.20.9) in Bioconductor. The association between clinical characteristics and MYO18B expression was evaluated by Chi-square test. Kaplan-Meier analysis with long-rank test and Cox regression analysis were used to determine the prognostic value of MYO18B in HCC patients. Student’s t-test was performed to determine the significance in cell experiments. P < 0.05 was considered statistically significant.
Results
MYO18B overexpression involves poor prognosis in HCC
We first analyzed the expression differences of MYO18B between tumor and normal tissues using TCGA cohort. Our work found that MYO18B was obviously upregulated in tumor tissues when compared with normal tissues (Fig. 1a, P < 0.05). To validate the result from TCGA, we thus determined the expression level of MYO18B in 80 pairs of tumor tissues and adjacent liver tissues by qRT-PCR. The result showed that MYO18B expression exhibited higher levels in tumor tissues than adjacent liver tissues (Fig. 1b, p < 0.05). These results suggested that MYO18B was overexpressed in HCC tumor tissues.
Fig. 1.
Relative MYO18B expression in tumor tissues and its clinical significance. a MYO18B was over-expressed in tumor tissues (n = 374) compared with normal tissues (n = 50), which was analyzed using TCGA database. b MYO18B was up-regulated in tumor tissues (n = 80) compared with adjacent liver tissues (ALT), which was analyzed by qRT-PCR. c Kaplan-Meier survival analysis showed that MYO18B over-expression was correlated with worse overall survival in patients with hepatocellular carcinoma (P = 0.001)
To further investigate whether MYO18B overexpression was involved in HCC progression, we measured the correlation between MYO18B levels and clinical pathological characteristics in HCC patients using chi-square test. Based on the median value of MYO18B, 80 HCC patients were divided into high and low expression groups. As shown in Table 1, statistical analysis revealed that no significant association was found in age, gender, grade, pathologic-N and pathologic-M and MYO18B expression (P > 0.05), while MYO18B expression was remarkably correlated with pathologic-stage (P = 0.025) and pathologic-T (P = 0.014), implying that high expression of MYO18B might be involved in the progression of HCC.
Additionally, the prognostic value of MYO18B in HCC was evaluated by Kaplan-Meier plotting with long-rank test for difference. The result showed that HCC patients with high expression of MYO18B displayed a significantly poorer prognosis than those with low expression (Fig. 1c, P = 0.001). Cox regression analysis was further performed to analyze the prognostic value of MYO18B in HCC. As shown in Table 2, univariate analysis showed that MYO18B expression (HR = 2.993, P = 0.002), pathologic-stage (HR = 3.708, p < 0.001) and pathologic-T (HR = 3.783, P < 0.001) were obviously related to overall survival in HCC, while age, gender, grade, pathologic-M and pathologic-N showed no significant association with overall survival (P > 0.05). Multivariate analysis revealed that MYO18B expression was an independent prognostic factor in HCC after adjusting for other clinical variables (Table 2, HR = 2.132, P = 0.039). The results reveal that MYO18B may be a potential marker for the prognosis and progression of HCC.
Table 2.
Cox regression analysis of overall survival in patients with hepatocellular carcinoma
| Variables | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| P value | HR | 95% CI | P value | HR | 95% CI | |
| MYO18B expression (high/low) | 0.002 | 2.993 | 1.505–5.952 | 0.039 | 2.132 | 1.038–4.380 |
| Age(<60/≥60) | 0.114 | 0.605 | 0.324–1.129 | |||
| Gender(female/male) | 0.324 | 1.373 | 0.731–2.576 | |||
| Grade(G1 + G2/G3 + G4) | 0.463 | 0.775 | 0.393–1.530 | |||
| Pathologic-Stage (I + II/III + IV) | 0.000 | 3.708 | 1.790–7.678 | 0.739 | 1.421 | 0.181–11.186 |
| Pathologic-T (T1 + T2/T3 + T4) | 0.000 | 3.783 | 1.855–7.718 | 0.461 | 2.133 | 0.285–15.989 |
| Pathologic-M (M0/M1) | 0.052 | 3.267 | 0.991–10.769 | |||
| Pathologic-N (N0/N1) | 0.080 | 2.351 | 0.902–6.130 | |||
HR hazard ratio, CI confidence interval
MYO18B knockdown inhibits the proliferation of HepG2 cells
We detected the mRNA expression of MYO18B in HCC cell line HepG2. Human normal liver cells HL-7702 were used as a control. qRT-PCR analysis found that MYO18B expression was significantly upregulated in HepG2 cells than that in HL-7702 cells (Fig. 2a, p < 0.05).
Fig. 2.
Relative MYO18B expression in hepatocellular carcinoma cells. a The expression levels of MYO18B in HepG2 and HL-7702 cell lines were analyzed by qRT-PCR. MYO18B expression displayed a higher level in HepG2 cells. b-d HepG2 cells were transfected with one specific siRNA (experimental group, si-MYO18B) or non-specific control siRNA (negative control group, si-con). MYO18B expression was determined by qRT-PCR (b) and western blot (c, d) respectively. *P < 0.05 versus si-con group
To investigate the effects of MYO18B on HCC, HepG2 cells were transfected with MYO18B siRNA or control siRNA, respectively. qRT-PCR and Western blot were utilized to determine the mRNA and protein levels of MYO18B, respectively. The result indicated that MYO18B expression was significantly downregulated by MYO18B siRNA in both mRNA (Fig. 2b, P < 0.05) and protein levels (Fig. 2c and d, P < 0.05).
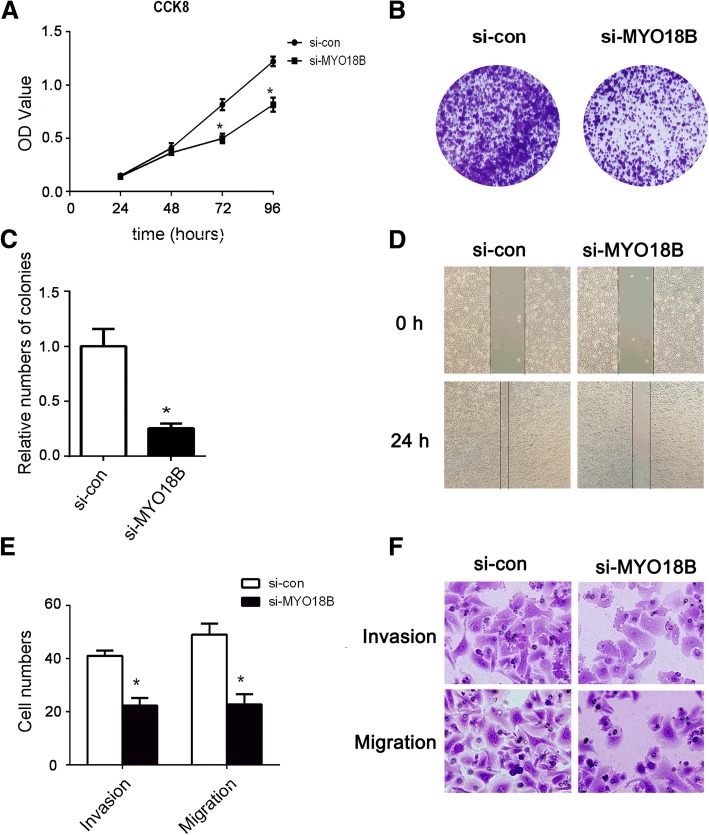
To study the effect of MYO18B on cell proliferation, the viability of HepG2 cells was detected by CCK-8 and colony formation assays. As shown in Fig. 3a, CCK-8 assay revealed that MYO18B knockdown significantly suppressed the proliferation of HepG2 cells after 72 h and 96 h of transfection when compared to control group (P < 0.05). In addition, Fig. 3b showed the representative images of the size and number of colonies per well. Colony formation assay showed that MYO18B knockdown significantly decreased the number of colony formation (Fig. 3c P < 0.05).
Fig. 3.
Knockdown of MYO18B inhibited the proliferation, migration and invasion of HepG2 cells. a CCK-8 assays were performed to measure the proliferation of the transfected HepG2 cells, which showed knockdown of MYO18B (si-MYO18B group) obviously inhibited the cell growth. b, c Colony formation assays were used to determined the proliferation of the transfected HepG2 cells, which also revealed knockdown of MYO18B (si-MYO18B group) obviously inhibited the cell growth. d After incubated the transfected cells for 24 h, knockdown of MYO18B (si-MYO18B group) significantly suppressed the migration of HepG2 cells analyzed by wound healing assays. e, f Transwell assays further demonstrated that knockdown of MYO18B (si-MYO18B group) obviously inhibited the migration and invasion of the transfected HepG2 cells. *P < 0.05 versus si-con group
MYO18B knockdown inhibits the migration and invasion of HepG2 cells
HCC cells are characterized by high migration ability. To evaluate the effect of MYO18B expression on the cell migration and invasion, wound healing and transwell assays were carried out. As shown in Fig. 3d, cell culture images were captured at 0 and 24 h and the wound gap was analyzed. The wound healing assay showed that the wound closure in si-MYO18B group was obviously slower compared with the control group, indicating that MYO18B knockdown suppressed the migration activity of HepG2 cells. Transwell results showed that the number of the invading and migrating cells in the si-MYO18B group was significantly decreased than that in the control group (Fig. 3e and f, P < 0.05), suggesting that MYO18B knockdown attenuated the migration and invasion of HepG2 cells.
Knockdown of MYO18B inhibited the activation of PI3K/AKT/mTOR pathway
PI3K/AKT/mTOR signaling pathway plays important roles in regulating multiple cellular functions, such as cell proliferation, differentiation and intracellular trafficking [24]. To explore the underlying mechanism of MYO18B in HCC, we analyzed the protein levels of PI3K/AKT/mTOR signaling pathway in HepG2 cells by Western blot. As shown in Fig. 4, the protein levels of PI3K, AKT and mTOR had no significant differences between si-MYO18B group and control group, but MYO18B knockdown significantly suppressed the phosphorylation of PI3K, AKT and mTOR, and led to an obvious decrease of p70S6K expression (P < 0.05). The results suggested that MYO18B knockdown might suppress the proliferation, migration and invasion of HepG2 cells by inhibiting the activation of PI3K/AKT/mTOR signaling pathway.
Fig. 4.
The effect of MYO18B on PI3K/AKT pathway. a Western blot assays were used to measure the expression of PI3K, p-PI3K, AKT, P-AKT, mTOR, p-mTOR and p-P70S6K proteins in the transfected HepG2 cells. b Quantification of PI3K, p-PI3K, AKT, P-AKT, mTOR, p-mTOR and p-P70S6K proteins was performed and normalized to the GAPDH a loading control. *P < 0.05 versus si-con group
Discussion
In this study, we investigated the expression and prognostic values of MYO18B in HCC using two independent cohorts, and found that MYO18B was overexpressed in HCC, and MYO18B high expression was involved in a poor prognosis in HCC. Silencing of MYO18B in HepG2 cells could inhibit cell proliferation, migration and invasion via attenuating the activation of PI3K/AKT/mTOR signaling pathway. Therefore, targeting MYO18B might represent a novel adjuvant clinical intervention for patients with HCC.
MYO18B is a novel unconventional myosin heavy chain that mainly expresses in human cardiac and skeletal muscles [25]. It is associated with the ATPase proteasome subunit Sug1 and is a substrate for proteasomal degradation [26]. Previous studies identified MYO18B as a tumor suppressor gene in several cancers [19–22]. Nishioka et al. [20] illustrated that MYO18B expression was decreased in 88% of nonsmall cell lung carcinoma and 47% of small cell lung carcinoma cell lines, and restoration of MYO18B expression can effectively suppress the growth of lung cancer cells. Nakano et al. [21] showed that 82% colorectal cancer cell lines presented reduced MYO18B expression, which was restored in all 9 by treatment. Yanaihara et al. [22] indicated that MYO18B expression was reduced in all 4 ovarian cancer cell lines and in 71% of primary ovarian cancers. Yokota et al. [27] demonstrated that MYO18B was involved in tumor suppression by regulating cell movement and maintaining cell structure. Nobutaka et al. [28] found that none of human malignant pleural mesothelioma cell lines expressed a detectable level of MYO18B, and a restored expression of MYO18B inhibited cell growth and increased cell apoptosis. Until now, MYO18B remains a mysterious player in HCC. Thus we attempted to investigate the specific effects of MYO18B on the progression of HCC. In the present study, we found that MYO18B expression was significantly upregulated in both HCC tumor tissues and HCC cell line, and high level of MYO18B was correlated with a poor prognosis for patients with HCC. Furthermore, MYO18B expression was an independent prognostic factor in HCC. In addition, experimental analysis showed that MYO18B promoted the proliferation, migration and invasion of HCC cells. Evidently, our results were inconsistent with the previous findings that MYO18B was a tumor suppressor gene and downregulated in cancer tissues. The occurrence of opposite results might partially due to the tissue-specific expression of MYO18B. Moreover, previous studies gave the conclusion based a relative small sample size. While, our finding was drawn using two large independent cohorts, suggesting a more reliable result. Even though, further studies should be performed to elucidate the true circumstance behind the inconsistence.
To explore the underlying mechanism of MYO18B in HCC, we evaluated the effect of MYO18B on the PI3K/AKT/mTOR signaling pathway. PI3K/AKT/mTOR signaling pathway is a major activated signaling pathway in human malignancies, and regulates multiple cellular processes, such as cell proliferation, apoptosis and migration [29–31]. The PI3K/Akt pathway is a key regulator of cell survival via inhibition of apoptosis in various types of human cancers [32–34]. Akt activation promotes metastasis and invasion of cancer cells, and phosphorylates mammalian target of rapamycin (mTOR) [35]. mTOR is an important downstream target of PI3K/Akt, and positively regulates the serine/threonine kinase p70 S6 kinase (p70S6K) [35]. Previous studies demonstrated frequent changes of the PI3K/Akt/mTOR pathway in HCC [24, 36, 37]. Components of this pathway are frequently deregulated in an extensive number of tumors, making PI3K/AKT/mTOR signaling pathway an attractive target for cancer therapy [30, 38, 39]. Increasing evidence indicates that inhibition of PI3K/Akt/mTOR pathway suppresses cell growth in many tumor types [24, 40–42]. In this report, we found that the phosphorylation levels of PI3K, Akt and mTOR were significantly suppressed by MYO18B knockdown in HepG2 cells, suggesting that MYO18B knockdown could inhibit the activation of PI3K/AKT/mTOR signaling pathway in HCC.
We acknowledge some limitations of our study. In particular, we determined the expression of MYO18B in HCC tumor tissues using two independent cohorts, one from TCGA and one from our clinical patients. In TCGA, the normal liver tissues were used for control, whereas the adjacent liver tissues were used for control in our clinical cohort. Most of HCCs develop in cirrhotic livers, therefore the tumor adjacent tissue is mostly not normal. A further limitation is that some clinical characteristics of these 80 HCC patients such as tumor size, tumor number, and lymphovascular invasion, background liver condition were not included, because the information was not available for a substantial proportion of cases. Thus, the significance and robustness of MYO18B as a biomarker requires further confirmation.
Conclusions
MYO18B was overexpressed in HCC and correlated with poor prognosis in HCC. MYO18B expression was an independent prognostic factor in HCC. MYO18B knockdown suppressed cell proliferation, migration and invasion via inhibiting the activation of PI3K/AKT/mTOR signaling pathway. Targeting MYO18B might represent a promising target for HCC treatment.
Acknowledgements
Not applicable.
Funding
This work is supported by the Joint Scientific Special foundation of Guangdong Provincial Department of Science and Technology-Academy of Traditional Chinese Medicine (NO: 2013A032500017).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- HCC
Hepatocellular carcinoma
- qRT-PCR
Quantitative real-time PCR
- TCGA
The Cancer Genome Atlas
Authors’ contributions
ZZ conceived the study, acquired the data and drafted the manuscript; JZ and YH acquired the data; WL analyzed the data; HC conceived the study and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The procedure was reviewed and approved by the ethics committee of Shantou University. Human tissue acquisition and usage in this study complied with the National Regulations on the Use of Clinical Samples in China. Written informed consent was obtained from all participants.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhenyu Zhang, Email: 179598450@qq.com.
Jinfeng Zhu, Email: 136605631@qq.com.
Yansong Huang, Email: 1393747899@qq.com.
Weibing Li, Email: L_w_bing@163.com.
Hongqiu Cheng, Phone: +86 0754 88915657, Email: 982115479@qq.com.
References
- 1.Marrero Jorge A., Kulik Laura M., Sirlin Claude B., Zhu Andrew X., Finn Richard S., Abecassis Michael M., Roberts Lewis R., Heimbach Julie K. Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology. 2018;68(2):723–750. doi: 10.1002/hep.29913. [DOI] [PubMed] [Google Scholar]
- 2.Bosetti C, Turati F, La Vecchia C. Hepatocellular carcinoma epidemiology. Best Pract Res Clin Gastroenterol. 2014;28(5):753–770. doi: 10.1016/j.bpg.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 3.Pinter M, Trauner M, Peck-Radosavljevic M, Sieghart W. Cancer and liver cirrhosis: implications on prognosis and management. ESMO Open. 2016;1(2):e000042. doi: 10.1136/esmoopen-2016-000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singal AG, Conjeevaram HS, Volk ML, et al. Effectiveness of hepatocellular carcinoma surveillance in patients with cirrhosis. Cancer Epidemiol Biomark Prev. 2012;21(5):793–799. doi: 10.1158/1055-9965.EPI-11-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391(10127):1301–1314. doi: 10.1016/S0140-6736(18)30010-2. [DOI] [PubMed] [Google Scholar]
- 6.Kulik LM, Chokechanachaisakul A. Evaluation and management of hepatocellular carcinoma. Clin Liver Dis. 2015;19(1):23–43. doi: 10.1016/j.cld.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Nishida N, Goel A. Genetic and epigenetic signatures in human hepatocellular carcinoma: a systematic review. Curr Genomics. 2011;12(2):130–137. doi: 10.2174/138920211795564359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niu ZS, Niu XJ, Wang WH. Genetic alterations in hepatocellular carcinoma: an update. World J Gastroenterol. 2016;22(41):9069–9095. doi: 10.3748/wjg.v22.i41.9069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou L, Liu J, Luo F. Serum tumor markers for detection of hepatocellular carcinoma. World J Gastroenterol. 2006;12(8):1175–1181. doi: 10.3748/wjg.v12.i8.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ajdarkosh H, Dadpay M, Yahaghi E, et al. Decrease expression and clinicopathological significance of miR-148a with poor survival in hepatocellular carcinoma tissues. Diagn Pathol. 2015;10:135. doi: 10.1186/s13000-015-0371-4. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Park SJ, Jang JY, Jeong SW, et al. Usefulness of AFP, AFP-L3, and PIVKA-II, and their combinations in diagnosing hepatocellular carcinoma. Medicine (Baltimore) 2017;96(11):e5811. doi: 10.1097/MD.0000000000005811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaiteerakij R, Addissie BD, Roberts LR. Update on biomarkers of hepatocellular carcinoma. Clin Gastroenterol Hepatol. 2015;13(2):237–245. doi: 10.1016/j.cgh.2013.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El-Serag HB, Marrero JA, Rudolph L, Reddy KR. Diagnosis and treatment of hepatocellular carcinoma. Gastroenterology. 2008;134(6):1752–1763. doi: 10.1053/j.gastro.2008.02.090. [DOI] [PubMed] [Google Scholar]
- 14.Reichl P, Mikulits W. Accuracy of novel diagnostic biomarkers for hepatocellular carcinoma: an update for clinicians (review) Oncol Rep. 2016;36(2):613–625. doi: 10.3892/or.2016.4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marrero JA, Feng Z, Wang Y, et al. Alpha-fetoprotein, des-gamma carboxyprothrombin, and lectin-bound alpha-fetoprotein in early hepatocellular carcinoma. Gastroenterology. 2009;137(1):110–118. doi: 10.1053/j.gastro.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi K, Kudo J, Ishibashi H, Hirata Y, Niho Y. Frequent loss of heterozygosity on chromosome 22 in hepatocellular carcinoma. Hepatology. 1993;17(5):794–799. doi: 10.1002/hep.1840170508. [DOI] [PubMed] [Google Scholar]
- 17.Guan XY, Fang Y, Sham JS, et al. Recurrent chromosome alterations in hepatocellular carcinoma detected by comparative genomic hybridization. Genes Chromosomes Cancer. 2000;29(2):110–116. doi: 10.1002/1098-2264(2000)9999:9999<::AID-GCC1022>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 18.Zhu GN, Zuo L, Zhou Q, et al. Loss of heterozygosity on chromosome 10q22-10q23 and 22q11.2-22q12.1 and p53 gene in primary hepatocellular carcinoma. World J Gastroenterol. 2004;10(13):1975–1978. doi: 10.3748/wjg.v10.i13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tani M, Ito J, Nishioka M, et al. Correlation between histone acetylation and expression of the MYO18B gene in human lung cancer cells. Genes Chromosomes Cancer. 2004;40(2):146–151. doi: 10.1002/gcc.20027. [DOI] [PubMed] [Google Scholar]
- 20.Nishioka M, Kohno T, Tani M, et al. MYO18B, a candidate tumor suppressor gene at chromosome 22q12.1, deleted, mutated, and methylated in human lung cancer. Proc Natl Acad Sci U S A. 2002;99(19):12269–12274. doi: 10.1073/pnas.192445899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakano T, Tani M, Nishioka M, et al. Genetic and epigenetic alterations of the candidate tumor-suppressor gene MYO18B, on chromosome arm 22q, in colorectal cancer. Genes Chromosomes Cancer. 2005;43(2):162–171. doi: 10.1002/gcc.20180. [DOI] [PubMed] [Google Scholar]
- 22.Yanaihara N, Nishioka M, Kohno T, et al. Reduced expression of MYO18B, a candidate tumor-suppressor gene on chromosome arm 22q, in ovarian cancer. Int J Cancer. 2004;112(1):150–154. doi: 10.1002/ijc.20339. [DOI] [PubMed] [Google Scholar]
- 23.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Q, Lui VW, Yeo W. Targeting the PI3K/Akt/mTOR pathway in hepatocellular carcinoma. Future Oncol. 2011;7(10):1149–1167. doi: 10.2217/fon.11.95. [DOI] [PubMed] [Google Scholar]
- 25.Salamon M, Millino C, Raffaello A, et al. Human MYO18B, a novel unconventional myosin heavy chain expressed in striated muscles moves into the myonuclei upon differentiation. J Mol Biol. 2003;326(1):137–149. doi: 10.1016/S0022-2836(02)01335-9. [DOI] [PubMed] [Google Scholar]
- 26.Inoue T, Kon T, Ajima R, et al. MYO18B interacts with the proteasomal subunit Sug1 and is degraded by the ubiquitin-proteasome pathway. Biochem Biophys Res Commun. 2006;342(3):829–834. doi: 10.1016/j.bbrc.2006.02.025. [DOI] [PubMed] [Google Scholar]
- 27.Yokota J, Kohno T. Molecular footprints of human lung cancer progression. Cancer Sci. 2004;95(3):197–204. doi: 10.1111/j.1349-7006.2004.tb02203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edakuni N, Ikuta K, Yano S, et al. Restored expression of the MYO18B gene suppresses orthotopic growth and the production of bloody pleural effusion by human malignant pleural mesothelioma cells in SCID mice. Oncol Res. 2006;16(5):235–243. doi: 10.3727/000000006783981062. [DOI] [PubMed] [Google Scholar]
- 29.Faes S, Dormond O. PI3K and AKT: unfaithful Partners in Cancer. Int J Mol Sci. 2015;16(9):21138–21152. doi: 10.3390/ijms160921138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porta C, Paglino C, Mosca A. Targeting PI3K/Akt/mTOR signaling in Cancer. Front Oncol. 2014;4:64. doi: 10.3389/fonc.2014.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ersahin T, Tuncbag N, Cetin-Atalay R. The PI3K/AKT/mTOR interactive pathway. Mol BioSyst. 2015;11(7):1946–1954. doi: 10.1039/C5MB00101C. [DOI] [PubMed] [Google Scholar]
- 32.Franke TF, Hornik CP, Segev L, Shostak GA, Sugimoto C. PI3K/Akt and apoptosis: size matters. Oncogene. 2003;22(56):8983–8998. doi: 10.1038/sj.onc.1207115. [DOI] [PubMed] [Google Scholar]
- 33.Altomare DA, Wang HQ, Skele KL, et al. AKT and mTOR phosphorylation is frequently detected in ovarian cancer and can be targeted to disrupt ovarian tumor cell growth. Oncogene. 2004;23(34):5853–5857. doi: 10.1038/sj.onc.1207721. [DOI] [PubMed] [Google Scholar]
- 34.Polivka J, Jr, Janku F. Molecular targets for cancer therapy in the PI3K/AKT/mTOR pathway. Pharmacol Ther. 2014;142(2):164–175. doi: 10.1016/j.pharmthera.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 35.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12(1):9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 36.Li W, Tan D, Zhang Z, Liang JJ, Brown RE. Activation of Akt-mTOR-p70S6K pathway in angiogenesis in hepatocellular carcinoma. Oncol Rep. 2008;20(4):713–719. [PubMed] [Google Scholar]
- 37.Ma L, Ji L, Yu Y, Wang J. Novel molecular targets for diagnosis and treatment of hepatocellular carcinoma. Discov Med. 2015;19(102):7–14. [PubMed] [Google Scholar]
- 38.Morgensztern D, McLeod HL. PI3K/Akt/mTOR pathway as a target for cancer therapy. Anti-Cancer Drugs. 2005;16(8):797–803. doi: 10.1097/01.cad.0000173476.67239.3b. [DOI] [PubMed] [Google Scholar]
- 39.Yap TA, Garrett MD, Walton MI, Raynaud F, de Bono JS, Workman P. Targeting the PI3K-AKT-mTOR pathway: progress, pitfalls, and promises. Curr Opin Pharmacol. 2008;8(4):393–412. doi: 10.1016/j.coph.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 40.Papadimitrakopoulou V, Adjei AA. The Akt/mTOR and mitogen-activated protein kinase pathways in lung cancer therapy. J Thorac Oncol. 2006;1(7):749–751. [PubMed] [Google Scholar]
- 41.Eiden AM, Zhang S, Gary JM, Simmons JK, Mock BA. Molecular pathways: increased susceptibility to infection is a complication of mTOR inhibitor use in Cancer therapy. Clin Cancer Res. 2016;22(2):277–283. doi: 10.1158/1078-0432.CCR-14-3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mabuchi S, Kawase C, Altomare DA, et al. mTOR is a promising therapeutic target both in cisplatin-sensitive and cisplatin-resistant clear cell carcinoma of the ovary. Clin Cancer Res. 2009;15(17):5404–5413. doi: 10.1158/1078-0432.CCR-09-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.