Abstract
Background
Emerging evidence indicate that miRNAs play an important role on gastric cancer (GC) progression via regulating several downstream targets, but it is still partially uncovered. This study aimed to explore the molecular mechanisms of GC by comprehensive analysis of mRNAs and miRNA expression profiles.
Methods
The mRNA and miRNA expression profiles of GSE79973 and GSE67354 downloaded from Gene Expression Omnibus were used to analyze the differentially expressed genes (DEGs) and DE-miRNAs among GC tissues and normal tissues. Then, targets genes of DE-miRNAs were predicted and the DE-miRNA–DEG regulatory network was constructed. Next, function enrichment analysis of the overlapped genes between the predicted DE-miRNAs targets and DEGs was performed and a protein–protein interactions network of overlapped genes was constructed. Finally, RT-PCR analysis was performed to detect the expression levels of several key DEGs and DE-miRNAs.
Results
A set of 703 upregulated and 600 downregulated DEGs, as well as 8 upregulated DE-miRNAs and 27 downregulated DE-miRNAs were identified in GC tissue. hsa-miR-193b-3p and hsa-miR-148a-3p, which targeted most DEGs, were highlighted in the DE-miRNA–DEG regulatory network, as well as hsa-miR-1179, which targeted KNL1, was newly predicted to be associated with GC. In addition, NCAPG, which is targeted by miR-193b-3p, and KNL1, which is targeted by hsa-miR-1179, had higher degrees in the PPI network. RT-qPCR results showed that hsa-miR-148a-3p, hsa-miR-193b-3p, and hsa-miR-1179 were downregulated, and NCAPG and KNL1 were upregulated in GC tissues; this is consistent with our bioinformatics-predicted results.
Conclusions
The downregulation of miR-193b-3p might contribute to GC cell proliferation by mediating the upregulation of NCAPG; as additionally, the downregulation of miR-193b-3p might contribute to the mitotic nuclear division of GC cells by mediating the upregulation of KNL1.
Keywords: Gastric cancer, Differentially expressed genes, Protein–protein interaction network, Regulatory network
Background
Gastric cancer (GC) is type of common malignant tumor that originates from gastric epithelial cells [1]. Early-stage GC patients may show the symptoms of epigastric pain and weight loss [2]. In 2015, it was evaluated that GC is the second-most common cancer in China, and the incidence rate for GC is two-fold higher in men than in women (320.8 vs 157.2 per 100,000), with the mean age being more than 50 [3]. In China, early-stage gastric cancer has a relatively low diagnosis rate (< 10%) [4]. In addition, although data show that during 1984–2013, the 5-year relative survival rates for GC patients have improved from 17.8 to 20.3 to 22.9% in each decade, the clinical prognosis of GC is still poor [5]. Thus, it is a great challenge to explore novel biomarkers for the early diagnosis and effective treatment of GC.
The pathogenesis of GC is complex, involving factors such as dietary habits and environmental risks [6]. However, genetic factors are believed to be predominant factors causing GC [7]. Recent progress in researches on miRNAs and gene alterations in GC has been reported [8–10]. Emerging evidence indicates that miRNAs play an important role on the progression of GC, via the regulation of several downstream targets [11]; however, detailed information is still unavailable. Reportedly, the overexpression of miR-223 can promote GC invasion and metastasis via the regulation of the downstream tumor suppressor, EPB41L3 [11]. In addition, upregulated miRNA-194 may also promote the proliferation and migration of GC cells by activating Wnt signaling by targeting the negative Wnt regulator, SUFU [12]. Moreover, decreased levels of miR-4317, which targets ZNF322, is related to GC cell proliferation and S-G2/M transition [13].
In recent years, more and more researchers have attempted to explore the therapeutic targets of GC by microarray analysis of genes and miRNA expression profiles [14–16], and many genes like ALDOB, MT1H, and KRT2, as well as miRNAs such as miR-495-3p, miR-421, and miR-658 have been shown to be differentially expressed in GC tissues, compared to healthy control tissues [14–16]. However, the comprehensive regulatory mechanisms between those miRNAs and genes in GC have not yet been studied comprehensively.
In the present study, we used the data of GSE79973 mRNA and GSE67354 miRNA datasets to analyze the differentially expressed miRNAs (DE-miRNAs) and differentially expressed genes (DEGs), and predicted both the regulatory pairs between those DE-miRNAs and DEGs, and the function of those genes. Finally, we validated the expression changes of several DE-miRNAs and DEGs by real-time RT-PCR. Our study might not only provide the potential regulatory relationships between miRNAs and genes, but also identify important biomarkers for GC diagnosis and treatment.
Results
DEGs and DE-miRNAs analyses
Based on the aforementioned cut-off criteria, a set of 1303 DEGs were identified in GC tissue samples, compared to normal adjacent non-tumor mucosa samples, among which 703 DEGs were upregulated and 600 DEGs were downregulated. In addition, a total of 35 DE-miRNAs were screened between GC and normal adjacent non-tumor mucosa tissue samples, including 8 upregulated DE-miRNAs and 27 downregulated DE-miRNAs. The number of downregulated DE-miRNAs and upregulated DEGs were higher than the number of upregulated DE-miRNAs and downregulated DEGs.
miRNA–miRNA network analysis
Among the 35 identified DE-miRNAs stated above, only 25 DE-miRNAs, which comprised 19 downregulated DE-miRNAs and 6 upregulated DE-miRNAs, were reported in the miRBase database. Of these, miRNA301b had no relevant targets. For the remaining 24 DE-miRNAs, a total of 2843 targets were found in the miRBase database. The top ten DE-miRNAs with more downstream targets (eg., hsa-miR-193b-3p, hsa-miR-148b-3p, and hsa-miR-193b-3p) and top ten targets regulated by more upstream miRNAs (eg., MYC, CDKN1B, and GATA6) are listed in Table 1, respectively.
Table 1.
Top ten DE-miRNAs with most downstream targets and top ten genes regulated by most upstream miRNAs
| miRNA | Degree | Gene | Degree |
|---|---|---|---|
| hsa-miR-193b-3p | 844 | MYC | 6 |
| hsa-miR-148b-3p | 374 | CDKN1B | 6 |
| hsa-miR-378a-5p | 370 | GATA6 | 5 |
| hsa-miR-196a-5p | 288 | IGF1R | 5 |
| hsa-miR-100-5p | 243 | DNMT1 | 5 |
| hsa-miR-148a-3p | 187 | STX16 | 5 |
| hsa-miR-140-3p | 174 | HMGB1 | 5 |
| hsa-miR-136-5p | 147 | NUFIP2 | 4 |
| hsa-miR-196b-5p | 136 | RCC2 | 4 |
| hsa-miR-99a-5p | 128 | FAM104A | 4 |
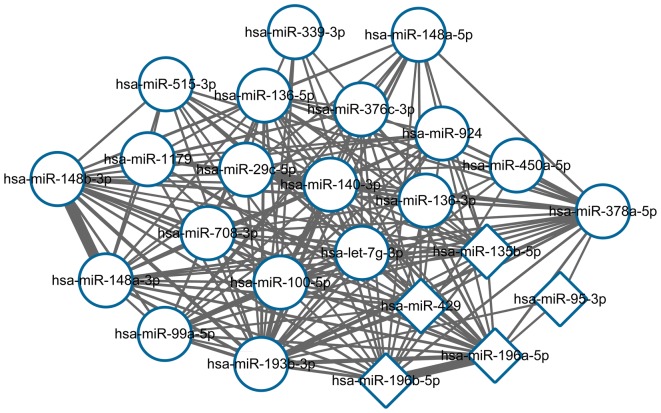
The co-regulated miRNA–miRNA target network included 24 DE-miRNAs and 171 interactions (Fig. 1). The top 15 miRNA–miRNA interactions that had more number of the same target genes are listed in Table 2; this includes hsa-miR-148a-3p and hsa-miR-148b-3p, hsa-miR-196a-5p and hsa-miR-196b-5p, and hsa-miR-100-5p and hsa-miR-99a-5p.
Fig. 1.
The co-regulated miRNA–miRNA target network. The circular nodes represent the downregulated miRNA, while the diamond-shaped nodes represent the upregulated miRNA. The lines indicate that the targeted genes between miRNAs are the same
Table 2.
The top 15 miRNA–miRNA interactions that had more same target genes
| miRNA1 | miRNA2 | Number of same target genes | miRNA1 | miRNA2 | Number of same target genes |
|---|---|---|---|---|---|
| hsa-miR-148a-3p | hsa-miR-148b-3p | 113 | hsa-miR-148b-3p | hsa-miR-193b-3p | 21 |
| hsa-miR-196a-5p | hsa-miR-196b-5p | 88 | hsa-miR-193b-3p | hsa-miR-99a-5p | 16 |
| hsa-miR-100-5p | hsa-miR-99a-5p | 42 | hsa-miR-148a-3p | hsa-miR-193b-3p | 13 |
| hsa-miR-100-5p | hsa-miR-193b-3p | 34 | hsa-miR-196a-5p | hsa-miR-140-3p | 11 |
| hsa-miR-196a-5p | hsa-miR-193b-3p | 28 | hsa-miR-196b-5p | hsa-miR-193b-3p | 11 |
| hsa-miR-429 | hsa-miR-193b-3p | 26 | hsa-let-7 g-3p | hsa-miR-193b-3p | 11 |
| hsa-miR-140-3p | hsa-miR-193b-3p | 24 | hsa-miR-136-5p | hsa-miR-193b-3p | 11 |
| hsa-miR-193b-3p | hsa-miR-378a-5p | 22 |
Pathways analysis of DE-miRNAs
In order to analysis the function of DE-miRNAs, pathways analysis based on DE-miRNA-targeted genes was performed. The results revealed that several miRNAs such as mir-196a-5p, mir-148a-3p, mir-148a-5p, mir-376c-3p, and mir-429 were closely associated with “Proteoglycans in cancer”. In addition, many miRNAs (eg., mir-100-5p, mir-148a-3p, and mir-193b-3p) were involved in the pathway of “cell cycle” (Fig. 2).
Fig. 2.
The functional enrichment result of miRNA-targeted genes. The size of each node represents the miRNA-targeted gene number ratio for the corresponding pathway, whereas the color change from blue to red indicates the p values from big to small for the corresponding pathway
DE-miRNA–DEG regulatory network analysis
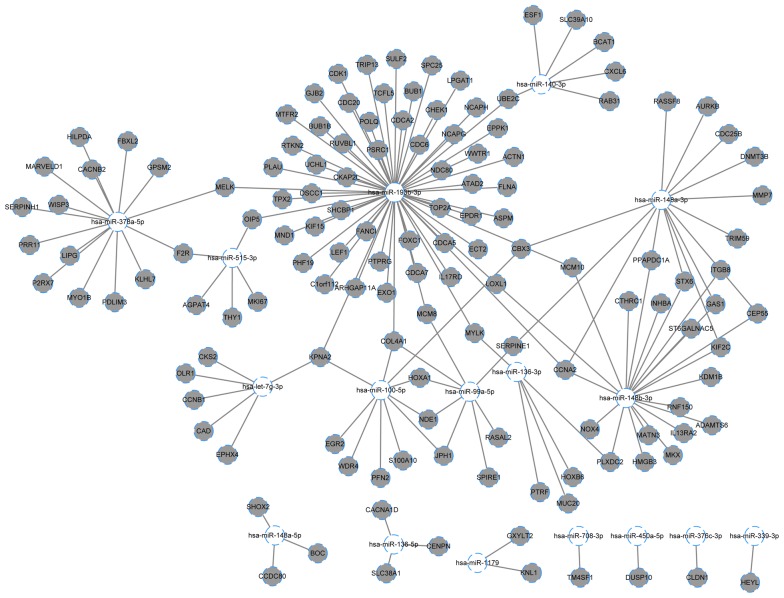
Based on the aforementioned methods, a total of 11 downregulated DEGs and 136 upregulated DEGs were overlapped between the targets of DE-miRNAs and all DEGs. The upregulated DE-miRNA–DEG regulatory network comprised of four upregulated DE-miRNAs, 11 downregulated DEGs, and 15 regulatory pairs, in which upregulated hsa-miR-196b-5p targeted more DEGs (eg., GLUL and GATA6) than other DE-miRNAs (Fig. 3). On the other hand, the downregulated DE-miRNA–DEG regulatory network involved 17 downregulated DE-miRNAs, 136 upregulated DEGs, and 162 regulatory pairs. In addition, hsa-miR-193b-3p and hsa-miR-148a-3p had more targeted DEGs than other miRNAs; hsa-miR-193b-3p had 59 targets (eg., NCAPG, CDK1, and CHEK1) and hsa-miR-148a-3p had 15 targets (eg., KIF2C and MYC) (Fig. 4).
Fig. 3.

Upregulated DE-miRNA-downregulated DEG regulatory network. The white rhombus-shaped nodes represent the upregulated DE-miRNA, and the gray rhombus-shaped nodes represent the downregulated DEGs. The lines stand for the interaction between DE-miRNA and their targeted DEGs. DEGs differentially expressed genes
Fig. 4.
Downregulated DE-miRNA-upregulated DEG regulatory network. The white circles represent the downregulated DE-miRNA, and the gray circles represent the upregulated DEGs. The lines stand for the interaction between DE-miRNA and their targeted DEGs. DEGs differentially expressed genes
Functional enrichment analysis of overlapped genes
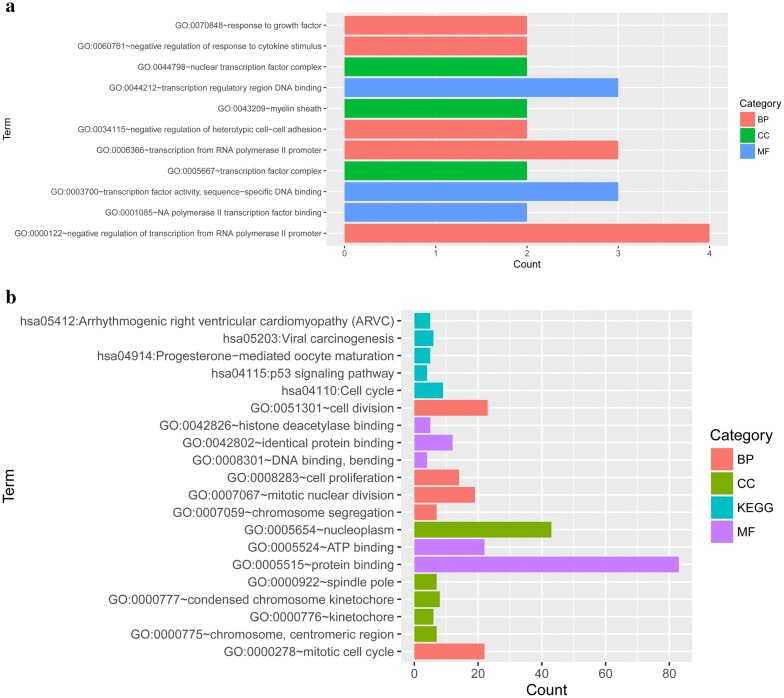
The functional enrichment analysis indicated that the downregulated DEGs were markedly related to functions such as “negative regulation of transcription from RNA polymerase II promoter” and “transcription regulatory region DNA binding’’ (Fig. 5a). However, no relevant pathways were predicted for the downregulated DEGs. In addition, most upregulated DEGs were significantly associated with functions like “cell division” (e.g., CCNB1 and KNL1), “cell proliferation” (e.g., BUB1), and “mitotic nuclear division” (e.g., KNL1), and with pathways such as “p53 signaling pathway” (e.g., CCNB1, CDK1, and CHEK1) and “cell cycle” (e.g., KNL1 and NCAPG) (Fig. 5b).
Fig. 5.
The top five enriched GO functions in the BP, CC, and MF categories, as well as the top five enriched KEGG pathways. a The functional enrichment results for downregulated DEGs targeted by upregulated DE-miRNAs. b The functional enrichment results for upregulated DEGs targeted by downregulated DE-miRNAs. MF molecular function, CC cellular component, BP biological process, KEGG Kyoto Encyclopedia of Genes and Genomes, DEGs differentially expressed genes
PPI network analysis
The interactions of the 147 aforementioned genes were investigated by constructing a PPI network (Fig. 6), which comprised of 95 nodes and 340 protein–protein interaction pairs. The top 20 nodes with high degrees are shown in Table 3, including CDK1, KNL1, NCAPG, and KIF2C.
Fig. 6.

The PPI network of the overlapped DEGs. The rhombus-shaped nodes represent the downregulated DEGs, and the circular nodes represent the upregulated DEGs. The lines stand for the interactions between genes. PPI protein–protein interaction, DEGs differentially expressed genes
Table 3.
The topological property scores for nodes in the PPI network (top 20)
| Node | Betweenness | Closeness | Degree |
|---|---|---|---|
| CDK1 | 898.30255 | 0.1 | 32 |
| TOP2A | 1276.175 | 0.09126214 | 31 |
| AURKB | 1021.4524 | 0.09978768 | 29 |
| CDC20 | 342.96075 | 0.10010649 | 29 |
| CCNB1 | 771.9478 | 0.1006424 | 27 |
| BUB1 | 206.33615 | 0.1 | 27 |
| CCNA2 | 1041.6703 | 0.09466264 | 24 |
| BUB1B | 90.71324 | 0.096114516 | 21 |
| CDC6 | 251.6078 | 0.084761046 | 20 |
| KIF2C | 70.66762 | 0.0939061 | 19 |
| CDCA5 | 130.50531 | 0.09108527 | 18 |
| NDC80 | 67.37463 | 0.09680741 | 18 |
| ECT2 | 224.30412 | 0.09989373 | 16 |
| CASC5 | 213.23108 | 0.09447236 | 14 |
| CHEK1 | 195.58467 | 0.092519686 | 14 |
| NCAPG | 187.03864 | 0.0985325 | 14 |
| TPX2 | 195.76343 | 0.096114516 | 13 |
| CEP55 | 68.46083 | 0.09791667 | 13 |
| MELK | 62.496788 | 0.091617934 | 13 |
PPI protein–protein interaction
Validation of gene expression
We used RT-qPCR to detect the expression of three DE-miRNAs (hsa-miR-148a-3p, hsa-miR-193b-3p, and hsa-miR-1179) and three DEGs (MYC, NCAPG, and KNL1). The results showed that the expression of NCAPG and KNL1 were obviously increased in GC tissue samples compared to normal controls (p < 0.05, Fig. 7a, b). On the contrary, the expression of hsa-miR-148a-3p, hsa-miR-193b-3p, and hsa-miR-1179 were significantly decreased in GC tissue samples, compared to the normal controls (p < 0.05, Fig. 7c–e). Notably, those experimental results were in accordance with our bioinformatics-predicted results in the GSE79973 and GSE67354 datasets. However, no significant difference in the expression level of MYC was detected between the experimental and control groups (p > 0.05, Fig. 7f).
Fig. 7.
The relative miRNA expressions of NCAPG (a), KNL1 (b), hsa-miR-148a-3p (c), hsa-miR-193b-3p (d), hsa-miR-1179 (e), and MYC (f) detected by RT-PCR in the gastric cancer tissues, compared to those in the normal controls. p < 0.05 was considered to be significantly significant
Discussion
In our study, a total of 1303 DEGs, including 703 upregulated and 600 downregulated genes, and 35 DE-miRNAs, comprising 8 upregulated and 27 downregulated miRNAs, were identified in GC tissues, compared to the normal adjacent non-tumor mucosa tissue samples. Importantly, hsa-miR-193b-3p, which targeted 59 DEGs (eg., NCAPG), and hsa-miR-148a-3p, which targeted 15 DEGs (eg., MYC), were highlighted in the DE-miRNA–DEG regulatory network; additionally, hsa-miR-1179, which targeted KNL1, was newly predicted to be associated with GC. In addition, NCAPG and KNL1 had higher degrees in the PPI network. Notably, overlapped DEGs were significantly associated with functions like “mitotic nuclear division” (e.g., KNL1) and with the pathway “cell cycle” (e.g., KNL1 and NCAPG). Moreover, our RT-qPCR results showed that hsa-miR-148a-3p, hsa-miR-193b-3p, and hsa-miR-1179 were downregulated, and NCAPG and KNL1 were upregulated in GC tissue, which were consistent with our bioinformatics-predicted results.
miR-148a-3p is a key regulatory factor to be involved in many cancers progression [28–30]. A study has reported that the downregulation of miR-148a-3p can promote cell migration and proliferation in patients with laryngeal squamous cell carcinoma [29]. In addition, miR-148a is detected to be downregulated in human breast cancer tissues, and its overexpression can inhibit the migration and invasion of breast cancer cells by targeting WNT-1, while inhibition of miR-148a-3p had the opposite effect [30]. Moreover, Wang et al. have suggested that the miR-148a-3p/ERBB3/AKT2/c-myc signaling axis has an important role in controlling bladder cancer progression. Consistently, we herein detected that miR-148a downregulation existed in GC tissue samples and that MYC was its target [28]. Furthermore, MYC has been suggested as a proto-oncogene; its expression is markedly high in GC tissue [31]. However, our RT-PCR results showed that there was no statistically significant difference in the expression of MYC between the GC tissue and control samples, and the difference with previous results might be caused due to the low number of tissue samples. Collectively, we suppose that the downregulation of miR-148a-3p might be closely associated with the development of GC, via the targeting of MYC.
miR-193b-3p, a tumor suppressor, is aberrantly expressed in several types of cancer. miR-193b is detected to be downregulated in ovarian cancer, and is associated with poor prognosis [32]. Similarly, Jin et al. have revealed that the reduction of miR-193b was detected in pancreatic cancer tissues and it can act as a cell-cycle brake in pancreatic cancer cells through the regulation of G1-phase arrest and fraction of cells in the S phase [33]. In addition, it has been suggested that miR-193b-3p functions as a tumor suppressor in T-cell acute lymphoblastic leukemia and can directly regulate the MYB oncogene [34]. Notably, our experimental results indicated that miR-193b-3p was significantly downregulated in GC tissues, and we predicted that NCAPG, as a target of miR-193b-3p, was upregulated and involved in the function of the cell cycle. Non-SMC Condensin I Complex Subunit G (NCAPG) encodes a subunit of the condensin complex I, which is associated with the proper segregation of sister chromatids in the condensation and fission of mitotic chromosomes, and is responsible for the stabilization of chromosomes during mitosis and meiosis [35]. Mitotic chromosome condensation plays a crucial role in cell proliferation and results in the reconstitution of chromosomes into rod-like mitotic chromosomes, ensuring the separation of sister chromatids during cell division. NCAPG has been reported to be a mitotic gene, and its overexpression is responsible for the cell proliferation and migration in hepatocellular carcinoma [36]. Consistent with our study, NCAPG is differentially expressed in GC tissues compared to normal control tissues, and is enriched in the cell cycle term [37]. Therefore, we speculated that the downregulation of miR-193b-3p might contribute to GC cell proliferation by mediating the upregulation of NCAPG.
Our study showed that miR-1179, a newly identified DE-miRNA, was related to the development of GC; RT-PCR analysis revealed that it was significantly downregulated in GC tissues. So far, there is no related report regarding the role of miR-1179 in GC. In 2017, Xu et al. had demonstrated that miR-1179 is downregulated in glioma tissues and it is associated with cell proliferation and cell cycle progression by targeting transcription factor 5 [38]. In our study, we predicted that KNL1, which is associated with mitotic nuclear division, is a direct target of miR-1179; experimental data showed that KNL1 was overexpressed in GC tissues. Kinetochore Scaffold 1 (KNL1, also named CASC5 or D40/AF15q14) encodes a component of the multiprotein assembly that regulates spindle assembly checkpoint and chromosome biorientation to promote accurate chromosome segregation during the cell cycle [39]. The normal expression of KNL1 contributes to multiple aspects of mitotic progression [40]. Evidence suggests that the overexpression of kinetochore components may lead to tumor progression by driving chromosome instability [41]. Reportedly, high D40 expression levels are detected in two human tumors cells lines: cervical cancer and lung cancer [42]. In addition, KNL1 is involved in cell growth and division and can interact with the tumor suppressor pRb to regulate cell proliferation in cancers [42]. We suggested that as a whole, the downregulation of miR-193b-3p might contribute to the mitotic nuclear division of GC cells by mediating the upregulation of KNL1.
Although the study has detected the expressions of NCAPG and KNL1 by RT-PCR, we fail to further validate these expressions by western blot due to no available samples and limited research funding. In future, an indepth study of regulatory mechanisms validations between genes and upstream miRNAs speculated in this study should be conducted.
Conclusion
In conclusion, we identified a set of 703 upregulated and 600 downregulated DEGs, as well as 8 upregulated DE-miRNAs and 27 downregulated DE-miRNAs in GC tissues, in total. In addition, our results revealed that the downregulation of miR-193b-3p might contribute to GC cell proliferation by mediating the upregulation of NCAPG. Additionally, the downregulation of miR-193b-3p might contribute to the mitotic nuclear division of GC cells by mediating the upregulation of KNL1. These results provide a theoretical direction for future research with regards to the molecular mechanisms of the progression of GC.
Methods
Date source and data processing
The datasets GSE79973 of mRNA and GSE67354 of miRNA used in the present study were both downloaded from Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/). The GSE79973 dataset was analyzed by Affymetrix Human Genome U133 Plus 2.0 Array platform, and it included 20 samples (10 GC tissue samples and 10 adjacent non-tumor mucosa samples) [14]. On the other hand, the GSE6735 dataset was analyzed by the Homo sapiens miRNA Ca_Hu_MiRNome_v2 platform. This dataset comprised data from five GC tissue samples and five adjacent non-tumor mucosa samples.
The raw data formatted as cel files and corresponding annotation files of those two datasets were obtained from GEO database. Data normalization was performed next, using the affy package (Version 1.48.0; http://www.bioconductor.org/packages/3.2/bioc/html/affy.html) in R [17], which included the background correction, quantile normalization, probe summarization, and translation of the probe ID to the gene symbol.
Identification of differentially expressed miRNAs and genes
The T test in limma package (Version 3.26.9; http://www.bioconductor.org/packages/3.2/bioc/html/limma.html) [18] in R was used to screen the DE-miRNAs and DEGs in GC tissue samples, compared to normal adjacent non-tumor mucosa samples. The threshold values for identifying DE-miRNAs in GSE67354 were set as |log2 fold change (FC)| > 0.8 and p-value < 0.05, while |log2 FC| > 1 and p value < 0.05 were selected as the cut-off criteria for defining DEGs.
Construction of co-regulated targets networks of DE-miRNAs
miRBase (http://www.mirbase.org/) is a public repository database, which contains all published microRNA sequences and annotation [19]. We compared the miRNA ID and mature miRNA sequence between DE-miRNAs identified above and miRNAs in miRBase database; only the DE-miRNAs with corresponding IDs in the miRBase database were reserved for following analysis. miRWalk2.0 (http://mirwalk.uni-hd.de) is a comprehensive database that provides the predicted and validated information of miRNA-target interaction [20]. The targets of DE-miRNAs were predicted from this database, and regulation pairs between the DE-miRNAs and their targeted genes were obtained. In addition, it was considered that DE-miRNAs with same target genes interacted with each other. Thus, co-regulated targets networks of miRNA–miRNA were constructed and presented using the cytoscape software (version: 3.2.0) [21].
Functional enrichment analysis of DE-miRNAs
clusterProfiler is a package of R that applies gene classification and enrichment analyses for gene cluster comparison [22]. In our study, the pathway enrichment analysis of miRNA-targeted genes was performed using clusterProfiler package, and the miRNA-related pathway was inferred from pathways associated with their targeted genes. Next, the p value of enriched pathways was revised by BH method [23], and the revised value of p < 0.01 was chosen as the cut-off criterion for significant pathway terms.
DE-miRNA–DEG regulatory network construction
First, we obtained the overlapped genes between the above predicted targets of DE-miRNAs and DEGs obtained from the GSE79973 dataset. Next, the overlapped upregulated genes and downregulated genes were divided. By acquiring the regulatory relationships between the upregulated DE-miRNAs and downregulated DEGs, and downregulated DE-miRNAs and upregulated DEGs, the upregulated DE-miRNAs-targeted DEG and downregulated DE-miRNAs-targeted DEG networks were constructed using the cytoscape software.
Functional enrichment analysis of overlapped genes
Gene ontology (GO) database offers functional annotations of genes from three aspects including biological process, molecular function, and cellular component [24]. The Kyoto Encyclopedia of Genes and Genomes (KEGG) is also an important database for genome annotation, which defines the functions of genes or proteins in several specific metabolic and regulatory pathways [25]. In the present study, the GO term and KEGG pathways analysis of overlapped genes was performed by using the biocloudservice platform (http://www.biocloudservice.com/). The cut-off criterion for significant GO terms and KEGG pathways was set as p < 0.05.
Protein–protein interactions (PPIs) network construction
The Search Tool for the Retrieval of Interacting Genes (STRING, https://string-db.org/) database provides the functional associations between proteins for more than 200 organisms [26]. In our study, the PPIs of overlapped DEGs were analyzed using STRING. Then, the obtained PPI pairs were used to construct the PPI network, which was visualized using Cytoscape [21]. The topological properties of each node in the PPI network were also analyzed.
Validation of gene expression by real-time RT-PCR analysis
RT-PCR analysis was performed to detect the expression levels of several key DEGs and DE-miRNAs that were predicted to be closely associated with GC. The experimental material, which was five normal gastric mucosa (Normal group) and five GC tissue samples (Experimental group), were collected from five gastric cancer patients, who had undergone radical gastrectomy at the Sino-Japanese Friendship Hospital of Jilin University. All the patients had signed the informed consent before participating in the study. The study has been approved by the Ethics Committee of the Sino-Japanese Friendship Hospital of Jilin University. Total RNA was extracted using the TriZol reagent (TAKARA, Dalian, China, Cat. No. 9109), and RNA was reversed to produce complementary DNA with 5× primeScript RT Master MIX (Takara, Dalian, China, Cat. No. RR036A). After cDNA synthesis, quantitative real-time PCR was conducted with Power SYBR Green PCR Master (Thermo Scientific, Waltham, MA, USA, Cat. No. 4367659). GAPDH and U6 were selected as reference genes for quantitating DEGs and miRNAs, respectively. The primer sequences of detected genes are listed in Table 4, and the relative expression of genes was calculated using the 2−ΔΔCt method [27]. All the experiments were repeated thrice.
Table 4.
The primer sequence for each validated gene
| Primer name | Primer sequence (5′-3′) |
|---|---|
| U6-F | CTCGCTTCGGCAGCACA |
| U6-R | AACGCTTCACGAATTTGCGT |
| Human-U6-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAATATG |
| GAPDH-F | TGACAACTTTGGTATCGTGGAAGG |
| GAPDH-R | AGGCAGGGATGATGTTCTGGAGAG |
| MYC-F | CCTGGTGCTCCATGAGGAGAC |
| MYC-R | CAGACTCTGACCTTTTGCCAGG |
| NCAPG-F | TTAAGGAGGCCTTTCGGCTG |
| NCAPG-R | TCCACAGCTGGTTCACGTTT |
| CASC5-F | AGAAATGGAAGAAACAGAAACAGG |
| CASC5-R | TGCATGTTTCCTTTCACGGG |
| hsa-miR-148a-3p-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACAAAG |
| JH-hsa-miR-148a-3p-F | GCGCTCAGTGCACTACAGAA |
| hsa-miR-193b-3p-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGCGGG |
| JH-hsa-miR-193b-3p-F | GCGCAACTGGCCCTCAAAGT |
| hsa-miR-1179-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCAACCA |
| JH-hsa-miR-1179-F | GCGCGCAAGCATTCTTTCAT |
Statistical analysis
All results are presented as the mean ± standard error of mean (SEM). Statistical analysis of differences between groups was performed using the SPSS 22.0 software, and p < 0.05 was considered to be significant. The graph software used was Graphpad prism 5 (Graphpad Software, San Diego, CA).
Authors’ contributions
JD carried out the conception and design of the research, participated in the acquisition of data, and drafted the manuscript. DS carried out the analysis and interpretation of data. JR participated in the statistical analysis. YF participated in the design of the study and performed the statistical analysis. BS conceived the study, and participated in its design and coordination, and helped to draft the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Not applicable. This study was only the primary research, and further studies have been in progress.
Consent for publication
Not applicable.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of the China-Japan Union Hospital, Jilin University.
Funding
None.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- GC
gastric cancer
- DE-miRNAs
differentially expressed miRNAs
- DEGs
differentially expressed genes
- GEO
Gene Expression Omnibus
- FC
fold change
- GO
gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- PPIs
protein–protein interactions
- STRING
Search Tool for the Retrieval of Interacting Genes
- SEM
mean ± standard error of mean
- NCAPG
Non-SMC Condensin I Complex Subunit G
- KNL1
kinetochore scaffold 1
Contributor Information
Bin Song, Email: BinSongbs@163.com.
Juan Du, Email: juandujdjd@163.com.
De-feng Song, Email: DefengSongdfs@163.com.
Ji-chen Ren, Email: jichenrenjcr@163.com.
Ye Feng, Phone: +86-0431-89876762, Email: bhc311@sina.com.
References
- 1.Nogueira A, Cabral M, Salles P, Araujo L, Rodrigues L, Rodrigues M, Oliveira C, Queiroz D, Rocha G, Oliveira A. Role of intestinal metaplasia and epithelial dysplasia in the pathogenesis of gastric carcinoma. Gastroenterology. 2000;118(4):A1404. doi: 10.1016/S0016-5085(00)81498-5. [DOI] [Google Scholar]
- 2.Fielding JWL, Brookes VS. Natural history of “early” gastric cancer: results of a 10-year regional survey. Br Med J. 1980;281(6246):965–967. doi: 10.1136/bmj.281.6246.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 4.Zou WB, Yang F, Li ZS. How to improve the diagnosis rate of early gastric cancer in China. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2015;44(1):9–14. doi: 10.3785/j.issn.1008-9292.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun F, Sun H, Mo X, Tang J, Liao Y, Wang S, Su Y, Ma H. Increased survival rates in gastric cancer, with a narrowing gender gap and widening socioeconomic status gap: a period analysis from 1984 to 2013. J Gastroenterol Hepatol. 2018;33(4):837–846. doi: 10.1111/jgh.14024. [DOI] [PubMed] [Google Scholar]
- 6.Compare D, Rocco A, Nardone G. Risk factors in gastric cancer. Eur Rev Med Pharmacol Sci. 2010;14(4):302–308. [PubMed] [Google Scholar]
- 7.Hudler P. Genetic aspects of gastric cancer instability. Sci World J. 2012;2012(4):761909. doi: 10.1100/2012/761909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hippo Y, Taniguchi H, Tsutsumi S, Machida N, Chong JM, Fukayama M, Kodama T, Aburatani H. Global gene expression analysis of gastric cancer by oligonucleotide microarrays. Cancer Res. 2002;62(1):233–240. [PubMed] [Google Scholar]
- 9.Qu Y, Dang S, Hou P. Gene methylation in gastric cancer. Clin Chim Acta. 2013;424:53–65. doi: 10.1016/j.cca.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Chang HK, Kim HK, Rettig RL, Kim J, Lee ET, Aprelikova O, Choi IJ, Munroe DJ, Green JE. miRNA signature associated with outcome of gastric cancer patients following chemotherapy. BMC Med Genom. 2011;4(1):79. doi: 10.1186/1755-8794-4-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X, Zhang Y, Zhang H, Liu X, Gong T, Li M, Sun L, Ji G, Shi Y, Han Z. miRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol Cancer Res. 2011;9(7):824–833. doi: 10.1158/1541-7786.MCR-10-0529. [DOI] [PubMed] [Google Scholar]
- 12.Peng Y, Zhang X, Ma Q, Yan R, Qin Y, Zhao Y, Cheng Y, Yang M, Wang Q, Feng X. MiRNA-194 activates the Wnt/β-catenin signaling pathway in gastric cancer by targeting the negative Wnt regulator, SUFU. Cancer Lett. 2017;385:117–127. doi: 10.1016/j.canlet.2016.10.035. [DOI] [PubMed] [Google Scholar]
- 13.Hu X, Zhang M, Miao J, Wang X, Huang C. miRNA-4317 suppresses human gastric cancer cell proliferation by targeting ZNF322. Cell Biol Int. 2017 doi: 10.1002/cbin.10870. [DOI] [PubMed] [Google Scholar]
- 14.He J, Jin Y, Chen Y, Yao HB, Xia YJ, Ma YY, Wang W, Shao QS. Downregulation of ALDOB is associated with poor prognosis of patients with gastric cancer. OncoTargets Ther. 2016;9:6099–6109. doi: 10.2147/OTT.S110203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eun J, Kim H, Shen Q, Yang H, Kim S, Yoon J, Park W, Lee J, Nam S. MicroRNA-495-3p functions as a tumour suppressor by regulating multiple epigenetic modifiers in gastric carcinogenesis. J Pathol. 2018;244:107–119. doi: 10.1002/path.4994. [DOI] [PubMed] [Google Scholar]
- 16.Guo J, Miao Y, Xiao B, Huan R, Jiang Z, Meng D, Wang Y. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol. 2009;24(4):652–657. doi: 10.1111/j.1440-1746.2008.05666.x. [DOI] [PubMed] [Google Scholar]
- 17.Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 18.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozomara A, Griffithsjones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42(Database issue):D68. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk—database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011;44(5):839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 21.Tang Y, Li M, Wang J, Pan Y, Wu FX. CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems. 2015;127:67–72. doi: 10.1016/j.biosystems.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 22.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benjamini YHY. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 1995;29(4):289–300. [Google Scholar]
- 24.Hulsegge I, Kommadath A, Smits MA. Globaltest and GOEAST: two different approaches for Gene Ontology analysis. BMC Proc. 2009;3(S4):S10. doi: 10.1186/1753-6561-3-s4-s10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Xiao W, Zhen L, Xin X, Li J, Yi Z, Shuai M, Li S, Song W, Bo X, Ji A. miR-148a-3p represses proliferation and EMT by establishing regulatory circuits between ERBB3/AKT2/c-myc and DNMT1 in bladder cancer. Cell Death Dis. 2016;7(12):e2503. doi: 10.1038/cddis.2016.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu T, Qu L, He G, Tian L, Li L, Zhou H, Jin Q, Ren J, Wang Y, Wang J. Regulation of laryngeal squamous cell cancer progression by the lncRNA H19/miR-148a-3p/DNMT1 axis. Oncotarget. 2016;7(10):11553–11566. doi: 10.18632/oncotarget.7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang Q, He M, Ma MT, Wu HZ, Yu ZJ, Guan S, Jiang LY, Wang Y, Zheng DD, Jin F. MicroRNA-148a inhibits breast cancer migration and invasion by directly targeting WNT-1. Oncol Rep. 2015;35(3):1425–1432. doi: 10.3892/or.2015.4502. [DOI] [PubMed] [Google Scholar]
- 31.de Souza CR, Leal MF, Calcagno DQ, Costa Sozinho EK, Borges BN, Montenegro RC, Dos Santos AK, Dos Santos SE, Ribeiro HF, Assumpção PP. MYC deregulation in gastric cancer and its clinicopathological implications. PloS ONE. 2013;8(5):e64420. doi: 10.1371/journal.pone.0064420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Xu Y, Qiu W, Zhao D, Zhang Y. Tissue miR-193b as a novel biomarker for patients with ovarian cancer. Med Sci Monit. 2015;21:3929–3934. doi: 10.12659/MSM.895407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin X, Sun Y, Yang H, Li J, Yu S, Chang X, Lu Z, Chen J. Deregulation of the MiR-193b-KRAS axis contributes to impaired cell growth in pancreatic cancer. PLoS ONE. 2015;10(4):e0125515. doi: 10.1371/journal.pone.0125515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mets E, Meulen JVD, Peer GV, Boice M, Mestdagh P, Walle IVD, Lammens T, Goossens S, Moerloose BD, Benoit Y. MicroRNA-193b-3p acts as a tumor suppressor by targeting the MYB oncogene in T-cell acute lymphoblastic leukemia. Leukemia. 2015;29(4):798–806. doi: 10.1038/leu.2014.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herzog S, Jaiswal SN, Urban E, Riemer A, Fischer S, Heidmann SK. Functional dissection of the Drosophila melanogaster condensin subunit Cap-G reveals its exclusive association with condensin I. PLoS Genet. 2013;9(4):e1003463. doi: 10.1371/journal.pgen.1003463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Q, Su R, Shan C, Gao C, Wu P. Non-SMC condensin I complex, subunit G (NCAPG) is a novel mitotic gene required for hepatocellular cancer cell proliferation and migration. Oncol Res. 2018;26(2):269–276. doi: 10.3727/096504017X15075967560980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang DG, Chen G, Wen XY, Wang D, Cheng ZH, Sun SQ. Identification of biomarkers for diagnosis of gastric cancer by bioinformatics. Asian Pac J Cancer Prev. 2015;16(4):1361–1365. doi: 10.7314/APJCP.2015.16.4.1361. [DOI] [PubMed] [Google Scholar]
- 38.Xu X, Cai N, Zhi T, Bao Z, Wang D, Liu Y, Jiang K, Fan L, Ji J, Liu N. MicroRNA-1179 inhibits glioblastoma cell proliferation and cell cycle progression via directly targeting E2F transcription factor 5. Am J Cancer Res. 2017;7(8):1680–1692. [PMC free article] [PubMed] [Google Scholar]
- 39.Vleugel M, Tromer E, Omerzu M, Groenewold V, Nijenhuis W, Snel B, Kops GJPL. Arrayed BUB recruitment modules in the kinetochore scaffold KNL1 promote accurate chromosome segregation. J Cell Biol. 2013;203(6):943–955. doi: 10.1083/jcb.201307016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caldas GV, DeLuca JG. KNL1: bringing order to the kinetochore. Chromosoma. 2014;123(3):169–181. doi: 10.1007/s00412-013-0446-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuen K, Montpetit B, Hieter P. The kinetochore and cancer: what’s the connection? Curr Opin Cell Biol. 2005;17(6):576–582. doi: 10.1016/j.ceb.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 42.Bogdanov KV, Takimoto M. Involvement of c-Abl and D40 (AF15Q14/CASC5) proteins in the regulation of cell proliferation and cancer. Tsitologiia. 2008;2(4):590–596. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable. This study was only the primary research, and further studies have been in progress.