ABSTRACT
The integrin-associated adaptor proteins integrin-linked kinase (ILK) and kindlin-2 play central roles in integrin signaling and control of cell morphology. A direct ILK–kindlin-2 interaction is conserved across species and involves the F2PH subdomain of kindlin-2 and the pseudokinase domain (pKD) of ILK. However, complete understanding of the ILK–kindlin-2 interaction and its role in integrin-mediated signaling has been impeded by difficulties identifying the binding site for kindlin-2 on ILK. We used conservation-guided mapping to dissect the interaction between ILK and kindlin-2 and identified a previously unknown binding site for kindlin-2 on the C-lobe of the pKD of ILK. Mutations at this site inhibit binding to kindlin-2 while maintaining structural integrity of the pKD. Importantly, kindlin-binding-defective ILK mutants exhibit impaired focal adhesion localization and fail to fully rescue the spreading defects seen in ILK knockdown cells. Furthermore, kindlin-2 mutants with impaired ILK binding are also unable to fully support cell spreading. Thus, the interaction between ILK and kindlin-2 is critical for cell spreading and focal adhesion localization, representing a key signaling axis downstream of integrins.
This article has an associated First Person interview with the first author of the paper.
KEY WORDS: Integrin-linked kinase, Kindlin-2, Integrin
Highlighted Article: We identify a binding site for kindlin-2 on ILK, and show that disrupting kindlin-2 binding diminishes localization to focal adhesions and impairs cell spreading.
INTRODUCTION
Cell adhesion to the extracellular matrix is a spatially and temporally regulated process governed by the dynamic assembly of complexes between transmembrane heterodimeric integrin adhesion receptors and cytoplasmic regulators (Morse et al., 2014). Hetero-dimerization of the 18 α- and 8 β-integrin subunits to form 24 αβ-integrin combinations drives ligand-binding specificity (Humphries, 2006) with signaling activity determined by the recruitment of specific binding partners to the cytoplasmic tails (Iwamoto and Calderwood, 2015). These complexes facilitate an essential link between the intracellular and extracellular environment, signaling bi-directionally through clusters of activated ligand-bound integrins that are associated with cytoplasmic regulators and cytoskeletal adaptors in foci known as focal adhesions (Anthis and Campbell, 2011).
Integrin-linked kinase (ILK) is a key component of focal adhesions, acting as a non-catalytic scaffold that serves as a hub for integrin-mediated signaling (Lange et al., 2009; Wickström et al., 2010). Loss of ILK is lethal at the embryonic stage in mice, and loss or depletion of ILK in cultured mammalian cells leads to defects in cell spreading and focal adhesion formation (Friedrich et al., 2004; Fukuda et al., 2003; Sakai et al., 2003). ILK comprises an N-terminal ankyrin repeat domain (ARD) and a C-terminal pseudokinase domain (pKD) (Chiswell et al., 2008; Fukuda et al., 2009). ILK forms a tripartite complex with particularly interesting new cysteine-histidine rich protein (PINCH, officially known as LIMS) through its ARD (Chiswell et al., 2008) and parvin through its pKD (Fukuda et al., 2009) that is known as the ILK–PINCH–parvin (IPP) complex. The IPP complex is important for stabilization of the three proteins as well as their cellular functions (Stiegler et al., 2013; Zhang et al., 2002). Mammals have two PINCH isoforms, PINCH1 and PINCH2 (LIMS1 and LIMS2, respectively), and three parvin isoforms, α-parvin, β-parvin and γ-parvin (PARVA, PARVB and PARVG, respectively), with PINCH1 and α-parvin being the most widely expressed across development and cell type (Nikolopoulos and Turner, 2000; Stanchi et al., 2005; Tu et al., 2001; Yamaji et al., 2001). The distinct signaling pathways downstream of ILK and the IPP complex remain largely un-elucidated, although the IPP complex has been shown to interact with many other signaling proteins such as paxillin (Nikolopoulos and Turner, 2002), Nck2, a regulatory adaptor of receptor tyrosine kinases (Velyvis et al., 2003), and kindlins (Fukuda et al., 2014; Huet-Calderwood et al., 2014; Mackinnon et al., 2002; Qadota et al., 2014).
Kindlins are a family of key cytoplasmic regulators of integrin activation, acting by direct interaction with the cytoplasmic tail of the β integrin subunit (Calderwood et al., 2013; Harburger et al., 2009; Li et al., 2017; Qadota et al., 2012). Kindlins are composed of an atypical 4.1, ezrin, radixin, moesin (FERM) domain that – in addition to the classic F1, F2 and F3 subdomains found in all FERM domains – is hallmarked by an N-terminal F0 lobe, a large flexible F1 insertion and a pleckstrin homology (PH) domain that splits the F2 subdomain to generate a module that we have termed F2PH (Calderwood et al., 2013; Li et al., 2017). The kindlin family consists of three isoforms (kindlin-1, kindlin-2 and kindlin-3) that exhibit restricted expression and non-overlapping functions. Kindlin-2 (officially known as FERMT2) is the ubiquitously expressed isoform and loss of kindlin-2 expression in mice is embryonic lethal (Montanez et al., 2008). In cultured murine fibroblasts, kindlin-2 deficiency impairs cell spreading and cell adhesion (Böttcher et al., 2017; Huet-Calderwood et al., 2014; Montanez et al., 2008; Theodosiou et al., 2016).
The interaction between ILK and kindlin-2 is conserved across species, and was first found in Caenorhabditis elegans (C. elegans) using a yeast two-hybrid screen in which the kindlin orthologue UNC-112, used as bait, interacted with PAT-4, the orthologue of ILK (Mackinnon et al., 2002). In mammals, ILK and kindlin-2 also form a complex (Fukuda et al., 2014; Huet-Calderwood et al., 2014; Montanez et al., 2008). Biochemical and biophysical studies have shown binding of highly conserved leucine residues – located at the hydrophobic face of an amphipathic helix within the F2PH subdomain of kindlin-2 – to an unidentified region on the pKD of ILK (ILK-pKD) (Fukuda et al., 2014; Huet-Calderwood et al., 2014). Overexpression studies have shown that mutation of one or more of the conserved leucine residues to alanines on this helix, which inhibits binding to the ILK-pKD, impairs cell spreading (Fukuda et al., 2014). However, difficulties in identifying the binding site for kindlin-2 on the ILK-pKD have hindered further understanding of the interaction and its role in signaling downstream of integrins.
Here we report the identification of a novel binding interface for kindlin-2 on the ILK-pKD, allowing us to investigate the functional significance of the ILK–kindlin-2 interaction. We show that mutations on helix-αH in the C-lobe of the ILK-pKD impair the interaction with kindlin-2 without perturbing α-parvin binding or structural integrity of the ILK-pKD. This binding site is distinct from a recently reported kindlin-2 binding interface in the N-lobe of the ILK-pKD (Guan et al., 2018), and we found that the reported double glycine for arginine substitutions (R243G and R334G) at that site destabilize the interaction of the ILK-pKD with α-parvin, potentially disrupting interaction with kindlin-2 indirectly. We demonstrate that mutations on helix-αH of the ILK-pKD impair the ability of GFP-tagged ILK (GFP–ILK) to localize to focal adhesions and disrupt the ability of GFP–ILK to fully rescue spreading defects observed in ILK-knockdown HeLa cells. Similarly, we show that a kindlin-2 mutant impaired in binding to ILK is unable to fully rescue spreading defects caused by depletion of kindlin-2, solidifying the role of the kindlin-2-ILK interaction in cell spreading and focal adhesion signaling.
RESULTS
Mutation of a highly conserved hydrophobic patch on the ILK-pKD impairs binding to the F2PH domain of kindlin-2
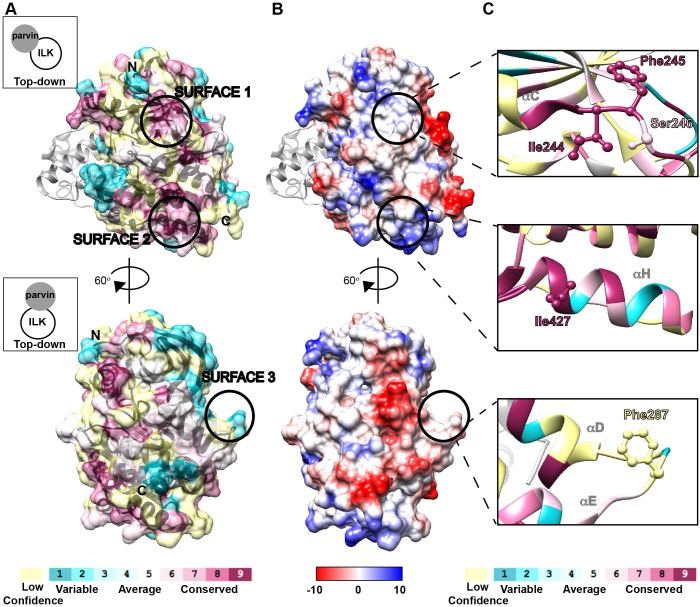
Previous work has demonstrated that the ILK-pKD binds to a series of highly conserved leucine residues, including L357 of human kindlin-2; these residues lie along the non-polar face of an amphipathic helix in the F2PH subdomain of kindlin-2 (Fukuda et al., 2014; Huet-Calderwood et al., 2014). Therefore, we hypothesized that kindlin-2 interacts with a conserved, hydrophobic patch on the ILK-pKD. To identify potential kindlin-2-binding surfaces on the ILK-pKD, we used ILK sequences from 37 species including C. elegans and Homo sapiens (Table S1), and the previously reported crystal structure of the human ILK-pKD in complex with the second calponin homology domain (CH2) of α-parvin (α-parvin-CH2) bound to MgATP (PDB ID: 3KMW) (Fukuda et al., 2009) to generate a conservation surface map with the ConSurf server (http://consurf.tau.ac.il; Landau et al., 2005). We initially identified two surfaces (surface 1 and 2) with clusters of highly conserved residues (Fig. 1A). We also selected a third, less well conserved surface on the lateral face of the ILK-pKD that may accommodate the helical fragment of the F2PH, which binds the ILK-pKD (surface 3) (Fig. 1A) (Fukuda et al., 2014). Next, we generated a map of the coulombic surface potential of the ILK-pKD to identify patches with neutral surface potential, a proxy for hydrophobicity, using Chimera software (https://www.cgl.ucsf.edu/chimera/; Pettersen et al., 2004) (Fig. 1B). We noticed that all three selected surfaces lie on hydrophobic patches. Importantly, none of the selected candidate kindlin-binding surfaces overlap with the binding interface for α-parvin or the ATP-binding site on the ILK-pKD (Fukuda et al., 2009). In order to disrupt the potential non-polar interaction with the kindlin-2 F2PH, we mutated selected non-polar, solvent-exposed residues on each surface to either an aspartic acid or glutamic acid (Fig. 1C). On surface 1, we generated substitution mutations of isoleucine, phenylanaline and serine (I244D, F245D and S246D) on a loop at the C-terminus of the αC helix. For surface 2, we replaced I427 with glutamic acid (I427E) on helix-αH and on surface 3 we replaced F287, which resides on a loop between helix-αD and helix-αE, with D (F287D).
Fig. 1.
Selection of highly conserved, hydrophobic patches on the ILK-pKD by surface mapping. (A) ConSURF (Landau et al., 2005) surface map generated from 37 species of ILK-pKD mapped onto the previously determined crystal structure of the ILK-pKD in complex with α-parvin-CH2 (gray ribbon) bound to MgATP (not visible in orientations shown), generated with Chimera software (Pettersen et al., 2004), and shown in two different orientations related by a 60° rotation as indicated (PDB ID: 3KMW). Schematic representing a top-down view of the complex to show the relative orientation of α-parvin-CH2 to the ILK-pKD (left). Color scale (bottom of panel), with positions for which the conservation score was assigned with low confidence indicated in light yellow. Color-coded surface is shown at 50% transparency, with ribbon structure in black. N- and C-termini are indicated. (B) Coulombic surface map indicating the electrostatic potential was generated by using Chimera software (Pettersen et al., 2004) for each orientation of the ILK-pKD–α-parvin-CH2 complex shown in Fig. 1A. Color scale (bottom of panel) is given in units of kcal mol−1 e−1 at 298 K. (C) Ribbon diagram of selected regions from the ConSURF map shown in A. Residues selected for mutagenesis are shown as ball-and-stick display.
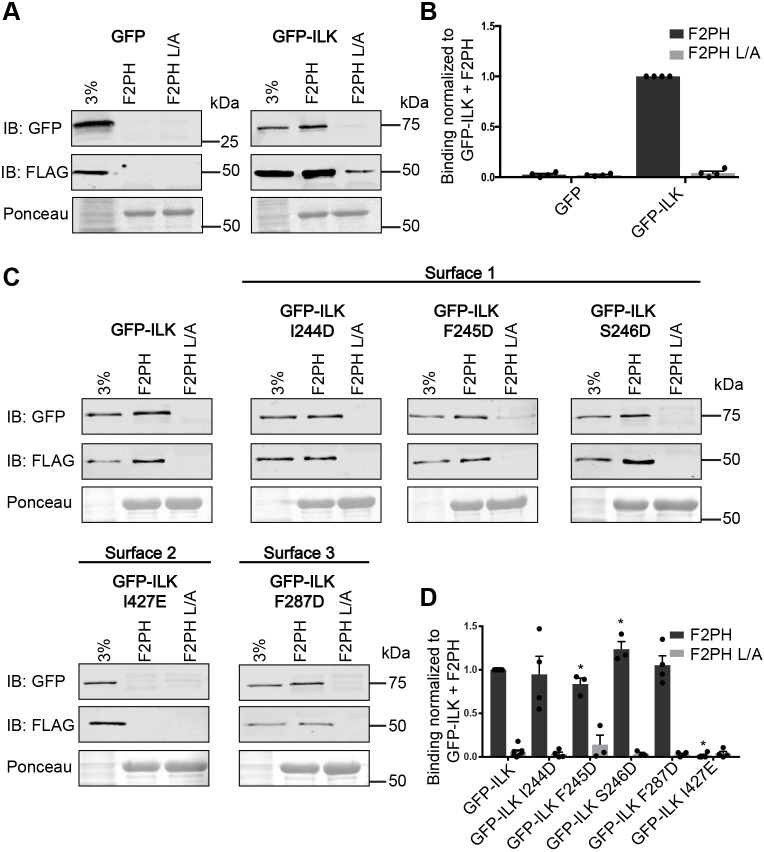
As ILK protein stability is crucially dependent on association with parvin (Fukuda et al., 2003) we co-expressed FLAG-tagged α-parvin (FLAG–α-parvin) and GFP–ILK or ILK mutants in chinese hamster ovary (CHO) cells and assessed the ability of purified recombinant glutathione S-transferase (GST) fused to kindlin-2 F2PH (GST–kindlin-2 F2PH) to pull down GFP–ILK from cell lysates. As previously reported (Huet-Calderwood et al., 2014), kindlin-2 F2PH bound GFP–ILK and FLAG–α-parvin in a specific ILK-dependent manner, as when GFP was co-expressed with FLAG-α-parvin, negligible GFP or FLAG–α-parvin binding was detected (Fig. 2A,B). Specificity was further confirmed using GST–kindlin-2 F2PH L357A (F2PH L/A), which contains a mutation in the previously identified ILK-binding site (Fukuda et al., 2014; Huet-Calderwood et al., 2014) as a negative control. All five ILK mutants expressed well at the expected molecular mass, and non-specific binding to GST–kindlin-2 F2PH L/A was minimal (Fig. 2C). While surface 1 and 3 mutants exhibited approximately wild-type levels of binding to kindlin-2 F2PH, the I427E mutant on surface 2 abolished binding (Fig. 2C,D). Importantly, the binding of FLAG–α-parvin to the GST–kindlin-2 F2PH only occurred concomitantly with the binding to GFP–ILK, consistent with the fact that the interaction is specifically mediated by ILK (Fukuda et al., 2014; Huet-Calderwood et al., 2014). We also tested a more conservative I427A mutation, in which the isoleucine side chain was removed but no negative charge was introduced and found that this also impaired binding to GST–kindlin-2 F2PH in pulldown assays (Fig. S1A,B). In summary, these data suggest that I427 on helix-αH of ILK-pKD is involved in the interaction between the ILK-pKD and kindlin-2.
Fig. 2.
I427E mutation of αH in surface 2 of ILK-pKD impairs binding of GFP–ILK to GST–kindlin-2 F2PH. (A) GFP or GFP–ILK co-expressed with FLAG–α-parvin in CHO cells bound to glutathione bead-immobilized GST–kindlin-2 F2PH or GST–kindlin-2 F2PH L357A (L/A) as a negative control detected by immunoblotting. One representative blot for each construct tested is shown. The lane labeled ‘3%’ indicates 3% of input lysate. Bead loading was visualized by Ponceau S staining. (B) Quantification of GFP or GFP–ILK binding to GST–kindlin-2 F2PH or GST–kindlin-2 F2PH L/A (mean±s.e.m.; n=4). (C) Representative immunoblots for pulldown of GFP–ILK mutants co-expressed with FLAG–α-parvin in CHO cell lysates by GST–kindlin-2 F2PH or GST–kindlin-2 F2PH L/A. The lane labeled ‘3%’ indicates 3% of input lysate. Bead loading was visualized by Ponceau S staining. (D) Quantification of binding of GFP–ILK and GFP–ILK mutants to GST–kindlin-2 F2PH or GST–kindlin-2 F2PH L/A (mean±s.e.m.; n≥3); *P<0.005 (Student's t-test). Pulldown quantification graphs are shown as bar charts with individual data points plotted (dots).
Additional mutations in helix-αH on surface 2 impair binding to kindlin-2
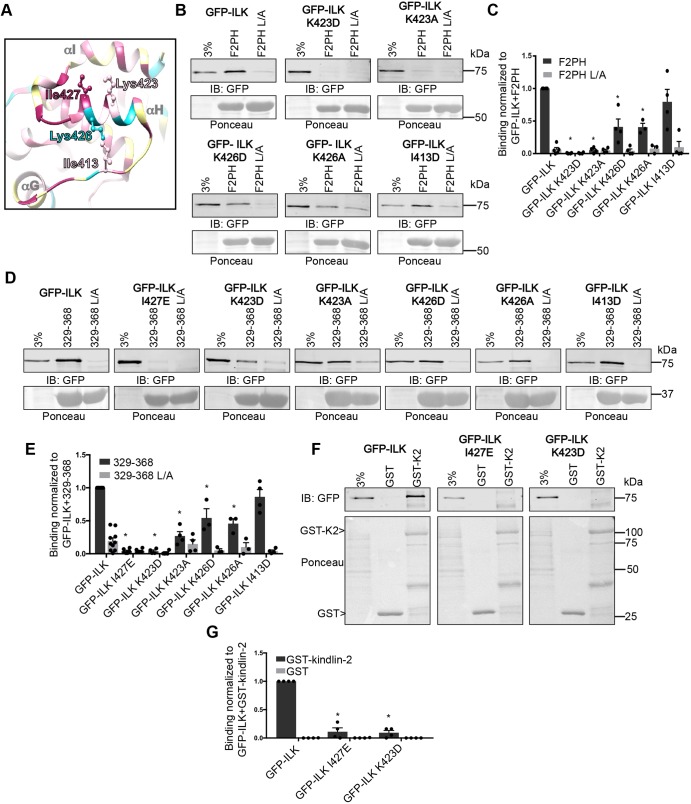
To further map the kindlin-binding surface on the ILK-pKD, we re-examined the ILK-pKD–α-parvin-CH2 co-crystal structure (Fukuda et al., 2009) to look for additional surface-exposed residues close to I427. We selected the well-conserved K423 and the poorly conserved K426 on helix-αH, as well as the more-distant but conserved hydrophobic residue I413 on the linker between helix-αG and helix-αH (Fig. 3A). We tested the effects of charge reversal and charge removal mutations at selected lysine residues (i.e. K423D, K423A, K426D or K426A), and the effect of replacing I413 with a charged residue (I413D). When co-expressed with FLAG-α-parvin, GFP–ILK K423D and GFP–ILK K423A were severely impaired in binding to GST–kindlin-2 F2PH, while the K426D and K426A mutations exhibited a more modest inhibition (Fig. 3B,C). The I413D mutation had no marked effect on binding to GST–kindlin-2 F2PH (Fig. 3B,C). This suggests that the highly conserved K423, which lies along the same face of helix-αH as I427, is also involved in the interaction between the ILK-pKD and the kindlin-2 F2PH, and that αH is a key mediator of the interaction with the kindlin-2 F2PH.
Fig. 3.
Additional residues on αH in the ILK-pKD are implicated in the interaction with kindlin-2. (A) Ribbon diagram of helix-αH and surrounding residues in the ILK-pKD–α-parvin-CH2 co-crystal structure (PDB ID: 3KMW) generated with Chimera software (Pettersen et al., 2004). Residues selected for mutation are labeled and shown as a ball-and-stick representation. Conservation coloring is indicated using the same color scale as shown in Fig. 1A. (B,C) Pulldown of GFP–ILK or GFP–ILK mutants by GST–kindlin-2 F2PH and GST–kindlin-2 F2PH L357A (L/A) from CHO cell lysate co-overexpressing FLAG–α-parvin assessed by representative immunoblots (B) and quantified (C); mean±s.e.m.; n≥3; *P<0.001 (Student's t-test). (D,E) Pulldown of GFP–ILK or GFP–ILK mutants from CHO cell lysate co-overexpressing FLAG–α-parvin using GST–kindlin-2 329-368 or GST–kindlin-2 329-368 L/A were assessed by representative immunoblots (D) and quantified (E); mean±s.e.m.; n≥3; *P≤0.0006. (F,G) Pulldown of GFP–ILK or GFP–ILK mutants from CHO cell lysate co-overexpressing FLAG–α-parvin using GST or GST–kindlin-2 were assessed in representative immunoblots (F) and quantified (G); mean±s.e.m.; n=4; *P≤0.0001 (Student's t-test). Pulldown quantification graphs are shown as bar charts with individual data points plotted (dots). GST- protein loading is indicated by Ponceau S staining.
We, and others, previously localized the ILK-binding site in the kindlin-2 F2PH to leucine residues in a helical region in the linker between the F2 and PH domains, spanning residues 329-368 (Fukuda et al., 2014; Huet-Calderwood et al., 2014). Therefore, to further confirm the specificity of our results, we performed pull-down assays with purified GST–kindlin-2 329-368. Consistent with our kindlin-2 F2PH data, mutants GFP–ILK I427E and GFP–ILK K423D were severely impaired in binding to GST–kindlin-2 329-368, while K426 mutants had a partial effect, and GFP–ILK I413D bound at levels comparable to those of GFP–ILK (Fig. 3D,E). Again, the L357A (L/A) mutations in the kindlin-2 protein strongly perturbed ILK binding. Finally, we also confirmed that GFP–ILK I427E and K423D were impaired in their ability to bind to purified full-length GST–kindlin-2 in pulldowns from cell lysate (Fig. 3F,G). Thus, the highly conserved surface-exposed ILK residues I427 and K423 are important for kindlin-2 binding, apparently by interacting with kindlin-2 residues 329-368 in the F2PH domain.
Prior studies have highlighted kindlin-2 residues L353 and L357 within the helical linker region as important for binding ILK (Fig. S2A) (Fukuda et al., 2014; Huet-Calderwood et al., 2014) but our data, showing the importance of ILK residue K423, suggest that charged kindlin residues are also important in the interaction with helix-αH of the ILK-pKD. We, therefore, re-examined the sequence of the helical region looking for negatively charged sidechains that might interact with ILK residue K423. We tested the importance of E354 and E358 in kindlin-2 by generating individual charge-reversal mutations (E354K and E358K) and an E354K/E358K double mutant (GST–kindlin-2 F2PH EE/KK). In pulldown assays with GST–kindlin-2 F2PH, we found that the E354K and E358K mutations partially impaired binding to GFP–ILK, and the EE/KK double mutation severely inhibited binding of GFP–ILK (Fig. S2B,C). Thus, kindlin-2 residues E354 and E358, in addition to L353 and L357, are important for the interaction between kindlin-2 and ILK. To test whether the effect of these charge-swap mutations could be reversed by a compensatory charge swap in ILK, we assessed binding of these GST–kindlin-2 F2PH mutants to GFP–ILK K423D (Fig. S2B,C). While GFP–ILK K423D was strongly impaired in binding to GST–kindlin-2 E358K and GST–kindlin-2 F2PH EE/KK, its binding to GST–kindlin-2 E354K was slightly enhanced (Fig. S2B,C). This raises the possibility of an interaction between K423 in ILK and E354 in kindlin-2, although a direct interaction between these residues cannot be confirmed without high-resolution structural information.
Kindlin-2-binding-deficient ILK-pKD mutants co-purify with α-parvin-CH2 and are structurally stable
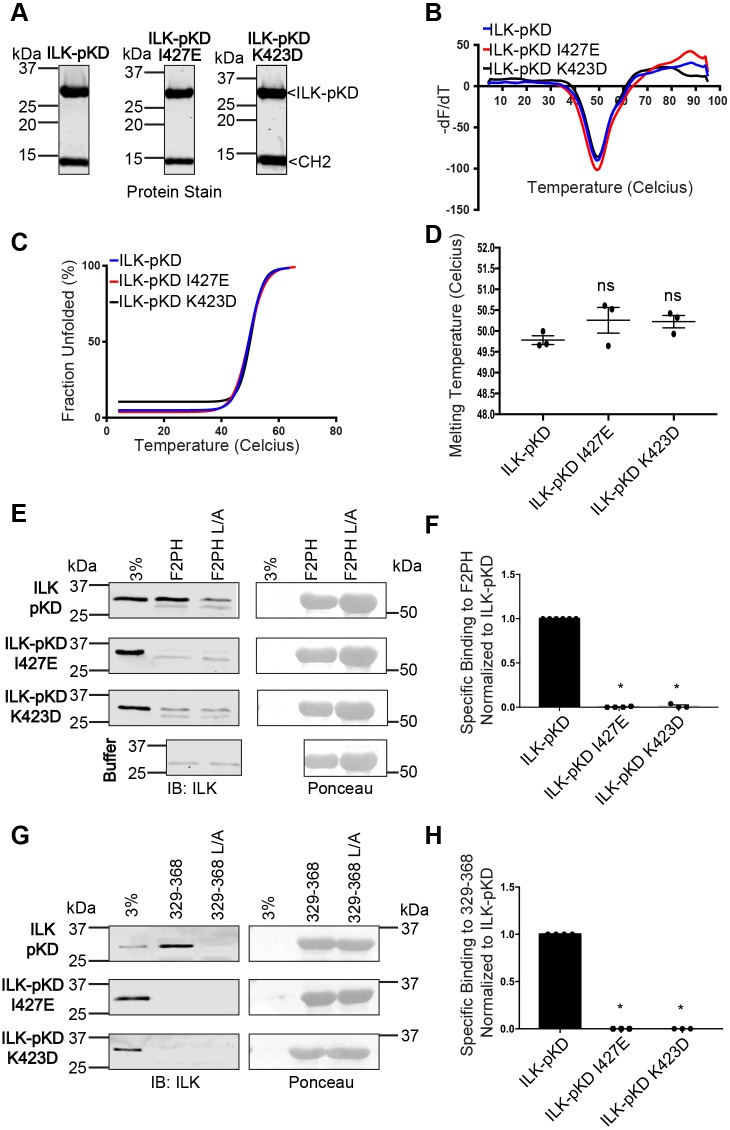
ILK stability and function require binding to the CH2 domain of α-parvin (Fukuda et al., 2009). To test whether the effect of ILK I427E and K423D mutations on kindlin binding might be mediated by destabilization of the ILK-pKD and/or disruption of the ILK-pKD–α-parvin-CH2 interaction, we co-purified recombinant GST-ILK-pKD (wild type or mutant) in complex with α-parvin-CH2 by co-expressing the GST-ILK-pKD and α-parvin-CH2 in Escherichia coli. The GST-fusion complex was purified by glutathione affinity chromatography and, after removal of the GST tag with tobacco etch virus protease (TEV), further purified by Resource S cation exchange chromatography. The wild-type ILK-pKD and the I427E and K423D point mutants each co-purified with α-parvin-CH2 through both chromatography steps (Fig. 4A). Thus, like wild-type ILK-pKD, the two helix-αH mutants are stably associated with α-parvin-CH2. To assess protein stability, we measured the apparent melting temperature of the purified protein complexes by differential scanning fluorimetry (DSF) using SYPRO orange fluorescence. Visual inspection of the first derivatives of fluorescence revealed similar melting profiles for the wild-type and mutant complexes (Fig. 4B), and non-linear fitting of the thermal denaturation data to identify the midpoint of unfolding (Tm) (Huynh and Partch, 2015) revealed similar Tm values for the wild-type, I427E and K423D ILK-pKD–α-parvin-CH2 complexes in multiple experiments (Fig. 4C,D). These values are comparable to the previously reported Tm for the ILK-pKD–α-parvin-CH2 complex (Murphy et al., 2014). Furthermore, by using the purified ILK-pKD–α-parvin-CH2 complexes described above, we assessed direct binding to GST–kindlin-2 constructs in pulldown assays. This showed that, as seen in the lysate pulldown assays (Fig. 2C,D and Fig. 3B-E), the I427E and K423D mutant complexes are severely impaired in binding to GST–kindlin-2 F2PH and GST–kindlin-2 329-368 (Fig. 4E-H). Thus, the inability of the ILK-pKD I427E and K423D mutants to bind to kindlin-2 F2PH cannot be explained by instability of the ILK-pKD or a defect in α-parvin binding and is, instead, likely to be due to direct disruption of a conserved binding surface on the C-lobe of ILK-pKD for a highly conserved helical fragment in the kindlin-2 F2PH.
Fig. 4.
ILK-pKD mutants that are defective in binding kindlin-2 co-purify with α-parvin-CH2 and are structurally stable. (A) Coomassie protein stain of purified non-mutant ILK-pKD, ILK-pKD I427E and K423D ILK-pKD–α-parvin-CH2 complexes. (B) Negative derivatives of SYPRO Orange fluorescence with respect to temperature for the non-mutant ILK-pKD, ILK-pKD I427E and K423D ILK-pKD–α-parvin-CH2 complexes from one representative experiment. (C) Normalized SYRPO Orange fluorescence data fitted to a Boltzmann sigmoid by using Graphpad Prism software for the purified ILK-pKD–α-parvin-CH2 complexes from one representative experiment. Statistical fit parameters: non-mutant ILK-pKD: r2=0.99, V50=49.7±0.2 (V50±s.e.m.); ILK-pKD I427E: r2=0.99, V50=49.7±0.2; ILK-pKD K423D: r2=0.95, V50=50.4±0.4. (D) Melting temperature extracted from the half-maximal point of the fitted fluorescence data (mean±s.e.m.; n=3); ns, not significant, P>0.05 (Student's t-test). (E,F) Pulldown of purified ILK-pKD–α-parvin-CH2 complexes by immobilized GST–kindlin-2 F2PH or GST–kindlin-2 F2PH L357A (L/A) assessed by representative immunoblots (E) and quantified (F) (mean±s.e.m.; n≥3); *P≤0.0001 (Student's t-test). (G,H) Pulldown of purified ILK-pKD–α-parvin-CH2 complexes by immobilized GST–kindlin-2 329-368 or GST–kindlin-2 329-368 L/A assessed by representative immunoblots (G) and quantified (H); mean±s.e.m.; n≥3; P≤0.0001 (Student's t-test). Pulldown quantification is shown as bar charts with individual data points plotted (dots). GST-protein loading for pulldowns is indicated by Ponceau S staining.
Mutations at a recently reported kindlin-2 binding interface on the ILK-pKD perturb α-parvin binding
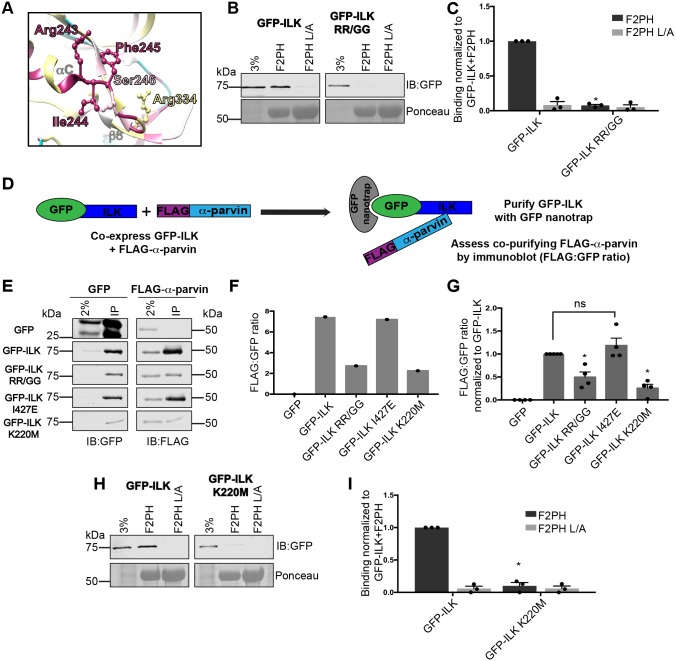
While our study was underway, it was reported that substitutions R243G and R334G in the N-lobe of the ILK-pKD inhibit binding to kindlin-2 F2PH (Guan et al., 2018). These residues are in the vicinity of our surface 1 mutants (Fig. 5A). While our surface 1 mutants had little impact on GFP–ILK binding to kindlin-2 F2PH (Fig. 2C,D), we confirmed that the R243G and R334G double mutant (GFP-ILK RR/GG) is strongly impaired in binding (Fig. 5B,C). However, when we attempted to purify recombinant GST-ILK-pKD RR/GG from bacteria co-expressing α-parvin-CH2, we were unable to obtain stable soluble material (data not shown). This prevented us from directly assessing the protein stability of the purified ILK-pKD RR/GG–α-parvin-CH2 mutant complex as we had done for the helix-αH mutants (Fig. 4B,C). We, therefore, utilized immunoprecipitation to assess association of co-overexpressed GFP–ILK RR/GG with FLAG–α-parvin and included GFP–ILK I427E as a control. We also included the additional control of the K220M substitution mutant of GFP–ILK (GFP–ILK K220M), originally generated as a kinase-dead mutant but now known to disrupt α-parvin binding (Lange et al., 2009). Using a GFP-nanotrap (Rothbauer et al., 2008), we precipitated GFP–ILK, assessed the amount of co-precipitating FLAG–α-parvin by immunoblotting and calculated the ratio of co-precipitated FLAG–α-parvin to precipitated GFP–ILK (FLAG:GFP ratio) (Fig. 5D). Fig. 5E shows representative immunoblots from one experiment, with quantification of the raw FLAG:GFP ratios shown in Fig. 5F. Pooled data from multiple experiments, where the FLAG:GFP ratio was normalized to the ratio of the GFP–ILK control in each experiment, are shown in Fig. 5G. Although this approach cannot provide the stoichiometry of ILK–kindlin-2 complexes, as expected from our experiments with purified proteins (Fig. 4A), GFP–ILK I427E co-precipitated an amount of FLAG–α-parvin that was comparable to that of wild-type GFP–ILK, while GFP–ILK K220M precipitated significantly less FLAG–α-parvin (Fig. 5E-G). Notably, GFP–ILK RR/GG also co-precipitated substantially less α-parvin than GFP–ILK wild type and GFP–ILK I427E (Fig. 5E-G). The crystallographically defined α-parvin-binding site, and ILK residues R243 and R334 lie on opposite sides of the ILK-pKD (Fukuda et al., 2009), yet our data clearly suggest that α-parvin binding is perturbed when these residues are mutated to glycines. Since α-parvin binding is required for ILK structural stability, it is possible that the impaired binding ability of the RR/GG mutant to kindlin-2 is the indirect result of its reduced ability to bind α-parvin. This is consistent with our inability to produce soluble ILK-pKD RR/GG in E. coli (data not shown). Notably, GFP–ILK K220M, another parvin-binding defective mutant (Lange et al., 2009), is also impaired in binding to GST–kindlin-2 F2PH in pulldown experiments (Fig. 5H,I), supporting the idea that disruption of the ILK–α-parvin interaction indirectly impairs kindlin binding, possibly by destabilization of the ILK-pKD.
Fig. 5.
R243G/R334G double mutation of GFP–ILK (GFP–ILK RR/GG) impairs binding of the ILK to α-parvin. (A) Ribbon diagram of selected regions in the ILK KD–α-parvin-CH2 complex co-crystal structure (PDB ID: 3KMW) surrounding I244, F245, and S246, generated with Chimera software (Pettersen et al., 2004). Residues selected for mutagenesis are labeled and shown as a ball-and-stick representation. Conservation coloring is indicated using the same color scale as shown in Fig. 1A. (B,C) Pulldown of GFP–ILK or GFP–ILK RR/GG from CHO cell lysates co-overexpressing FLAG–α-parvin using GST–kindlin-2 F2PH or GST–kindlin-2 F2PH L357A (L/A) assessed by representative immunoblots (B) and quantified (C); mean±s.e.m.; n=3; *P≤0.0001 (Student's t-test). (D) Diagram of the GFP nanotrap experiment. GFP–ILK was purified from lysate of cells co-expressing GFP–ILK and FLAG-α-parvin. The amount of co-purifying FLAG–α-parvin was assessed by immunoblotting and the FLAG:GFP ratio for each construct was calculated. (E,F) GFP-nanotrap co-purification of GFP–ILK constructs co-expressed FLAG–α-parvin in CHO cells was assessed by immunoblot from one experiment (E) and quantified (F) as a raw FLAG:GFP ratio. Dots represent single data points for each construct tested. The lane labeled ‘2%’ indicates the 2% input of lysate. (G) The FLAG:GFP ratio for each GFP or GFP–ILK construct tested is expressed relative to the FLAG:GFP ratio of the GFP–ILK control within each experiment, which is set to 1. The dataset includes the experiment shown in E and F (mean±s.e.m.; n≥4); *P≤0.0015; ns, statistically not significant, P>0.05 (Student's t-test). (H,I) Pulldown of GFP–ILK K220M from CHO cell lysate co-overexpressing FLAG–α-parvin using GST–kindlin-2 F2PH or GST–kindlin-2 F2PH L/A assessed by representative immunoblot (H) and quantified (I); mean±s.e.m.; n=3; *P≤0.0001. Pulldown and co-purification quantification graphs are shown as bar charts with individual data points plotted (dots). GST-protein loading control for pulldown experiments is indicated by Ponceau S staining.
Kindlin-2 binding facilitates ILK localization to focal adhesions
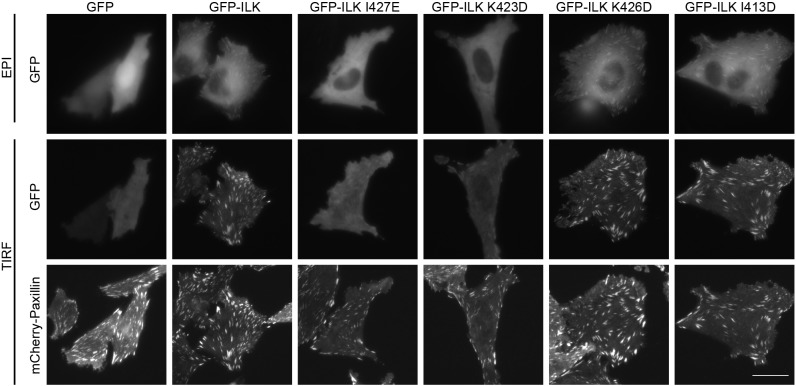
To test whether the interaction with kindlin-2 is required for the localization of ILK to focal adhesions, we utilized total internal reflection fluorescence (TIRF) microscopy to visualize the localization of GFP–ILK in live cells. We generated stable CHO cell lines expressing mCherry-paxillin as a marker for focal adhesions. We transiently overexpressed GFP–ILK or GFP–ILK mutants in these cells concomitantly with FLAG-α-parvin, plated them on fibronectin-coated glass-bottom dishes and examined the localization of GFP–ILK mutants to paxillin-containing focal adhesions by TIRF and epifluorescence microscopy. Although GFP–ILK localized to focal adhesions when conducting epifluorescence microscopy, GFP, GFP–ILK I427E and GFP–ILK K423D exhibited barely detectable focal adhesion targeting (Fig. 6). When the same cells were imaged by TIRF microscopy to enhance visualization at the cell-matrix interface, GFP–ILK was very clearly detected in focal adhesions, while GFP alone was not; and although GFP–ILK I427E and GFP–ILK K423D were observed in some focal adhesions, they appeared to target much more weakly than wild-type GFP–ILK (Fig. 6). GFP–ILK K426D, which only partially inhibited binding to kindlin-2 in pulldown experiments, and GFP–ILK I413D, which had no strong effect on kindlin-2 binding, localized to focal adhesions when visualized by either epifluorescence or TIRF microscopy (Fig. 6). The kindlin-binding defective I427A mutant is also strongly impaired in its localization to focal adhesions in these cells (Fig. S3A), suggesting that changes in charge distribution along helix-αH do not explain the defect in focal adhesion targeting imparted by surface 2 mutations. Consistent with prior reports, GFP–ILK RR/GG, which is impaired in binding to parvin and kindlin, exhibits strongly impaired focal adhesion localization (Fig. S3B and Guan et al., 2018). In addition, GFP–ILK K220M is also impaired in focal adhesion targeting (Fig. S3B). Together, these results suggest that binding of kindlin-2 to ILK facilitates the localization of ILK to focal adhesions.
Fig. 6.
GFP–ILK mutants that are impaired in kindlin-2 binding localize poorly to focal adhesions. CHO cells stably expressing mCherry-paxillin were transiently co-transfected with FLAG–α-parvin and either GFP alone, GFP–ILK or one of the GFP–ILK mutants. Six hours after replating on fibronectin-coated glass-bottom dishes, live cells were imaged by epifluorescence (EPI) and/or TIRF microscopy as indicated. Images in each channel were linearly and uniformly adjusted, and cropped for clarity. Scale bar: 20 µm.
Interaction between kindlin-2 and ILK is required for spreading of HeLa cells
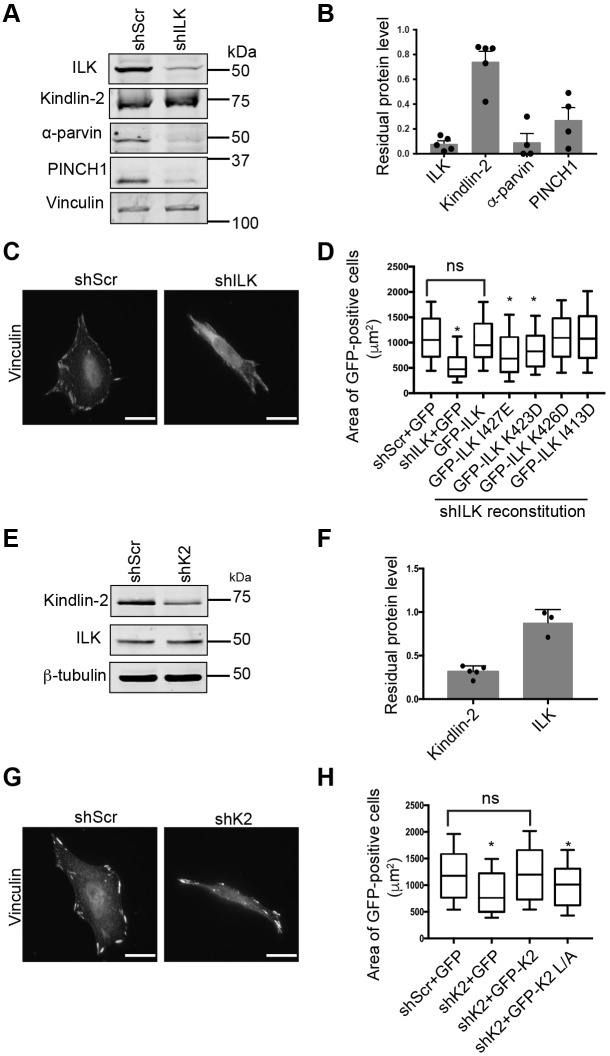
Both kindlin-2 and ILK have been implicated in cell spreading (Friedrich et al., 2004; Montanez et al., 2008; Sakai et al., 2003; Stanchi et al., 2009; Theodosiou et al., 2016). We, therefore, evaluated the role of the ILK–kindlin-2 interaction in cell spreading. To generate ILK-deficient cells, we infected HeLa cells with a lentivirally delivered shRNA construct targeting human ILK (shILK) or a scrambled shRNA (shScr) control. Immunoblotting showed only ∼10% residual ILK protein in shILK-infected ILK knockdown HeLa (shILK cells) and, as expected, protein levels of α-parvin and PINCH1– components of the IPP complex – were also substantially diminished (Fig. 7A,B). Kindlin-2 levels were only modestly impacted (Fig. 7A,B). Morphologically, shILK cells spread poorly on fibronectin and formed few, small vinculin-containing focal adhesions (Fig. 7C). This was not simply due to delayed spreading of shILK cells as the phenotype was still evident 18 h after cell plating (Fig. 7C). To investigate the importance of interactions between ILK and kindlin for the spreading of HeLa cells, we transiently re-expressed shILK-resistant GFP–ILK or our surface 2 GFP–ILK mutants (I427E, K423D, K426D and I413D) in shILK cells, and plated the cells on fibronectin-coated coverslips.
Fig. 7.
The ILK–kindlin-2 interaction is important for normal cell spreading in HeLa cells. (A) Immunoblotting of shScr and shILK HeLa cells to show protein levels of ILK, kindlin-2, α-parvin, PINCH1 and vinculin. (B) Bar graph showing residual protein levels in shILK cells calculated relative to those in shScr cells (mean±s.e.m.); individual data points are indicated (dots; n≥4). (C) Immunofluorescence staining of endogenous vinculin in fixed shScr or shILK HeLa cells spread on fibronectin-coated glass coverslips and acquired by epifluorescence microscopy. Scale bars: 20 µm. (D) CellProfiler quantification of GFP-positive cell areas pooled across three independent experiments shown as box and whiskers plots indicating 10th and 90th percentile range. n=155 shScr+GFP cells, 169 shILK+GFP cells, 154 shILK+GFP–ILK cells, 162 shILK+I427E cells, 143 shILK+K423D cells, 137 shILK+K426D cells, 125 shILK+I413D cells; *, significantly different from shScr+GFP calculated using one-way ANOVA and Tukey's correction for multiple comparisons (P≤0.01); ns, statistically not significant; P>0.05. (E) Immunoblotting of shScr and shK2 HeLa cells to show protein levels of kindlin-2, ILK, and β-tubulin. (F) Bar graph showing residual kindlin-2 or ILK protein levels in shK2 cells calculated relative to those in shScr cells (mean±s.e.m.); individual data points are indicated (dots; n≥3). (G) Immunofluorescence staining of endogenous vinculin in fixed shScr or shK2 HeLa cells spread on fibronectin-coated glass coverslips and acquired by epifluorescence microscopy. Scale bars: 20 µm. (H) CellProfiler quantification of GFP-positive cells areas pooled across three independent experiments; n=143 shScr+GFP cells, 121 shK2+GFP cells, 145 shK2+GFP-K2 cells, 130 shK2+GFP-K2 LA cells. Data are shown as box and whiskers plot, with whiskers indicating the 10th and 90th percentile range. *, significantly different from shScr+GFP calculated using one-way ANOVA and Tukey's correction for multiple comparisons (P≤0.015); ns, statistically not significant. Microscopy images were linearly and uniformly adjusted for clarity.
Cells were fixed and stained with phalloidin 18 h after plating and CellProfiler version 2.0 (Kamentsky et al., 2011) was used to calculate areas of GFP-positive cells across multiple fields of view and multiple replicate experiments using the phalloidin stain to identify the cell outline. Immunoblotting confirmed that all constructs expressed at their expected molecular mass (Fig. S4A). We clearly observed a profound spreading defect in shILK cells expressing GFP, which was fully reversed in shILK cells expressing GFP–ILK (Fig. 7D). Surface 2 mutant GFP–ILK I413D, for which kindlin-2 binding was not significantly altered, and GFP–ILK K426D, for which only partial inhibition of kindlin-2 binding was produced, were also able to fully rescue shILK cell spreading (Fig. 7D). Notably, neither GFP–ILK I427E nor GFP–ILK K423D, which both strongly impair binding to kindlin-2, were able to fully rescue the spreading defect of shILK cells (Fig. 7D), suggesting that kindlin binding contributes to ILK function in cell spreading. Notably, when we examined the localization of GFP–ILK in live shILK HeLa cells using mCherry-Paxillin as a marker for focal adhesions, GFP–ILK K423D and GFP–ILK I427E clearly localized to focal adhesions in these cells, although targeting appeared weaker compared to that of GFP–ILK (Fig. S5). This is consistent with the notion that, under conditions of reduced competition from endogenous ILK, GFP–ILK can be recruited to focal adhesions in a kindlin-2-independent manner (Huet-Calderwood et al., 2014). Nonetheless, despite targeting to focal adhesions, the kindlin-binding defective ILK was unable to fully support cell spreading.
To further test the importance of kindlin-ILK interactions in cell spreading, we knocked down kindlin-2 in HeLa cells (shK2 cells). Despite screening five shRNAs that target kindlin-2, the best specific knockdown we achieved was ∼70% depletion and this hairpin was used in further experiments (Fig. 7E,F). Kindlin knockdown had minimal effect on ILK expression levels (Fig. 7E,F). Nonetheless, similar to shILK cells, shK2 cells spread poorly on fibronectin, and formed only small vinculin-containing adhesions (Fig. 7G). We expressed GFP or knockdown-resistant GFP-kindlin-2 or GFP–kindlin-2 L357A (L/A) in these cells and used Cell Profiler 2.0 (Kamentsky et al., 2011) to quantify the area of GFP-positive cells across multiple biological replicates. We found that shK2 cells transiently expressing GFP were smaller than shScr cells; this defect was fully rescuable with GFP-kindlin-2 but not with the ILK-binding defective GFP–kindlin-2 L/A (Fig. 7H), despite both constructs migrating at their expected size in immunoblots (Fig. S4B). Together, with our results in reconstituted ILK knockdown cells, this strongly supports the conclusion that kindlin-2-ILK interactions are essential for normal cell spreading.
DISCUSSION
Kindlins and ILK are key components of cell-matrix adhesions with essential roles in cell adhesion and spreading (Calderwood et al., 2013; Li et al., 1999). Kindlins make direct interactions with the β-integrin cytoplasmic tail (Calderwood et al., 2013) and the recognition that kindlins also directly bind ILK (Fukuda et al., 2014; Huet-Calderwood et al., 2014) highlighted a potential kindlin-mediated linkage between integrins and the IPP complex. However, a thorough understanding of the kindlin-ILK interaction and its importance in integrin-mediated signaling has been hindered by an inability to identify the binding site for kindlin-2 on ILK and, hence, to generate mutations that selectively impair kindlin binding. In this study, we used a combination of conservation-guided surface mapping of the ILK-pKD, biochemical analysis of the kindlin-2-ILK interaction, and functional studies in knockdown cells to show that (1) kindlin-2 interacts with highly conserved residues on helix-αH in the C-lobe of the ILK-pKD, (2) efficient localization of ILK to focal adhesions requires binding to kindlin-2 and, (3) the ILK–kindlin-2 interaction is important for normal cell spreading.
Focusing on highly conserved hydrophobic patches on the surface of the ILK-pKD, we identified three candidate kindlin-binding sites that we termed surface 1, 2 and 3. Mutagenesis localized the interaction to surface 2 on the C-lobe of the pKD and revealed key residues on helix-αH, i.e. I427 and K423, with the adjacent K426 playing a less important role. We demonstrate that helix-αH mutations that disrupt binding to kindlin-2 did not impair the association of ILK with α-parvin, a requirement for stability (Fukuda et al., 2009), and maintained structural integrity of the ILK-pKD–α-parvin-CH2 complex when assessed by DSF. Furthermore, even somewhat conservative ILK I427A mutations perturbed kindlin-2 binding, indicating that the effect is not simply due to changing the surface charge in this area. Notably, helix-αH has been implicated in protein–protein interactions in other kinase domains. For example, an important component of activation of the epidermal growth factor receptor (EGFR) involves formation of an asymmetric dimer through hydrophobic interactions between the N-lobe of one kinase domain and residues in the C-lobe of its partner kinase domain that includes residues on helix-αH (Zhang et al., 2006). ILK is a catalytically incompetent scaffold (Fukuda et al., 2009); thus, we propose that helix-αH is important not for activation but for binding kindlin-2.
Previous studies have proposed other potential kindlin-2 interaction surfaces on the ILK-pKD. Most recently, a binding interface for the ILK-binding helical fragment in the kindlin-2 F2PH (Fukuda et al., 2014; Huet-Calderwood et al., 2014) was identified on the N-lobe of the ILK-pKD by rigid-body docking (Guan et al., 2018). The reported binding surface is formed by the highly conserved R243 and the less-well conserved R334 (Guan et al., 2018). These residues lie near the surface 1 residues that we found to have a minimal effect on kindlin-2 binding; but we confirmed that the reported GFP-ILK RR/GG double mutant (Guan et al., 2018) impaired kindlin-2 binding in our pulldown assays and ILK targeting to focal adhesions in cells. Unfortunately, we were unable to obtain soluble purified GST-ILK RR/GG in complex with α-parvin-CH2 from E. coli, preventing us from assessing the stability of the mutant ILK-pKD (data not shown). Co-immunoprecipitation assays from transfected mammalian cells did however indicate that the GFP-ILK RR/GG mutation significantly impaired association with FLAG-α-parvin. R243 and R334 are located far from the crystallographically defined parvin-binding site (Fukuda et al., 2009) but we suggest that the double RR/GG mutation alters ILK confirmation or stability indirectly, resulting in impaired parvin and kindlin binding. In support of this idea, we found that a K220M mutation known to indirectly disrupt ILK–α-parvin association (Lange et al., 2009) is also severely impaired in binding to kindlin-2. Thus, although we cannot definitively rule out the possibility that the interface formed by R243 and R334 is involved in the interaction with another part of the kindlin-2 F2PH, we clearly show that mutations on helix–αH selectively impair binding to the conserved helical fragment in the kindlin-2 F2PH, while maintaining association with α-parvin and integrity of the ILK-pKD. Furthermore, spatial separation of the helix-αH residues that we identified as being important for binding kindlin-2, and R243 and R334, make it unlikely that the ILK-binding helical fragment of kindlin-2 could engage both sites simultaneously. We also note that, although not discussed or tested further, in the prior docking study two additional models showed the kindlin-2 helical fragment binding to helix-αH in the ILK-pKD (Guan et al., 2018), further supporting a role for helix-αH in kindlin binding.
Notably, studies with the C. elegans orthologues of kindlin (UNC-112) and ILK (PAT-4) have also investigated the ILK-kindlin interface (Mackinnon et al., 2002; Qadota et al., 2012, 2014). A yeast two-hybrid screen of UNC-112 (kindlin) mutants defective in PAT-4 (ILK) binding identified a D382V mutant in the linker between the F2 and PH domains of UNC-112 (Qadota et al., 2012). This region contains the ILK-binding site in mammalian kindlins (Fukuda et al., 2014; Huet-Calderwood et al., 2014) but D382 corresponds to highly conserved S351 in mammalian kindlins. However, a S351V mutation in kindlin-2 does not inhibit ILK binding (Huet-Calderwood et al., 2014), indicating differences in the mammalian and C. elegans interactions. A yeast two-hybrid suppressor mutation screen for PAT-4 mutants that restore binding to UNC-112 D382V suggested two large potential interaction surfaces for UNC-112 (Qadota et al., 2014). The first maps to a region in the N-lobe of the ILK-pKD overlapping with our surface 1 and includes the C. elegans PAT-4 residues I261 and F262 that correspond to human ILK residues I244 and F245. However, as in our pulldown assays – where I244D or F245D mutants of ILK bound kindlin-2 at near wild-type levels – PAT4 I261N or F262L mutants still bound UNC-112 (Qadota et al., 2014). The second interface maps to the C-lobe of the ILK-pKD and includes two residues on helix-αH: M440 (human residue M425) and A433 (human residue S418). In human ILK, M425 is located on the buried face of αH, and S418 lies on the N-terminal cap of helix-αH oriented away from the key kindlin-binding residues K423 and I427 identified in our study, which are conserved in PAT-4 (R438 and I442 respectively). While the mechanisms by which suppressor mutations facilitate PAT-4 binding to mutant UNC-112 are unclear, their localization supports our conclusion that the ILK helix-αH is important for binding kindlin. The role of residues in surface 1 appears more complex, as no surface 1 mutations that selectively impair kindlin binding have been identified. However, the fact that this region has been identified as a potential kindlin-binding site in independent studies (Guan et al., 2018; Qadota et al., 2014) suggests that residues in surface 1 may be important for overall pseudokinase domain conformation.
Our observation that ILK mutants impaired in binding to kindlin-2 are hindered in their ability to localize to focal adhesions is consistent with a linear model where integrin β subunit cytoplasmic tails bind and recruit kindlin, which in turn recruits ILK and the IPP complex. However, additional data indicate that this model is overly simplistic as our data, and that of others, shows that kindlin and ILK are co-dependent because kindlin mutations that inhibit ILK binding also impair kindlin accumulation at adhesions in mammalian cells and in C. elegans muscle cells in vivo (Fukuda et al., 2014; Huet-Calderwood et al., 2014; Qadota et al., 2012). It has been proposed that ILK binding induces conformational changes in kindlin that expose the integrin-binding site allowing kindlin to recruit ILK to adhesions (Qadota et al., 2012) which may explain the reliance of both kindlin and ILK on binding for recruitment to adhesions, but direct evidence of such a conformational change is currently not available. Understanding kindlin and ILK targeting to adhesions is further complicated by the observation that kindlins (through the F0 and F2-PH domains) and the IPP complex (through the CH2 domain of parvin) both bind paxillin, and that these interactions are also important for their targeting to adhesions (Gao et al., 2017; Stiegler et al., 2012; Theodosiou et al., 2016; Wang et al., 2008). Nonetheless, our observation that mutants in ILK or kindlin-2 that disrupt the ILK-kindlin complex also fail to fully rescue spreading defects in ILK or kindlin-2 knockdown cells further underscores the importance of this complex. Dissection of the specific downstream signaling along the ILK–kindlin-2 axis that promotes normal cell spreading will be the subject of future studies.
In summary, we have mapped the interaction between kindlin-2 and ILK to highly conserved residues on helix-αH of the ILK-pKD, and established that the interaction between kindlin-2 and ILK is important for their correct localization to focal adhesions and for normal cell spreading.
MATERIALS AND METHODS
Antibodies
Primary antibodies against GFP (Rockland, catalog #601-101-215; Limerick, PA) (used at 1:1000), ILK (Cell Signaling, catalog #3862; Danvers, MA) (used at 1:1000), FLAG (Sigma, catalog #F1804; St. Louis, MO) (used at 1:1000), vinculin (Sigma, catalog #V9131) (used at 1:10,000), kindlin-2 (Proteintech, catalog #11453-1-AP; Rosemont, IL) (used at 1:1000), α-parvin (Cell Signaling, catalog #8190) (used at 1:1000), PINCH1 (Proteintech, catalog #55336-1-AP) (used at 1:1000), carbonyl reductase (Santa Cruz Biotechnology, catalog #sc-70212; Dallas, Texas) (used at 1:1000) and β-tubulin (Developmental Studies Hybridoma Bank, catalog #E7; Iowa City, Iowa) (used at 1:1000) as well as IRDye-conjugated secondary antibodies (Li-Cor; Lincoln, NE) (used at 1:10,000) were purchased from commercial sources.
Constructs
Vectors encoding N-terminally GFP-tagged human ILK and kindlin-2, GFP alone, FLAG-tagged human α-parvin, GST-tagged human ILK-pKD, and His-FLAG-tagged human α-parvin-CH2 were as previously described (Huet-Calderwood et al., 2014; Stiegler et al., 2012, 2013). Point mutations were generated by QuikChange site directed mutagenesis following the manufacturer's instructions (Stratagene; La Jolla, CA). A pLENTI CMV Hygro lentiviral paxillin-mCherry expression vector was previously described (Huet-Calderwood et al., 2017). All constructs were verified by DNA sequencing. pLKO expression vectors containing shRNAs (Scramble: catalog no. SHC002; human ILK: TRCN0000199983, target sequence 5′-GCAATGACATTGTCGTGAAGG-3′; human kindlin-2: TRCN0000127493, target sequence 5′-CCAGGGCTTAACCTATATGAA-3′ were purchased from Sigma.
Cell culture and transfection
HeLa cells were cultured in Dulbecco's modified Eagle’s medium (DMEM) with 9% fetal bovine serum (FBS), sodium pyruvate, and non-essential amino acids (NEAA) (Gibco Laboratories; Gaithersburg, MD) at 37°C in 5% CO2, and were obtained from colleagues at Yale University. CHO cells, which have been engineered to stably express integrin αIIbβ3 and described previously (O'Toole et al., 1994), were cultured in DMEM with 9% FBS, sodium pyruvate, and NEAA at 37°C in 5% CO2. CHO cells were transiently transfected with linear polyethylenimine (PEI, MW 25,000) (Polysciences, Inc.; Warrington, PA), and HeLa cells were transiently transfected with Lipofectamine 2000 (Invitrogen; Waltham, MA). Cells were regularly tested for mycoplasma using the MycoAlert mycoplasma detection kit (Lonza; Basel, Switzerland).
Recombinant protein production and purification
GST-ILK-pKD in complex with His-FLAG–α-parvin-CH2 was produced in BL21(De3) E. coli cells (Millipore Sigma) as previously described (Huet-Calderwood et al., 2014). The complex was co-purified by glutathione affinity chromatography using glutathione-sepharose 4b (GE Healthcare; Chicago, IL), and eluted with reduced glutathione. The GST- and His-tags were removed by cleavage with purified tobacco etch virus protease (TEV). Following affinity tag removal, the ILK-pKD–α-parvin-CH2 complex was analyzed and purified by Resource S cation exchange chromatography (GE Healthcare). Fractions containing the complex were pooled, concentrated using Amicon Ultra centrifugal filter units (Sigma), snap frozen in liquid nitrogen, and stored at –80°C for subsequent usage. Recombinant GST–kindlin-2 F2PH and GST–kindlin-2 F2PH 329-368 were purified as previously described (Huet-Calderwood et al., 2014).
GFP nanotrap purification
GFP binding protein, derived from a llama single-chain antibody (Rothbauer et al., 2008), was produced in BL21 RIPL competent E. coli (Millipore Sigma). Cultures (1 l) were induced with 0.1 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG) at 16°C for 20 h. Bacterial pellets were harvested by centrifugation, and re-suspended in lysis buffer consisting of 20 mM HEPES (pH 7.2), 250 mM NaCl, 10% glycerol, cOmplete EDTA-free protease inhibitor (Roche; Indianapolis, IN), and 1 mg ml−1 lysozyme. The lysate was incubated on a rotary mixer for 1 h at 4°C and then sonicated. After clarification by centrifugation, the lysate was loaded onto a pre-equilibrated Ni-NTA column (Millipore Sigma), and bound protein was eluted with 20 mM HEPES (pH 7.2), 250 mM NaCl, 5% glycerol, and 200 mM imidazole. Fractions containing protein were pooled and dialyzed into PBS at 4°C. After dialysis, the material was fractionated using a HiLoad Superdex S200 preparative grade size-exclusion chromatography column (GE Healthcare). Appropriate fractions were pooled, and snap frozen in liquid nitrogen. Covalent coupling was done according to manufacturer instructions. In brief, aliquots were thawed on ice, and 1 M NaHCO3 (pH 8.3) added to a final concentration of 200 mM. After clarification by centrifugation, soluble protein was incubated with NHS-activated FastFlow sepharose (GE Healthcare) for 24 h at 4C. The reaction was quenched with 0.1 M Tris-HCl (pH 8.5) for 4 h at room temperature. Beads were washed with 0.1 M sodium acetate (pH 4.5), 500 mM NaCl and PBS and re-suspended in a 50% slurry in PBS with 0.04% NaN3 and cOmplete EDTA-free protease inhibitor (Roche), and subsequently stored at 4°C.
Lentiviral knockdown and overexpression
Lentiviruses were produced by transfecting HEK293T cells with packaging vectors psPAX2 (viral proteins Gag and Rev under the SV40 promoter; Addgene plasmid #12260, a gift from Didier Trono (École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland) and pMD2.G (viral protein VSV-G expressed under the CMV promoter; Addgene plasmid #12259, a gift from D. Trono) together with the pLKO shRNA or pLenti-Hygro mCherry-paxillin constructs. Viral supernatant was harvested 48 and 72 h after transfection, and filtered with a 0.45-µm low-protein-binding filter. Cell lines were transduced by incubating cells with viral supernatant diluted 1:5 in cell culture media supplemented with 8 µg ml−1polybrene (Sigma) for 16 h. pLKO-infected HeLa cells were selected with 1 µg ml−1 puromycin for ∼72 h, until non-infected control cells were dead. pLenti-Hygro-infected HeLa cells and CHO cells were, respectively, selected with 250 µg ml−1 and 350 µg ml−1 hygromycin B for 5 days, until non-infected control cells were dead.
To calculate the residual protein level in ILK and kindlin-2 knockdown cells, the densitometric signal obtained from immunoblots was normalized to an internal loading control (β-tubulin or vinculin) for each protein; this ratio was then normalized to that of shScramble cells to calculate the residual protein level.
GST-pulldown and GST-binding experiments
For pulldown experiments from cell lysate performed with GST–kindlin-2 F2PH, GST–kindlin-2 329-368, and GST–kindlin-2, GFP–ILK was overexpressed together with FLAG–α-parvin in CHO cells by transient transfection with PEI. Cells were lysed in buffer X (1 mM NaVO4, 50 mM NaF, 40 mM sodium pyrophosphate, 50 mM NaCl, 150 mM sucrose, 10 mM PIPES (pH 6.8) containing 0.5% Triton X-100, 0.2% deoxycholic acid and cOmplete EDTA-free protease inhibitor cocktail (Roche) for 15 min at 4°C. After clarification by centrifugation, lysate was diluted in buffer X-T (buffer X with 0.05% Triton X-100), and incubated with GST–kindlin-2 F2PH, GST–kindlin-2 329-368 or GST–kindlin-2 coupled to Glutathione Sepharose 4b. Lysate was also incubated with GST–kindlin-2 F2PH L/A or GST–kindlin-2 329-368 L/A, or GST only as a negative control. Incubations were carried out for 2 h while rocking at 4°C. Beads were then washed three times with buffer X-T, and bound proteins fractionated by reducing SDS-PAGE and analyzed by immunoblotting. Immunoblots were imaged on an Odyssey Infrared Imaging System (Li-Cor), and analyzed using Image Studio Lite (Li-Cor). Ponceau S stains were imaged using a UVP EpiChemI II Darkroom equipped with a CCD camera (UVP; Upland, CA). For quantification of binding from immunoblots, the fluorescence intensity of the band corresponding to bound material was quantified as fraction of the fluorescence of the 3% input material band for each condition. The binding of the internal positive control (e.g. GFP–ILK binding to GST–kindlin-2 F2PH, GST–kindlin-2 329-368 or GST–kindlin-2) was set to 1, and the binding in all other conditions was expressed relatively within each experiment. Bead loading was verified by staining the nitrocellulose membrane with Ponceau S.
For pulldowns performed with recombinant ILK-pKD–α-parvin-CH2, the complex was diluted in buffer X-T to a final concentration of ∼27 nM. Diluted protein was incubated with Glutathione-bead immobilized GST–kindlin-2 F2PH or GST–kindlin-2 F2PH 329-368 (and relevant negative control) for 2 h rocking at 4°C. Beads were washed three times with buffer X-T, and bound proteins fractionated by reducing SDS-PAGE and analyzed by immunoblotting in the same fashion. Specific binding was calculated from immunoblots by subtracting the fluorescence intensity for binding to the negative control beads from that of the positive control (e.g. ILK-pKD binding to GST–kindlin-F2PH or GST–kindlin-2 329-368). This value was then quantified as fraction of the fluorescence of the 3% input material band for each condition. The binding of the internal positive control (e.g. ILK-pKD binding to GST–kindlin-2 F2PH or GST–kindlin-2 329-368) was set to 1, and the binding in all other conditions was expressed relatively within each experiment. Bead loading was verified by staining the nitrocellulose membrane with Ponceau S.
Differential scanning fluorimetry (DSF)
Recombinant purified ILK-pKD-α-parvin-CH2 complex was diluted in 50 mM Tris, 150 mM NaCl pH 8.0, to a final concentration of 4 µM in a PCR multi-plate (BioRad; Hercules, CA). 5000× SYPRO orange protein gel stain (Invitrogen) was then diluted to a final concentration of 2.5× into the protein-buffer mixture immediately before the assay. The multi-plate was inserted into a BioRad CFX384 Touch Real-time PCR Machine (BioRad), and emission monitored using the FAM filter. Following a short step at 4°C, temperature steps of 1°C/min were performed from 5°C to 95°C. Raw data, as well as the negative first derivatives of fluorescence emission, were exported into Microsoft Excel (Microsoft, Redmond, WA).
Non-linear fitting of DSF melting curves
Raw fluorescence data were exported from the BioRad CFX384 Touch real-time PCR Machine into a Microsoft Excel spreadsheet. The maximum numerical fluorescence for each condition was identified, and the data truncated beginning two points after this maximum point (Huynh and Partch, 2015). The data were exported into GraphPad Prism (GraphPad Software; La Jolla, CA), and data within each condition were normalized to a value between 0–100, where 0 represents the smallest value in the dataset and 100 the largest. These normalized data were then fitted to a Boltzmann sigmoid function by non-linear regression, using a least-squares-based method (ordinary fit) in GraphPad Prism (GraphPad Software).
Total internal reflection fluorescence microscopy
For total internal reflection fluorescence (TIRF) microscopy, 35 mm glass bottom microwell dishes with a 14 mm microwell diameter (MatTek Corporation; Ashland, MA) were coated with 5 µg ml−1 bovine plasma fibronectin (Sigma) for 18 h at 37°C. Cells (CHO cells stably expressing mCherry-paxillin and transiently transfected with GFP–ILK or GFP–ILK mutant and FLAG–α-parvin or shILK HeLa cells stably expressing mCherry-paxillin and transiently transfected with GFP, GFP–ILK or GFP–ILK mutant) were plated on the coated glass bottom dishes in DMEM without glutamine and Phenol Red (Gibco Laboratories) but supplemented with 9% FBS, sodium pyruvate, NEAA, and GlutaMAX supplement (Gibco Laboratories). Cells were live imaged 6 h (CHO) or 18 h (shILK HeLa) later in a temperature- and CO2-controled environment OkoLab (OkoLab; Burlingame, CA) chamber mounted onto a Nikon Ti-2 Eclipse microscope (Nikon; Tokyo, Japan) equipped with a motorized Ti-LA-HTIRF module with LUN4 488 and 561 nm lasers (15 mW), using a CFI Plan Apo Lambda 100× Oil TIRF objective and a Prime95B RoHS cMOS Camera (pixel size=110 nm) (Photometrics; Tuscon, AZ). Images were acquired and processed with the NIS-Elements AR software and ImageJ.
Immunofluorescence and quantification of the cell area
For cell spreading experiments, transiently transfected HeLa cells were plated on glass coverslips that had been coated with 5 µg ml−1 bovine plasma fibronectin (Sigma) for 2 h at 37°C, and allowed to spread for 18 h at 37°C in 5% CO2. Cells were then washed with PBS, fixed with 4% paraformaldehyde in PBS for 5 min, permeabilized with PBS containing 0.1% Triton X-100 for 3 min, quenched and blocked with 50 mM NH4Cl, 0.2% BSA and 0.1% Triton X-100 in PBS (PMZ-T) for 30 min at room temperature. Coverslips were stained with Alexa Fluor 647 phalloidin at a 1:500 dilution in PMZ-T for 30 min (Invitrogen). Coverslips were washed three times with PBS, rinsed in milliQ H2O and mounted in FluorSave Reagent (Millipore Sigma).
To quantify cell areas, multiple fields of view containing GFP-positive cells were imaged on each coverslip across multiple biological replicates using a Nikon Ti inverted microscope equipped with a ×40 immersion oil objective. Cell Profiler version 2.0 (Kamentsky et al., 2011) was used to measure cell areas. Phalloidin staining was used to identify cells and measure cell area, and the daa were pooled across multiple biological replicates.
Statistics
Statistical tests to calculate P values were performed using Prism software. Two-tailed Student's t-test or one-way ANOVA using Tukey's correction for multiple comparisons were performed as indicated in figure legends.
Supplementary Material
Acknowledgements
The authors thank Amy Stiegler for her help with purification of the ILK-pKD–α-parvin-CH2 complexes. We also thank Josie Bircher for her contributions at the beginning of this work, and members of the Calderwood lab for helpful discussions and comments.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Y.A.K. designed and performed experiments, analyzed data, and wrote the paper. C.H.-C. performed experiments and provided expertise. B.S. provided essential reagents and expertise. D.A.C. designed experiments, analyzed data, and wrote the paper.
Author contributions metadata
Conceptualization: Y.A.K., D.A.C.; Investigation: Y.A.K., C.H.-C., B.S.; Writing - original draft: Y.A.K.; Writing - review & editing: Y.A.K., C.H.-C., B.S., D.A.C.; Visualization: Y.A.K.; Supervision: D.A.C.; Project administration: D.A.C.; Funding acquisition: D.A.C.
Funding
This work was supported by the National Institutes of Health grants R01-GM068600 and R01-NS085078 (to D.A.C). Y.A.K is supported by a National Science Foundation Graduate Research Fellowship (DGE1122492). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.221184.supplemental
References
- Anthis N. J. and Campbell I. D. (2011). The tail of integrin activation. Trends Biochem. Sci. 36, 191-198. 10.1016/j.tibs.2010.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher R. T., Veelders M., Rombaut P., Faix J., Theodosiou M., Stradal T. E., Rottner K., Zent R., Herzog F. and Fässler R. (2017). Kindlin-2 recruits paxillin and Arp2/3 to promote membrane protrusions during initial cell spreading. J. Cell Biol. 216, 3785-3798. 10.1083/jcb.201701176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood D. A., Campbell I. D. and Critchley D. R. (2013). Talins and kindlins: partners in integrin-mediated adhesion. Nat. Rev. Mol. Cell Biol. 14, 503-517. 10.1038/nrm3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiswell B. P., Zhang R., Murphy J. W., Boggon T. J. and Calderwood D. A. (2008). The structural basis of integrin-linked kinase-PINCH interactions. Proc. Natl. Acad. Sci. USA 105, 20677-20682. 10.1073/pnas.0811415106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich E. B., Liu E., Sinha S., Cook S., Milstone D. S., MacRae C. A., Mariotti M., Kuhlencordt P. J., Force T., Rosenzweig A. et al. (2004). Integrin-linked kinase regulates endothelial cell survival and vascular development. Mol. Cell. Biol. 24, 8134-8144. 10.1128/MCB.24.18.8134-8144.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T., Chen K., Shi X. and Wu C. (2003). PINCH-1 is an obligate partner of integrin-linked kinase (ILK) functioning in cell shape modulation, motility, and survival. J. Biol. Chem. 278, 51324-51333. 10.1074/jbc.M309122200 [DOI] [PubMed] [Google Scholar]
- Fukuda K., Gupta S., Chen K., Wu C. and Qin J. (2009). The pseudoactive site of ILK is essential for its binding to alpha-Parvin and localization to focal adhesions. Mol. Cell 36, 819-830. 10.1016/j.molcel.2009.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda K., Bledzka K., Yang J., Perera H. D., Plow E. F. and Qin J. (2014). Molecular basis of kindlin-2 binding to integrin-linked kinase pseudokinase for regulating cell adhesion. J. Biol. Chem. 289, 28363-28375. 10.1074/jbc.M114.596692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Huang M., Lai J., Mao K., Sun P., Cao Z., Hu Y., Zhang Y., Schulte M. L., Jin C. et al. (2017). Kindlin supports platelet integrin αIIbβ3 activation by interacting with paxillin. J. Cell Sci. 130, 3764-3775. 10.1242/jcs.205641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan S.-Y., Chng C.-P., Ong L.-T., Tan H.-F., Alex Law S. K. and Tan S.-M. (2018). The binding interface of kindlin-2 and ILK involves Asp344/Asp352/Thr356 in kindlin-2 and Arg243/Arg334 in ILK. FEBS Lett. 592, 112-121. 10.1002/1873-3468.12938 [DOI] [PubMed] [Google Scholar]
- Harburger D. S., Bouaouina M. and Calderwood D. A. (2009). Kindlin-1 and -2 directly bind the C-terminal region of β integrin cytoplasmic tails and exert integrin-specific activation effects. J. Biol. Chem. 284, 11485-11497. 10.1074/jbc.M809233200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet-Calderwood C., Brahme N. N., Kumar N., Stiegler A. L., Raghavan S., Boggon T. J. and Calderwood D. A. (2014). Differences in binding to the ILK complex determines kindlin isoform adhesion localization and integrin activation. J. Cell Sci. 127, 4308-4321. 10.1242/jcs.155879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet-Calderwood C., Rivera-Molina F., Iwamoto D. V., Kromann E. B., Toomre D. and Calderwood D. A. (2017). Novel ecto-tagged integrins reveal their trafficking in live cells. Nat. Commun. 8, 570 10.1038/s41467-017-00646-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries J. D. (2006). Integrin ligands at a glance. J. Cell Sci. 119, 3901-3903. 10.1242/jcs.03098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh K. and Partch C. L. (2015). Analysis of protein stability and ligand interactions by thermal shift assay. Curr. Protoc. Protein Sci. 79, 28.9.1-28.9.14. 10.1002/0471140864.ps2809s79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto D. V. and Calderwood D. A. (2015). Regulation of integrin-mediated adhesions. Curr. Opin. Cell Biol. 36, 41-47. 10.1016/j.ceb.2015.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamentsky L., Jones T. R., Fraser A., Bray M.-A., Logan D. J., Madden K. L., Ljosa V., Rueden C., Eliceiri K. W. and Carpenter A. E. (2011). Improved structure, function and compatibility for cellprofiler: modular high-throughput image analysis software. Bioinformatics 27, 1179-1180. 10.1093/bioinformatics/btr095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau M., Mayrose I., Rosenberg Y., Glaser F., Martz E., Pupko T. and Ben-Tal N. (2005). ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 33, 299-302. 10.1093/nar/gki370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange A., Wickström S. A., Jakobson M., Zent R., Sainio K. and Fässler R. (2009). Integrin-linked kinase is an adaptor with essential functions during mouse development. Nature 461, 1002-1006. 10.1038/nature08468 [DOI] [PubMed] [Google Scholar]
- Li F., Zhang Y. and Wu C. (1999). Integrin-linked kinase is localized to cell-matrix focal adhesions but not cell-cell adhesion sites and the focal adhesion localization of integrin-linked kinase is regulated by the PINCH-binding ANK repeats. J. Cell Sci. 112, 4589-4599. [DOI] [PubMed] [Google Scholar]
- Li H., Deng Y., Sun K., Yang H., Liu J., Wang M., Zhang Z., Lin J., Wu C., Wei Z. et al. (2017). Structural basis of kindlin-mediated integrin recognition and activation. Proc. Natl. Acad. Sci. USA 114, 9349-9354. 10.1073/pnas.1703064114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackinnon A. C., Qadota H., Norman K. R., Moerman D. G. and Williams B. D. (2002). C. elegans PAT-4/ILK functions as an adaptor protein within integrin adhesion complexes. Curr. Biol. 12, 787-797. 10.1016/S0960-9822(02)00810-2 [DOI] [PubMed] [Google Scholar]
- Montanez E., Ussar S., Schifferer M., Bosl M., Zent R., Moser M. and Fassler R. (2008). Kindlin-2 controls bidirectional signaling of integrins. Genes Dev. 22, 1325-1330. 10.1101/gad.469408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse E. M., Brahme N. N. and Calderwood D. A. (2014). Integrin cytoplasmic tail interactions. Biochemistry 53, 810-820. 10.1021/bi401596q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy J. M., Zhang Q., Young S. N., Reese M. L., Bailey F. P., Eyers P. A., Ungureanu D., Hammaren H., Silvennoinen O., Varghese L. N. et al. (2014). A robust methodology to subclassify pseudokinases based on their nucleotide-binding properties. Biochem. J. 457, 323-334. 10.1042/BJ20131174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolopoulos S. N. and Turner C. E. (2000). Actopaxin, a new focal adhesion protein that binds paxillin LD motifs and actin and regulates cell adhesion. J. Cell Biol. 151, 1435-1448. 10.1083/jcb.151.7.1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolopoulos S. N. and Turner C. E. (2002). Molecular dissection of actopaxin-integrin-linked kinase-Paxillin interactions and their role in subcellular localization. J. Biol. Chem. 277, 1568-1575. 10.1074/jbc.M108612200 [DOI] [PubMed] [Google Scholar]
- O'Toole T. E., Katagiri Y., Faull R. J., Peter K., Tamura R., Quaranta V., Loftus J. C., Shattil S. J. and Ginsberg M. H. (1994). Integrin cytoplasmic domains mediate inside-out signal transduction. J. Cell Biol. 124, 1047-1059. 10.1083/jcb.124.6.1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C. and Ferrin T. E. (2004). UCSF Chimera - A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605-1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Qadota H., Moerman D. G. and Benian G. M. (2012). A molecular mechanism for the requirement of PAT-4 (Integrin-linked Kinase (ILK)) for the localization of UNC-112 (kindlin) to integrin adhesion sites. J. Biol. Chem. 287, 28537-28551. 10.1074/jbc.M112.354852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota H., Luo Y., Matsunaga Y., Park A. S., Gernert K. M. and Benian G. M. (2014). Suppressor mutations suggest a surface on PAT-4 (integrin-linked kinase) that interacts with UNC-112 (kindlin). J. Biol. Chem. 289, 14252-14262. 10.1074/jbc.M114.556308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbauer U., Zolghadr K., Muyldermans S., Schepers A., Cardoso M. C. and Leonhardt H. (2008). A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell. Proteomics 7, 282-289. 10.1074/mcp.M700342-MCP200 [DOI] [PubMed] [Google Scholar]
- Sakai T., Li S., Docheva D., Grashoff C., Sakai K., Kostka G., Braun A., Pfeifer A., Yurchenco P. D. and Fässler R. (2003). Integrin-linked kinase (ILK) is required for polarizing the epiblast, cell adhesion, and controlling actin accumulation. Genes Dev. 17, 926-940. 10.1101/gad.255603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanchi F., Bordoy R., Kudlacek O., Braun A., Pfeifer A., Moser M. and Fässler R. (2005). Consequences of loss of PINCH2 expression in mice. J. Cell Sci. 118, 5899-5910. 10.1242/jcs.02686 [DOI] [PubMed] [Google Scholar]
- Stanchi F., Grashoff C., Nguemeni Yonga C. F., Grall D., Fässler R. and Van Obberghen-Schilling E. (2009). Molecular dissection of the ILK-PINCH-parvin triad reveals a fundamental role for the ILK kinase domain in the late stages of focal-adhesion maturation. J. Cell Sci. 122, 1800-1811. 10.1242/jcs.044602 [DOI] [PubMed] [Google Scholar]
- Stiegler A. L., Draheim K. M., Li X., Chayen N. E., Calderwood D. A. and Boggon T. J. (2012). Structural basis for paxillin binding and focal adhesion targeting of β-Parvin. J. Biol. Chem. 287, 32566-32577. 10.1074/jbc.M112.367342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiegler A. L., Grant T. D., Luft J. R., Calderwood D. A., Snell E. H. and Boggon T. J. (2013). Purification and SAXS analysis of the integrin linked kinase, PINCH, parvin (IPP) heterotrimeric complex. PLoS ONE 8, e55591 10.1371/journal.pone.0055591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodosiou M., Widmaier M., Böttcher R. T., Rognoni E., Veelders M., Bharadwaj M., Lambacher A., Austen K., Müller D. J., Zent R. et al. (2016). Kindlin-2 cooperates with talin to activate integrins and induces cell spreading by directly binding paxillin. Elife 5, 1-24. 10.7554/eLife.10130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y., Huang Y., Zhang Y., Hua Y. and Wu C. (2001). A new focal adhesion protein that interacts with integrin-linked kinase and regulates cell adhesion and spreading. J. Cell Biol. 153, 585-598. 10.1083/jcb.153.3.585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velyvis A., Vaynberg J., Yang Y., Vinogradova O., Zhang Y., Wu C. and Qin J. (2003). Structural and functional insights into PINCH LIM4 domain-mediated integrin signaling. Nat. Struct. Biol. 10, 558-564. 10.1038/nsb938 [DOI] [PubMed] [Google Scholar]
- Wang X., Fukuda K., Byeon I.-J., Velyvis A., Wu C., Gronenborn A. and Qin J. (2008). The structure of α-Parvin CH2-paxillin LD1 complex reveals a novel modular recognition for focal adhesion assembly. J. Biol. Chem. 283, 21113-21119. 10.1074/jbc.M801270200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickström S. A., Lange A., Montanez E. and Fässler R. (2010). The ILK/PINCH/parvin complex: the kinase is dead, long live the pseudokinase! EMBO J. 29, 281-291. 10.1038/emboj.2009.376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji S., Suzuki A., Sugiyama Y., Koide Y. I., Yoshida M., Kanamori H., Mohri H., Ohno S. and Ishigatsubo Y. (2001). A novel integrin-linked kinase-binding protein, affixin, is involved in the early stage of cell-substrate interaction. J. Cell Biol. 153, 1251-1264. 10.1083/jcb.153.6.1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen K., Tu Y., Velyvis A., Yang Y., Qin J. and Wu C. (2002). Assembly of the PINCH-ILK-CH-ILKBP complex precedes and is essential for localization of each component to cell-matrix adhesion sites. J. Cell Sci. 115, 4777-4786. 10.1242/jcs.00166 [DOI] [PubMed] [Google Scholar]
- Zhang X., Gureasko J., Shen K., Cole P. A. and Kuriyan J. (2006). An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell 125, 1137-1149. 10.1016/j.cell.2006.05.013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.