Abstract
The mechanisms restricting regeneration and maintaining cell identity following injury are poorly characterized in higher vertebrates. Upon β-cell loss, 1–2% of the glucagon-producing α-cells spontaneously engage in insulin production in mice. Here we explore the mechanisms inhibiting α-cell plasticity. We show that the adaptive α-cell identity changes are constrained by intra-islet Insulin- and Smoothened-mediated signaling, among others. The combination of β-cell loss, or insulin signaling inhibition, with Smoothened inactivation in α- or δ-cells, stimulates insulin production in more α-cells. These findings suggest that removing constitutive “brake signals” is crucial for neutralizing the refractoriness to adaptive cell-fate changes. It appears that cell identity maintenance is an active process mediated by repressive signals, released by neighbor cells, curbing an intrinsic trend of differentiated cells to change.
Half-century of research into cell identity determination and maintenance revealed that adult cells are not terminally differentiated but maintain some plasticity potential even in higher organisms1–3. Spontaneous adult cell type interconversion is considered a rare event, highly regulated, often activated exclusively after injury, and whose efficiency correlates with mechanisms preserving a specific cell identity2,4. Conversely, our knowledge of the intricate mechanisms maintaining adult cell identity is still limited.
Cell-fate allocation and maintenance1,5,6 result from the activity of transcriptional regulators and epigenetic modifiers controlling the constitutive expression of identity genes that become ‘locked’ due to autoregulatory feed-back loops, stable chromatin modifications7–9 or through the action of regulatory signals from the microenvironment in which cells reside. The plasticity potential of a given cell depends on the level of redundancy in which these complex mechanisms operate and on the physiological needs of the corresponding tissue.
Changes in adult cell identity, especially if triggered by stress responses, are a basis for in situ regenerative medicine10,11. In the pancreas of adult mice, following near total β-cell ablation, 1–2% of glucagon-expressing α-cells and somatostatin-expressing δ-cells spontaneously express insulin, leading to significant β-cell mass regeneration and normoglycemia12,13. The mechanisms controlling this insulin expression are unknown. We have previously shown that in α-cells inhibition of the transcription factor Arx and the dimethyltransferase Dnmt1 causes transdifferentiation into insulin+ cells irrespective of β-cell loss14. However, nothing is known about the control of α-cell identity by extrinsic signals. Here we define the cellular mechanisms that regulate the expression of insulin in glucagon+ α-cells after near-total β-cell ablation or insulin action inhibition. We focus on local signals that act as constitutive brakes limiting cell reprogramming. We identified Smoothened and Insulin signaling pathways in α-cells, and surprisingly also in δ-cells, as regulators of α-cell identity and conversion into insulin-producing cells.
RESULTS
α-cell conversion is driven by local signals
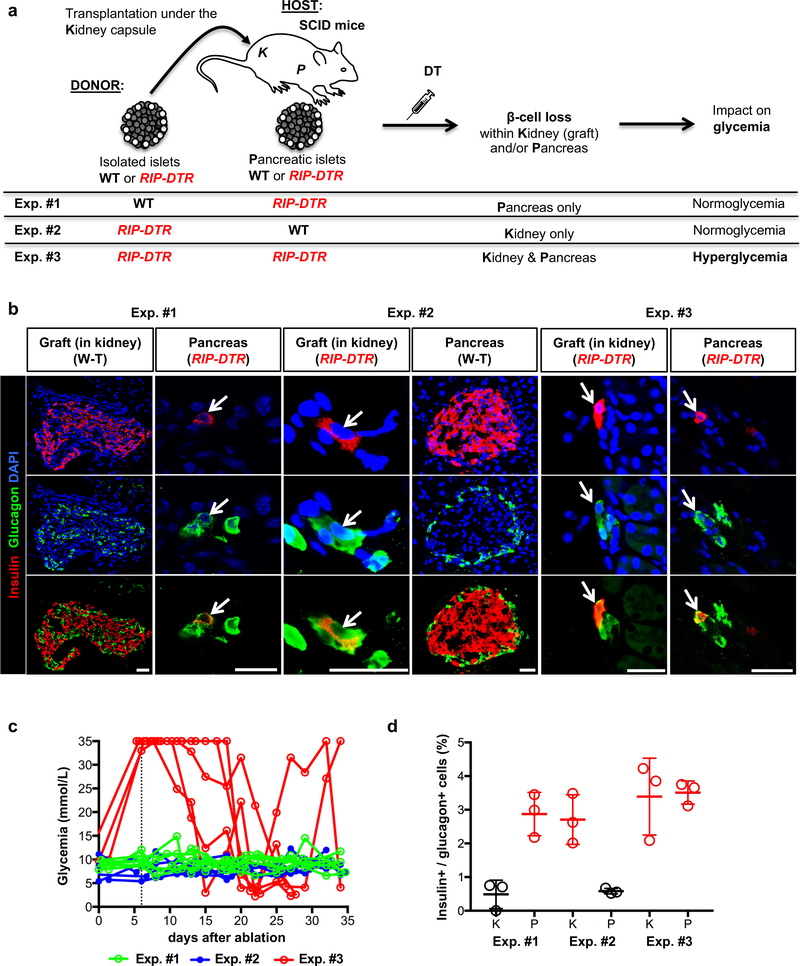
To elucidate the signals leading to insulin production in α-cells upon β-cell loss in mice, we set up a series of islet transplantation experiments (Fig. 1a). To prevent allograft rejection, we used immunodeficient SCID mice15 as hosts receiving islets isolated from immunocompetent donor mice. In different experimental conditions, islet donor and/or recipient mice also bore the RIP-DTR transgene, allowing for β-cell ablation upon diphtheria toxin (DT) administration, in either engrafted or pancreatic islets, or both13. As a readout for α-cell conversion toward insulin production, we assessed the fraction of α-cells coexpressing glucagon and insulin in the engrafted and/or host’s pancreatic islets. Indeed, cell lineage tracing experiments have shown that these bihormonal cells appearing after β-cell loss are reprogrammed α-cells13.
Figure 1. α-cells engage insulin production after β-cell ablation in islets transplanted under the kidney capsule.
(a) Experimental design of islet transplantation underneath the kidney capsule of immunocompromised host mice (SCID). Exp. #1. WT islets are transplanted into RIP-DTR hosts; upon DT administration, β-cell ablation occurs in pancreatic islets, while transplanted islets remain unaffected and maintain normoglycemia. Exps. #2 & #3. Islets isolated from RIP-DTR donors are transplanted into either WT or RIP-DTR hosts; DT only ablates β-cells in transplanted islets (in WT hosts) or in both transplanted and endogenous pancreatic islets (in RIP-DTR hosts). (b) Immunofluorescence staining of insulin (red) and glucagon (green) in Exps. #1 to #3. Arrows indicate bihormonal glucagon+ insulin+ cells. Scale bars: 20 μm. Experiment repeated independently 3 times. (c) Random-fed blood glycemia after DT-treatment in 3 experimental conditions tested. The vertical dash line indicates the administration of one insulin pellet to the hyperglycemic mice. Experiment performed once. (d) Proportion of glucagon+ cells coexpressing insulin. Red: RIP-DTR islets; black: WT islets. n=3 biologically independent animals per condition. Data shown as mean ± s.d.; see Supplementary Table 1 for source data.
We first transferred wild type (WT) islets (~700) under the kidney capsule of RIP-DTR SCID hosts to ensure normoglycemia after DT-induced pancreatic β-cell loss (Fig. 1a, Exp. #1). The reverse experiment, transferring RIP-DTR islets into WT normoglycemic SCID host mice, was also performed (Fig. 1a, Exp. #2). β-cells were efficiently ablated in engrafted RIP-DTR islets 10 days post-DT (Supp. Fig. 1a). We also transplanted islets from RIP-DTR mice into RIP-DTR SCID hosts (Fig. 1a, Exp. #3). In this case, DT treatment led to hyperglycemia, because of β-cell loss in both host and grafted islets (Fig. 1c, red lines). One month after β-cell ablation, insulin production in α-cells was observed in all 3 experiments, but only in β-cell-ablated islets (Fig. 1b,d), irrespective of their location (pancreatic or extrapancreatic; Supp. Table 1a) and glycemia. α-cell conversion was also observed after β-cell ablation in RIP-DTR islets engrafted on the iris (anterior chamber of the eye) of normoglycemic WT mice (not shown).
Noticeably, the fraction of α-cells producing insulin in β-cell-ablated islets (regardless their location) was 2-fold higher in RIP-DTR SCID mice (1–2% vs. 3–4% in SCIDs, and ref.12,13). This suggests that the genetic background (SCID) somewhat influences α-cell plasticity.
To explore the effect of global 50% β-cell mass loss, we cotransplanted WT and RIP-DTR islets in a 1:1 ratio into WT SCID hosts, either at two separate places or mixed together at one single location (Supp. Fig. 1b, Exp. #4). To distinguish the two types of engrafted islets, we labeled: i) α-cells with YFP in RIP-DTR islets (using Glucagon-rtTA; TetO-Cre; R26-YFP; RIP-DTR mice as donors, in which doxycycline (DOX) triggers the irreversible expression of YFP in α-cells13), and ii) β-cells with mCherry in WT islets (using islets from Insulin-mCherry mice, whose insulin-producing cells constitutively express mCherry). In the two-spot transplantation experiment, after DT treatment, no glucagon+ cell was insulin+ in unablated WT mCherry+ islets, while 4% of YFP+ α-cells produced insulin in β-cell-ablated RIP-DTR (Supp. Fig. 1c; Supp. Table 1a, Exp. #4). This further confirms that α-cells start insulin expression only in massively ablated islets. Similarly, no evidence of α-cell conversion after DT-mediated β-cell ablation was found in WT islets when mixed with RIP-DTR islets, while the proportion of YFP-labeled α-cells producing insulin in RIP-DTR islets was like in the “two-spot” experiment (Supp. Fig. 1c-d; Supp. Table 1a, Exp. #4). This suggests that no signals act locally between adjacent β-cell-ablated (RIP-DTR) and unablated (WT) islets: neither from β-cell-ablated RIP-DTR islets that would promote conversion in WT islets with an intact β-cell mass, nor from WT islets that would restrict α-cell conversion in nearby RIP-DTR islets.
We next explored the effect of 50% β-cell ablation taking place in every single individual islet, either in pancreas or after transplantation. We used hemizygous RIP-DTR females, in which about 50% of the β-cells express DTR13. In these settings (Exp. #5, Supp. Table 1a), mice remained normoglycemic after DT, and no evidence of α-cell conversion was observed (Supp. Table 1a). Partial β-cell loss is thus insufficient to trigger α-cell conversion, even in the more permissive genetic backgrounds (i.e. SCID mice).
In summary, these experiments reveal that α-cell conversion occurs only in islets undergoing massive β-cell loss, in an autonomous manner, irrespective of their location or the glycemia. The existence of signals from a putative systemic “β-cell mass sensor” (Exps. #1, #2 and #3) or acting at short distance (Exp. #4) is not supported, since bihormonal cells were detected in massively β-cell-ablated islets only, in hyper- and normo-glycemic mice, but not in islets retaining ~50% of their β-cell mass (Exp. #5). Instead, our findings suggest that massive β-cell injury may lead to the release of local signals acting as modulators of α-cell change or maintenance.
β-cell loss facilitates insulin expression in α-cells
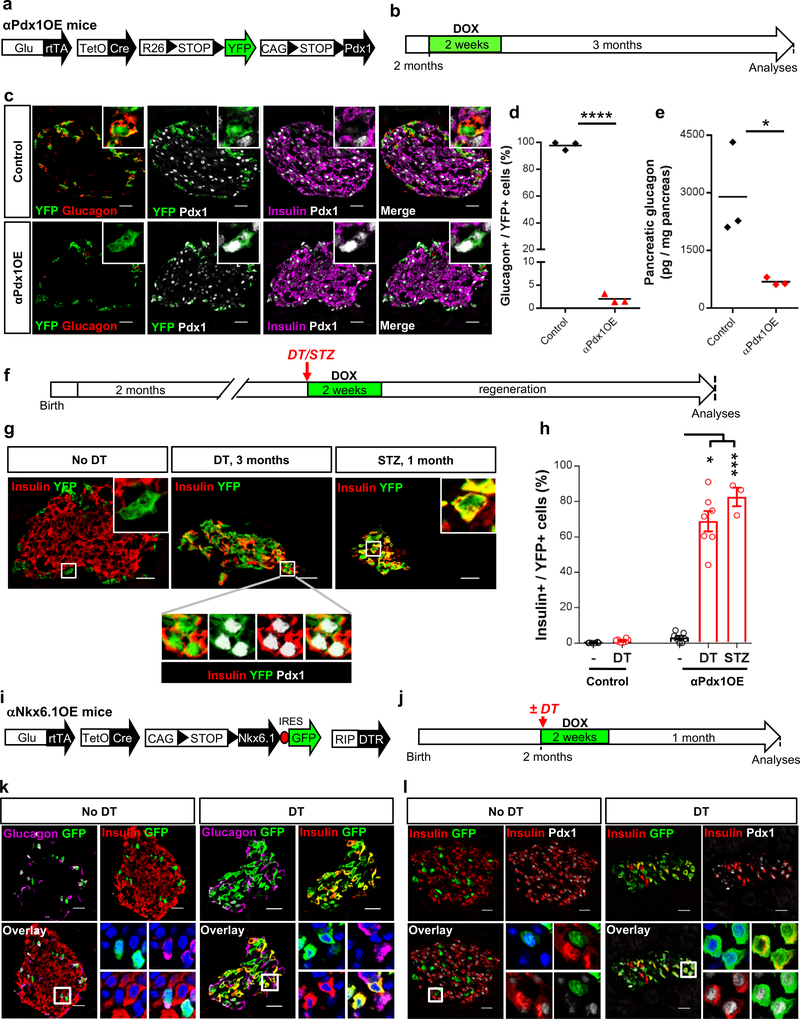
Why most α-cells do not express insulin after massive β-cell loss? To explore whether all α-cells can produce insulin, we induced Pdx1 (a β-cell-specific transcription factor) in “αPdx1OE” mice. In this quintuple transgenic line (Fig. 2a) DOX administration causes the irreversible expression of Pdx1 and YFP in α-cells (Fig. 2b). Pdx1 activity in α-cells in vivo leads to suppression of glucagon16 (Fig. 2c-e and Suppl. Table 1b,c) and to insulin expression in a small subset (3%) (Supp. Table 1d). Yet most Pdx1-expressing α-cells produce insulin after β-cell loss induced by treatment with DT or streptozotocin (STZ, a glucose analogue toxic for β-cells) (Fig. 2f-h, Supp. Table 1d), suggesting that all α-cells can produce insulin. αPdx1OE mice remained hyperglycemic after β-cell loss, implying that the insulin-producing α-cells are likely not fully functional (not shown).
Figure 2. Pdx1 expression inhibits glucagon production in adult α-cells.
(a) Transgenes required for α-cell tracing and ectopic Pdx1 expression. (b) Experimental design. (c) α-cells are specifically and efficiently YFP-labeled upon DOX administration in controls (upper panel). Pdx1-expressing YFP+ α-cells cease glucagon expression (bottom panel). The experiment was performed once. (d) Percentage of YFP-traced α-cells expressing glucagon after Pdx1 expression. Two-tailed unpaired t-test P<0.0001. n=3 control mice and 3 α-PdxOE mice. (e) Pancreatic glucagon content is decreased in αPdx1OE mice. Two-tailed unpaired t-test P=0.0362. n=3 control mice and 3 α-PdxOE mice. (f) Experimental design for Pdx1 induction in α-cells, and DT- or STZ-triggered β-cell loss. (g) A vast fraction of YFP+ α-cells (i.e. expressing Pdx1) starts insulin production upon DT- or STZ-mediated β-cell loss. The experiment was performed once with mice treated asynchronously according to their availability. (h) Fraction of YFP+ α-cells expressing insulin in αPdx1OE mice upon β-cell loss. Two-tailed Mann Whitney test, P=0.0167 for STZ+Pdx1 vs Pdx1 and P=0.0006 for DT+Pdx1 vs Pdx. n=6 and 8 untreated and DT-treated control mice, respectively and n=7, 7, 3 untreated, DT-treated and STZ treated α-PdxOE mice. (i) Transgenes required for α-cell tracing, DT-mediated β-cell ablation and ectopic Nkx6.1 expression. (j) Experimental design. (k) Nkx6.1 expression (GFP+ cells) does not induce insulin production in α-cells in presence of a normal β-cell mass. Glucagon expression persists in Nkx6.1OE α-cells (upper lane). After DT-mediated β-cell loss, most Nkx6.1-expressing α-cells start insulin expression and stop glucagon production (bottom lane). The experiment was performed once with all animals treated asynchronously according to their availability. (l) Pdx1 is not expressed in Nkx6.1OE α-cells when the β-cell mass is normal (upper lane) but is induced after DT (bottom lane). All centers indicate the mean. Scale bars: 20 μm. See Supplementary Tables 1b,c,d as source data.
We also used DOX to induce another β-cell-specific transcription factor, Nkx6.1, in α-cells (Fig. 2i-l): this led to Pdx1 induction, glucagon inhibition and insulin expression in most α-cells, yet only after β-cell loss (Fig. 2k,l).
In summary, in intact islets, β-cell transcription factor activity in α-cells is insufficient to trigger insulin protein production. However, it reveals an intrinsic ability of α-cells to produce insulin when combined with β-cell injury. Thus, β-cell loss furthers the capacity of α-cells to produce insulin.
Antagonistic response of α-cells to β-cell loss
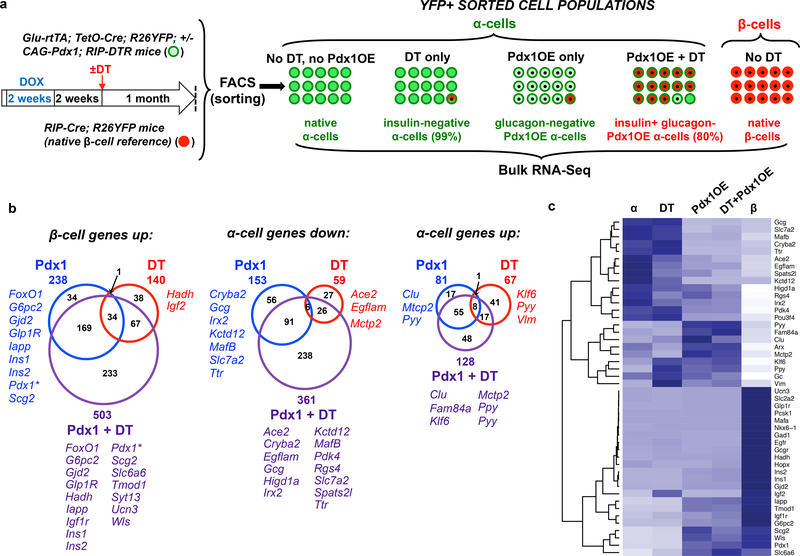
To understand how massive β-cell loss influences the α-cell population, we performed RNA-Seq on: i) native α-cells, ii) α-cells 30 days after DT-induced β-cell ablation (“αDT”), and iii) native β-cells. We also profiled α-cells expressing Pdx1: iv) α-cells from Pdx1OE mice (“αPdx1”), that lack glucagon expression, and v) α-cells from Pdx1OE mice 30 days after DT-induced β-cell loss (“αPdx1+DT”), coexpressing glucagon and insulin (Fig. 3a and Supp. Fig. 2). Principal component analyses revealed that αDT, αPdx1, and αPdx1+DT cells retain a strong α-like gene signature, indicating that they are significantly different from native β-cells (Supp. Fig. 3a). We filtered all differentially expressed genes between native α- and β-cells (α: 1682 genes and β: 1258 genes, FDR<0.01, FC>2; Supp. Table 2a), assessed how they were impacted in α-cells in all conditions (αDT, αPdx1 and αPdx1+DT, Supp. Table 2) and categorized them according to their modulation (up- or downregulated compared to native α-cells; Fig. 3b,c; Supp. Fig. 3b,c). 59 α-cell genes were downregulated and 140 β-cell genes were upregulated in the α-cells not expressing insulin after DT (Fig. 3b; Supp. Fig. 3c), indicating that upon β-cell loss α-cells acquire some β-cell identity traits. Concomitantly, 67 α-cell-enriched genes were further upregulated, reinforcing the α-cell signature. This dual response was also observed in α-cells expressing Pdx1 in islets with intact β-cell mass: 238 β-cell genes were induced (Ins1, Ins2, Gjd2…), 153 α-cell genes were downregulated (Gcg, MafB, …) and 81 α-cell genes were upregulated. Some of the induced β-cell genes (Ins1 and G6pc2) are direct Pdx1 targets. 35 β-cell-enriched genes were induced by DT or Pdx1 (Fig. 3b).
Figure 3. DT-mediated β-cell loss facilitates β-cell gene expression and elicits dual responses in α-cells.
(a) Experimental design for RNA-Seq. Transgenic mice allowing α- and β-cell lineage tracing were used to sort by FACS the following cell populations: i) native control α-cells (“native α”, n=6 biologically independent samples), ii) α-cells 1 month after DT-induced β-cell ablation (“DT”, n=3 biologically independent samples), iii) α-cells overexpressing Pdx1 (“Pdx1OE”, n=3 biologically independent samples), iv) α-cells overexpressing Pdx1 combined with β-cell ablation (“Pdx1OE+DT”, n=5 biologically independent samples). Native β-cells were also collected (“native β”, n=5 biologically independent samples) and analyzed to identify differentially-expressed genes (DEGs) between native α- and β-cells as reference gene sets (α-/β-cell genes). Note that insulin protein was detected in only 1 % of α-cells in the DT group, and 2% in Pdx1OE group (immunofluorescence). Experiment (RNA-Seq) performed once. (b) Venn diagrams showing the regulation of α-/β-cells enriched genes upon Pdx1OE, DT and Pdx1OE+DT. DEGs were calculated for each condition (DT, Pdx1OE, Pdx1OE+DT) compared to native α-cells and intersected with α-/β-cell enriched genes to identify α-/β-cell signature changes. DEGs in left and middle panels are upregulated β-cell genes and downregulated α-cell genes, respectively, which could be considered as an increased β-cell signature in α-cells. Conversely, all genes in the right panel are up-regulated α-cell genes, which could represent a resistance to reprogramming. For every category, only representative genes with FDR < 0.05 are shown. The entire gene lists are reported in Table S2. (c) Heatmap showing scaled expression (blue, high; white, low) of representative α-/β-cell genes in Fig. 3b and Fig. S3c. Gene clustering shown by dendrogram indicates separated gene clusters with different modulation patterns in each condition analyzed, as seen in Fig. 3b and Fig. S4c. See Suppl. Table 2 for source data.
Although not expressed at levels comparable to native β-cells, 40% of the β-cell-enriched genes (503 out of 1258) were upregulated in α-cells expressing Pdx1 when β-cells were ablated (Fig. 3b, “αPdx1+DT”). Interestingly, Pdx1 was expressed in these α-cells at levels similar to those observed in β-cells (Fig. 3c). Out of these 503 upregulated β-cell genes, 233 (46%) were not significantly induced by DT or Pdx1 alone (Fig. 3d). Similarly, 238 (66%) of the 361 downregulated α-cell genes in “αPdx1+DT” cells, were not significantly modulated by Pdx1 activity alone or after β-cell loss (Fig. 3b-c; Supp. Fig. 3c).
These results indicate that β-cell loss facilitates the expression of β-cell-enriched genes in α-cells. This correlates with the strong induction of insulin in most αPdx1 cells after β-cell ablation (Supp. Fig. 3). Massive β-cell ablation decreases the α-cell expression of some α-cell-enriched genes; this is accentuated when combined to Pdx1 induction. Also, other α-cell-enriched genes were further upregulated, indicating that these conditions alter α-cell identity in antagonistic ways.
Insulin deprivation leads to α-cell identity changes
Our results suggest that massive β-cell loss is a requisite for spontaneous α-cell conversion. We therefore hypothesized that active insulin signaling in α-cells may prevent their commitment to produce insulin. We assessed whether insulin signaling is compromised in α-cells after β-cell depletion on sorted Venus+ α-cells from Glucagon-Venus; RIP-DTR mice, 5 days after β-cell ablation induction (Supp. Fig. 3d,e). Massive β-cell death triggered a rapid downregulation of insulin signaling genes in α-cells (Supp. Fig. 3e). Interestingly, insulin-like growth factor 1 (Igf1), an activator of the insulin-signaling pathway, was upregulated in β-cell-depleted islets (Supp. Fig. 3f), likely as a compensatory attempt at maintaining insulin/IGF1 signaling in islets. This confirms that insulin signaling is active in α-cells under physiological conditions and is blunted after β-cell loss. Along this line, gene set enrichment analyses from RNA-Seq indicate that insulin receptor signaling pathways (PKC activity and PI3K binding) are modulated in α-cells upon β-cell ablation (Supp. Fig. 3g).
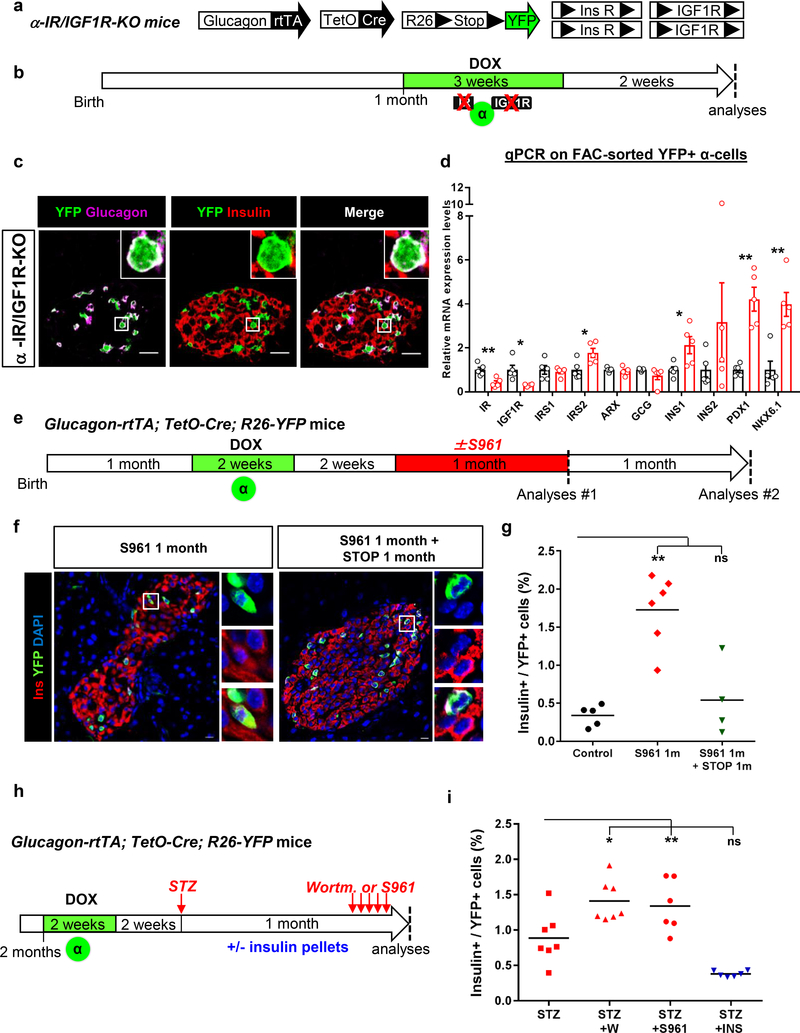
To investigate whether local insulin deprivation alters α-cell identity, we impaired insulin/IGF1 receptor signaling genetically or pharmacologically in healthy mice. Through DOX administration to α-IR/IGF1R-KO mice, insulin and IGF1 receptors (IR and IGF1R) were downregulated and YFP was activated in α-cells (Fig. 4a,b). This led to Ins1, Nkx6.1 and Pdx1 transcript upregulation in α-cells, although insulin was not detected at the protein level (Fig. 4c,d). Therefore, the downregulation of insulin/IGF1 signaling specifically in α-cells initiates insulin gene transcription. We speculate that a more efficient inactivation of IR/IGF1R genes in α-cells, and maybe in all non-β islet cell-types, would lead to stronger upregulation of β-cell genes. Also, the observed upregulation of IRS2 likely compensates in part the effects of IR/IGF1R downregulation.
Figure 4. Decreased insulin signaling predisposes α-cells to insulin production in islets with an intact β-cell mass.
(a) Transgenes for α-cell tracing and insulin (IR) and IGF-1 receptor (IGF1R) downregulation in adult α-cells. (b) Experimental design. (c) Immunofluorescence on islets and (d) RT-qPCR on purified α-cells from αIR/IGF1R KO mice. Impaired insulin/IGF1 signaling does not lead to insulin protein production but induces insulin, Pdx1 and Nkx6.1 gene expression. Data shown as mean ± s.e.m.; n=5 independent biological samples (i.e. one mouse or pool of mice/sample). Experiment performed once. Two-tailed Mann Whitney test (P=0.0079, IR; P=0.0159, IGF1R; P=0.0317, IRS2; P=0.0317 INS1; P=0.0079 PDX1; P=0.0079 NKX6.1) (e) Experimental design for α-cell tracing and insulin signaling blockade with S961 in mice with intact β-cell mass. Islets were analyzed either immediately after stopping S961 (Analyses #1) or 1 month later (Analyses #2). (f) Immunofluorescence of islets of S961 treated mice. YFP-traced α-cells expressing insulin are present only in islets of mice analyzed during S961 treatment (Analyses #1). IF were performed at on 3–5 consecutive sections per animal with similar results. The experiment was performed once, with mice treated asynchronously according to their availability. (g) % of YFP-traced α-cells expressing insulin after ± S961 treatment (analyses #1 and #2). Center indicates the mean. n=5, 6 and 4 animals for control (no treatment), S961 1month and S961 1 month + STOP 1 month, respectively. Two-tailed Mann Whitney test, P=0.0025. (h) Experimental design for α-cell tracing and STZ-induced β-cell ablation followed either by blockade of residual insulin signaling with Wortmannin or S961, or insulin signaling enhancement through insulin administration (subcutaneous pellets). (i) Fraction of converted α-cells after β-cell loss and inhibition or enhancement of residual insulin signaling. n=7 mice for STZ and STZ+WORT groups and n=6 mice for STZ+S961 and STZ+INS groups. Center indicates the mean. Two-tailed Mann Whitney test, P=0.0070, STZ+WORT; P=0.0350, STZ+S961; P=0.0047, STZ+INS. Scale bars: 10 μm. See Supplementary Table 1e for source data.
In parallel, we transiently blocked insulin action using the IR antagonist S961 (Novo Nordisk)17–19, which induces hyperglycemia and insulin resistance. Healthy Glucagon-rtTA; TetO-Cre; R26-YFP mice were treated with DOX to tag α-cells, and then with S961 (Fig. 4e). Mice were either analyzed during S961 treatment (analyses #1) or after stopping it (analyses #2). Under the inhibition of insulin action 13,20, ~2% of the YFP-traced α-cells expressed insulin (Fig. 4f,g, analyses #1; Supp. Table 1e), Pdx1 and Nkx6.1 (Supp. Fig. 4a-c). Insulin (Ins1 & Ins2) transcripts were upregulated in α-cells sorted from S961-treated Glucagon-Venus mice (Supp. Fig. 4d,e). In contrast, when islets were analyzed 1 month after stopping S961 treatment (Fig. 4e, analyses #2; Supp. Table 1e), insulin protein became undetectable in α-cells, suggesting that its production had ceased (Fig. 4f,g).
We next combined STZ-induced partial β-cell loss with the pharmacological inhibition of insulin20,21. One month after STZ, Glucagon-rtTA; TetO-Cre; R26-YFP mice received either Wortmannin (a PI3K inhibitor) or S961, to inhibit the residual insulin signaling (Fig. 4h). The proportion of α-cells producing insulin increased upon drug treatment compared to mice treated with STZ only (Fig. 4i; Supp. Table 1e). Conversely, α-cell reprogramming was abrogated when STZ was followed by insulin therapy (Fig. 4i; Supp. Table 1e), confirming that insulin negatively modulates α-cell plasticity.
When combined with Pdx1 overexpression in αPdx1OE mice, the number of insulin-producing α-cells were greatly increased in all conditions described above (Supp. Fig. 4f-k; Supp. Table 1d). Importantly, the number of insulin-negative YFP+ α-cells increased again after stopping S961 (Supp. Fig. 4j), indicating that α-cells are maintained after discontinuing insulin production.
These observations suggest that β-cell death per se is not required for insulin gene expression in α-cells. Insulin signaling deprivation promotes insulin production in α-cells in a reversible manner. Therefore, in homeostatic conditions, islet insulin signaling helps maintaining α-cell identity.
Constitutive Smo-mediated signaling restricts α-cell plasticity
Since the vast majority of α-cells is apparently unaffected after β-cells loss, we hypothesized that signaling pathways less perturbed after β-cell injury could convey molecular cues restricting α-cell plasticity, even when insulin signaling is compromised.
Two observations suggest that Smoothened (Smo)-mediated Hedgehog (Hh) signaling (SmoHh) could act as an α-cell identity keeper: (i) SmoHh components are expressed in α-, β-, and δ-cells, and active signaling was reported in intact islets (Supp. Fig. 5a-c and ref.22,23); (ii) SmoHh is linked to cell differentiation and maintenance in many organs, including pancreas23–26, where it regulates Pdx1 and Ins expression27,28.
To test whether SmoHh controls α-cell plasticity, we generated Glucagon-rtTA; TetO-Cre; R26-YFP; Smoothenedfl/fl; RIP-DTR mice (“α-Smo-KO”) (Fig. 5a). One-month-old α-Smo-KO mice were DOX-treated to downregulate Smo in α-cells and tag them with YFP (Supp. Fig. 5e,f). α-cell-Smo downregulation triggered the downregulation of Gcg and Arx, 2 α-cell specific markers (Supp. Fig. 5g), suggesting that active SmoHh in α-cells maintains α-cell identity.
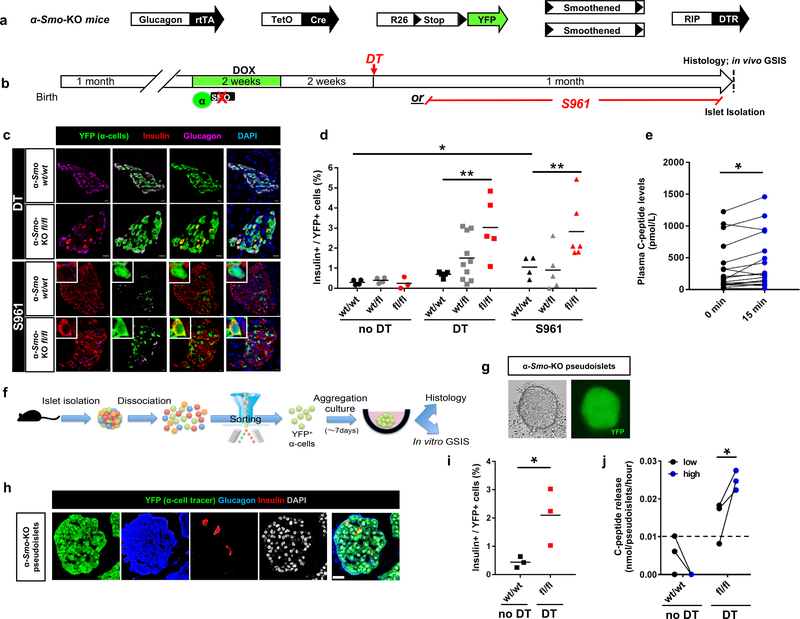
Figure 5. Smoothened (Smo) inactivation in α-cells facilitates their engagement into insulin production.
(a) Transgenes required for simultaneous α-cell lineage tracing, Smo co-receptor downregulation and DT-induced β-cell ablation. (b) Experimental design. (c) Smo inactivation in α-cells leads to insulin production when combined with β-cell loss (upper panels) or insulin receptor antagonism (bottom panels). IF were performed at on 3–5 consecutive sections per animal with similar results. The experiment was performed once, with mice treated asynchronously according to their availability. (d) Fraction of YFP+ cells producing insulin upon inactivation of Smo in α-cells combined with DT or S961. n=4,4,3 mice for Smowt/wt, Smofl/fl and Smofl/fl noDT, respectively; n=4,10,5 mice for Smowt/wt, Smofl/fl and Smofl/fl DT, respectively; n=4,5,6 mice for Smowt/wt, Smofl/fl and Smofl/fl S961, respectively. Center indicates the mean. Two-tailed Mann Whitney test, P=0.0079 Smofl/fl DT vs Smowt/wt DT, P=0.0095 Smofl/fl DT vs Smowt/wt S961 and P=0.0286 Smowt/wt vs Smowt/wt S961. Scale bars: 10 μm. (e) In vivo glucose challenge in α-Smo-KO mice. n=18 mice, P=0.012, Wilcoxon test, two-tailed. (f) Pipeline for α-cell sorting, in vitro pseudoislet reconstruction and functional tests. (g) Live imaging of 7-day-cultured pseudoislet reconstituted using α-cells from α-Smo-KO mice. Representative images from 3 independent experiments. (h) α-Smo-KO pseudoislet at day 7 of aggregation culture (immunofluorescence). Representative images from 3 independent experiments. Scale bar: 25 μm. (i) Percentage of YFP+ cells producing insulin in pseudoislets from control α-Smo-WT animals (no β-cell ablation) or α-Smo-KO mice after cell ablation (DT). n = 3 independent cohorts from 18 α-Smo-WT mice, and 3 independent cohorts from 18 α-Smo-KO mice, P=0.049, unpaired t-test, two-tailed. Center indicates the mean. (j) Glucose-stimulated C-peptide secretion. α-Smo-KO cells secrete C-peptide in response to glucose in vitro, while α-Smo-WT (no DT control) cells have no measurable secretion. n = 3 independent cohorts obtained from 18 α-Smo-Wt mice, n=3 independent cohorts obtained from 18 α-Smo-KO mice, P=0.048, paired t test, one-tailed. The dash line indicates the detection threshold documented by manufacturer (0.0127 nmole/pseudoislet/hour). All values are means ± s.e.m. See Supplementary Tables 1f-l for source data.
α-cell-Smo downregulation did not trigger insulin production (Supp. Fig. 5h). Conversely, Smo inactivation along with β-cells loss or S961 (Fig. 5b), increased the fraction of insulin-expressing α-cells (Fig. 5c,d; Supp. Table 1f). Hence, downregulation of SmoHh facilitates α-cell reprogramming when combined with insulin signaling inhibition.
We next examined whether insulin-producing α-cells secrete insulin in response to glucose. As these cells are rare, we took advantage of their increased number in the islets of DT-treated α-Smo-KO mice. C-peptide was released in a glucose-responsive manner in the blood of α-Smo-KO mice, 1 month after β-cell ablation (Fig. 5e; Supp. Table 1g). To determine the precise contribution of converted α-cells and of escaping β-cells (< 0.5%), we assessed insulin secretion specifically from α-cells in vitro. We isolated islets from α-Smo-KO fl/fl and α-Smo-KOwt/wt mice after β-cell loss (Supp. Table 1h), sorted YFP+ α-cells and reaggregated them to reconstitute highly purified “monotypic pseudoislets” (Fig. 5f,g). We confirmed by immunofluorescence the increased insulin production in these α-cells (Fig. 5h,i; Supp. Table 1i), and performed glucose-stimulated insulin secretion tests in vitro. α-Smo-KO pseudoislets secreted C-peptide in a glucose-dependent manner (Fig. 5j; Supp. Table 1l). These results suggest that insulin-producing α-cells are functional and naturally secrete insulin in response to glucose.
δ-cells restrict α-cell plasticity
A fraction of δ-cells produce insulin upon β-cell loss12. As we observed that SmoHh-mediated signaling is active in δ-cells (Supp. Fig. 5a-c), we investigated its putative role in blocking δ-cell plasticity by downregulating it in Somatostatin-rtTA; TetO-Cre; R26-YFP; Smoothenedfl/fl; RIP-DTR mice (“δ-Smo-KO”) (Fig. 6a; Supp. Fig. 5i,j).
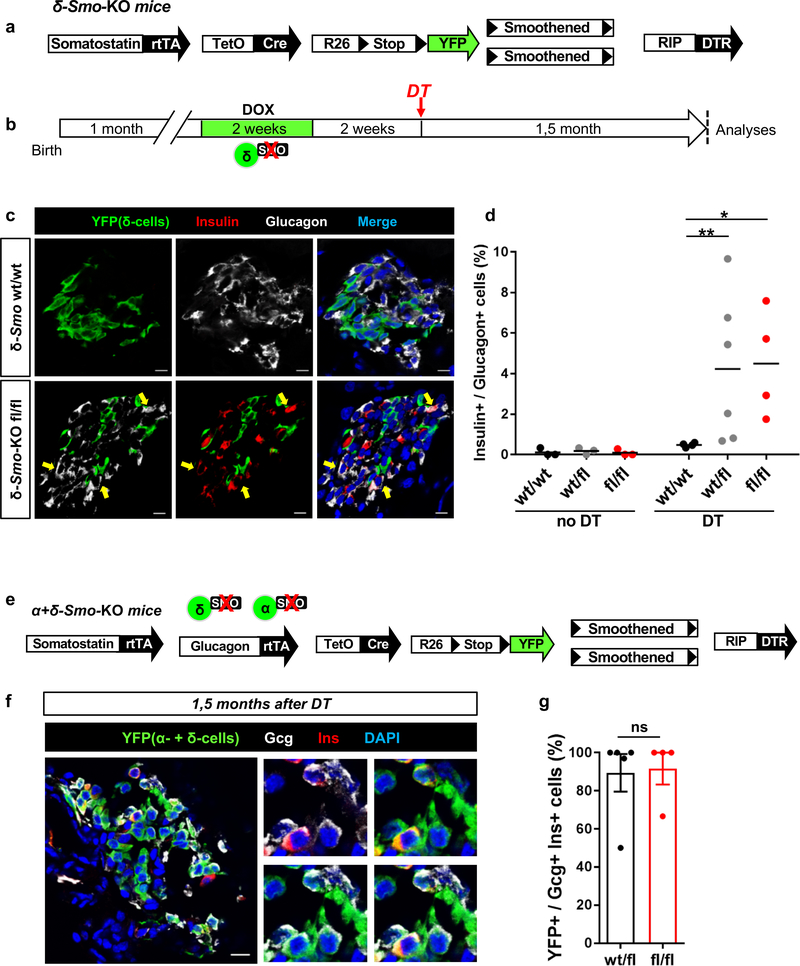
Figure 6. Smo inactivation in δ-cells leads to enhanced α-to-β cell conversion.
(a) Transgenes required for simultaneous δ-cell lineage tracing, Smo co-receptor downregulation and DT-induced β-cell ablation. (b) Experimental design (c) Immunofluorescence staining of islets from δ-Smo-KO mice 1,5 month after DT-induced β-cell loss. δ-Smo-KO mice display increased numbers of insulin+/glucagon+ coexpressing cells as compared to controls. (d) Percentage of cells coexpressing insulin and glucagon in δ-Smo-KO mice. n=3 mice wt/wt, wt/fl, fl/fl no DT; n =4, 6, 4 mice in wt/wt, wt/fl, fl/fl DT respectively. Two-tailed Mann Whitney test, P=0.0095 wt/fl vs wt/wt and P=0.0286 fl/fl vs wt/wt. Center indicates the mean. (e) Transgenes for simultaneous inactivation of Smo in α- and δ-cells along with their lineage tracing, and for DT-induced β-cell ablation. (f) Immunofluorescence staining of islets from α+δ-Smo-KO mice 1,5 month after DT. (g) Fraction of insulin+/glucagon+ coexpressing cells traced with YFP. n=5 wt/fl mice, n=4 fl/fl mice. Two-tailed Mann Whitney test, P=0.84. Center indicates the mean. Experiments in c-f were repeated two times independently with similar results. Scale bars: 10 μm. See Supplementary Tables 1n-p for source data.
In δ-Smo-KO mice, DOX induces δ-cells Smo downregulation along with YFP-labeling (Supp. Fig. 5i,j; Supp. Table 1m).
Smo downregulation did not affect δ-cell-specific gene expression nor induced insulin production (Supp. Fig. 5j; Supp. Table 1n). After β-cell ablation, the fraction of δ-cells engaging insulin expression was similar in control and δ-Smo-KO mice (Supp. Fig. 5k; Supp. Table 1n). Yet, surprisingly, the number of glucagon+/insulin+ bihormonal cells was 10-fold higher in mice with Smo downregulated in δ-cells (Fig. 6b-d; Supp. Table 1o). These bihormonal cells were not YFP-traced, indicating that they were not reprogrammed δ-cells (Fig. 6c). This suggests a potent δ-cell-mediated non-α-cell-autonomous regulation of α-cell plasticity.
To further explore this non-cell-autonomous regulation of α-cell identity, we generated “[α+δ]-Smo-KO” mice (Glucagon-rtTA; Somatostatin-rtTA; TetO-Cre; R26-YFP; Smoothenedfl/fl; RIP-DTR), to inactivate Smo in α- and δ-cells (Fig. 6e). In this situation, the glucagon+ & insulin+ bihormonal cells were YFP-traced (Fig. 6f, g; Supp. Table 1p), thus suggesting their α-cell origin.
Pharmacological inhibition of SmoHh with GANT61 also increased the number of insulin-producing α-cells after β-cell loss (Supp. Fig. 5l-m).
These observations suggest that active SmoHh-mediated signaling in δ-cells is a non-cell-autonomous intra-islet inhibitory signal that restricts α-cell plasticity in β-cell-ablated islets.
δ- and β-cell co-ablation enhances α-cell conversion
To further confirm that δ-cells act as negative regulators of α-cell plasticity, we generated Somatostatin-Cre; R26-YFP; R26-iDTR; RIP-DTR mice, to co-ablate β- and δ-cells simultaneously (Supp. Fig. 5n). β- + δ-cells loss doubled the proportion of glucagon+/insulin+ bihormonal cells (Supp. Fig. 5n; Supp. Table 1q). This result is compatible with a δ-cell-mediated restriction of α-cell plasticity.
DISCUSSION
Maintenance of adult cell identity is a highly dynamic process depending on the tight convergence of diverse signals whose complexity might be correlated with tissue regenerative capacity. The pressure to preserve cell identity is likely stronger in highly specialized cells implicated in vital metabolic processes. Perhaps endodermal cells are somewhat intrinsically different, always in a “regenerative state”, because they are exposed to external insults1. Any uncontrolled phenotypic instability could be detrimental and result in disease onset.
Our results show that the near-total loss of β-cells triggers simultaneous signals with antagonistic effects on α-cells: they activate insulin production and favor regeneration while also increasing α-cell marker expression, seemingly enforcing the α-cell fate and opposing identity changes.
We have identified paracrine repressive signals that maintain α-cell identity. We show here that α-cell identity is tightly maintained under physiological conditions through the constant repressive influence of local insulin and SmoHh signaling, coming from proximate β- and δ-cells and whose inhibition leads to substantially increased insulin+ α-cell numbers. Thus, beyond regulating α-cell function through somatostatin and insulin, δ- and β-cells also ensure α-cell fate maintenance in a non-α-cell-autonomous manner. Even with dual Smo-Ins downregulation, α-cell conversion is only partially improved, suggesting that α-cell conversion is restricted by the synergistic influence of multiple signals. Recent studies on transcription factors expressed in β-cells (such as Pdx1, Nkx6.1, Nkx2.2, Pax6) suggest their direct involvement in repressing non-β-cell genes29–35. Perhaps the extracellular repressive signals that lock α-cells in their state are transmitted via such transcriptional repressors.
The cell identity maintenance detected in the critical physiologically relevant islets may be much more widespread in differentiated cells, like a natural “tendency” or “capacity” of adult mature cells, and appear to be subtler than the complete iPSC reprogramming, with several levels of control of switching of cell phenotype.
This has outstanding implications regarding our comprehension of how the cell identity / differentiation equilibrium is established.
Our results indicate that spontaneous insulin production in α-cells is not simply due to uncontrolled stress-induced insulin gene dysregulation but is dynamically regulated, representing a meaningful compensatory response to cope with situations of insulin insufficiency.
Noteworthy, Lee et al. reported the appearance of insulin-resistant α-cells during diabetes36. We propose that these insulin-resistant α-cells would be more susceptible to changes in their identity, facilitating their coping with insulin deficiency. In agreement with such a speculation, bihormonal cells have been reported in the pancreas of diabetic patients37,38. The similarity of insulin-producing α-cells to native β-cells and their level of maturity were not addressed in this study and must be investigated.
In the context of this study, α-cell recruitment into insulin production, encompassing reduction of glucagon expression, would also be beneficial for diabetics by limiting glucagon secretion and hepatic glucose mobilization, without major metabolic defects caused by α-cell deficit39,40.
In conclusion, we have found that the stability of cell identity is heavily context-dependent, not “carved in stone”, with several levels of control of switching of cell phenotype. A physiological input from signaling pathways inside the islet keeps the identity of the cells in homeostasis. The endocrine-cell plasticity detected in this critical organ may be much more widespread in differentiated cells, like a natural “tendency” of adult mature cells. Maintaining cell identity is therefore an active process of repressive signals released from surrounding neighbor cells, blocking an intrinsic tendency of specialized differentiated cells to modify their phenotype and functional characteristics (Suppl. Fig. 6)49.
METHODS
Mice
Transgenic mice were previously described12,13,16,41-44. SstrtTA knock-in mice were newly generated as follows: a SstrtTALCA allele was made using a targeting vector made by BAC recombineering starting with BAC clone RP23-274H19 (purchased from CHORI, Oakland, CA). Both the general strategy and vectors were previously described45. The targeting vector, pSst.rtTA.LCA, contained a 5’ homology arm of 7.3 kb and a 3’ homology arm of 3.6 kb flanking a Lox66 site, a modified region of the Sst gene in which the reverse tetracycline TransActivator (rtTA), a cassette containing both a PGK-puromycinΔthymidine kinase and EM7-kanamycin selectable markers flanked by 2 tandemly-oriented FRT sites, and a Lox2272 site. After electroporation of the targeting vector into 129S6-derived mouse ES cells, 192 puromycin-resistant clones were obtained, 24 of which had undergone the desired homologous recombination event. Clone 1A8 was used to derive chimeras that were then bred with C57BL/6J mice to obtain germline transmission. The FRT-flanked selectable markers were removed by interbreeding with a FlpE-expressing transgenic mouse46.
Male and female mice were used for all experiments, except for transplantation experiments, where only males were used as hosts. Animals were treated with care and respect. The study is compliant with all relevant ethical regulations regarding animal research and all experiments have been approved and performed according to the guidelines of the Direction générale de la santé du Canton de Genève, state of Geneva (license numbers: GE/103/14; GE/111/17; GE/121/17). The number of mice used was limited by the availability of the required complex genotype. Mice were randomly selected for treatments and control. Sample size is in the range of the published literature and exclusion criteria were transgene set, general health status and occasional spontaneous death during the experiments.
Diphtheria toxin (DT), doxycycline (DOX), streptozotocin (STZ), S961, wortmannin and insulin treatments
DT (Sigma) was administrated by intra-peritoneal (i.p.) injections as previously described13. DOX (1mg/ml; Sigma) was added in the drinking water. STZ was injected i.p. as 1 single dose of 200 mg/kg body weight. S961 (Novo-Nordisk) was given via ALZET osmotic pumps implanted subcutaneously (40 nmol/1week). Wortmannin (Sigma) was i.p. injected daily for 5 consecutive days (1 mg/kg body weight). Mice received a subcutaneous insulin pellet (Linbit) when glycemia exceeded 25 mM.
Islet isolation, FACS, RNA extraction and qPCR
Islet isolation, cell sorting, RNA preparation and qPCR were performed as described12. All qPCRs were performed in triplicate. Primers are listed in Supplementary Table 3. Components of the insulin signaling pathway were evaluated with the PAMM-030Z and qPCR were performed as previously reported12.
Non-quantitative RT-PCR
Total RNA was isolated from liver, uterus and duodenum and used as controls. Tissue RNA was prepared using the Qiagen RNeasy Mini Kit. Islets cells were FAC-sorted as previously described12 and RNA prepared using the Qiagen RNeasy Micro Kit. cDNA was prepared using the Qiagen QuantiTect Reverse Transcription Kit starting from 1μg of RNA. Amplification conditions and primers were as previously reported47,48. Primers for actin were unpublished and are reported in Table 3.
RNA sequencing and quality control
Islet isolation and cell isolation by FACS were performed using previously described protocols12. The gating strategy used for FACS sorting is provided in Supplementary Figure 2. Experimental design for sample collection is reported in Fig. 2a. Briefly, purified cells were obtained as follows: i) native α-cells from 3-month-old Glucagon-rtTA; TetO-Cre; Rosa26YFP; RIP-DTR mice, ii) αDT: α-cells 1 month after DT from Glucagon-rtTA; TetO-Cre; Rosa26YFP; RIP-DTR mice, iii) αPdxlOE: α-cells ectopically expressing PDX1 from 3-month-old Glucagon-rtTA; TetO-Cre; R26YFP; CAG-Pdx1; RIP-DTR mice, iv) αPdx1OE+DT: α-cells expressing PDX1 one month after DT from Glucagon-rtTA; TetO-Cre; R26YFP; CAG-Pdx1; RIP-DTR mice, and v) native β-cells from 3-month-old RIP-Cre; Rosa26YFP mice.
Extracted RNA was assessed for quality by Agilent bioanalyzer. Libraries were prepared (according to Illumina's standard protocols), multiplexed and sequenced on an Illumina platform with paired- end 100-bp reads. The sequencing quality control was done with FASTQC v.0.10.1, followed by sequence alignment to the mouse reference genome (UCSC mm10) using the TopHat v2.0.9 (default parameters). Biological quality control and summarization were done with the RSeQC v2.4, the PicardTools v1.92 and the SamTools v0.1.18. In brief, RNA sequencing quality for each sample was assessed for overall coverage, base composition, presence of adaptors and 3’ bias, among other factors. A subset of samples were excluded from further analyses due to poor sequencing quality, 3’ bias, contamination, or outlier status for technical or biological reasons, including outliers on the first principal component analysis (PCA) across the entire sample set or samples with PDX-1 expression levels exceeding 3 SD from the mean of each respective experimental group. Finally, 22 samples were included for all downstream analyses (n=6 for native α-cells, n=3 for αDT, n=3 for αPdx1OE, n=5 for αPdx1OE+DT and n=5 for native β-cells). The raw count data was prepared with HTSeq v.0.6.1p1 (htseq-count, defaults parameters). The RNA-Seq data generated in this study have been deposited in the NCBI GEO database under accession number GSE109285.
Transcriptomic data analyses
The normalization and differential expression analysis was performed with the R/Bioconductor package edgeR package v.3.20.9. Briefly, very lowly expressed genes were filtered out and genes achieved 10 counts in at least 5 samples were kept. The filtered data were normalized by the library size and DEGs were estimated with the negative binomial general model statistics. To identify differentially expressed genes (DEGs), pairwise comparison was performed (edgeR, GLM approach for the design matrix setup, the factors were combined together). To obtain reference gene expression profile datasets for native α- and β-cell phenotypes, DEGs between sorted α- and β-cells were analyzed (FC>2 FDR<0.01). The 2940 DEGs were used as reference set of genes to separate α-cell- and β-cell-specific expression patterns (1682 DEGs as α-cell genes, 1258 DEGs as β-cell genes). Three additional comparisons were generated with respect to the reference α-cells: αDT vs α, αPdx1OE vs α, and αPdx1OE+DT vs α (FDR < 0.05). All DEGs we identified are provided in Supp. Table 5.
We considered upregulated β-cell genes or downregulated α-cell genes in the DEGs list as genes inducing a β-cell signature (“induced β-cell signature” category), and, reciprocally, upregulated α-cell genes as enhancing α-cell signature (“enhanced α-cell signatureβ category). DEGs from each condition were intersected with α-/β-cell reference genes, and shown in Fig. 2b and Supp. Table 5. The output data were displayed graphically as either a PCA-plot, heatmap, dendrogram, or Venn diagrams.
Pathway analysis
The pathway analyses were performed with gene set enrichment analysis (GSEA, http://software.broadinstitute.org/gsea/index.isp). Gene sets with significant enrichment in GSEA were identified among mm_GO of Gene Set Knowledgebase (GSKB). All significant gene sets were shown in Supp. Table 5.
Physiological studies
Pancreatic glucagon was measured as previously described49.
C-peptide measurements and pseudoislets experiments
In vivo glucose challenge tests, islets isolation and FACS-sorting of YFP+ cells were previously described12,13. The gating strategy used for FACS sorting is provided in Supplementary Figure 2.
For reaggregation into pseudoislets, sorted islet cells from 3-8 mice were pooled and seeded on 96- well ultra-low adherent culture plates for 5-7 days (1,000 cells/well) at 37°C in 5% CO2 incubator, in the following culture medium: Advanced DMEM/F12 (12634-010, Invitrogen) supplemented with penicillin/streptomycin, 10 mM HEPES (15630056, Invitrogen), 2 mM GlutaMAX (35050038, Invitrogen), 10% FBS, 10 mM Nicotinamide (N0636, Sigma), and 1 mM N-acetyl-L-cysteine (A9165, Sigma). Every other day of culture medium was changed.
For live imaging of cultured cells, images were captured manually at culture day 7 using Nikon Eclipse TE300 microscope (Nikon).
For evaluation of reprogramming events in pseudoislets, histology of pseudoislets were examined in cryo-sections as described previously12,13.
Pseudoislets were handpicked for each assay replicate and washed by incubation for 1 hour at 37°C in RPMI medium (11879020, Invitrogen) and then equilibrated by incubation for 1 hour at 37°C in Krebs-Ringer Bicarbonate buffer (KRB) containing 3 mM glucose (Sigma). Samples were then transferred into fresh KRB containing 3 mM (Low) glucose for 1 hour followed by incubation for another hour in KRB containing 20 mM (Hi) glucose at 37°C. Medium was collected after 1-hour incubation at each glucose concentration and stored at −80°C for subsequent analyses. Mouse C-peptide concentration was quantified using Mouse C-peptide ELISA kit (80-CPTMS-E01, Alpco).
For the independent 3 cohorts of experiments in each control and α-Smo-KO group, sorted α-cells (90,000-225,000 α-cells from 3-8 mice) were pooled and reaggregated into pseudoislets. In glucose-stimulated C-peptide secretion test, the maximum number of pseudoislets (90-225 pseudoislets) that we could generate from the pooled α-cells were tested as a cohort and then pseudoislet number normalized C-peptide release.
Transplantations
Islet transplantations under the kidney capsule were performed as described50.
Immunofluorescence
Cryostat sections were 10 μm-thick. The antibodies used were: rabbit and guinea pig anti-Pdx1 (C.W. Wright, 1/5000 and 1/750 respectively), rabbit anti-Nkx6.1 (BCBC AB1069, 1/800), guinea pig anti-porcine insulin (DAKO, 1/400), mouse anti-glucagon (Sigma, 1/1000), mouse antisomatostatin (BCBC Ab1985, 1/200) or goat anti-somatostatin (Santa Cruz 7918 1:200), rabbit anti- GFP (Molecular Probes, 1/400), mouse anti-mCherry (Abcam ab125096, 1/500), goat anti-Ihh (Santa Cruz sc-1196 1:50). Secondary antibodies were coupled to Alexa 405, 488, 647 (Molecular Probes), Cy3, Cy5 (Jackson Immunoresearch), or TRITC (Southern Biotech). All antibodies are listed in Suppl. Table 4. Sections were examined with a confocal microscope (Leica TCS SPE). In all experiments cells were considered bihormonal (glucagon+insulin+; somatostatin+insulin+) or coexpressing markers (i.e. insulin+YFP+) when one nucleus was clearly surrounded by both hormone / reporter staining.
Statistics & Reproducibility
Error bars represent standard error of the mean (s.e.m.) or standard deviation (s.d.), as indicated in the figure legends. One representative biological replicate of an experiment is presented in the figures. All experiments were performed three or more times independently under identical or similar conditions, except when differently indicated in the figure legends. Statistical analyses were performed using the Prism v6.0 software, and either unpaired t-test or Mann-Whitney test were applied for sample comparisons. Glycemia was measured once on multiple time-points for each animal (Figure 1c). The RNA-Seq experiment/reaction was performed once (Figure 3, Suppl. Figure 3a-c). qPCRs were performed once, using 3-5 individual biological samples as indicated in the figure legends; each biological sample was run in triplicate (Fig. 4, Suppl. Fig. 3-5). Immunofluorescence for a certain antibody cocktail was performed once for each mouse with >3 cryosections/animal being stained at once and analyzed (Figure 2 c, g, k and i; Figure 4 c, f; Figure 5 c; Fig. 6 c,f; Suppl. Fig. 4 b, g, j; Suppl. Fig. 5 h, j, k, l). The immunofluorescence reaction was repeated twice for Suppl. Figure 5b).
Data availability
The RNA-Seq data generated in this study have been deposited in the NCBI GEO database under accession number GSE109285. Source data for Figures 1, 2, 3, 4, 5 and 6 and Supplementary Figures 1, 2, 4, 5, 6 have been provided as supplementary table 1, and RNA analyses have been provided in Supplementary Table 2. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Yuval Dor and, very specially, to Marcos González Gaitán, for carefully reading and discussing the manuscript; to O. Fazio, M. Urwyler, C. Gysler, B. Polat and R. Gangula for their technical help, and to J-P. Aubry-Lachainaye for FACS assistance. We acknowledge C. Delucinge-Vivier, M. Docquier A. Efanov, P. Ebert, J. Calley, H. Wu, S.K. Syed and T. Wei for technical and expert assistance supporting RNA sequencing, quality control, statistical and bioinformatics analyses. M.S. was supported by R01DK068471. SC is supported by grants from the Research Council of Norway (NFR n° 247577) and the Novo Nordisk Foundation (n° NNF15OC0015054). M.A.M. was supported by U01DK072473 (Beta Cell Biology Consortium). Work was funded with grants from the National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Disease (Beta Cell Biology Consortium, U01DK089566 and Human Islet Research Network, UC4-DK104209 and UC4-DK108132), the Juvenile Diabetes Research Foundation (in part through joint research funding with Lilly Research Laboratories, 17–2011-276, and 2-SRA-2015–67-Q-R), the Innovative Medicines Initiative Joint Undertaking under grant agreement n° 155005 (IMIDIA), resources of which are composed of financial contribution from the European Union’s Seventh Framework Programme (FP7/2007–2013) and EFPIA companies’ in kind contribution, the Fondation privée des Hôpitaux Universitaires de Genève – Confirm, the Fondation Aclon, and the Swiss National Science Foundation (NRP63, 310030_152965, and the BONUS OF EXCELLENCE #310030B_173319) to P.L.H.
Footnotes
COMPETING INTEREST
The authors declare no financial and non-financial competing interests.
REFERENCES
- 1.Holmberg J & Perlmann T Maintaining differentiated cellular identity. Nat Rev Genet 13, 429–439, doi: 10.1038/nrg3209 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Thowfeequ S, Myatt EJ & Tosh D Transdifferentiation in developmental biology, disease, and in therapy. Dev Dyn 236, 3208–3217, doi: 10.1002/dvdy.21336 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Chera S & Herrera PL Regeneration of pancreatic insulin-producing cells by in situ adaptive cell conversion. Curr Opin Genet Dev 40, 1–10, doi: 10.1016/j.gde.2016.05.010 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pasque V, Jullien J, Miyamoto K, Halley-Stott RP & Gurdon JB Epigenetic factors influencing resistance to nuclear reprogramming. Trends Genet 27, 516–525, doi: 10.1016/j.tig.2011.08.002 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Natoli G Maintaining cell identity through global control of genomic organization. Immunity 33, 12–24, doi: 10.1016/j.immuni.2010.07.006 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Barrero MJ, Boue S & Izpisua Belmonte JC Epigenetic mechanisms that regulate cell identity. Cell Stem Cell 7, 565–570, doi: 10.1016/j.stem.2010.10.009 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Szabat M et al. Maintenance of beta-cell maturity and plasticity in the adult pancreas: developmental biology concepts in adult physiology. Diabetes 61, 1365–1371, doi: 10.2337/db11-1361 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eade KT & Allan DW Neuronal phenotype in the mature nervous system is maintained by persistent retrograde bone morphogenetic protein signaling. J Neurosci 29, 3852–3864, doi: 10.1523/JNEUROSCI.0213-09.2009 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopez-Coviella I, Berse B, Krauss R, Thies RS & Blusztajn JK Induction and maintenance of the neuronal cholinergic phenotype in the central nervous system by BMP-9. Science 289, 313–316 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Jessen KR, Mirsky R & Arthur-Farraj P The Role of Cell Plasticity in Tissue Repair: Adaptive Cellular Reprogramming. Dev Cell 34, 613–620, doi: 10.1016/j.devcel.2015.09.005 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Knapp D & Tanaka EM Regeneration and reprogramming. Curr Opin Genet Dev 22, 485–493, doi: 10.1016/j.gde.2012.09.006 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Chera S et al. Diabetes recovery by age-dependent conversion of pancreatic delta-cells into insulin producers. Nature 514, 503–507, doi: 10.1038/nature13633 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thorel F et al. Conversion of adult pancreatic alpha-cells to beta-cells after extreme beta-cell loss. Nature 464, 1149–1154, doi: 10.1038/nature08894 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chakravarthy H et al. Converting Adult Pancreatic Islet alpha Cells into beta Cells by Targeting Both Dnmt1 and Arx. Cell Metab 25, 622–634, doi: 10.1016/j.cmet.2017.01.009 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bosma MJ & Carroll AM The SCID mouse mutant: definition, characterization, and potential uses. Annu Rev Immunol 9, 323–350, doi: 10.1146/annurev.iy.09.040191.001543 (1991). [DOI] [PubMed] [Google Scholar]
- 16.Yang YP, Thorel F, Boyer DF, Herrera PL & Wright CV Context-specific alpha- to-beta-cell reprogramming by forced Pdx1 expression. Genes Dev 25, 1680–1685, doi: 10.1101/gad.16875711 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaffer L et al. A novel high-affinity peptide antagonist to the insulin receptor. Biochem Biophys Res Commun 376, 380–383, doi: 10.1016/j.bbrc.2008.08.151 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Vikram A & Jena G S961, an insulin receptor antagonist causes hyperinsulinemia, insulin-resistance and depletion of energy stores in rats. Biochem Biophys Res Commun 398, 260–265, doi: 10.1016/j.bbrc.2010.06.070 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Yi P, Park JS & Melton DA Betatrophin: a hormone that controls pancreatic beta cell proliferation. Cell 153, 747–758, doi: 10.1016/j.cell.2013.04.008 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Cigliola V, Thorel F, Chera S & Herrera PL Stress-induced adaptive islet cell identity changes. Diabetes Obes Metab 18 Suppl 1, 87–96, doi: 10.1111/dom.12726 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Damond N et al. Blockade of glucagon signaling prevents or reverses diabetes onset only if residual beta-cells persist. Elife 5, doi: 10.7554/eLife.13828 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grieco FA et al. Delta-cell-specific expression of hedgehog pathway Ptch1 receptor in murine and human endocrine pancreas. Diabetes Metab Res Rev 27, 755–760, doi: 10.1002/dmrr.1247 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Lau J & Hebrok M Hedgehog signaling in pancreas epithelium regulates embryonic organ formation and adult beta-cell function. Diabetes 59, 1211–1221, doi: 10.2337/db09-0914 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawahira H et al. Combined activities of hedgehog signaling inhibitors regulate pancreas development. Development 130, 4871–4879, doi: 10.1242/dev.00653 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Cervantes S, Lau J, Cano DA, Borromeo-Austin C & Hebrok M Primary cilia regulate Gli/Hedgehog activation in pancreas. Proc Natl Acad Sci U S A 107, 10109–10114, doi: 10.1073/pnas.0909900107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landsman L, Parent A & Hebrok M Elevated Hedgehog/Gli signaling causes beta-cell dedifferentiation in mice. Proc Natl Acad Sci U S A 108, 17010–17015, doi: 10.1073/pnas.1105404108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas MK, Lee JH, Rastalsky N & Habener JF Hedgehog signaling regulation of homeodomain protein islet duodenum homeobox-1 expression in pancreatic beta-cells. Endocrinology 142, 1033–1040, doi: 10.1210/endo.142.3.8007 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Thomas MK, Rastalsky N, Lee JH & Habener JF Hedgehog signaling regulation of insulin production by pancreatic beta-cells. Diabetes 49, 2039–2047 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Schaffer AE et al. Nkx6.1 controls a gene regulatory network required for establishing and maintaining pancreatic Beta cell identity. PLoS Genet 9, e1003274, doi: 10.1371/journal.pgen.1003274 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao T et al. Pdx1 maintains beta cell identity and function by repressing an alpha cell program. Cell Metab 19, 259–271, doi: 10.1016/j.cmet.2013.12.002 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gutierrez GD et al. Pancreatic beta cell identity requires continual repression of non-beta cell programs. J Clin Invest 127, 244–259, doi: 10.1172/JCI88017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swisa A et al. PAX6 maintains beta cell identity by repressing genes of alternative islet cell types. J Clin Invest 127, 230–243, doi: 10.1172/JCI88015 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Papizan JB et al. Nkx2.2 repressor complex regulates islet beta-cell specification and prevents beta-to-alpha-cell reprogramming. Genes Dev 25, 2291–2305, doi: 10.1101/gad.173039.111 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gauthier BR, Gosmain Y, Mamin A & Philippe J The beta-cell specific transcription factor Nkx6.1 inhibits glucagon gene transcription by interfering with Pax6. Biochem J 403, 593–601, doi: 10.1042/BJ20070053 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schisler JC et al. The Nkx6.1 homeodomain transcription factor suppresses glucagon expression and regulates glucose-stimulated insulin secretion in islet beta cells. Proc Natl Acad Sci U S A 102, 7297–7302, doi: 10.1073/pnas.0502168102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee Y et al. Hyperglycemia in rodent models of type 2 diabetes requires insulin-resistant alpha cells. Proc Natl Acad Sci U S A 111, 13217–13222, doi: 10.1073/pnas.1409638111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Butler AE et al. Adaptive changes in pancreatic beta cell fractional area and beta cell turnover in human pregnancy. Diabetologia 53, 2167–2176, doi: 10.1007/s00125-010-1809-6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White MG et al. Expression of mesenchymal and alpha-cell phenotypic markers in islet beta-cells in recently diagnosed diabetes. Diabetes Care 36, 3818–3820, doi: 10.2337/dc13-0705 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thorel F et al. Normal glucagon signaling and beta-cell function after near-total alpha-cell ablation in adult mice. Diabetes 60, 2872–2882, doi: 10.2337/db11-0876 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hayashi Y et al. Mice deficient for glucagon gene-derived peptides display normoglycemia and hyperplasia of islet {alpha}-cells but not of intestinal L-cells. Mol Endocrinol 23, 1990–1999, doi: 10.1210/me.2009-0296 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyatsuka T et al. Persistent expression of PDX-1 in the pancreas causes acinar-to-ductal metaplasia through Stat3 activation. Genes Dev 20, 1435–1440, doi: 10.1101/gad.1412806 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dietrich P, Dragatsis I, Xuan S, Zeitlin S & Efstratiadis A Conditional mutagenesis in mice with heat shock promoter-driven cre transgenes. Mamm Genome 11, 196–205 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Bruning JC et al. A muscle-specific insulin receptor knockout exhibits features of the metabolic syndrome of NIDDM without altering glucose tolerance. Mol Cell 2, 559–569 (1998). [DOI] [PubMed] [Google Scholar]
- 44.Long F, Zhang XM, Karp S, Yang Y & McMahon AP Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation. Development 128, 5099–5108 (2001). [DOI] [PubMed] [Google Scholar]
- 45.Chen SX et al. Quantification of factors influencing fluorescent protein expression using RMCE to generate an allelic series in the ROSA26 locus in mice. Dis Model Mech 4, 537–547, doi: 10.1242/dmm.006569 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodriguez CI et al. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet 25, 139–140, doi: 10.1038/75973 (2000). [DOI] [PubMed] [Google Scholar]
- 47.Russell MC, Cowan RG, Harman RM, Walker AL & Quirk SM The hedgehog signaling pathway in the mouse ovary. Biol Reprod 77, 226–236, doi: 10.1095/biolreprod.106.053629 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Li Z et al. Reduced white fat mass in adult mice bearing a truncated Patched 1. Int J Biol Sci 4, 29–36 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strom A et al. Unique mechanisms of growth regulation and tumor suppression upon Apc inactivation in the pancreas. Development 134, 2719–2725, doi: 10.1242/dev.02875 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Mathe Z et al. Tetracycline-regulated expression of VEGF-A in beta cells induces angiogenesis: improvement of engraftment following transplantation. Cell Transplant 15, 621–636 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-Seq data generated in this study have been deposited in the NCBI GEO database under accession number GSE109285. Source data for Figures 1, 2, 3, 4, 5 and 6 and Supplementary Figures 1, 2, 4, 5, 6 have been provided as supplementary table 1, and RNA analyses have been provided in Supplementary Table 2. All other data supporting the findings of this study are available from the corresponding author on reasonable request.