Abstract
Little is known of the structural and functional properties of abnormal brain networks associated with neurological disorders. We used a social network approach to characterize the properties of the Parkinson’s disease (PD) metabolic topography in 4 independent patient samples and in an experimental non-human primate model. The PD network exhibited distinct features. Dense, mutually facilitating functional connections linked the putamen, globus pallidus, and thalamus to form a metabolically active core. The periphery was formed by weaker connections linking less active cortical regions. Notably, the network contained a separate module defined by interconnected, metabolically active nodes in the cerebellum, pons, frontal cortex, and limbic regions. Exaggeration of the small-world property was a consistent feature of disease networks in parkinsonian humans and in the non-human primate model; this abnormality was only partly corrected by dopaminergic treatment. The findings point to disease-related alterations in network structure and function as the basis for faulty information processing in this disorder.
Keywords: FDG PET, graph theory, metabolic brain networks, Parkinson’s disease
Introduction
Pattern detection techniques are currently being used to identify abnormal functional topographies in resting-state brain scans from patients with neurodegenerative disorders (Eidelberg 2009; Habeck et al. 2010; Vo et al. 2017). Disease-related spatial covariance patterns obtained using principal component analysis (PCA) and related multivariate approaches have proved to be helpful as functional imaging biomarkers for differential diagnosis (Tang, Poston, Eckert et al. 2010; Niethammer et al. 2014; Tripathi et al. 2016) and for tracking the progression of neurodegenerative processes and their response to treatment (Feigin et al. 2007; Tang, Poston, Dhawan et al. 2010; Tang et al. 2013; Ko, Feigin et al. 2014). In Parkinson’s disease (PD), for instance, a significant disease-related metabolic covariance pattern (PDRP) has been identified, which is characterized by increased activity in the putamen/globus pallidus/ventral thalamus, dorsal pons, and cerebellum, with relatively reduced activity in the lateral premotor and parieto-occipital cortex (Eidelberg 2009; Spetsieris and Eidelberg 2011). Subject scores for this pattern, which quantify its expression in individual brain scans, have been found to correlate with clinical motor disability ratings (Niethammer and Eidelberg 2012), intraoperative ratings of basal ganglia output (Lin et al. 2008; Eidelberg 2009), as well as independent PET measurements of nigrostriatal dopaminergic input in PD patients (Holtbernd et al. 2015; Ko et al. 2017). In addition to its excellent reproducibility in individual subjects and across populations (Eidelberg 2009; Niethammer and Eidelberg 2012; Ko, Feigin et al. 2014), the PDRP topography has been found to be disease specific, as revealed by its accuracy as a diagnostic marker in patients with parkinsonism (Tang, Poston, Eckert et al. 2010; Tripathi et al. 2016) and in individuals with preclinical changes (Holtbernd et al. 2014; Wu et al. 2014). Importantly, PDRP expression increases with advancing disease (Huang et al. 2007; Tang, Poston, Dhawan et al. 2010; Ko, Feigin et al. 2014), while declining toward normal in response to effective treatment (Asanuma et al. 2006; Hirano et al. 2008; Niethammer and Eidelberg 2012).
In spite of the clinical utility of the PDRP—and its experimental homolog, the parkinsonism-related pattern (PRP) in the non-human primate model (Ma et al. 2012, 2015; Peng et al. 2016)—the biological basis of these abnormal topographies is only partly understood. Indeed, it is not known whether such characteristic disease-related topographies as PDRP and PRP represent distinct brain networks composed of interconnected regional elements (nodes) with defined spatial structure (topology). Moreover, the computation of individual expression values (subject scores) for a given pattern, while integral to its role as a disease biomarker, may entail the sacrifice of critical data needed to delineate underlying network architecture.
To address this issue, we explored the topographical relationship between the PDRP and corresponding disease networks defined using graph theory. In particular, we determined whether nodal centrality, a graph theoretic measure of a region’s importance to the network, correlated with corresponding region weights on established parkinsonism-related metabolic covariance patterns in PD patients and parkinsonian non-human primates. Additionally, graph theory allowed us to examine the topology of the disease network in a manner not possible with spatial covariance mapping. The 2 representations, that is, PDRP region weights in PCA (Spetsieris and Eidelberg 2011) and eigenvector centrality in graph theory (Bullmore and Sporns 2009), describe different characteristics of the disease network. The PDRP as a whole-brain anatomical representation assigns regional values (weights, PC loadings) according to each area’s functional significance to the abnormal PD topography. The graph theoretical representation, by contrast, is more of a topographical schematic based on functional inter-relationships between major regions. Using a novel computational approach borrowed from social network analysis (Correa et al. 2009, 2012), we considered how small perturbations in the centrality of one network node influence the centrality of the others. Thus, for each pair of regions—that is, for each graphical edge connecting them—we computed the partial derivative of centrality with respect to the number of incoming connections. The resulting value, termed the centrality sensitivity (Correa et al. 2009), was used to rank the edges of the graph by the strength of its node-to-node interactions (Correa et al. 2012). By additionally taking into account the sign of the centrality derivatives, this approach also allowed us to visualize mutually enhancing (“friendship”) versus competing (“enmity”) functional interactions between pairs of nodes. Indeed, we found that the PD network was characterized by a distinct core-periphery architecture. The disease network was additionally associated with abnormal information processing. The small-world property, defined by shortened communication distance (path length) between nodes and increased connectivity between nearest neighbors (clustering), normally serves to optimize the efficiency of information transfer with biological networks (Watts and Strogatz 1998; Bassett and Bullmore 2016). The PD network, by contrast, was characterized by a pathological increase in “small-worldness”, which was only partially reversed by dopamine treatment.
Materials and Methods
Participants
Human Subjects
We studied metabolic brain images acquired using 18F-fluorodeoxyglucose (FDG) positron emission tomography (PET) in 4 independent PD (PD1–PD4) and 2 normal (NL1 and NL2) samples (Supplementary Table 1). Graph theory analysis was performed on resting-state scan data from each group. For the 3 samples PD1–PD3, imaging was conducted in the “off-state” (OFF), 12 h after cessation of antiparkinsonian medications. Scans from PD1 (n = 33), a mixed patient sample with mild to moderate motor symptoms, were used in combination with NL1 (n = 33), an age-matched healthy sample, for spatial covariance analysis to identify a region-of-interest (ROI)-based PDRP topography for correlation with corresponding centrality measures. (Scans from these subjects were originally used to identify the current voxel-based PDRP biomarker (Ma et al. 2007).) PD2 (n = 33) and PD3 (n = 33), independent patient samples with mild and more advanced motor symptoms, respectively, and NL2 (n = 33), a separate healthy control group, were age- and gender-matched to PD1; scan data from these samples were used for validation. PD4 (n = 21), a separate age- and gender-matched sample, was comprised of patients with mild-moderate motor symptoms who were scanned twice with FDG PET: once in the baseline “off-state” (OFF) (as above, off medication for 12 h), and again in the “on-state” (ON) during an individually titrated intravenous levodopa infusion without dyskinesia (Jourdain et al. 2016). Scan data from the PD4 subjects were used to assess the effects of clinically effective levodopa administration on the network parameters.
In all disease subjects, a diagnosis of PD was made according to the United Kingdom Parkinson’s Disease Society Brain Bank criteria (Hughes et al. 1992). Patients were excluded if Mini-Mental State Examination score was less than 27. Further exclusion criteria included severe hypertension, cardiovascular disease, diabetes mellitus, and past or current psychiatric history. Routine anatomical magnetic resonance imaging (MRI) disclosed no evidence of structural brain abnormalities or visually discernible atrophy. Ethical permission for all studies was obtained from the Institutional Review Board of Northwell Health. Written consent was obtained from each subject after detailed explanation of the procedures.
Non-human Primates
We additionally studied non-human primates with experimental parkinsonism induced by chronic low-dose systemic 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) administration (Porras et al. 2012). We compared FDG PET scans from the parkinsonian monkeys (MPTP, n = 11) with their normal counterparts (NL, n = 12) as described previously (Ma et al. 2012). (These scans were originally used to identify the PRP, a validated marker of parkinsonism in non-human primates (Ma et al. 2012, 2015; Peng et al. 2016).) MPTP and NL monkeys were similar in age (12.4 ± 4.6 vs. 10.0 ± 3.9 years; P = 0.20) and weight (9.8 ± 2.2 vs. 8.8 ± 2.6 kg; P = 0.31). Demographic information concerning these animals and the details of the experimental protocol have been provided elsewhere (Ma et al. 2012). All non-human primate experiments were conducted in accordance with the relevant guidelines and regulations of Canadian federal government on animal welfare and were approved by the Committee on Animal Care at the University of British Columbia.
Positron Emission Tomography
Human Subjects
In this study, human subjects were scanned with FDG PET in 3D mode using a GE Advance tomograph (General Electric, Milwaukee, WI, USA). The performance characteristics of this instrument have been provided elsewhere (Eckert et al. 2005). PET scans were conducted in a dimly lit room with eyes open and minimal auditory stimulation. The resulting metabolic images were normalized and smoothed using SPM5 software (www.fil.ion.ucl.ac.uk/spm/). The details of image processing are described elsewhere (Spetsieris and Eidelberg 2011). Local metabolic activity normalized to global activity was measured in prespecified 95 ROIs according to the automated anatomical labeling (AAL) atlas (Tzourio-Mazoyer et al. 2002). In addition to 90 ROIs from the AAL, we measured cerebellar activity in 3 discrete ROIs corresponding to the cerebellar hemispheres and vermis. Similarly, activity in the left and right pons was averaged for further analysis.
Non-human Primates
Non-human primates in the normal (NL) and parkinsonian (MPTP) groups were scanned using the high-resolution PET tomograph (ECAT HRRT; CPS Innovations, Knoxville, TN, USA) at the University of British Columbia. The details of these procedures are provided elsewhere (Ma et al. 2012). In these studies, the animal stayed awake in a quiet dimly lit room for ~40 min after FDG injection. The monkeys were rapidly sedated (ketamine 10 mg/kg intramuscularly) at the end of the uptake period. They were then brought to the PET suite where they were intubated and scanned for 30 min under isoflurane anesthesia. The resulting FDG PET scans were normalized and smoothed using SPM5 software after alignment using the INIA19 macaque brain template (Rohlfing et al. 2012). From this template, we selected 53 ROIs corresponding to primate regions homologous to those used in the analysis of the human scan data. All regions contained at least 30 contiguous voxels (voxel size: 2×2×2 mm) with gray matter probability of 50% or more. Local metabolic activity was quantified in each ROI as described elsewhere (Ma et al. 2012). Several of the animals underwent dopamine cell implantation in the right putamen (Peng et al. 2016). We therefore restricted the analysis of the non-human primates to unoperated left hemispheres.
Network Analysis
Globally normalized metabolic activity in each of the ROIs was used to generate a region × region correlation matrix for each of the 6 human samples (95×95 for the PD1–PD4 patient and NL1 and NL2 control groups) and for the 2 non-human primate samples (53×53 for the MPTP and NL groups). For PD4, metabolic data acquired on and off levodopa, termed the ON and OFF treatment conditions, were analyzed separately. A correlation matrix based on absolute values of pairwise regional correlation coefficients was constructed for each sample using ROI values for local FDG uptake determined in that particular group. FDG PET signal in a given region reflects afferent synaptic activity (Raichle and Mintun 2006; Lin et al. 2008). In this context, metabolic correlations between network nodes (either in the same or opposite directions) denote interneuronal information transfer that is more efficient than between arbitrarily chosen region pairs (Matsui et al. 2011). Thus, absolute values of the correlation matrix elements were sorted and adjacency matrices were constructed at varying cost thresholds (1–50%) (Hosseini et al. 2012), with cost defined as the graph’s connection density, that is, the ratio of the actual to the maximum number of possible edges in the graph. Thus, at the 25% cost threshold, an adjacency matrix corresponding to an undirected graph was generated by setting the top 25% of the absolute r-values to 1 and the rest including the diagonal elements to 0. The graphs were determined to be fully connected at minimum cost of 18% (NL1), 11% (NL2), 14% (PD1), 12% (PD2), 10% (PD3), 12% (PD4-OFF), 8% (PD4-ON) for the human data and 24% (NL) and 17% (MPTP) for the non-human primate data. For each dataset, we computed the following network metrics: “Clustering Coefficient” (C), a measure of network density represented by the average probability that the nearest neighbors of given node are themselves nearest neighbors of one another; “Characteristic Path Length” (L), a measure of network efficiency represented by the average shortest path length on the graph; “Small-worldness coefficient” (S), the C/L ratio for the graph divided by the analogous measure for a random graph with the same wiring cost (Humphries and Gurney 2008); and “Edges”, an estimate of the relative number of connected edges within a subgraph. These measures were computed at varying thresholds, at a cost range of 18–50% for the comparisons between NL1 and PD1, 12–50% for NL2, PD2, PD3 and PD4, and 24–50% for the NL and MPTP monkey groups. The details of these calculations are provided in Supplementary Note A. In the human data, differences in the analytical parameters were evaluated separately for the PDRP and non-PDRP spaces of the relevant samples (PD1 vs. NL1, PD2 and PD3 vs. NL2, and PD4-OFF vs. PD4-ON), as well as corresponding group × subspace interaction effects. This was done by performing 10 000 permutations of random samples of the combined subject pool at each cost threshold (Hosseini et al. 2012). An analogous approach was used to evaluate these parameters in the PRP and non-PRP spaces of the MPTP and normal non-human primate samples. When the subgraphs were not fully connected, path length (L) values were estimated after removing the unconnected nodes.
For each sample, we used graph theory to compute “eigenvector centrality” (EC), an index of the relative importance of the component regions (nodes) that make up the dominant pattern vector (brain network) (Newman 2010). In addition, we used finite-difference methods to estimate the partial derivatives of EC with respect to nodal degree for each incident edge (connections) of the graph (Correa et al. 2009) (see Supplementary Note B for details). The measure of “centrality sensitivity”, introduced above, gauges the impact of a given node on the centrality of another. Centrality sensitivity can be used to simplify the visualization of large, complex networks by retaining only the most influential graph edges (Van Ham and Wattenberg 2008; Correa et al. 2012). Edge ranking also provides an index of “centralization”, that is, the ways centrality is distributed across a graph (Wasserman and Faust 1994), which in turn can be used to uncover the latent community structure of the network (Girvan and Newman 2002; Van Ham and Wattenberg 2008).
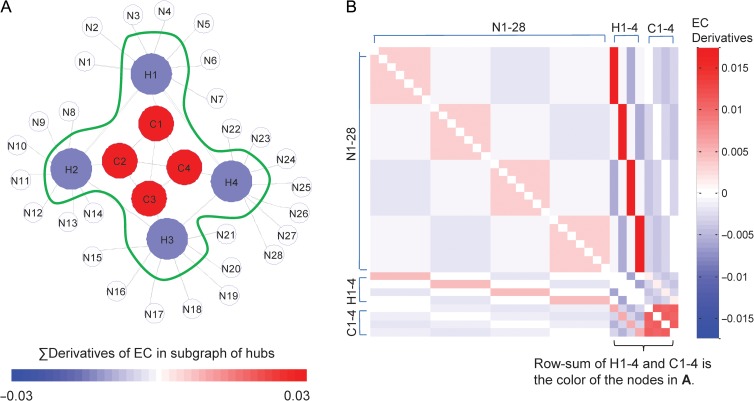
In this study, edge ranking was based upon centrality sensitivity for 2 reasons: 1) centrality sensitivity preserves the centrality distribution of the original graph; and 2) it distinguishes mutually facilitating node-to-node interactions (“collaboration”/“friendship”) from inhibitory nodal relationships (“competition”/“enmity”) on the basis of the sign of the corresponding partial derivatives (Correa et al. 2012). The utility of this approach in probing network structure is illustrated in Figure 1. In this model network, local hubs exhibit the highest degree (= 10) and EC (= 0.0746), central hubs have moderate degree (= 4) and EC (= 0.0553), while peripheral nodes exhibit low degree (= 1) and small EC (= 0.0172). Nonetheless, in the subgraph of connected hubs (green outline), EC derivatives for the edges connecting the central (red) core hubs have higher average values (0.0284) than those connecting the isolated (blue), more peripheral hubs (−0.0137). This implies that strengthening the connectivity between core hubs increases EC and facilitates information flow within the subgraph.
Figure 1.
Derivatives of eigenvector centrality in a model graph with core-periphery structure. (A) Model network composed of a central hub (C1-4) that connects the local hub (H1-4), and set of peripheral nodes (N1-28). The gray lines represent edges that connect nodes. Local hubs showed the highest degree (= 10) and eigenvector centrality (EC = 0.0746). Central hubs showed moderate degree (= 4) and EC (= 0.0553). Peripheral nodes had the lowest degree (= 1) and small EC (= 0.0172). (B) Matrix of sensitivity values. The entries of this square, asymmetric matrix are defined by the partial derivatives of EC with respect to the number of edges incident upon (pointing to) a given node (see text). To measure the impact of adding 1° to the subgraph of hubs (C1-4 and H1-4; green outline in A), EC partial derivatives for edges incident upon subgraph nodes were row-summed; values for self-nodes were set to 0. Central hubs had higher averaged EC derivatives for incident subgraph edges than local hubs (= −0.0137). The greater centrality sensitivity of the central hubs implies that strengthening its connectivity will further facilitate information flow (i.e., increasing EC) within the hub subgraph. [Partial derivatives of EC were computed using the variational method described by Correa et al. (2012). In the model network presented in A, we assessed the effect of increasing the degree of the hub subgraph (C1-4 and H1-4) by 1. This was done by row-summing the entries for the hub regions in the partial derivative matrix. The resulting values were color-coded to match the corresponding nodes depicted in A. See Supplementary Notes for details.]
Relationship Between Spatial Covariance Topography and Eigenvector Centrality
FDG PET scans from the combined PD1 and NL1 sample were analyzed using the Scaled Subprofile Model (SSM) and principal component analysis (PCA) to identify a significant PDRP topography as described elsewhere (Eidelberg 2009; Spetsieris and Eidelberg 2011). In this study, we used a regional approach analogous to that described previously (Spetsieris et al. 2013) in which spatial covariance analysis was applied to measurements of metabolic activity from the prespecified ROIs (n = 95) determined for individual subjects in both groups using an automated computerized algorithm. This ROI-based PDRP topography (Supplementary Fig. 1A) was represented by the first principal component (PC1) in the combined group analysis, accounting for 37.9% of the subject × ROI variance. PDRP region weights were z-scored based upon the mean and standard deviation of all regions as described elsewhere (Spetsieris et al. 2013). Subject scores for the PDRP, denoting pattern expression in individual subject scores (Eidelberg 2009; Spetsieris et al. 2013), were elevated in PD1 relative to NL1 subjects (P < 0.001; Student’s t-test). Region weight absolute values on this pattern were correlated with corresponding nodal EC values for each group by computing Pearson product-moment correlation coefficients. In this analysis, 19 ROIs had PDRP region weights that were greater than +1.0; these network regions were considered metabolically “active”. Eighteen ROIs showed region weights on the PDRP that were lower than −1.0, and were considered metabolically “underactive”.
All the examined graphs were undirected and fully connected, indicating that the corresponding adjacency matrices were irreducible. Because these non-negative matrices had positive diagonal entries, a unique, strictly positive principal eigenvector was guaranteed by the Perron Frobenius Theorem (Meyer 2000, p. 678). As a result, the EC vectors obtained for the various samples in this study had strictly positive components, which were correlated with absolute values for the corresponding PDRP region weights.
For further analysis, we divided the brain into PDRP and non-PDRP subspaces based upon metabolic data from 37 and 58 ROIs, corresponding respectively to ROIs with high (absolute region weight ≥1.0) and low (absolute region weight < 1.0) local contributions to overall PDRP activity. Graph theoretic metrics from each subspace were permuted 10 000 times for purposes of group comparison (Hosseini et al. 2012). A similar approach was used in the analysis of the corresponding metrics from the non-human primate scans. Computations were performed using the Brain Connectivity Toolbox (Rubinov and Sporns 2010) and in-house programs running on MATLAB 8.3.0 (MathWorks, Inc., Natick, MA, USA).
Statistical Analysis
Topographical correlations between the EC vectors for each sample and corresponding PDRP/PRP region weights were evaluated by computing Pearson product-moment correlation coefficients at varying cost. For regional comparison of the EC sensitivity measures, values from the 95 ROIs were divided into 4 categories based upon the magnitude and sign of the corresponding PDRP region weights (i.e., PDRP < −1, −1 < PDRP < 0, 0 < PDRP < 1 and PDRP > 1). Differences across categories were assessed separately for each sample using 1-way ANOVA and post hoc Bonferroni tests. The number of edges, and C and L values for the PDRP and non-PDRP subgraphs and the differences between the subgraphs were compared across samples using permutation tests (Hosseini et al. 2012). All statistical tests were performed using MATLAB 8.3.0 run on a Windows platform. Values were considered significant for P < 0.05, 2-tailed.
Results
Eigenvector Centrality Measures Correlate with Abnormal Disease Topographies
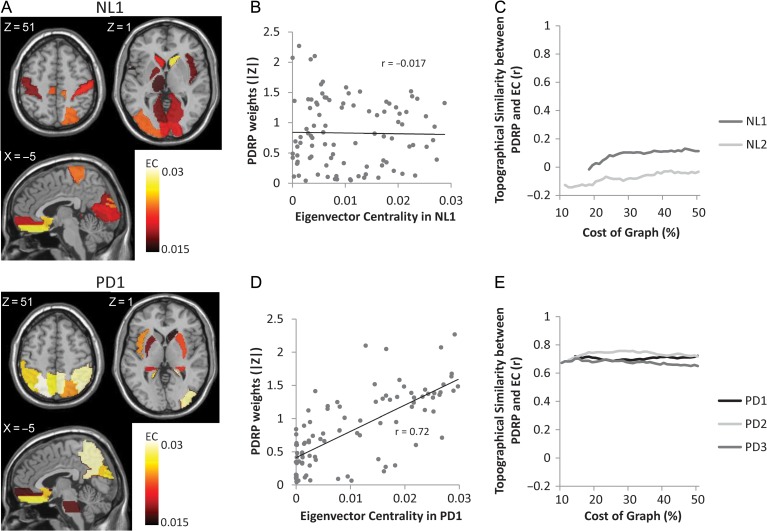
Distinct relationships were identified between the dominant regional covariance patterns identified in the NL1 and PD1 groups and corresponding EC values measured at the 18% cost threshold, which was the minimum necessary for the 2 graphs to be fully connected. For NL1, nodal EC values (Fig. 2A, top) did not correlate (Fig. 2B) with region weights on the ROI-based PDRP identified in the spatial covariance analysis (R2 = 0.0003, P = 0.88; Pearson correlations). By contrast, for PD1, the correlation between EC values (Fig. 2A, bottom) and absolute PDRP region weights (Fig. 2D) was significant at the 18% cost threshold (R2 = 0.51, P < 0.001). Indeed, in PD1, EC correlations with absolute PDRP region weights were significant (Fig. 2E) across the cost range of 18–50% (R2 > 0.51). Similar EC correlations were seen in the PD2 and PD3 testing sets (R2 > 0.42; Fig. 2E). Indeed, EC values for PD2 and PD3 correlated significantly with one another (R2 > 0.65) and with corresponding PD1 values (R2 > 0.54) across the cost range. The close relationship between PDRP region weights and corresponding EC values for the various PD samples suggests that the disease topography is a prominent component of overall resting network activity in patients but not necessarily in healthy subjects.
Figure 2.
Correlations between eigenvalue centrality (EC) and region weights on metabolic covariance patterns. (A) Display of EC maps identified in metabolic brain images from the NL1 normal group (top) and the PD1 patient group (bottom). (B) In NL1, there was no relationship (r = −0.017) between local EC values and absolute region weights on the Parkinson’s disease-related covariance pattern (PDRP). (C) EC correlations with PDRP region weights are presented for NL1 and NL2, over the range of cost thresholds on the graph (see Methods). (D) Local EC values for PD1 network (A, bottom panel) correlated significantly with absolute PDRP region weights (r = 0.72, P < 0.001). (E) Significant EC correlations with absolute PDRP region weights were also present in the PD2 and PD3 groups (r > 0.65, P < 0.001). Correlations in all 3 PD groups were maintained across the range of cost thresholds.
Analogous findings were observed in non-human primates with and without MPTP-induced parkinsonism (Supplementary Fig. 2). A significant topographic correlation was observed (R2 = 0.27, P < 0.001) between the EC vector identified in the MPTP group and corresponding absolute region weights on the ROI-based PRP topography (Supplementary Fig. 2B, bottom); the correlation between these measures was not significant (R2 = 0.021, P = 0.30) in the NL monkey group (Supplementary Fig. 2B, top). Indeed, significant EC correlations with absolute PRP region weights (Supplementary Fig. 2C) were evident in MPTP monkeys as varying cost thresholds (r > 0.47, P < 0.001; cost 24–50%), whereas such correlations were not found in the NL group despite varying cost (r > −0.18, P > 0.30; cost 24–50%). In summary, these findings suggest that a consistent relationship exists between graph theoretic disease networks, represented by the EC vectors identified in scans from human PD patients and parkinsonian macaques, and the corresponding disease-related covariance patterns.
PD-related Metabolic Network: Core-periphery Structure
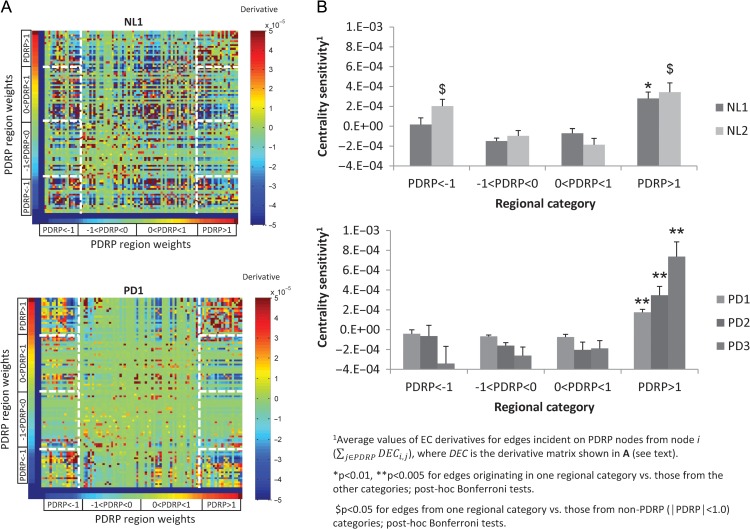
We next examined the nodal architecture of the disease network using a graph visualization approach described elsewhere (Correa et al. 2012). This method, used previously in the analysis of social networks, ranks the edges of a graph by the sensitivity of EC for a given node to small changes in the number of edges that connect it to the others (see Supplementary Notes). Thus, using a variational approach, we computed partial derivatives of EC with respect to nodal degree for each incident edge. The resulting sensitivity values for each region (Fig. 3A) were sorted by PDRP region weight. In this way, we analyzed the relationship between the sensitivity of a given node to perturbation in the number of afferent connections (incident edges) and local metabolic contribution to the network, that is, the corresponding region weight. Regions contributing to the PDRP, that is, those with absolute weights ≥ 1.0 (z-scale) were classified as being either metabolically active (region weight ≥ +1.0) or underactive (region weight ≤ −1.0). Regions were considered to be non-contributing to the pattern for absolute region weight < 1.0; weights of these regions on the PDRP were likewise dichotomized by sign.
Figure 3.
Eigenvector centrality sensitivity: relationship to region weight category. (A) Partial derivatives of eigenvector centrality (EC) computed for each edge in graphs constructed from the NL1 (top) and PD1 (bottom) scan data. The partial derivative of EC (i,j) represented the effect on node j’s centrality when node i’s degree is minimally increased (see text). Spectrum plots of EC derivative values for each group were sorted by the sign of the corresponding PDRP region weights. Values corresponding to edges between salient PDRP nodes (absolute region weights ≥ 1.0) are outlined by white dashed lines. [Eigenvector centrality sensitivity for PDRP edges was computed by averaging EC derivatives incident upon network nodes. This was done by taking row means of values delineated by the vertical dashed lines on the far left and right of the array. The resulting values were grouped for further analysis by the magnitude and sign of the corresponding PDRP region weights (horizontal dashed lines).] (B) Centrality sensitivity values (mean ± SE) for edges originating from the salient PDRP nodes with brain regions grouped according to the magnitude and sign of their respective pattern weight. In both NL1 and NL2, the 2 groups of healthy subjects (top), centrality sensitivity differed significantly across the region weight categories (P < 0.001; 1-way ANOVA). Higher sensitivity values were associated with edges incident upon metabolically active network regions (i.e., salient PDRP nodes with positive region weights; see text). Analogous differences in centrality sensitivity across regional categories (P < 0.001) were observed for PD1, PD2, and PD3, the 3 patient groups (bottom). In each of the patient samples, sensitivity values were greater for edges incident on nodes in the metabolically active category as compared to those in the other regional categories (P < 0.005; post hoc Bonferroni tests). See text for details.
Average sensitivity values for nodes in the 4 region weight categories are displayed in Figure 3B. Significant differences in sensitivity were observed across the categories in both the NL1 and NL2 groups (NL1: F(3,91) = 13.1, P < 0.001; NL2: F(3,91) = 12.35, P < 0.001; 1-way ANOVA). In NL1 (Fig. 3B, top) sensitivity was high for the active PDRP regions compared to the remaining categories (P < 0.01; post hoc Bonferroni test). In NL2, nodal sensitivity was high in both active and underactive PDRP regions (|region weight| > 1.0) compared to the non-contributing regions (P < 0.05; post hoc Bonferroni tests). No significant difference in sensitivity was observed between the active and underactive PDRP regions (P = 1.0; post hoc Bonferroni test) in NL2.
Significant differences in sensitivity across metabolic weight categories were also present in the 3 PD samples (PD1: F(3,91) = 16.6, P < 0.001; PD2: F(3,91) = 10.4, P < 0.001; PD3: F(3,91) = 16.4, P < 0.001). In each patient group, increased nodal sensitivity was evident only in the metabolically active PDRP regions (P < 0.005; post hoc Bonferroni tests), while significant differences were not seen in the remaining categories (P = 1.0; post hoc Bonferroni tests).
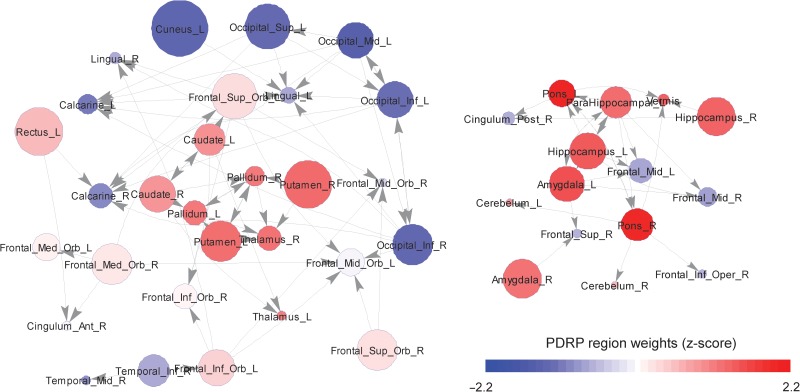
The significantly high sensitivity of the edges connecting metabolically active PDRP nodes suggests the presence of central hubs (see Fig. 1A) and possible core-periphery organization. To visualize this aspect of network architecture, we restricted the graph to edges representing only the top 1% of their computed sensitivity values (see Supplementary Note B). This approach revealed the presence of 2 nodal communities. The major nodal cluster (Fig. 4, left), representing basal ganglia-thalamocortical connections, was populated almost exclusively by metabolically active (red) PDRP nodes. Members of this nodal community were connected via edges that were not only highly sensitive (i.e., had large magnitude centrality derivatives), but also reinforcing (i.e., the centrality derivatives had positive values). Interestingly, the periphery was populated to a large extent by metabolically underactive (blue) PDRP nodes. While these nodes tended to have high EC values (represented in Fig. 4 by disc radius), the lower centrality sensitivity values denoted weaker node-to-node interactions between metabolically underactive PDRP regions compared to the stronger interactions exhibited by their metabolically active counterparts (Fig. 3B, bottom). Thus, overall, this subgraph (Fig. 4, left) exhibited a discrete core comprised of metabolically active PDRP nodes in the caudate, putamen, globus pallidus, and thalamus, with relatively underactive cortical nodes in the periphery.
Figure 4.
Graph visualization based on the top 1% of centrality derivatives. In this radial graph display of the PD1 connectivity data, nodes connected by edges with high centrality sensitivity were positioned close to the center, while those with lower sensitivity were positioned in the periphery. In this directed graph, incident edges are represented by arrows; the radius of each node is proportioned to local EC. For each network node, corresponding PDRP region weights were color-coded such that metabolically active regions (PDRP weights ≥1.0) were depicted in red while relatively underactive regions (PDRP weights ≤−1.0) were depicted in blue. This display revealed the presence of 2 distinct nodal clusters: a prominent basal ganglia-thalamocortical subnetwork with a distinct core-periphery mesostructure (left), and a smaller discrete subnetwork involving primarily ponto-cerebellar and limbic interconnections (right). Both clusters were centered around cores defined by high magnitude mutually reinforcing node-to-node interactions (i.e., edges with relatively high, positive centrality derivative values). Interestingly, the core nodes identified in this display corresponded almost exclusively to metabolically active (red) PDRP regions, whereas the underactive (blue) regions were localized mainly to the network periphery.
A minor nodal cluster (Fig. 4, right) was defined by a set of high sensitivity, mutually reinforcing correlations linking the pons, cerebellum, amygdala, and hippocampus. While core-periphery structure was not as well-defined in this subgraph, these nodes were influential (high EC) and also metabolically active. The remaining members of this community, represented mainly by areas of the frontal cortex and cingulate gyrus, were less influential (lower EC) and also relatively underactive. We additionally note that despite the similarity between the PD1 and NL1 partial derivative matrices, core-periphery nodal organization was not evident in the latter group.
Parkinson’s Disease is Characterized by Excessive Small-worldness
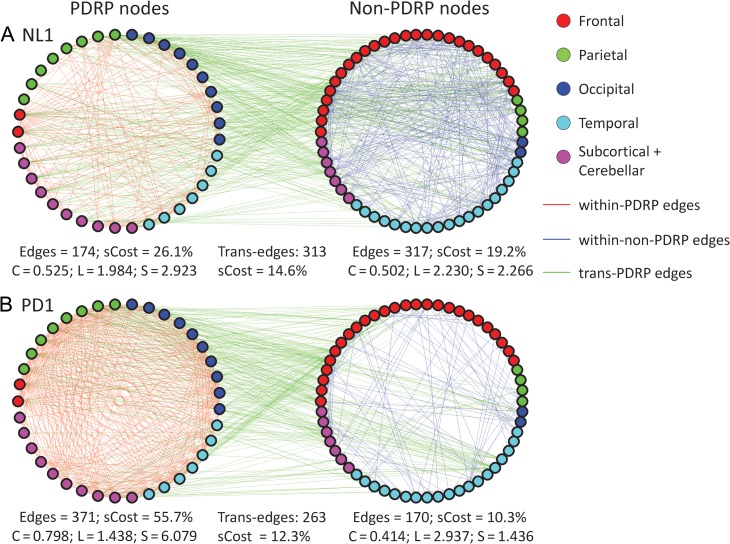
We next considered the potential impact of the disease network on brain information processing. Many biological networks exhibit the small-world property as a means of maintaining efficient information processing in response to perturbation (Bassett and Bullmore 2016). To determine whether this feature differed for PD and healthy subjects, we partitioned the graph data from each group into 2 discrete subspaces. As above, the PDRP subspace was defined by the influential network regions (absolute weights ≥ 1.0); the non-PDRP subspace was composed of the remaining brain regions (absolute weights < 1.0). Analysis of graph data from PD1 and NL1 (Fig. 5) disclosed increases in the clustering coefficient (C) and number of edges for the PDRP subspace in the patient sample (PD1: C = 0.798, edges = 371; NL1: C = 0.525, edges = 174). By contrast, the characteristic path length (L) for the PDRP subspace was reduced in the same group (PD1: L = 1.438; NL1: L = 1.984), with corresponding increases in the small-worldness coefficient (S) (PD1: S = 6.079; NL1: S = 2.923). Indeed, permutation analysis (Supplementary Fig. 3) revealed that the observed group differences in the PDRP subspace, as well as the corresponding group × subspace interaction effects, were significant over the cost range (P < 0.05; 10 000 permutations). Group differences were not observed, however, in the non-PDRP subspace or over the brain as a whole. Differences between groups in the number of trans-edges connecting PDRP and non-PDRP regions were likewise not significant (P > 0.05; 10 000 permutations at cost 18–50%).
Figure 5.
Node-to-node functional connections in the NL1 (A) and PD1 (B) groups in the PDRP and non-PDRP spaces. The PDRP space (left) is defined by nodes (n = 37) with absolute region weights greater than or equal to 1, whereas the non-PDRP space (right) is defined by nodes (n = 58) with absolute region weights below 1. The edges that connect PDRP nodes to PDRP nodes are shown in red. The edges that connect non-PDRP nodes to non-PDRP nodes are blue. The edges that connect PDRP nodes and non-PDRP nodes (trans-edges) are shown in green. The total number of edges and the number of edges divided by the number of possible edges (the subgraph wiring cost, sCost) are shown at the bottom of each subgraph along with corresponding values for the clustering coefficient (C) and characteristic path length (L). Enhancement of the small-worldness (S) with increased clustering and reduced path length is seen in the PDRP space of the PD1 graph. Analogous changes were also seen in the PD2 and PD3 graphs (Supplementary Fig. 4). [In each subgraph, the small circles represent vertices (ROIs defined by the AAL atlas; see Methods); brain location is represented by color (red: frontal, green: parietal, blue: occipital, cyan: temporal, pink: subcortical).]
Similar differences between patient and control network parameters (Supplementary Fig. 4) were also seen in the PD2 and PD3 testing samples relative to corresponding NL2 values. As above, permutation analysis (Supplementary Figs. 5 and 6) disclosed increases in PDRP clustering and number of edges, and corresponding path length reductions, in each of the patient validation samples. PDRP small-worldness was likewise abnormally increased in both patient groups. Group differences in these measures and related group × subspace interaction effects were significant over most of the cost range (P < 0.05; 10 000 permutations). Group differences in network parameters were not consistently different for the non-PDRP subspace or for the whole brain.
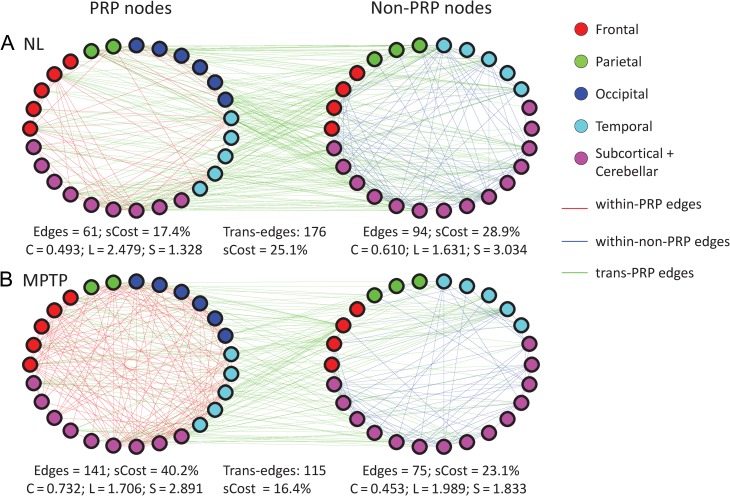
Analogous network differences (Fig. 6) were also seen in graph data from non-human primates. As in the human, MPTP primates (Supplementary Fig. 7) exhibited significant increases in clustering within the PRP subspace (MPTP: C = 0.732; NL: C = 0.493; P < 0.05; 10 000 permutations at cost 24–42%). Small-worldness was likewise increased within the PRP subspace (MPTP: S = 2.891; NL: S = 1.328; P < 0.05 at cost 24–29, 31–33%). Group differences in the non-PRP subspace were overall not significant.
Figure 6.
Node-to-node connections in normal and parkinsonian macaques. By analogy to the human, a specific network space (left) was defined by the nodes (n = 27) with absolute region weights on the primate parkinsonism-related covariance pattern (PRP, see text) greater than 0.8. Likewise, a non-PRP testing space (right) was defined by the nodes (n = 26) with absolute region weights below this threshold. The edges that connect PRP nodes to other PRP nodes are shown in red. The edges that connect PRP nodes and non-PRP nodes (trans-edges) are shown in green. The total number of edges and the number of possible edges (the subgraph wiring cost, sCost) are shown at the bottom of each graph, along with corresponding values for clustering coefficient (C) and characteristic path length (L). An increase in small-worldness (S) is seen in the PRP space of the parkinsonian macaque. [In each graph, the small circles represent ROIs defined according to the INIA19 template (Rohlfing et al. 2012); brain location is represented by color (red: frontal, green: parietal, blue: occipital, cyan: temporal, pink: subcortical).]
Discussion
This report highlights the topographic similarity that exists between the absolute value of PDRP region weights and corresponding nodal EC measures. Indeed, this relationship was identified in the PD subjects whose metabolic scan data were used in the original PDRP derivation (Ma et al. 2007), and then replicated in 2 additional independent cohorts comprised of early and more advanced stage patients, as well as in non-human primates with experimental parkinsonism. EC for a given node represents the probability that the node receives random traffic through the network after a sufficient number of steps have passed (Borgatti 2005). Thus, the current study shows that the brain regions with high magnitude loadings (region weights) on the disease-related spatial covariance pattern are also the sites of high information flow through the network in the equilibrium state.
Organizational Structure of the PD Metabolic Network
A natural disparity exists between EC vector components (i.e., nodal centrality values), which are positive by virtue of the Perron Frobenius Theorem (Meyer 2000), and signed region weights on the corresponding spatial covariance patterns. Indeed, significant correlations with the graph theoretic centrality measures were observed only with region weight magnitude (absolute value) but not with the corresponding signed values. EC provides information concerning the relative importance of a given network node. Nodal centrality is, however, a positive measure that correlates with region weights of comparable magnitude (absolute value), whether positive or negative, on the corresponding spatial covariance pattern. What functional characteristics, if any, distinguish metabolically active from underactive network regions? To answer this question, we considered the possibility that nodal metabolic activity within a specific network is determined to at least some degree by its topology. Indeed, based upon the heuristic in Figure 1A, we predicted that synaptic activity, and concomitantly local glucose consumption, would be relatively increased in core regions with dense, mutually facilitating nodal interactions. By contrast, local metabolic activity was expected to be lower in weakly connected peripheral nodes.
To test this hypothesis, we computed partial derivatives of EC at each node with respect to the number of incident edges as a measure of centrality sensitivity. The resulting values allowed us to rank relationships between pairs of nodes by the impact that one node has on the centrality of the other (Correa et al. 2012). According to this approach, positive sensitivity values denoted mutually facilitating nodal interactions, whereas negative sensitivity values denoted inhibitory or competitive relationships between nodes. A striking observation in our study was that in node-to-node facilitation within the PDRP subspace (i.e., edges with positive EC derivatives pointing to PDRP nodes) was detected almost exclusively in metabolically active (“red”) PDRP regions, while the underactive (“blue”) PDRP regions were associated with low-magnitude nodal interactions that had summed derivatives that were either 0 or slightly negative. We note that relatively active subcortical regions also contribute to the previously characterized healthy aging-related covariance topography (Moeller et al. 1996). The presence of such nodes in both networks may explain why significant increases in positive centrality sensitivity values were also seen in healthy subjects (Fig. 3B), albeit of smaller magnitude than in PD.
Based upon the sensitivity measurements, we suggested that the subgraph defined by high sensitivity, that is, mutually facilitating nodal interactions within the PDRP space, can be viewed as a compact, densely connected core zone. The periphery, by contrast, is defined by node-to-node interactions that are either substantially weaker or altogether absent. Region weights on a given covariance pattern represent the activity of a specific regional component relative to the mean for the overall topography. We found that correlations of region weights with EC were valid only for absolute values—an observation that we confirmed (data not shown) using EC vectors from adjacency matrices constructed based on mutual information, an intrinsically positive functional distance metric (Muskulus et al. 2009). This indicates that EC values do not relate to nodal metabolic activity within the network as represented by the sign of the corresponding region weight. The data suggested, however, that while region weight sign had no clear influence on the centrality of a given node, it did relate its sensitivity to small perturbations in input from other nodes. This in turn was reflected in the core property (“coreness”) of the node within the overall network.
The PDRP was derived from a PCA-based algorithm (Eidelberg 2009; Spetsieris and Eidelberg 2011). As with other multivariate approaches, PCA-based covariance topographies are defined by the loadings (region weights) on the resulting covariance patterns which may have positive or negative (or 0) values. It is noteworthy that PCA inherently does not impose an orientation on the eigenvector read-out. Thus, region weight sign can be reversed without altering the intrinsic relational topography (although in that instance the associated subject scores would have to be multiplied by −1). Moreover, the biological significance of positive (“red”) versus negative (“blue”) region weights in PCA-based disease topographies has been questioned with regard to the global normalization procedure performed as part of the model (Borghammer et al. 2008, 2009). The idea that the disease-related covariance topographies identified using PCA reflect normalization artifacts has been discounted based upon theoretical as well as empiric considerations (Ma et al. 2009; Spetsieris and Eidelberg 2011; Dhawan et al. 2012). In this study, graph theory independently identified the metabolically overactive (“red”) PDRP components as the facilitatory network core, while the relatively underactive (“blue”) components were identified as its periphery. In this context, graph theory provides a deeper understanding of the organization of the disease network than routine PCA.
This is illustrated by the structure of the graph that we created based upon edges defined by the top 1% of the measured sensitivity values (see Supplementary Notes). We used a standard display package to visualize the disease network by ranking the graph edges by the corresponding sensitivity values (Fig. 4). To simplify the visualization, we limited the display to the top 1% of measured sensitivity values, omitting any unconnected (“dangling”) nodes. The chosen layout thus positioned the most sensitive facilitating connections centrally, whereas lower ranked interactions were placed at varying distances from center based upon their relative sensitivity values. Importantly, the graph was constructed using only centrality derivative data; information regarding nodal EC values or the magnitude and sign of the corresponding PDRP region weights was not used to shape the topology of the display. Even so, the resulting graph was substantially “centralized”, with an uneven distribution of sensitivity values across the network space (Wasserman and Faust 1994).
The visualized graph exhibited a number of interesting organizational features. Our visualization approach revealed two discrete nodal clusters as part of the overall network topology: a major module corresponding to the cortico-striatal-pallido-thalamo-cortical (CSPTC) system and a minor module involving ponto-cerebello-limbic pathways. Each module is likely to possess distinct functional correlates. Increased metabolic activity in the core nodes of the major module (Fig. 4, left) is a stereotyped feature of the PDRP, which consistently correlates with akinetic-rigid clinical manifestations (Eidelberg 2009; Niethammer and Eidelberg 2012). The minor module (Fig. 4, right), by contrast, involved abnormal ponto-cerebello-limbic nodal interactions that have been linked to other clinical manifestations of the disorder. Indeed, parkinsonian tremor and the placebo response have been associated with functional changes in the pathways that connect these nodes (Mure et al. 2011; Ko, Feigin et al. 2014). It is noteworthy that there was virtually no spatial overlap between these metabolic patterns (r = 0.03; topographical similarity test (Ko, Spetsieris et al. 2014)). Nonetheless, a significant correlation (r = 0.41, P = 0.05; Pearson’s correlation) was observed between expression levels for these 2 topographies measured at baseline in 23 PD subjects who received sham surgery as part of a randomized double-blind gene therapy trial (LeWitt et al. 2011; Ko, Feigin et al. 2014; Niethammer et al. 2017). Moreover, expression values for the 2 patterns trended together (r = 0.39, P = 0.07) in these subjects when they were rescanned under blinded conditions 6 months following sham surgery. This suggests that interactions between discrete nodal subpopulations within the module can occur under specific conditions. The clinical correlates of such interactions are not known.
From a topological standpoint, the 2 network modules exhibited similarities as well as differences. Both contained centralized clusters of interconnected mutually facilitating nodes, which, in aggregate, defined the network core. As demonstrated above, the major module exhibited a well-defined core versus periphery structure: a core defined by a dense, metabolically active collection of nodes with strong functional interactions involving the basal ganglia, thalamus, and frontal cortex. This module also contained a periphery that was defined by a discrete ring of metabolically underactive cortical nodes with relatively weak functional interactions. It remains nonetheless unclear whether the minor module possesses a defined peripheral zone, and whether its nodes interact with the core of the major module. The answers to these questions may impact the design of novel network-based treatment strategies for PD, particularly those targeting non-motor manifestations of the disorder.
The disease network was thus characterized by 2 independent core clusters, each populated largely by metabolically active PDRP regions. Regional glucose metabolism measured in the resting state with FDG PET is known to be an index of afferent synaptic activity (Sokoloff 1999; Lin et al. 2008). Because of this relationship, it is perhaps not surprising that densely connected mutually facilitating core nodes exhibit concomitant increases in local metabolic activity. We note that a close topographic correlation was present in all samples between the vectors of nodal degree and EC (r > 0.90, P < 0.001 for PD1–PD4 and NL1–NL2), corresponding respectively to the first and last iterations of the power method (ƙ = 1 and ƙ = 7) employed to extract the principal eigenvector of the adjacency matrix (see Supplementary Fig. 8). Even so, the core-periphery structure displayed in Figure 4 was not discerned until the fourth power interaction (ƙ = 4, Supplementary Fig. 8E). Indeed, the segregation of metabolically active nodes into a discrete core cluster did not become apparent until the final 3 power iterations (ƙ ≥ 5). This suggests that the core-periphery structure of the PD network is dependent more on the pattern of node-to-node interactions than on the number of connections (i.e., degree centrality) per se.
The PD Metabolic Network: Pathological Exaggeration of the Small-world Phenomenon
As with many biological networks, information processing in the PDRP exhibits small-world properties. Indeed, small increases in clustering and reductions in path length were consistently evident with the PDRP space of healthy subjects, relative to corresponding values for the rest of the brain (i.e., the “non-PDRP” space). These changes are compatible with “small-world” processing within the PDRP space in healthy individuals. Given that cost is relatively conserved in the PDRP and non-PDRP spaces of healthy subjects, it is likely that the small-world property of the normal PDRP subspace represents an adaptation to the high information processing requirements of these brain regions in the resting state. That said, measures of small-worldness differed when independently evaluated in the patient samples. In each PD group, there was a substantial increase in clustering in the PDRP relative to non-PDRP space. Moreover, clustering in the PDRP space was also significantly greater in PD compared to healthy subjects, with analogous reductions in path length. Clustering in the non-PDRP space was similar for the patient and control groups, whereas corresponding path length was relatively greater than normal in this “non-disease” space. In aggregate, these findings point to a substantial increase in the small-world property of the PDRP network space in the disease brain.
Several additional features accompanied the increase in small-worldness that was seen in the PDRP space. In the normal samples, there was little difference in wiring cost for information processing in the PDRP space relative to the rest of the brain (difference in wiring cost: NL1 7%, NL2 2.5%). In PD, by contrast, the increase in subgraph wiring cost was strikingly large for the PDRP space (difference: PD1 45%, PD2 30.1%, PD3 23.6%). The escalation of wiring cost within the PDRP space is compatible with the presence of a dense metabolically active core zone, as was demonstrated for this network. It is likely, therefore, that under pathological conditions the increased small-world property of the PDRP becomes a liability by increasing the energetic cost of information processing at critical core nodes. Interestingly, changes in these descriptors were not limited to the space occupied by the disease network. For example, all 3 patient samples exhibited relative increases in path length in the “non-disease” space, with reduced edge numbers and conserved clustering. While these changes were associated with reductions in overall wiring cost outside the PDRP space, the loss of connections and increased path length in this subgraph points to dysfunction in other brain networks such as those associated with cognitive functioning at more advanced disease stages (Niethammer and Eidelberg 2012; Meles et al. 2015; Mattis et al. 2016).
It is also noteworthy that analogous increases in clustering with diminished path length were seen within the PRP, the abnormal metabolic pattern identified in the spatial covariance analysis of scan data from MPTP and control non-human primates (Ma et al. 2012, 2015; Peng et al. 2016). As in humans with PD, the parkinsonian monkeys exhibited concurrent increases in connected edges and overall wiring cost within the PRP subgraph, as well as a relative increase in path length in the rest of the brain. This suggests that in both species, the small-world property of the disease network is potentiated by dopaminergic loss. That said, in both human PD and experimental parkinsonism, increased small-worldness comes at the price of high wiring cost and diminished processing efficiency within the network space.
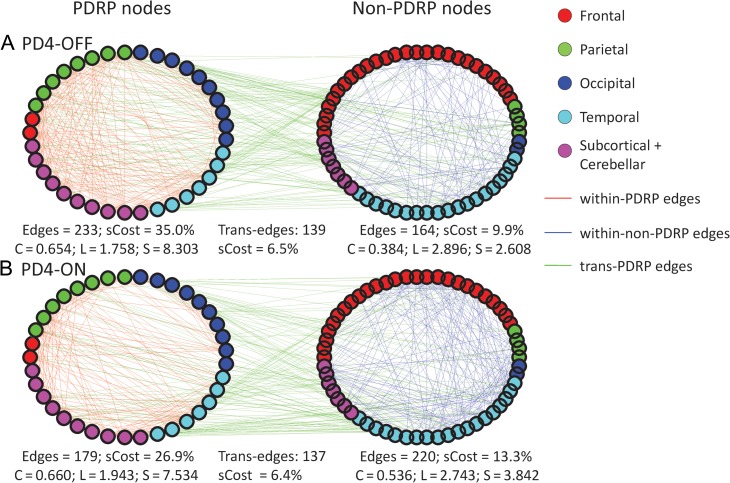
The current findings accord with a previous study showing detrimental effects of dopamine D2 receptor blockade on normal small-world networks (Achard and Bullmore 2007). Moreover, the exaggerated small-worldness that characterized information processing within the PDRP space was only partially corrected by levodopa treatment (Table 1). During clinically effective levodopa infusion, PD4 subjects (Fig. 7) increased characteristic path length toward normal and reduced subgraph wiring cost within the PDRP subspace. Like the other patient samples, PD4 subjects also exhibited significant increases in PDRP clustering and small-worldness when measured in the baseline unmedicated condition (Supplementary Fig. 9). However, unlike characteristic path length, PDRP clustering was unaltered by levodopa, with only minimal change in small-worldness in the treated condition (Table 1; Supplementary Fig. 10). The findings thus suggest that the clinically beneficial effects of the drug relate to the improved efficiency and lower cost of information processing within the network space. Pharmacological dopamine repletion, however, likely has minimal impact on the underlying changes in clustering and small-worldness that typify the disease network.
Table 1.
Small-worldness (S) of the study groups estimated within the PDRP network space, non-PDRP network space and the whole brain. The wiring costs were at minimum values that produced the fully connected graphs, i.e., 18% for PD1 and NL1, 12% for PD2, PD3, PD4 and NL2, and 24% for MPTP and NL (for details, see text).
| PD1 | PD2 | PD3 | PD4 [OFF] | PD4 [ON] | MPTP | NL1 | NL2 | NL | |
|---|---|---|---|---|---|---|---|---|---|
| PDRP | 6.079** | 8.477** | 8.109** | 8.303** | 7.534* | 2.891* | 2.923 | 5.286 | 1.328 |
| Non-PDRP | 1.436* | 2.385* | 2.453 | 2.608* | 3.842 | 1.833* | 2.266 | 3.849 | 3.034 |
| Whole brain | 2.416 | 4.002 | 3.589 | 3.833 | 4.016 | 1.986 | 2.457 | 3.837 | 1.952 |
Small-worldness coefficients (S) for each group and subspace. Significant differences in S for the PD groups relative to corresponding control values (P < 0.05, permutation test) at minimum cost (*) or throughout most of the cost range (**). See Supplementary Figures 3, 5–7, 9, and 10 for group differences in S and in the other network parameters, plotted over the cost range.
Figure 7.
Node-to-node functional connections in the PD4 group shown in the off-medication condition (A) and during and intravenous levodopa infusion titrated to achieve maximal motor benefit (B, see text) for the PDRP (left) and non-PDRP (right) spaces. The data suggest that levodopa treatment was associated with an increase in characteristic path length (L) toward normal levels in the PDRP space from the shortened values seen in the PD groups. Nonetheless, treatment had no effect on PDRP space clustering values, which remained elevated compared to normal. However, in the PDRP space, small-worldness (S), sCost and the number of edges declined with levodopa infusion, while concurrently these measures increased in the non-PDRP space. Thus, in the treated state, values for these parameters matched closely those for the NL1 graph (Fig. 5A).
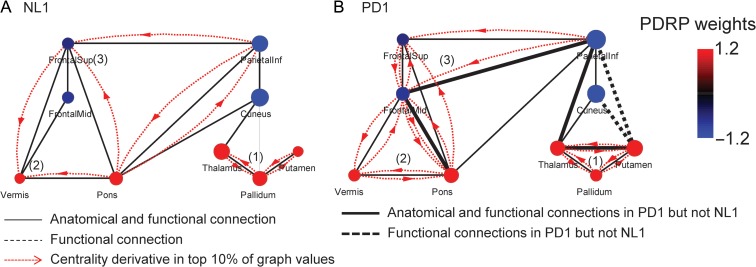
In general terms, the network changes observed in PD can be construed in economic terms. The healthy brain utilizes sparse small-world networks to achieve efficient information processing at reduced energetic cost (Bullmore and Sporns 2009). PD, by contrast, is associated with an abnormal network in which the small-world property is exaggerated with attendant increases in metabolic cost. Indeed, contrary to neurodegenerative disorders such as Alzheimer’s disease, in which extensive loss of “small-worldness” lends to profound behavioral deficits (Stam et al. 2007), PD is associated with an excess of this otherwise favorable network characteristic. Graph visualization (Fig. 8) linking this abnormality to the presence of “closed triples” (triangles) within the disease space reflects anomalous, mutually facilitating functional interconnections between nearest neighbors. This pathological form of information transfer is likely to incur high energetic cost, as indicated by the localization of these pathological links to metabolically active core zones within the disease network.
Figure 8.
Abnormal network-level clustering in Parkinson’s disease. Centrality derivatives can be useful in identifying the location of closed triples, i.e., triangles formed when a node’s nearest neighbors are connected, within a particular subgraph. In the case of the PDRP, in which overall subgraph clustering is increased, functional connections between pairs of network nodes can be assigned based upon the top 10% of the derivative values. (A) In the healthy NL1 group, 3 discrete sets of interconnected nodes (open triples) are evident involving (1) the putamen, globus pallidus, and the thalamus, (2) the pons, cerebellar vermis, and premotor/prefrontal cortex, and (3) superior and middle frontal gyri, and inferior parietal lobule. (B) In the PD1 group, additional edges (heavy lines) were formed sealing each of the triples off as a discrete triangle. These edges denote specific node-to-node functional interactions present in PD but not in healthy subjects. Notably, the closed triples (triangles) in (1) and (2) were located within the core zones identified in the structural analysis of the PD network (Fig. 4). These were formed by abnormal functional connections linking the nearest neighbors of core nodes through bidirectional mutually facilitating interactions (red arrows). [Edges conforming to known anatomical connections between nodes are represented by black solid lines. Edges corresponding to functional links between nodes “without” corresponding direct anatomical correlations are represented by dotted lines. Corresponding edge values are represented by red curved arrows.]
Notably, analogous core-periphery topologies were not discerned in graphs from either of the normal NL1 or NL2 samples. Indeed, the data suggest that these changes constitute a distinct maladaptive response to the underlying pathological process, which parallel the progression of the illness. Increases in the small-world property are associated with synchronization of nodal discharges and spontaneous oscillatory behavior in experimental systems (Arenas et al. 2008), phenomena that are also characteristically seen in electrophysiological studies of non-human primates with experimental parkinsonism and in recordings from human PD subjects undergoing deep brain stimulation (DBS) surgery (Wichmann and Dostrovsky 2011; Guridi and Alegre 2017). Although refractory to acute dopaminergic pharmacotherapy, this feature of the PD functional pathology may be amenable to interventions that directly interrupt such anomalous pathways within the network core. Indeed, knowledge of local network architecture may help in the design of new therapies for PD and other circuit disorders (Ko, Choi et al. 2014).
Finally, we emphasize that our data were derived from an analysis of metabolic brain data using a standardized ROI approach. Together, the combination of the relatively high resting signal-to-noise of FDG PET and the association of local radiotracer uptake with afferent synaptic activity (Feigin et al. 2007; Lin et al. 2008; cf. Patel et al. 2014; Dienel 2017) proved conducive to the detection of abnormal nodal interactions at the group level. That being said, voxel-based EC maps and disease-related functional topographies can be generated with resting-state fMRI techniques (Lohmann et al. 2010; Vo et al. 2017). Relationships between nodal EC values and region weights on fMRI-based disease patterns need to be rigorously examined, as well as similarities and differences in their respective graph theoretic topologies.
Supplementary Material
Notes
The authors wish to thank Dr Vijay Dhawan (The Feinstein Institute for Medical Research, Manhasset, NY, USA) for overseeing the human PET studies. We thank Dr Doris Doudet (University of British Columbia, Vancouver, BC, Canada) for providing the non-human primate scans, and Drs Yilong Ma and Shichun Peng (The Feinstein Institute for Medical Research, Manhasset, NY, USA) for their valuable contributions to the regional analysis of these data. Special thanks to Mr Peter G. Brown for critical editorial support. Lastly, we are grateful to Ms Yoon Young Choi and Ms Toni Fitzpatrick for their indispensable help in manuscript preparation. Conflict of Interest. Dr Eidelberg serves on the scientific advisory board and has received honoraria from The Michael J. Fox Foundation for Parkinson’s Research; is listed as coinventor of patents re: Markers for use in screening patients for nervous system dysfunction and a method and apparatus for using same, without financial gain; and has received research support from the NIH (NINDS, NIDCD, NIAID) and the Dana Foundation.
Funding
Aspects of this work were supported by the National Institute of Neurological Disorders and Stroke (P50 NS 071675 [Morris K. Udall Center of Excellence for Parkinson’s Disease Research at The Feinstein Institute for Medical Research] to D.E.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the National Institute of Neurological Disorders and Stroke. The sponsor did not play a role in study design, collection, analysis and interpretation of data, writing of the report or in the decision to submit the paper for publication.
References
- Achard S, Bullmore E. 2007. Efficiency and cost of economical brain functional networks. PLoS Comput Biol. 3:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas A, Diaz-Guilera A, Kurths J, Moreno Y, Zhou C. 2008. Synchronization in complex networks. Phy Rep. 469:93–153. [Google Scholar]
- Asanuma K, Tang C, Ma Y, Dhawan V, Mattis P, Edwards C, Kaplitt MG, Feigin A, Eidelberg D. 2006. Network modulation in the treatment of Parkinson’s disease. Brain. 129:2667–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Bullmore ET. 2016. Small-world brain networks revisited. Neuroscientist. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgatti SP. 2005. Centrality and network flow. Soc Netw. 27:55–71. [Google Scholar]
- Borghammer P, Cumming P, Aanerud J, Forster S, Gjedde A. 2009. Subcortical elevation of metabolism in Parkinson’s disease—a critical reappraisal in the context of global mean normalization. Neuroimage. 47:1514–1521. [DOI] [PubMed] [Google Scholar]
- Borghammer P, Jonsdottir KY, Cumming P, Ostergaard K, Vang K, Ashkanian M, Vafaee M, Iversen P, Gjedde A. 2008. Normalization in PET group comparison studies—the importance of a valid reference region. Neuroimage. 40:529–540. [DOI] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. 2009. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 10:186–198. [DOI] [PubMed] [Google Scholar]
- Correa CD, Crnovrsanin T, Ma KL. 2012. Visual reasoning about social networks using centrality sensitivity. IEEE Trans Vis Comput Graph. 18:106–120. [DOI] [PubMed] [Google Scholar]
- Correa CD, Crnovrsanin T, Ma KL, Keeton K. 2009. The derivatives of centrality and their applications in visualizing social networks. Citeseer.
- Dhawan V, Tang CC, Ma Y, Spetsieres P, Eidelberg D. 2012. Abnormal network topographies and changes in global activity: absence of a causal relationship. Neuroimage. 63:1827–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dienel GA. 2017. Lack of appropriate stoichiometry: strong evidence against an energetically important astrocyte-neuron lactate shuttle in brain. J Neurosci Res. 95:2103–2125. [DOI] [PubMed] [Google Scholar]
- Eckert T, Barnes A, Dhawan V, Frucht S, Gordon MF, Feigin AS, Eidelberg D. 2005. FDG PET in the differential diagnosis of parkinsonian disorders. Neuroimage. 26:912–921. [DOI] [PubMed] [Google Scholar]
- Eidelberg D. 2009. Metabolic brain networks in neurodegenerative disorders: a functional imaging approach. Trends Neurosci. 32:548–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feigin A, Kaplitt MG, Tang C, Lin T, Mattis P, Dhawan V, During MJ, Eidelberg D. 2007. Modulation of metabolic brain networks after subthalamic gene therapy for Parkinson’s disease. Proc Natl Acad Sci USA. 104:19559–19564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girvan M, Newman ME. 2002. Community structure in social and biological networks. Proc Natl Acad Sci USA. 99:7821–7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guridi J, Alegre M. 2017. Oscillatory activity in the basal ganglia and deep brain stimulation. Mov Disord. 32:64–69. [DOI] [PubMed] [Google Scholar]
- Habeck C, Stern Y, Alzheimer’s Disease Neuroimaging I . 2010. Multivariate data analysis for neuroimaging data: overview and application to Alzheimer’s disease. Cell Biochem Biophys. 58:53–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano S, Asanuma K, Ma Y, Tang C, Feigin A, Dhawan V, Carbon M, Eidelberg D. 2008. Dissociation of metabolic and neurovascular responses to levodopa in the treatment of Parkinson’s disease. J Neurosci. 28:4201–4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtbernd F, Gagnon JF, Postuma RB, Ma Y, Tang CC, Feigin A, Dhawan V, Vendette M, Soucy JP, Eidelberg D, et al. 2014. Abnormal metabolic network activity in REM sleep behavior disorder. Neurology. 82:620–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtbernd F, Ma Y, Peng S, Schwartz F, Timmermann L, Kracht L, Fink GR, Tang CC, Eidelberg D, Eggers C. 2015. Dopaminergic correlates of metabolic network activity in Parkinson’s disease. Hum Brain Mapp. 36:3575–3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini SM, Hoeft F, Kesler SR. 2012. GAT: a graph-theoretical analysis toolbox for analyzing between-group differences in large-scale structural and functional brain networks. PLoS One. 7:e40709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Tang C, Feigin A, Lesser M, Ma Y, Pourfar M, Dhawan V, Eidelberg D. 2007. Changes in network activity with the progression of Parkinson’s disease. Brain. 130:1834–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A, Daniel S, Kilford L, Lees A. 1992. Accuracy of clinical diagnosis of idiopathic Parkinson’s disease: a clinico-pathological study of 100 cases. J Neurol Neurosurg Psychiatry. 55:181–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries MD, Gurney K. 2008. Network ‘small-world-ness’: a quantitative method for determining canonical network equivalence. PLoS One. 3:e0002051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdain VA, Tang CC, Holtbernd F, Dresel C, Choi YY, Ma Y, Dhawan V, Eidelberg D. 2016. Flow-metabolism dissociation in the pathogenesis of levodopa-induced dyskinesia. JCI Insight. 1:e86615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JH, Choi YY, Eidelberg D. 2014. Graph theory-guided transcranial magnetic stimulation in neurodegenerative disorders. Bioelect Med. 1:15–18. [Google Scholar]
- Ko JH, Feigin A, Mattis PJ, Tang CC, Ma Y, Dhawan V, During MJ, Kaplitt MG, Eidelberg D. 2014. Network modulation following sham surgery in Parkinson’s disease. J Clin Invest. 124:3656–3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JH, Lee CS, Eidelberg D. 2017. Metabolic network expression in parkinsonism: clinical and dopaminergic correlations. J Cereb Blood Flow Metab. 37:683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JH, Spetsieris P, Ma Y, Dhawan V, Eidelberg D. 2014. Quantifying significance of topographical similarities of disease-related brain metabolic patterns. PLoS One. 9:e88119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeWitt PA, Rezai AR, Leehey MA, Ojemann SG, Flaherty AW, Eskandar EN, Kostyk SK, Thomas K, Sarkar A, Siddiqui MS, et al. 2011. AAV2-GAD gene therapy for advanced Parkinson’s disease: a double-blind, sham-surgery controlled, randomised trial. Lancet Neurol. 10:309–319. [DOI] [PubMed] [Google Scholar]
- Lin TP, Carbon M, Tang C, Mogilner AY, Sterio D, Beric A, Dhawan V, Eidelberg D. 2008. Metabolic correlates of subthalamic nucleus activity in Parkinson’s disease. Brain. 131:1373–1380. [DOI] [PubMed] [Google Scholar]
- Lohmann G, Margulies DS, Horstmann A, Pleger B, Lepsien J, Goldhahn D, Schloegl H, Stumvoll M, Villringer A, Turner R. 2010. Eigenvector centrality mapping for analyzing connectivity patterns in fMRI data of the human brain. PLoS One. 5:e10232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Johnston TH, Peng S, Zuo C, Koprich JB, Fox SH, Guan Y, Eidelberg D, Brotchie JM. 2015. Reproducibility of a Parkinsonism-related metabolic brain network in non-human primates: a descriptive pilot study with FDG PET. Mov Disord. 30:1283–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Peng S, Spetsieris PG, Sossi V, Eidelberg D, Doudet DJ. 2012. Abnormal metabolic brain networks in a nonhuman primate model of parkinsonism. J Cereb Blood Flow Metab. 32:633–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Tang C, Moeller JR, Eidelberg D. 2009. Abnormal regional brain function in Parkinson’s disease: truth or fiction? Neuroimage. 45:260–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Tang C, Spetsieris P, Dhawan V, Eidelberg D. 2007. Abnormal metabolic network activity in Parkinson’s disease: test-retest reproducibility. J Cereb Blood Flow Metab. 27:597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Tamura K, Koyano KW, Takeuchi D, Adachi Y, Osada T, Miyashita Y. 2011. Direct comparison of spontaneous functional connectivity and effective connectivity measured by intracortical microstimulation: an fMRI study in macaque monkeys. Cereb Cortex. 21:2348–2356. [DOI] [PubMed] [Google Scholar]
- Mattis PJ, Niethammer M, Sako W, Tang CC, Nazem A, Gordon ML, Brandt V, Dhawan V, Eidelberg D. 2016. Distinct brain networks underlie cognitive dysfunction in Parkinson and Alzheimer diseases. Neurology. 87:1925–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meles SK, Tang CC, Teune LK, Dierckx RA, Dhawan V, Mattis PJ, Leenders KL, Eidelberg D. 2015. Abnormal metabolic pattern associated with cognitive impairment in Parkinson’s disease: a validation study. J Cereb Blood Flow Metab. 35:1478–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer CD. 2000. Matrix analysis and applied linear algebra. Philadelphia, PA: Society for Industrial and Applied Mathematics. [Google Scholar]
- Moeller JR, Ishikawa T, Dhawan V, Spetsieris P, Mandel F, Alexander GE, Grady C, Pietrini P, Eidelberg D. 1996. The metabolic topography of normal aging. J Cereb Blood Flow Metab. 16:385–398. [DOI] [PubMed] [Google Scholar]
- Mure H, Hirano S, Tang CC, Isaias IU, Antonini A, Ma Y, Dhawan V, Eidelberg D. 2011. Parkinson’s disease tremor-related metabolic network: characterization, progression, and treatment effects. Neuroimage. 54:1244–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muskulus M, Houweling S, Verduyn-Lunel S, Daffertshofer A. 2009. Functional similarities and distance properties. J Neurosci Methods. 183:31–41. [DOI] [PubMed] [Google Scholar]
- Newman MEJ. 2010. Networks: an introduction. Oxford, UK: Oxford University Press. [Google Scholar]
- Niethammer M, Eidelberg D. 2012. Metabolic brain networks in translational neurology: concepts and applications. Ann Neurol. 72:635–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niethammer M, Tang CC, Feigin A, Allen PJ, Heinen L, Hellwig S, Amtage F, Hanspal E, Vonsattel JP, Poston KL, et al. 2014. A disease-specific metabolic brain network associated with corticobasal degeneration. Brain. 137:3036–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niethammer M, Tang CC, LeWitt PA, Rezai AR, Leehey MA, Ojemann SG, Flaherty AW, Eskandar EN, Kostyk SK, Sarkar A, et al. 2017. Long-term follow-up of a randomized AAV2-GAD gene therapy trial for Parkinson’s disease. JCI Insight. 2:e90133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AB, Lai JC, Chowdhury GM, Hyder F, Rothman DL, Shulman RG, Behar KL. 2014. Direct evidence for activity-dependent glucose phosphorylation in neurons with implications for the astrocyte-to-neuron lactate shuttle. Proc Natl Acad Sci USA. 111:5385–5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S, Ma Y, Flores J, Cornfeldt M, Mitrovic B, Eidelberg D, Doudet DJ. 2016. Modulation of abnormal metabolic brain networks by experimental therapies in a nonhuman primate model of Parkinson’s disease: an application to human retinal pigment epithelial (hRPE) cell implantation. J Nucl Med. 57:1591–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porras G, Li Q, Bezard E. 2012. Modeling Parkinson’s disease in primates: the MPTP model. Cold Spring Harb Perspect Med. 2:a009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME, Mintun MA. 2006. Brain work and brain imaging. Annu Rev Neurosci. 29:449–476. [DOI] [PubMed] [Google Scholar]
- Rohlfing T, Kroenke CD, Sullivan EV, Dubach MF, Bowden DM, Grant KA, Pfefferbaum A. 2012. The INIA19 template and NeuroMaps atlas for primate brain image parcellation and spatial normalization. Front Neuroinform. 6:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinov M, Sporns O. 2010. Complex network measures of brain connectivity: uses and interpretations. Neuroimage. 52:1059–1069. [DOI] [PubMed] [Google Scholar]
- Sokoloff L. 1999. Energetics of functional activation in neural tissues. Neurochem Res. 24:321–329. [DOI] [PubMed] [Google Scholar]
- Spetsieris P, Ma Y, Peng S, Ko JH, Dhawan V, Tang CC, Eidelberg D. 2013. Identification of disease-related spatial covariance patterns using neuroimaging data. J Vis Exp. 76:e50319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spetsieris PG, Eidelberg D. 2011. Scaled subprofile modeling of resting state imaging data in Parkinson’s disease: methodological issues. Neuroimage. 54:2899–2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam CJ, Jones BF, Nolte G, Breakspear M, Scheltens P. 2007. Small-world networks and functional connectivity in Alzheimer’s disease. Cereb Cortex. 17:92–99. [DOI] [PubMed] [Google Scholar]
- Tang CC, Feigin A, Ma Y, Habeck C, Paulsen JS, Leenders KL, Teune LK, Van Oostrom JC, Guttman M, Dhawan V, et al. 2013. Metabolic network as a progression biomarker of premanifest Huntington’s disease. J Clin Invest. 123:4076–4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang CC, Poston KL, Dhawan V, Eidelberg D. 2010. Abnormalities in metabolic network activity precede the onset of motor symptoms in Parkinson’s disease. J Neurosci. 30:1049–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang CC, Poston KL, Eckert T, Feigin A, Frucht S, Gudesblatt M, Dhawan V, Lesser M, Vonsattel JP, Fahn S, et al. 2010. Differential diagnosis of parkinsonism: a metabolic imaging study using pattern analysis. Lancet Neurol. 9:149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi M, Tang CC, Feigin A, De Lucia I, Nazem A, Dhawan V, Eidelberg D. 2016. Automated differential diagnosis of early parkinsonism using metabolic brain networks: a validation study. J Nucl Med. 57:60–66. [DOI] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M. 2002. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 15:273–289. [DOI] [PubMed] [Google Scholar]
- Van Ham F, Wattenberg M. 2008. Centrality based visualization of small world graphs. Com Graphics Forum. 27:975–982. [Google Scholar]
- Vo A, Sako W, Fujita K, Peng S, Mattis PJ, Skidmore FM, Ma Y, Ulug AM, Eidelberg D. 2017. Parkinson’s disease-related network topographies characterized with resting state functional MRI. Hum Brain Mapp. 38:617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman S, Faust K. 1994. Social network analysis: methods and applications. Cambridge: Cambridge University Press. [Google Scholar]
- Watts DJ, Strogatz SH. 1998. Collective dynamics of ‘small-world’ networks. Nature. 393:440–442. [DOI] [PubMed] [Google Scholar]
- Wichmann T, Dostrovsky JO. 2011. Pathological basal ganglia activity in movement disorders. Neuroscience. 198:232–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Yu H, Peng S, Dauvilliers Y, Wang J, Ge J, Zhang H, Eidelberg D, Ma Y, Zuo C. 2014. Consistent abnormalities in metabolic network activity in idiopathic rapid eye movement sleep behaviour disorder. Brain. 137:3122–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.