Abstract
Structural rearrangements of the genome can drive lung tumorigenesis through the generation of fusion genes with oncogenic properties. Advanced genomic approaches have identified the presence of a genetic fusion between fibroblast growth factor receptor 3 (FGFR3) and transforming acidic coiled-coil 3 (TACC3) in non-small cell lung cancer (NSCLC), providing a novel target for FGFR inhibition. To interrogate the functional consequences of the FGFR3-TACC3 fusion in the transformation of lung epithelial cells, we generated a novel transgenic mouse model that expresses FGFR3-TACC3 concomitant with loss of the p53 tumor suppressor gene. Intra-nasal delivery of an Ad5-CMV-Cre virus promoted seromucinous glandular transformation of olfactory cells lining the nasal cavities of FGFR3-TACC3 (LSL-F3T3) mice, which was further accelerated upon loss of p53 (LSL-F3T3/p53). Surprisingly, lung tumors failed to develop in intra-nasally infected LSL-F3T3 and LSL-F3T3/p53 mice. In line with these observations, we demonstrated that intra-nasal delivery of Ad5-CMV-Cre induces widespread Cre-mediated recombination in the olfactory epithelium. Intra-tracheal delivery of Ad5-CMV-Cre into the lungs of LSL-F3T3 and LSL-F3T3/p53 mice however, resulted in the development of lung adenocarcinomas. Taken together, these findings provide in vivo evidence for an oncogenic function of FGFR3-TACC3 in respiratory epithelium.
Introduction
Lung cancer represents the most common malignancy worldwide, and the leading cause of cancer-related death (1). Next-generation sequencing technologies have provided an in-depth characterization of small cell lung cancer (SCLC) (2) and non-small cell lung cancer (NSCLC) subtypes, including adenocarcinoma (ADC) (3) and squamous cell carcinoma (SCC) (4). Combined, these studies have highlighted the highly complex genomic landscape of lung cancer, represented by high rates of somatic alterations, copy number changes and genomic rearrangements (5). Structural genomic rearrangements can lead to the development of fusion genes with oncogenic properties. In the respiratory system, the best characterized of these is the EML4-ALK gene fusion, which is detected in 1-5% of ADC (6). Characterization of the EML4-ALK fusion gene has led to more successful targeted treatments for these gene rearrangement patients (7).
The FGFR3-TACC3 fusion gene has been described in 3-4% of lung SCC (8, 9) and also in nasopharyngeal carcinoma (NPC) (10), glioblastoma (11), cervical cancer (12) and bladder cancer (5, 13). The chromosomal translocation between fibroblast growth factor receptor 3 (FGFR3) and transforming acidic coiled-coiled 3 (TACC3) generates a fusion of the tyrosine kinase domain of FGFR3, upstream of the coiled-coil domain of TACC3, however the exact position of the breakpoint is variable between patients (8, 11, 12, 14). The transforming activity of FGFR3-TACC3 has been suggested to result from alterations in canonical signaling activity (13), loss of microRNA regulation (15), or from loss of maintenance of the mitotic spindle (11). However, the oncogenic potential of the FGFR3-TACC3 fusion in lung epithelium has not yet been assessed in vivo.
Genetically engineered mouse models (GEMMs) of human lung cancer genomic alterations are a powerful in vivo platform to dissect molecular mechanisms underlying tumor initiation and progression (16). Moreover, mouse models that use strategic gene targeting to both amplify and accelerate candidate pathogenic mechanisms have an especially high utility because they allow drug sensitivity and resistance studies to be performed in a timely, systematic and definitive manner (17). This is exemplified in mouse models of EML4-ALK, where therapeutic response mimicked that observed in human patients (18, 19). FGFR signaling is implicated in the etiology of lung pathogenesis, and to date several GEMMs have been generated based on Cre-inducible expression of a mutant active form of FGFR1 (K656E) (20) and FGFR9 (21). However, to date no GEMMs of FGFR3-TACC3 exist, representing a significant gap in our ability to mechanistically understand treatments to combat this subset of lung cancer patients.
To interrogate the functional significance of the FGFR3-TACC3 gene fusion in lung tumorigenesis, we generated a novel GEMM driven by the Cre-inducible expression of the FGFR3-TACC3 fusion oncoprotein. Expression of FGFR3-TACC3 (F3T3) was activated in the respiratory tract through intra-nasal inhalation of Adenovirus-Cre, mediating recombination in both lung epithelium (22) and olfactory and respiratory epithelia of the nasal cavity (23). Strikingly, in contrast to lung epithelium, the nasal olfactory epithelium was exquisitely sensitive to the oncogenic effects of F3T3. Tumorigenesis was accelerated by additional loss of p53 with a median survival of 6-months for LSL-F3T3/p53 mice due to obstruction of the nasal airway. Intra-tracheal administration of recombinant Ad5-Cre viruses effectively bypassed the nasal cavity, allowing lung tumors to develop. FGFR3-TACC3 promoted the transformation of respiratory cells through augmentation of the mitogen-activated protein kinase (MAPK) pathway characterized by elevated levels of phosphorylated-ERK1/2 but did not affect phosphoinositide 3-kinase (PI3K) pathway activity. Taken together, these findings indicate that the FGFR3-TACC3 fusion protein acts as an oncogene in the respiratory tract.
Results and Discussion
FGFR3-TACC3 is an oncogenic fusion protein in human lung NSCLC cells
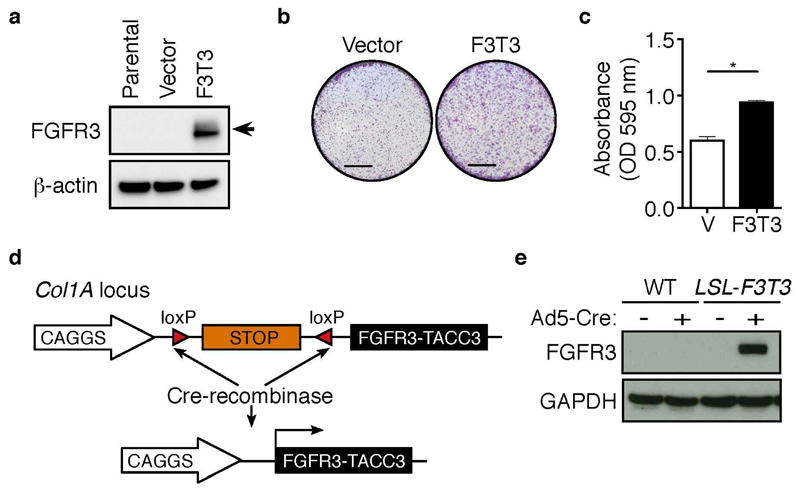
To determine whether FGFR3-TACC3 acts as an oncogenic fusion protein in NSCLC, we stably expressed the human fusion protein FGFR3-TACC3 (FGFR3 exon 18 – TACC3 exon 10) (9) (hereafter F3T3) in H2170 cells (Figure 1a). H2170 cells are a human lung SCC cell line expressing normal FGFR1 copy number (24). Colony formation assays were performed using H2170 stable cell lines to investigate whether the F3T3 fusion protein can enhance the transformation capacity of lung SCC cells. As shown in Figure 1b, F3T3-expressing H2170 cells formed significantly more colonies compared to the empty vector control cells. Quantification of crystal violet staining confirmed these observations (p<0.01; Figure 1c). Taken together, these results suggest that F3T3 acts as a chimeric oncogene in lung SCC.
Figure 1. Generation of conditional FGFR3-TACC3 fusion protein.
(a) Western blot analysis of FGFR3 (Santa Cruz, Dallas, TX, USA; sc-13121, 1:700) in parental, vector and FGFR3-TACC3-expressing H2170 cells. β-actin (Sigma-Aldrich, Saint Louis, MO, USA; A5441, 1:1000) served as a loading control. H2170 cells (ATCC, Manassas, VA, USA; CRL-5928) were maintained in RPMI 1640 supplemented with 10% fetal calf serum, 100 Units/ml penicillin and 100 μg/ml streptomycin (Life Technologies, Carlsbad, CA, USA). cDNA encoding the human FGFR3-TACC3 fusion (exon 18 – exon 10) was PCR amplified from FGFR3-TACC3 pCDH510B, a kind gift from Dr Chinnaiyan (9), using primers; 5’-GCGGTCGACGCCACCATGGGCGCC-3’ and 5’-GCGAGAATTCTCAGATCTTCTCCATCTTGGA-3’, and sub-cloned into the pMSCV-GFP retrovirus. To generate retroviral particles, pMSCV-FGFR3-TACC3 or pMSCV-GFP were transfected into 293T cells with calcium chloride, together with the packaging plasmids gag-pol and VSV. Culture supernatants were harvested after 32 and 48 hours, filtered through a 0.45 μm filter and used to transduce H2170 cells with 1 μg/ml polybrene. Forty-eight hours post transduction, GFP+ transduced cells were sorted on the BD FACS Aria (BD Biosciences), before replating for in vitro analysis. Western blot analysis was performed according to standard procedures. (b) Representative images of crystal violet stained colony assays of vector and F3T3 H2170 cells. Scale, 5 mm. Cells were plated in 12 well plates at a density of 10,000 cells/well. After 72 hours, cells were fixed with 1:1 ice-cold acetone/methanol for 1 minute and incubated with 0.1 % crystal violet for 20 minutes. Wells were imaged with Zeiss Stemi 2000-C using Zen software (Zeiss). (c) Quantification of colony assays were performed as described (55). Mean ± SEM. p<0.01. (d) Schematic representation of the Col1a1 locus targeted with the LSL-FGFR3-TACC3 transgene before and following Cre-mediated recombination. cDNA encoding the human FGFR3-TACC3 fusion (exon 18 – exon 10) (9) was PCR amplified using primers; 5’-GCGGGCGCGCCACCATGGGCGCCCCTGCCTGC-3’ and 5’-GCGATCTAGATCAGATCTTCTCCATCTTGGA-3’, and sub-cloned into the pCAGGS-LSL vector and KH2 ES cells were targeted as previously described (56). (e) Western blot of FGFR3 (Santa Cruz, Dallas, TX, USA; sc-13121, 1:700) protein in mouse embryonic fibroblasts (MEFs) generated from wildtype (WT) and LSL-FGFR3-TACC3 (LSL-F3T3) E14.5 embryos, 72 hours following Ad5-CMV-Cre infection (+). Uninfected (−) cells served as a control, while GAPDH (Sigma-Aldrich, Saint Louis, MO, USA; G8795, 1:5000) provides the loading control. MEFs were generated as previously described (57).
Generation of LSL-FGFR3-TACC3 conditional mice
To directly assess the effects of FGFR3-TACC3 fusion expression in the respiratory system, we engineered a novel mouse model harboring the human FGFR3-TACC3 fusion cDNA. The transgene cassette was inserted into the Col1A locus, with temporal expression achieved utilizing the control of lox-stop-lox (LSL) recombination under a strong CAGGs promoter (Figure 1d), an approach that allows for robust transgene expression (25). To validate a successful recombination event, targeted embryonic stem (ES) cell clones were infected with Ad5-CMV-Cre and protein lysates were prepared 48 hours following infection. Western blot analysis using an antibody that recognizes the intact FGFR3 portion of the fusion protein, confirmed restricted expression of the FGFR3-TACC3 fusion protein in ES cell clones following Ad5-CMV-Cre infection. No expression was detected in uninfected clones (Supplementary Figure 1a), validating that the LSL system is not leaky. To rule out the possibility of rearrangement of the locus in vivo, mouse embryonic fibroblasts (MEFs) were isolated from embryonic day (E) 14.5 LSL-FGFR3-TACC3 embryos. Human FGFR3 protein expression was detected in F3T3 transgenic MEFs analyzed 72 hours following infection with an Ad5-CMV-Cre virus (Figure 1e). No expression of FGFR3 was detected in uninfected LSL-F3T3 MEFs (Figure 1e), confirming stable expression of the transgene in vivo. Next, to validate recombination in cells of the respiratory tract, tracheal epithelial cells were isolated from LSL-F3T3 adult mice according to established protocols (26) and infected in vitro with Ad5-CMV-Cre (Supplementary Figure 1b). Consistent with our findings in ES cells (Supplementary Figure 1a) and MEFs (Figure 1c), expression of the FGFR3-TACC3 fusion protein was restricted to Cre-recombined LSL-F3T3 tracheal epithelial cells (Supplementary Figure 1c).
The FGFR3-TACC3 fusion is highly oncogenic in nasal olfactory epithelium
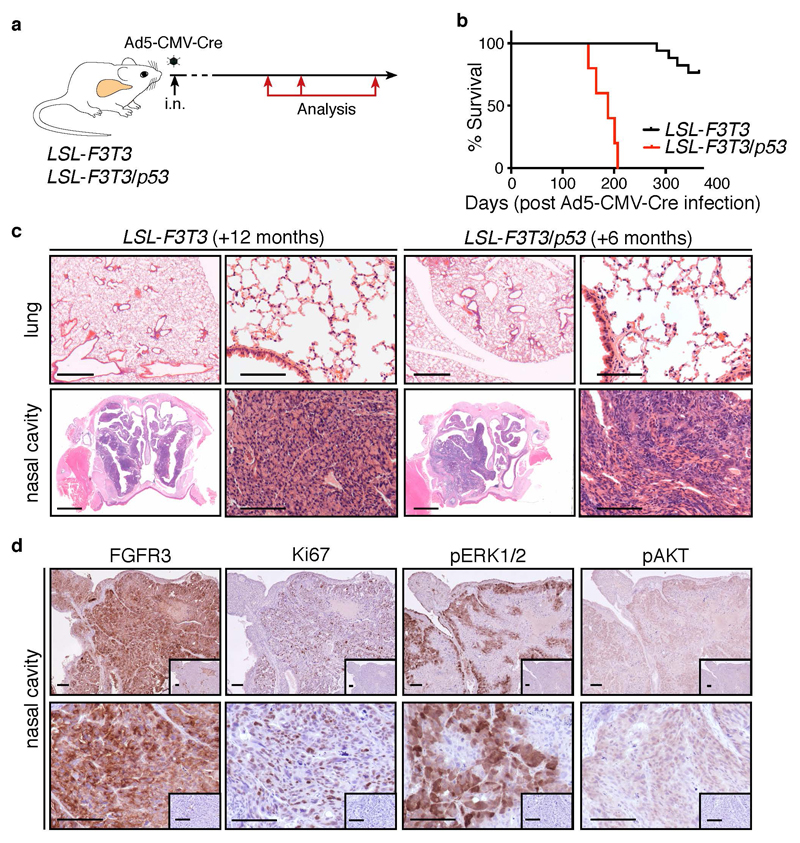
To confirm that FGFR3-TACC3 acts as an oncogenic fusion protein in lung epithelium, LSL-F3T3 mice were intra-nasally (i.n.) infected with an Ad5-CMV-Cre virus. Given that TP53 mutations occur in >90 % of human lung SCC (4) and 46 % lung ADC (3), LSL-F3T3 mice were also crossed with p53f/f mice (27) (hereafter LSL-F3T3/p53). Tumor initiation and progression was monitored at defined time points (3, 6, 9 months) post Ad5-CMV-Cre-infection or analyzed when LSL-F3T3 and LSL-F3T3/p53 mice displayed signs of morbidity (breathing difficulties, weight loss) (Figure 2a). Consistent with observations made in glioblastoma (11), enforced expression of FGFR3-TACC3 in combination with p53 loss reduced the survival latency of LSL-F3T3 mice (undefined vs. 188 days; p<0.0001) (Figure 2b). Surprisingly, histological analysis of lung tissue from LSL-F3T3 and LSL-F3T3/p53 Ad5-CMV-Cre-infected mice revealed the complete absence of epithelial hyperplasia and gross tumor formation (Figure 2c, Supplementary Table 1). Interestingly however, extensive seromucinous (glandular) hyperplasia of epithelial cells lining the posterior nasopharynx was observed in both LSL-F3T3 and LSL-F3T3/p53 mice (Figure 2c, Table 1), and was already apparent in LSL-F3T3/p53 mice, 3 months following intra-nasal Ad5-CMV-Cre infection (Supplementary Table 1; Supplementary Figure 2). Therefore, given that mice are obligate nose breathers (28), the cause of mortality in LSL-F3T3/p53 mice was confirmed to be restricted airway capacity in the nasal cavity.
Figure 2. FGFR3-TACC3 fusion protein is a potent oncogene in nasal epithelium.
(a) Schematic outline of experimental strategy. Animal experiments were conducted according to the regulatory standards approved by the Walter and Eliza Hall Animal Ethics Committee (AEC 2016.024). p53f/f mice were obtained from Jackson Laboratories and genotyped as previously described (27). LSL-F3T3 mice were genotyped using primers: 5’-CCAGTTCAATCATCCCAGGT-3’, 5’- GGCCATCGGAATAGGAACTT-3’ and 5’-CAACCTGGTCCTCCATGTCT-3’, according to standard procedures. Seven-to-eight-week old conditional mice were intra-nasally (i.n.) infected with 20 μl of 1x1010 PFU/ml Ad5-CMV-Cre virus (University of Iowa Gene Transfer Core Facility) according to standard procedures (22). Lungs and nasal cavities were harvested at defined time points following Ad5-CMV-Cre administration, or when mice showed signs of morbidity (breathing difficulties and weight loss) for Kaplan-Meier analysis. (b) Kaplan-Meier survival analysis of LSL-F3T3 (n = 17) and LSL-F3T3/p53 (n = 5) mice following i.n. of Ad5-CMV-Cre virus. ****p<0.0001. Kaplan-Meier analysis was performed using the Mantel-Cox test. (c) Representative hematoxylin and eosin (H&E) stained sections of the nasal cavities and lungs LSL-F3T3 and LSL-F3T3/p53 mice analyzed 12 and 6 months, respectively, following Ad5-CMV-Cre infection. Scale, low power: 1 mm; high power: 100 μm. Lungs were perfused and fixed with 4% paraformaldehyde for 24 hours at 4 °C and embedded in paraffin. Nasal cavities were fixed in 4% paraformaldehyde for 24 hours at 4 °C, de-calcified using Fast-Cal and paraffin embedded. Sections 2 μm thick were stained with H&E. (d) Representative immunohistochemistry of FGFR3 (Santa Cruz, Dallas, TX, USA; sc-13121, 1:100), Ki67 (Cell Signaling, Beverly, MA, USA; #12202, 1:400), p-ERK1/2 (Cell Signaling, Beverly, MA, USA; #9101, 1:200) and p-AKT (Cell Signaling, Beverly, MA, USA; #4060, 1:200) in hyperplastic nasal epithelium of LSL-F3T3/p53 mice, analyzed 6-months following Ad5-CMV-Cre infection. Inset, isotype control. Scale, 100 μm. Immunohistochemistry was performed as described (58).
To confirm that the nasal olfactory hyperplasia observed resulted from enforced expression of F3T3, lesions were immunostained using an FGFR3 antibody. As expected, high FGFR3 expression was specifically detected in hyperplastic cells (Figure 2d), with no expression observed in adjacent normal epithelium (data not shown). Moreover, hyperplastic F3T3-expressing cells stained positive for Ki67 (Figure 2d), indicating that the lesions are highly proliferative. Given that canonical FGFR3 kinase activity signals through the MAPK and PI3K pathways (29, 30), we investigated the activity of these signaling pathways in hyperplastic nasal epithelium. Hyperplastic lesions detected in LSL-F3T3/p53 and LSL-F3T3 nasal cavities exhibited strong, albeit heterogeneous pERK1/2 staining, while pAKT staining was uniformly weak (Figure 2d; data not shown). Thus, consistent with previous reports in human cancer cells (15, 31, 32), the oncogenic capacity of the FGFR3-TACC3 fusion is mediated through the enhanced activity of the MAPK signaling pathway.
The exquisite sensitivity of olfactory epithelial cells to enforced expression of F3T3 is consistent with the identification of the FGFR3-TACC3 gene fusion in 2.5% of NPCs (10). Indeed, NPC cell lines engineered to overexpress FGFR3-TACC3 were found to display growth-promoting qualities, most likely through the functionality of the FGFR3 kinase portion of the fusion protein (10). The relationship between the hyperplasia observed in LSL-F3T3 and LSL-F3T3/p53 mice and NPC remains unclear, as mice succumb to breathing difficulties before advanced disease develops.
Intranasal adenovirus infection results in widespread nasal epithelial cell infection
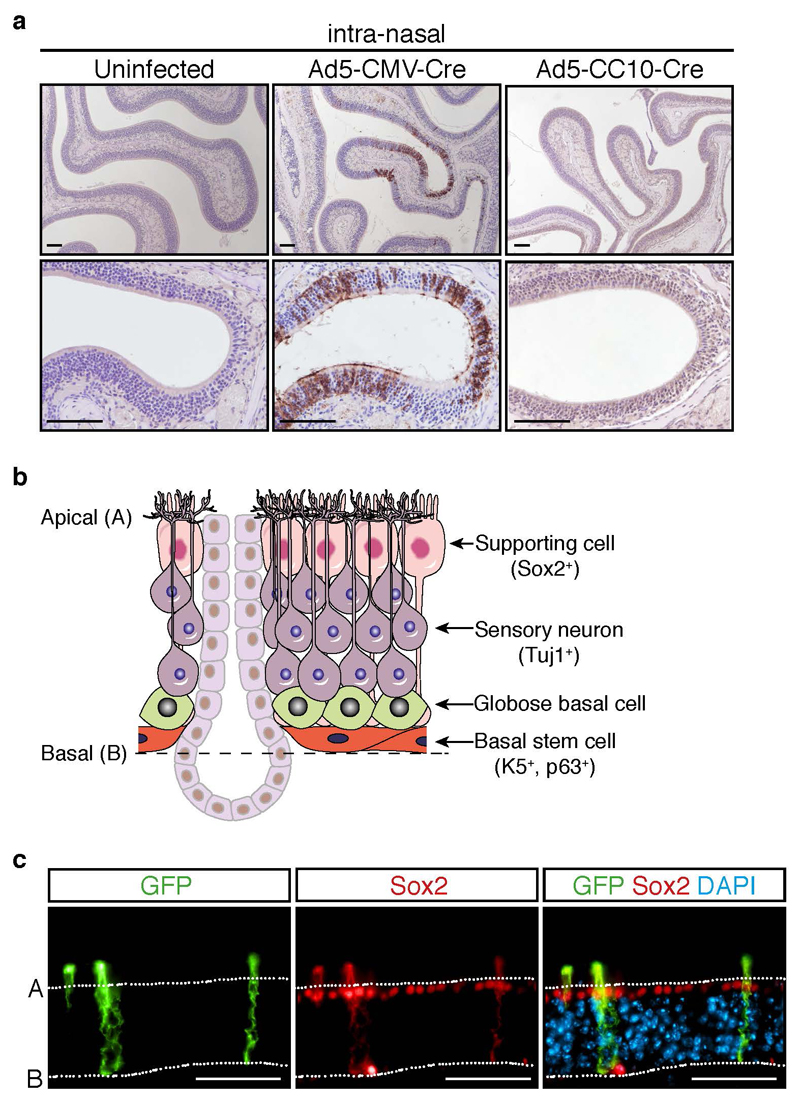
To gain greater insight into the efficiency and repertoire of cell types infected in the nasal cavity following intranasal delivery of Ad5-Cre viruses, we utilized the mT/mG reporter mouse strain (33). In mT/mG mice, all cells express membrane-bound Tomato (mT) prior to Cre- recombination and are induced to express membrane-bound GFP (mG) upon Cre-mediated excision. To investigate the presence of Cre-infected cells in both the proximal and distal nasal cavity (34), mT/mG mice were intra-nasally infected with either a ubiquitous Ad5-CMV-Cre virus or Ad5-CC10-Cre virus, with the latter targeting Cre-recombinase expression specifically to club cells lining the large and small airways of the mouse lung (35). Immunostaining performed on nasal cavity tissue harvested two weeks following infection revealed extensive sporadic GFP expression in the nasal cavities of Ad5-CMV-Cre-infected mT/mG mice (Figure 3a). Conversely, no GFP-positive cells were detected in the nasal cavities of Ad5-CC10-Cre-infected and mT/mG uninfected control mice (Figure 3a).
Figure 3. Adenovirus-mediated Cre delivery effectively infects nasal epithelium.
(a) Representative immunostaining of GFP (Abcam, Cambridge, UK; ab6556, 1:200 (20)) in the nasal cavity of mT/mG reporter mice two weeks following i.n infection of Ad5-CMV-Cre (n = 3) or Ad5-CC10-Cre (n = 3) adenoviruses. Uninfected mT/mG mice (n = 3) served as controls. Scale, 100 μm. (b) Schematic representation of the olfactory epithelium (adapted from (37)). (c) Immunofluorescence co-staining of GFP and Sox2 (Santa-Cruz, Santa Cruz, CA, USA; sc-17320, 1:1000) in the olfactory epithelium of mT/mG mice 2 weeks following Ad5-CMV-Cre infection. Scale, 100 μm. For immunofluorescence, fluorescently labeled secondary antibodies (Thermo Scientific) were incubated with 1 μg/ml DAPI (Sigma-Aldrich, Saint Louis, MO, USA; D9542) in the dark for 1 hour at room temperature. After washing, slides were mounted in Fluoromount-G (Southern Biotech, Chicago, IL, USA; 0100-01), cover-slipped and imaged on the Zeiss AxioObserver microscope using Zen software (Zeiss).
The olfactory sensory epithelium is composed of three major cell types: neurons, supporting cells and basal stem cells, responsible for generating olfactory neurons throughout life (36) (Figure 3b). To determine which of these cell types are most susceptible to Ad5-CMV-Cre infection, we performed fluorescence co-staining for GFP together with Sox2, a marker of supporting cells (37). GFP-positive “switched” cells had long projections that spanned the sensory neuron layer, with cell bodies overlapping with Sox2 staining. (Figure 3c), indicating that Cre is specifically activated in supporting cells. It has been observed, and we clearly demonstrate, that intra-nasal inhalation of adenovirus infects the olfactory and respiratory epithelium (23, 38–40), although the degree of infection and potential functional consequences associated have been far less documented. Interestingly, the incidence of sinonasal adenocarcinomas observed in K-rasLSL-G12D/+/p53f/f mice following intra-nasal delivery of Ad5-CMV-Cre, was significantly potentiated when oncogenic activation of KrasG12D was combined with expression of mutant p53 (p53R270H or p53R175H) (41). Moreover, tumors exhibiting a squamous phenotype were observed in the nasal cavities of mice engineered to overexpress Sox2 in combination with Pten and Cdkn2ab inactivation, following administration of an adenovirus expressing Cre-recombinase under the control of the keratin 5 (K5) gene promoter (20). Thus, together with results from this study, we would argue that caution should be taken in the design of new conditional mouse models for lung tumor studies, to avoid the preferential transformation of epithelial cells lining the nasal cavity.
FGFR3-TACC3 expression promotes tumorigenesis in lung epithelium
We hypothesized that the presence of hyperplasia in the nasal cavities of LSL-F3T3 and LSL-F3T3/p53 mice at early time points following intra-nasal delivery of Ad5-CMV-Cre, may restrict the time required for lung tumor formation. Thus, to bypass the infection of nasal epithelium, we delivered the Ad5-CMV-Cre virus via intra-tracheal instillation, a procedure routinely used to generate autochthonous lung tumors in genetically modified mice (42–44). Firstly, we needed to confirm that intra-tracheal administration of Ad5-CMV-Cre would bypass the infection of nasal epithelium observed following intranasal delivery of Cre-recombinase. To directly address this question, mT/mG mice were intra-tracheally infected with Ad5-CMV-Cre virus, and nasal cavities and lung tissue were harvested 2 weeks following infection. In contrast to the GFP staining pattern observed following intra-nasal infection (Figure 3a), intra-tracheal delivery of Ad5-CMV-Cre dramatically reduced the number of GFP-positive cells in the olfactory epithelium (Supplementary Figure 3a). The presence of GFP-positive “switched” cells lining the bronchiolar and alveolar airways of the lung validated the efficiency of the intra-tracheal procedure (Supplementary Figure 3a).
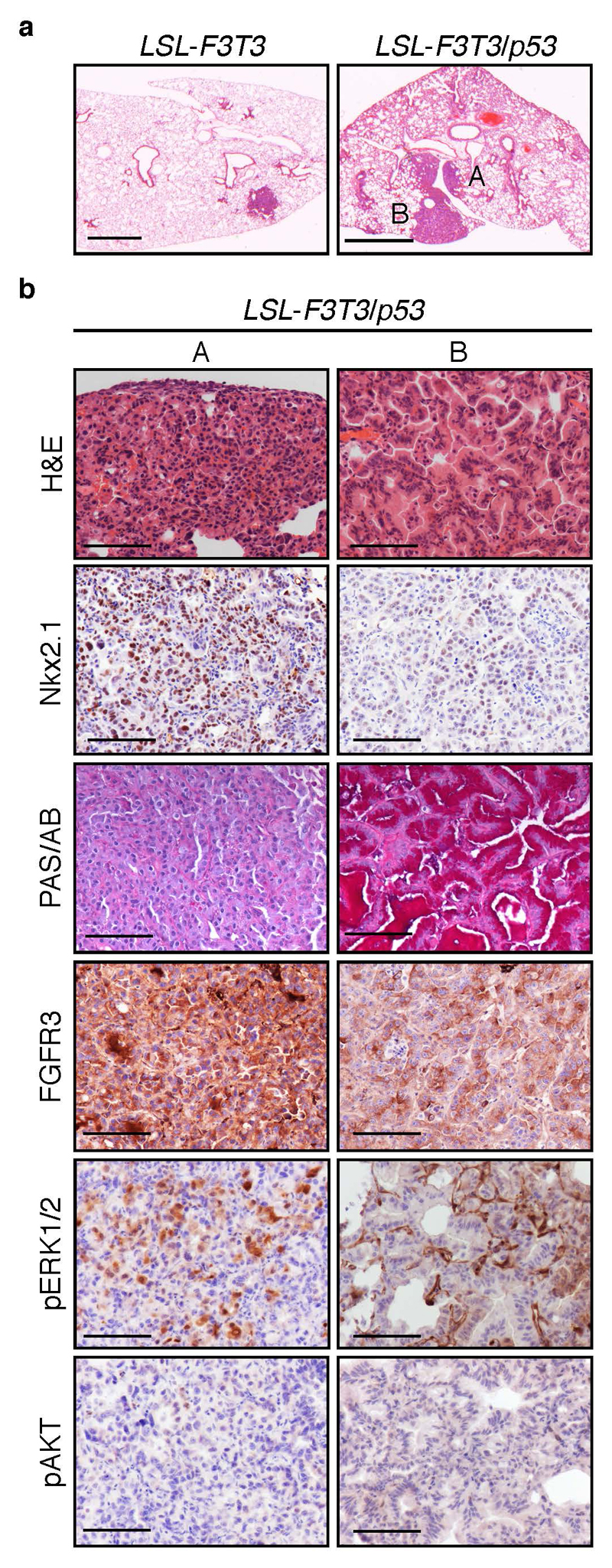
Consistent with a significant reduction in the infection of nasal epithelium, the presence of seromucinous hyperplasia was less frequently observed in LSL-F3T3 and LSL-F3T3/p53 mice analyzed 12 months following intra-tracheal Ad5-CMV-Cre infection (Supplementary Table 1). Moreover, the degree of hyperplasia was also less pronounced (Supplementary Figure 3b) enabling LSL-F3T3/p53 mice to survive past 7 months. Interestingly, focal lung tumors were observed in the lungs of 2/13 LSL-F3T3 mice, which further increased in penetrance following the additional loss of p53 (Figure 4a; Supplementary Table 1). The tumors detected in LSL-F3T3 mice were classified as adenocarcinomas, with tumors exhibiting a homogeneous papillary-like morphology and displaying high expression levels of Nkx2.1/TTF-1, a clinical marker of adenocarcinomas (Supplementary Figure 3c). Moreover, the tumors co-expressed FGFR3, confirming that these tumors were driven by expression of the FGFR3-TACC3 transgene (Supplementary Figure 3c). In comparison, the lung tumors that developed in 4/8 of LSL-F3T3/p53 mice displayed two distinct pathologic characteristics (Figure 4a-b). One subset of tumors displayed a well-differentiated papillary growth pattern expressing high levels of Nkx2.1 (Figure 4b, left panel A), while the remaining tumors showed signs of mixed lipidic, acinar and papillary growth (Figure 4b, right panel B). Lipidic pattern lesions (Type B) displayed an abundance of neutral-mucin PAS-positive cells, reduced Nkx2.1 expression, and sporadic regions expressing the squamous markers p63 and Sox2 (Figure 4b, Supplementary Figure 4a, right panel B). The mucinous transdifferentiation however, appears not to be driven by transcription factor HNF4α expression (45) (Supplementary Figure 4a, b). Consistent with observations made in the nasal cavity (Figure 2d), overexpression of FGFR3 correlated with increased MAPK, but not PI3K signaling in both tumor subsets (Figure 4b). While these findings confirm that enforced expression of the FGFR3-TACC3 fusion in vivo can promote the transformation of lung epithelium, its capacity as an oncogenic driver is weaker than what is observed with other lung cancer gene fusions, such as EML4-ALK (18, 19).
Fig. 4. Intra-tracheal mediated Adenovirus-Cre delivery enables lung tumor formation.
(a) Representative H&E stained sections of LSL-F3T3 and LSL-F3T3/p53 12 months following intra-tracheal (i.t.) delivery of Ad5-CMV-Cre. Briefly, seven-to-eight-week old conditional mice were infected (i.t.) with 20 μl of 1x1010 PFU/ml Ad5-CMV-Cre virus (University of Iowa Gene Transfer Core Facility) as described (59). A represents Type A lesion, box B represents a Type B lesion. Scale, 100 μm. (b) Representative H&E and immunostaining of Nkx2.1 (TTF-1; DAKO, Santa Clara, CA, USA; #M3575, 1:200), PAS/Alcian Blue (PAS/AB), FGFR3, p-ERK1/2, p-AKT on Type A and Type B lung tumors from LSL-F3T3/p53 mice, 12 months following intra-tracheal Ad5-CMV-Cre infection. Scale, 100 μm.
Although the FGFR3-TACC3 fusion is also observed in lung SCC (8, 9), the lung tumors that developed in LSL-F3T3 and LSL-F3T3/p53 mice were classified as lung adenocarcinomas (Figure 4a). To determine whether F3T3 gene fusions are also found in human lung ADC, we interrogated publicly available cancer datasets including TCGA (3) and MSK-IMPACT (46, 47), accessed via cBioPortal (48, 49). While no fusion events were identified in the TCGA dataset consistent with a recent report (5), five lung ADC patients harboring F3T3 gene fusions were identified in the MSK-IMPACT clinical sequencing cohort (46, 47). Critically, this analysis revealed the presence of F3T3 gene fusions in lung ADC, consist with our findings and highlighting the clinical relevance of our GEMM.
Interestingly, markers of squamous cell differentiation, Sox2 and p63 were observed in a subset of ADC cells, suggesting that additional genetic events, such as Sox2 overexpression, may be required to drive a fully-penetrant squamous cell phenotype (20, 50). Moreover, we cannot rule out that directing F3T3 expression to lung basal progenitor cells may also promote the formation of lung SCC tumors, given that basal cells are the putative cell-of-origin of lung SCC (20, 51, 52). Interestingly, perturbations in PI3K pathway components are more prevalent in lung SCC (53), where Pten inactivation has been shown to play an important role in driving lung SCC formation characterized by elevated p-AKT activity (54). The preferential activation of the MAPK signal transduction pathway downstream of FGFR3-TACC3 may have therefore influenced the formation of lung ADC over lung SCC. Interestingly, expression of the FGFR3-TACC3 oncoprotein in head and neck squamous cell carcinoma cells was recently shown to be responsible for acquired resistance to combined blockade of EGFR/ERBB3 therapies by reactivating MAPK signaling (32). Together, our findings highlight the importance of our novel transgenic model as a tool to better understand the behavior of the F3T3 fusion and how it could be therapeutically exploited in the clinic.
Supplementary Material
Aknowledgements
We are grateful to C. Alvarado, L. Scott, H. Johnson, K. Birchall and L. Mackiewicz for animal husbandry, E. Tsui and C. Tsui in the WEHI Histology Facility for expert support and P. Maltezos for preparation of the nasal cavity graphic image. The authors thank T. Wilson, J. Mighall and J. Pasquet for technical support and W. Alexander for useful discussions and critically reading the manuscript. This work was supported in part by a Worldwide Cancer Research Project Grant (14-0433). S.A.B. is supported by a Victorian Cancer Agency (VCA) Early Career Seed Grant (ECSG16001); M.L A-L is supported by a Viertel Foundation Senior Medical Research Fellowship; K.D.S. is supported by the Peter and Julie Alston Centenary Fellowship. This work was made possible through Victorian Government Operational Infrastructure Support and Australian Government.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Society AC. Cancer Facts & Figures 2016. Atlanta: American cancer Society; 2016. [Google Scholar]
- 2.George J, Lim JS, Jang SJ, Cun Y, Ozretic L, Kong G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524(7563):47–53. doi: 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–50. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research N. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao Q, Liang WW, Foltz SM, Mutharasu G, Jayasinghe RG, Cao S, et al. Driver Fusions and Their Implications in the Development and Treatment of Human Cancers. Cell reports. 2018;23(1):227–38 e3. doi: 10.1016/j.celrep.2018.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soda M, Young Lim C, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4–ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–6. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 7.Solomon BJ, Mok T, Kim D-W, Wu Y-L, Nakagawa K, Mekhail T, et al. First-Line Crizotinib versus Chemotherapy in ALK-Positive Lung Cancer. The New England Journal of Medicine. 2014;371(23):2167–77. doi: 10.1056/NEJMoa1408440. [DOI] [PubMed] [Google Scholar]
- 8.Majewski IJ, Mittempergher L, Davidson NM, Bosma A, Willems SM, Horlings HM, et al. Identification of recurrent FGFR3 fusion genes in lung cancer through kinome-centred RNA sequencing. The Journal of pathology. 2013;230(3):270–6. doi: 10.1002/path.4209. [DOI] [PubMed] [Google Scholar]
- 9.Wu YM, Su F, Kalyana-Sundaram S, Khazanov N, Ateeq B, Cao X, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer discovery. 2013;3(6):636–47. doi: 10.1158/2159-8290.CD-13-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan L, Liu Z-H, Lin Z-R, Xu L-H, Zhong Q, Zeng M-S. Recurrent FGFR3-TACC3 fusion gene in nasopharyngeal carcinoma. Cancer Biology and Therapy. 2014;15(12):1613. doi: 10.4161/15384047.2014.961874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh D, Chan JM, Zoppoli P, Niola F, Sullivan R, Castano A, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337(6099):1231–5. doi: 10.1126/science.1220834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carneiro BA, Elvin JA, Kamath SD, Ali SM, Paintal AS, Restrepo A, et al. FGFR3-TACC3: A novel gene fusion in cervical cancer. Gynecologic Oncology Reports. 2015;13:53–6. doi: 10.1016/j.gore.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams SV, Hurst CD, Knowles MA. Oncogenic FGFR3 gene fusions in bladder cancer. Human Molecular Genetics. 2013;22(4):795–803. doi: 10.1093/hmg/dds486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capelletti M, Dodge ME, Ercan D, Hammerman PS, Park S-I, Kim J, et al. Identification of recurrent FGFR3-TACC3 fusion oncogenes from lung adenocarcinoma. Clinical Cancer Research: An Official Journal Of The American Association For Cancer Research. 2014;20(24):6551–8. doi: 10.1158/1078-0432.CCR-14-1337. [DOI] [PubMed] [Google Scholar]
- 15.Parker BC, Annala MJ, Cogdell DE, Granberg KJ, Sun Y, Ji P, et al. The tumorigenic FGFR3-TACC3 gene fusion escapes miR-99a regulation in glioblastoma. The Journal Of Clinical Investigation. 2013;123(2):855–65. doi: 10.1172/JCI67144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwon MC, Berns A. Mouse models for lung cancer. Molecular oncology. 2013;7(2):165–77. doi: 10.1016/j.molonc.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharpless NE, Depinho RA. The mighty mouse: genetically engineered mouse models in cancer drug development. Nature reviews Drug discovery. 2006;5(9):741–54. doi: 10.1038/nrd2110. [DOI] [PubMed] [Google Scholar]
- 18.Chen Z, Sasaki T, Tan X, Carretero J, Shimamura T, Li D, et al. Inhibition of ALK PI3K/MEK, and HSP90 in murine lung adenocarcinoma induced by EML4-ALK fusion oncogene. Cancer research. 2010;70(23):9827–36. doi: 10.1158/0008-5472.CAN-10-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soda M, Takada S, Takeuchi K, Choi YL, Enomoto M, Ueno T, et al. A mouse model for EML4-ALK-positive lung cancer. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(50):19893–7. doi: 10.1073/pnas.0805381105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferone G, Song JY, Sutherland KD, Bhaskaran R, Monkhorst K, Lambooij JP, et al. SOX2 Is the Determining Oncogenic Switch in Promoting Lung Squamous Cell Carcinoma from Different Cells of Origin. Cancer Cell. 2016;30(4):519–32. doi: 10.1016/j.ccell.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin Y, Betsuyaku T, Garbow JR, Miao J, Govindan R, Ornitz DM. Rapid induction of lung adenocarcinoma by fibroblast growth factor 9 signaling through FGF receptor 3. Cancer research. 2013;73(18):5730–41. doi: 10.1158/0008-5472.CAN-13-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nature protocols. 2009;4(7):1064–72. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arimoto Y, Nagata H, Isegawa N, Kumahara K, Isoyama K, Konno A, et al. In vivo expression of adenovirus-mediated lacZ gene in murine nasal mucosa. Acta Otolaryngol. 2002;122:627–33. doi: 10.1080/000164802320396303. [DOI] [PubMed] [Google Scholar]
- 24.Dutt A, Ramos AH, Hammerman PS, Mermel C, Cho J, Sharifnia T, et al. Inhibitor-sensitive FGFR1 amplification in human non-small cell lung cancer. PloS one. 2011;6(6):e20351. doi: 10.1371/journal.pone.0020351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huijbers IJ, Bin Ali R, Pritchard C, Cozijnsen M, Kwon MC, Proost N, et al. Rapid target gene validation in complex cancer mouse models using re-derived embryonic stem cells. EMBO molecular medicine. 2014;6(2):212–25. doi: 10.1002/emmm.201303297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(31):12771–5. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jonkers J, Meuwissen R, Gulden Hvd, Peterse H, Valk Mvd, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breat cancer. Nature Genetics. 2001;29:418–25. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 28.Hatch TF, Gross P. Pulmonary Deposition and Retention of Inhaled Aerosols. Academic Press; 1964. [Google Scholar]
- 29.Ong SH, Hadari YR, Gotoh N, Guy GR, Schlessinger J, Lax I. Stimulation of phosphatidylinositol 3-kinase by fibroblast growth factor receptors is mediated by coordinated recruitment of multiple docking proteins. PNAS. 2001;98(11):6074–9. doi: 10.1073/pnas.111114298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kouhara H, Hadari YR, Spivak-Kroizman T, Schilling J, Bar-Sagi D, Lax I, et al. A Lipid-Anchored Grb2-Binding Protein That Links FGF-Receptor Activation to the Ras/MAPK Signaling Pathway. Cell. 1997;89(5):693–702. doi: 10.1016/s0092-8674(00)80252-4. [DOI] [PubMed] [Google Scholar]
- 31.Nelson KN, Meyer AN, Siari A, Campos AR, Motamedchaboki K, Donoghue DJ. Oncogenic Gene Fusion FGFR3-TACC3 is Regulated by Tyrosine Phosphorylation. Molecular Cancer Research. 2016 doi: 10.1158/1541-7786.MCR-15-0497. [DOI] [PubMed] [Google Scholar]
- 32.Daly C, Castanaro C, Zhang W, Zhang Q, Wei Y, Ni M, et al. FGFR3-TACC3 fusion proteins act as naturally occurring drivers of tumor resistance by functionally substituting for EGFR/ERK signaling. Oncogene. 2016:1–11. doi: 10.1038/onc.2016.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A Global Double-Fluorescent Cre Reporter Mouse. genesis. 2007;45(9):593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 34.Johnson VJ, Yucesoy B, Reynolds JS, Fluharty K, Wang W, Richardson D, et al. Inhalation of toluene diisocyanate vapor induces allergic rhinitis in mice. Journal of immunology. 2007;179(3):1864–71. doi: 10.4049/jimmunol.179.3.1864. [DOI] [PubMed] [Google Scholar]
- 35.Sutherland KD, Proost N, Brouns I, Adriaensen D, Song J-Y, Berns A. Cell of Origin of Small Cell Lung Cancer: Inactivation of Trp53 and Rb1 in Distinct Cell Types of Adult Mouse Lung. Cancer Cell. 2011;19:754–64. doi: 10.1016/j.ccr.2011.04.019. [DOI] [PubMed] [Google Scholar]
- 36.Leung C, Coulombe P, Reed R. Contribution of olfactory neural stem cells to tissue maintenance and regeneration. Nature Neuroscience. 2007;10(6):720–6. doi: 10.1038/nn1882. [DOI] [PubMed] [Google Scholar]
- 37.Duggan C, Ngai J. Scent of a stem cell. Nature Neuroscience. 2007;10(6):673–4. doi: 10.1038/nn0607-673. [DOI] [PubMed] [Google Scholar]
- 38.Damjanovic D, Zhang X, Mu J, Fe Medina M, Xing Z. Organ distribution of transgene expression following intranasal mucosal delivery of recombinant replication-defective adenovirus gene transfer vector. Genetic Vaccines and Therapy. 2008;6(5) doi: 10.1186/1479-0556-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doi K, Nibu K, Ishida H, Okado H, Terashima T. Adenovirus-mediated gene transfer in olfactory epithelium and olfactory bulb: a long-term study. Annals of Ontology, Rhinology & Larygology. 2005;114(8):629–33. doi: 10.1177/000348940511400808. [DOI] [PubMed] [Google Scholar]
- 40.Zhao H, Otaki J, Firestein S. Adenovirus-mediated gene transfer in olfactory neurons in vivo. Journal of Neurobiology. 1996;30(4):521–30. doi: 10.1002/(SICI)1097-4695(199608)30:4<521::AID-NEU7>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 41.Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, et al. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer research. 2005;65(22):10280–8. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- 42.Mollaoglu G, Guthrie MR, Bohm S, Bragelmann J, Can I, Ballieu PM, et al. MYC Drives Progression of Small Cell Lung Cancer to a Variant Neuroendocrine Subtype with Vulnerability to Aurora Kinase Inhibition. Cancer Cell. 2017;31(2):270–85. doi: 10.1016/j.ccell.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sutherland KD, Proost N, Brouns I, Adriaensen D, Song JY, Berns A. Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell. 2011;19(6):754–64. doi: 10.1016/j.ccr.2011.04.019. [DOI] [PubMed] [Google Scholar]
- 44.Sutherland KD, Song JY, Kwon MC, Proost N, Zevenhoven J, Berns A. Multiple cells-of-origin of mutant K-Ras-induced mouse lung adenocarcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(13):4952–7. doi: 10.1073/pnas.1319963111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Snyder EL, Watanabe H, Magendantz M, Hoersch S, Chen TA, Wang DG, et al. Nkx2-1 represses a latent gastric differentiation program in lung adenocarcinoma. Mol Cell. 2013;50(2):185–99. doi: 10.1016/j.molcel.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jordan EJ, Kim HR, Arcila ME, Barron D, Chakravarty D, Gao J, et al. Prospective Comprehensive Molecular Characterization of Lung Adenocarcinomas for Efficient Patient Matching to Approved and Emerging Therapies. Cancer discovery. 2017;7(6):596–609. doi: 10.1158/2159-8290.CD-16-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23(6):703–13. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2(5):401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mukhopadhyay A, Berrett KC, Kc U, Clair PM, Pop SM, Carr SR, et al. Sox2 cooperates with Lkb1 loss in a mouse model of squamous cell lung cancer. Cell reports. 2014;8(1):40–9. doi: 10.1016/j.celrep.2014.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sutherland KD, Berns A. Cell of origin of lung cancer. Molecular oncology. 2010;4(5):397–403. doi: 10.1016/j.molonc.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weeden CE, Chen Y, Ma SB, Hu Y, Ramm G, Sutherland KD, et al. Lung Basal Stem Cells Rapidly Repair DNA Damage Using the Error-Prone Nonhomologous End-Joining Pathway. PLoS Biol. 2017;15(1):e2000731. doi: 10.1371/journal.pbio.2000731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet. 2016;48(6):607–16. doi: 10.1038/ng.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu C, Fillmore CM, Koyama S, Wu H, Zhao Y, Chen Z, et al. Loss of Lkb1 and Pten leads to lung squamous cell carcinoma with elevated PD-L1 expression. Cancer Cell. 2014;25(5):590–604. doi: 10.1016/j.ccr.2014.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Best SA, Nwaobasi AN, Schmults CD, Ramsey MR. CCAR2 Is Required for Proliferation and Tumor Maintenance in Human Squamous Cell Carcinoma. The Journal of investigative dermatology. 2017;137(2):506–12. doi: 10.1016/j.jid.2016.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44(1):23–8. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- 57.Leong HS, Chen K, Hu Y, Lee S, Corbin J, Pakusch M, et al. Epigenetic regulator Smchd1 functions as a tumor suppressor. Cancer research. 2013;73(5):1591–9. doi: 10.1158/0008-5472.CAN-12-3019. [DOI] [PubMed] [Google Scholar]
- 58.Best SA, De Souza DP, Kersbergen A, Policheni AN, Dayalan S, Tull D, et al. Synergy between the KEAP1/NRF2 and PI3K Pathways Drives Non-Small-Cell Lung Cancer with an Altered Immune Microenvironment. Cell Metab. 2018;27(4):935–43 e4. doi: 10.1016/j.cmet.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 59.Best SA, Kersbergen A, Asselin-Labat ML, Sutherland KD. Combining Cell Type-Restricted Adenoviral Targeting with Immunostaining and Flow Cytometry to Identify Cells-of-Origin of Lung Cancer. Methods Mol Biol. 2018;1725:15–29. doi: 10.1007/978-1-4939-7568-6_2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.