Summary
Comprehensive genomic and transcriptomic analysis demonstrates that tumor-infiltrating T lymphocytes that react to mutated neo-epitopes could be identified in recurrent ovarian cancer. Two of these T cell populations reacted against TP53 hotspot missense mutations that are present in a wide variety of malignancies.
In this issue of Clinical Cancer Research, Deniger and colleagues demonstrate that T cells reacting to specific neoantigens, including mutant p53, can be identified in relapsed ovarian cancer (1).
The immune microenvironment is crucially important in ovarian cancer, particularly in the commonest histological type, high grade serous carcinoma (HGSC). Specific immune populations have both positive and negative prognostic influence; high density intra-epithelial tumor-infiltrating CD8+ lymphocytes (TILs) are associated with increased overall survival as is the presence of an immunoreactive gene expression signature. Conversely, intra-tumoral immunosuppressive macrophages are associated with poor survival. Recent data have also identified an inverse relationship between clonal diversity and immune cell infiltration in HGSC, and demonstrated that T cell clones spatially track tumor clones, implying a close relationship between genotype and immune response (2).
These observations suggest that patients with HGSC could benefit from immunotherapy. Clinical trials in which ovarian cancer patients were infused with ex vivo-expanded autologous TILs were first conducted in the 1990s. However, sample sizes were small and the studies were not randomized (3). Multiple trials of PD-1 or PD-L1 inhibition are currently being undertaken in ovarian cancer, in which some durable responses are occasionally observed. However, overall response rates are only 10 – 15%, with little correlation between response and expression of PD-L1 within tumors.
Although these studies and others reaffirm the idea that immunological therapies may have benefit in ovarian cancer, critical key questions remain unanswered, in particular how to identify the subset of patients best suited to immunotherapy. Certainly, the tumor-autonomous features driving immune cell infiltration into tumors remain unclear and it is uncertain which epitopes most frequently induce TIL responses.
It is against this background that Deniger and colleagues from the Rosenberg lab have set out to identify whether there are T cell responses to neo-epitopes in recurrent ovarian cancer. They studied tumors from women with recurrent ovarian cancer of multiple histological types enrolled in an NCI-sponsored trial of autologous TIL therapy (NCT01174121).
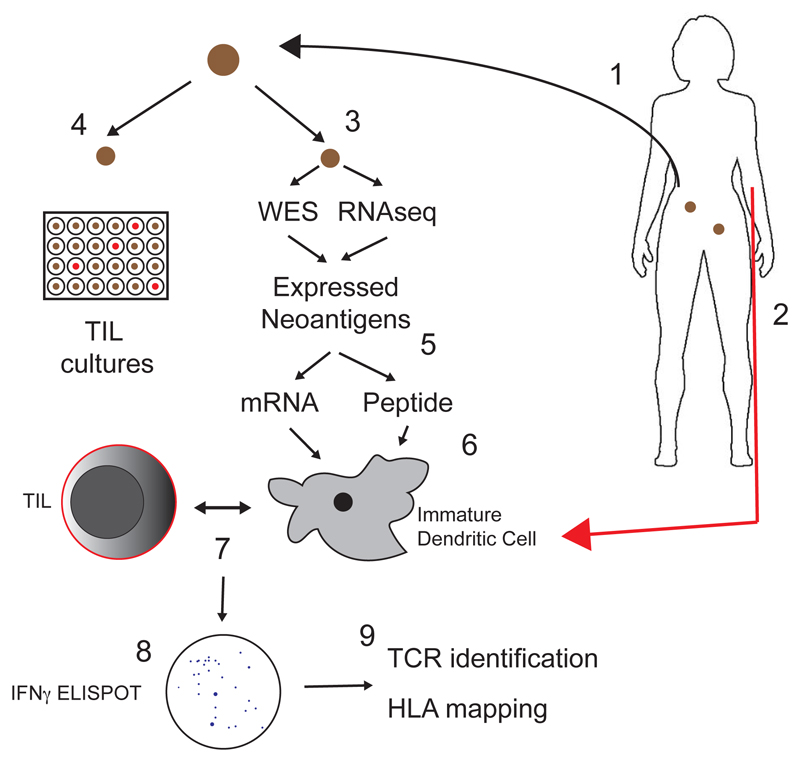
The study methodology was comprehensive and complex (Figure). A key trial inclusion criterion was the presence of at least two measurable tumor deposits, one of which was excised. Patients also underwent leukapheresis. The excised tumor sample was subjected to whole exome sequencing to identify missense mutations predicted to generate neo-epitopes, and RNA sequencing to assess their transcription. Putative neo-epitopes were then expressed both as minigenes, each encoding 25 amino acids, and as pools of 25-mer peptides. Tandem minigene constructs (TMG) containing up to 18 minigenes in parallel were then transcribed ex vivo. Autologous immature dendritic cells (DC) isolated from the leukapheresis sample were either transfected with TMG mRNA or pulsed with peptide pools, thus allowing presentation via both intra- and extracellular pathways on HLA class I and II molecules. Each resected tumor was also dissected into 24 pieces and cultured with high dose IL-2 to allow TIL growth. The pulsed or transfected DC were then co-cultured with immature TIL microcultures to identify immunoreactive populations via interferon-γ ELISPOT. Finally, the HLA mapping of epitopes was calculated, and putative T cell receptors identified.
Figure.
Schematic of the protocol utilized by Deniger et al. 1. A tumor deposit was excised from women with recurrent ovarian cancer. 2. The patient also underwent leukapheresis to isolate peripheral blood lymphocytes. 3. Whole exome sequencing (WES) and RNA sequencing (RNAseq) was performed on the exised nodule to identify expressed neoantigens. 4. Tumors were also cut into 24 pieces and cultured in high-dose IL-2 to allow TIL expansion. 5. Expressed neoantigens were in-vitro transcribed into mRNA or expressed as peptide pools. 6. Immature dendritic cells (DC) from leukapheresis sample were transfected with neoantigen mRNA or pulsed with peptide pools and 7. co-cultured with autologous TIL (from step 4). 8. TIL reactivity was assessed by interferon-γ (IFNγ) ELISPOT assay. 9. Putative T cell receptors were identified, and epitope HLA binding mapped.
The results are simultaneously fascinating and sobering. A total of 1714 putative neoantigens were identified, a median of 228 per patient, suggesting that there are indeed many potentially immunogenic mutant proteins in ovarian cancer. However, only 8 of these neoantigens, identified in five of the seven patents, were immunoreactive. All immunoreactive mutations were unique to each patient, but overall mutational burden did not correlate with T cell responsiveness. Thus, the results can be viewed in two ways: a large proportion of patients (c.70%; 5/7) with recurrent ovarian cancer are likely to have identifiable immunoreactive neoantigens. Conversely, the rate of neoantigen positivity is very low (<1%; 8/1714) and hard to predict using current algorithms.
The most noteworthy results relate to p53 missense mutations. TP53 mutation is near-universal in HGSC and is the key initiating event. In patients with metastatic disease, the same TP53 mutation is found across deposits at both diagnosis and relapse. In HGSC, approximately two-thirds of TP53 mutations are missense, with several hotspot residues (R175, Y220, G245, R248 and R273) that collectively comprise c.20% mutations (see p53.iarc.fr/TP53SomaticMutations.aspx). Deniger and colleagues identified mutation-specific TIL populations directed against two hotspot mutants (G245S and Y220C). Interestingly, both neoepitopes, although distinct and non-overlapping, were presented in association with HLA-DRB3*02:02:010.
Beyond p53, however, the neoantigens did not involve genes thought to be critical drivers in ovarian cancer biology and appeared to occur at random. One mutation, USP9XY2009C, generated three separate epitopes each capable of binding a single HLA and forming interactions with a specific T cell receptor, whilst a mutant histone, H1.5A71D, generated two neo-epitopes restricted by different HLA class II molecules.
The final example was identified in a patient who had multiple tumor samples, separated both temporally and spatially. CD4+ TIL reacting to a RAPTORD654G epitope were isolated from one deposit, and were only reactive to tumor cells containing the RAPTORD654G mutation. The mutant allele fraction in the index deposit was only 0.15, suggesting that immunogenic mutations can be subclonal and/or appear later in disease evolution. The clinical implication for this patient is that targeting RAPTORD654G would, at best, eliminate only a fraction of the tumor population, reflecting the therapeutic challenges of a disease with profound clonal diversity.
Two immediate points arise from this paper. The first and more important is that ovarian cancer is not a single disease. Here, patients with multiple different histological types were enrolled, including one case each of clear cell, endometrioid and low grade serous carcinoma (the latter, unusually, contained a TP53 mutation). It will be important to undertake studies in larger and more histologically homogeneous populations to assess whether these results are generalized across all ovarian cancer patients. Second, ovarian cancer, regardless of histological type, is rarely a disease of high mutational burden, and rates of mismatch repair are generally very low (4). Rather, HGSC is a disease of aberrant copy number, and is thus unlikely to be a prolific generator of neoantigens.
It is also important to frame these results in the wider context. It is now thirty years since Rosenberg and colleagues first demonstrated durable responses to re-infused TIL in melanoma, and their recent data suggest that clinical responses to TIL therapy can be achieved in breast cancer (5). However, the practical logistics of autologous cell harvest, expansion and re-infusion continue to preclude widespread clinical use.
Thus, these data may have more immediate impact upon vaccination protocols or chimeric antigen receptor cell therapy strategies. There is renewed interest in vaccination strategies in ovarian cancer – indeed, two presentations at the 2018 American Society of Clinical Oncology annual meeting suggested that vaccination against shared tumor-associated antigens (including survivin, MUC-1, NY-ESO-1 and WT1) may have genuine clinical utility in ovarian cancer when given following conventional carboplatin/paclitaxel chemotherapy or in association with low dose cyclophosphamide and an IDO-1 inhibitor. The results presented here suggest that future vaccination approaches in ovarian cancer could be widened to include shared mutations, such as the common p53 hotspot mutants, and potentially also unique mutations, such as the USP9XY2009C epitope identified here. Excitingly, the challenges of turning somatic sequence data into patient-specific vaccines in a clinically-relevant timeframe are already being addressed, thus allowing rapid translation of these results into clinical application.
Acknowledgements
Iain McNeish receives grant support from Ovarian Cancer Action (grant reference 006), Cancer Research UK (grant reference A15973), the NIHR Imperial Biomedical Research Centre and the Imperial Experimental Cancer Medicine Centre.
Footnotes
Conflicts of interest: No relevant conflicts of interest to declare
References
- 1.Deniger DC, Pasetto A, Robbins PF, Gartner JJ, Prickett TD, Paria BC, et al. T-cell responses to TP53 "hotspot" mutations and unique neoantigens expressed by human ovarian cancers. Clin Cancer Res. 2018 doi: 10.1158/1078-0432.Ccr-18-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang AW, McPherson A, Milne K, Kroeger DR, Hamilton PT, Miranda A, et al. Interfaces of Malignant and Immunologic Clonal Dynamics in Ovarian Cancer. Cell. 2018 doi: 10.1016/j.cell.2018.03.073. [DOI] [PubMed] [Google Scholar]
- 3.Ikarashi H, Fujita K, Takakuwa K, Kodama S, Tokunaga A, Takahashi T, et al. Immunomodulation in patients with epithelial ovarian cancer after adoptive transfer of tumor-infiltrating lymphocytes. Cancer Res. 1994;54(1):190–6. [PubMed] [Google Scholar]
- 4.Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357(6349):409–13. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zacharakis N, Chinnasamy H, Black M, Xu H, Lu YC, Zheng Z, et al. Immune recognition of somatic mutations leading to complete durable regression in metastatic breast cancer. Nat Med. 2018;24(6):724–30. doi: 10.1038/s41591-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]