SUMMARY
In a prior study, baseline mutational load (ML) predicted progression to high-grade dysplasia (HGD) or esophageal adenocarcinoma (EAC) in Barrett's esophagus (BE) with an area under the curve (AUC) of 0.95. We aimed to validate the test characteristics of this predictive biomarker panel using crude DNA lysates in a larger well-characterized cohort. We performed a nested case-control study of BE patients from three tertiary referral centers in the Netherlands. Cases had baseline nondysplastic BE (NDBE) and developed HGD/EAC ≥ 2 years later. Controls were matched 2:1, had baseline NDBE, and no progression. Polymerase chain reaction (PCR)-based mutational analysis was performed on crude lysates from formalin-fixed, paraffin-embedded tissue. ML was calculated from loss of heterozygosity (LOH) and microsatellite instability (MSI) at 10 genomic loci. Receiver operator characteristic (ROC) curves were created to assess the diagnostic utility of various cutoffs of ML for progression. Of 159 subjects, 58 were progressors and 101 were nonprogressors, there was no difference in mean ML in preprogression tissue in progressors and nonprogressors (ML = 0.73 ± 0.69 vs. ML = 0.74 ± 0.61, P = 0.93). ROC curves showed poor discrimination of ML in predicting progression with AUC of 0.50 at ML ≥ 1. AUC did not vary with different ML cut-points. The utility of the ML to stratify BE patients for risk of progression was not confirmed in this study. The etiology for discrepancies between this and prior studies showing high predictiveness is likely due to the use of crude lysates in this study, but requires further investigation.
Keywords: Barrett's esophagus, biomarkers, esophageal adenocarcinoma
INTRODUCTION
The goal of endoscopic surveillance in Barrett's esophagus (BE) is for early detection of dysplasia and esophageal adenocarcinoma (EAC). Early detection and treatment of malignant and premalignant lesions is associated with improved survival in EAC patients.1–3 However, current surveillance reliant on histologic interpretation of endoscopic biopsies have several shortcomings. Histological evaluation has a high inter- and intraobserver variability and random biopsies are associated with sampling error, resulting in an inaccurate prediction of progression to EAC.4–8 The need for better risk stratification has prompted the investigation of potential biomarkers 9–14 to predict which BE patients are at an increased risk of progression to EAC, so that surveillance and more aggressive treatment could be targeted to this group.
In a previous case-control study,15 we assessed a panel of genetic markers comprised of 10 genomic loci to calculate a mutational load (ML) score that could be used to predict progression to high-grade dysplasia (HGD) or EAC. We found that mean ML was significantly higher in 23 BE subjects who progressed to HGD or EAC compared to 46 who did not progress over a mean time of 4 years. The ML test at a threshold of ≥0.5 was 100% sensitive at predicting progression and the area under the curve (AUC) for the assay at a cut-off of ≥1 was 0.95.
Given that these preliminary data showed that ML could potentially be a robust biomarker for risk stratification in BE, we aimed to validate these findings in a larger, stringently selected community-based population to determine whether ML in preprogression BE tissue predicted risk of progression to HGD or EAC. We hypothesized that study participants with ML ≥ 1 at baseline would be at an increased risk of progression to HGD or EAC.
MATERIALS and METHODS
Study design and setting
We performed a nested case-control study assessing ML in preprogression tissue of nondysplastic BE (NDBE) patients at baseline, who progressed to HGD or EAC (progressors or cases) and those who did not progress (nonprogressors or controls) on follow-up endoscopy. All subjects were recruited as part of the ReBus biorepository at the Academic Medical Centre, Amsterdam (AMC).
Progressors were identified at three tertiary referral centers in the Netherlands between 2000 and 2012. All subsequent surveillance endoscopies were identified through the nationwide network and registry of histology and cytopathology in the Netherlands (PALGA database).16 Progressors were defined as those with initial NDBE who subsequently developed HGD or T1 disease on a resection specimen or on two separate biopsy sessions. Progressors were required to have at least 2 years of surveillance before the diagnosis of HGD/EAC, and to have at least two surveillance exams demonstrating NDBE prior to progression.
Nonprogressors were identified from a BE surveillance registry in 10 community hospitals in the Amsterdam region, comprising the same catchment area from which progressors originated. Nonprogressors were included if they underwent at least two surveillance endoscopies with a minimum surveillance interval of two years and did not develop HGD/EAC. For both progressor and nonprogressor patients, baseline samples were required to contain solely NDBE histology.
Controls were matched to cases in up to a 2:1 ratio by age, sex, BE segment length, and duration of surveillance. Subject demographics, number of surveillance endoscopies, and duration of surveillance were obtained from patient records. Duration of surveillance was defined as time from baseline biopsy to time of progression in cases, and baseline to last surveillance endoscopy with biopsies for controls.
Tissue processing and mutational load analysis
Laboratory personnel blinded to the case-control status assessed ML in baseline tissue of progressors and nonprogressors. To do this, hematoxylin and eosin (H&E)-stained histology slides from formalin fixed, paraffin embedded (FFPE) biopsies were microscopically evaluated to identify BE-related targets that were microdissected from consecutively cut unstained slides.
Mutational analysis was performed on crude lysates of all microdissected histological targets. Briefly, microdissected targets were placed into a Tris/Tween buffer. All samples underwent proteinase K digestion at 56°C for at least 48 hours and 1 round of freeze-thawing at −8°C. An aliquot of the resulting crude lysate was analyzed by quantitative polymerase chain reaction (qPCR) to assess the amount of PCR amplifiable DNA in each microdissected sample. A previously established15,17 qPCR cycle threshold (CT) value (CT < 32 = sufficient; 32–34 = borderline; ≥34 = insufficient) was used to measure amplifiable DNA to assess for loss of heterozygosity (LOH) or microsatellite instability (MSI) mutations. Samples with CT values ≥34 were excluded from the analysis. All mutations were confirmed in triplicate for ML analysis, with confirmation of mutations in all three runs, in order to help avoid PCR-related errors due to contaminates and low template DNA.
ML is a previously determined15,17 summary score of genetic aberration, based on the presence of LOH and MSI mutations at 10 genomic loci (with associated tumor suppressor genes): 1p(CMM1, L-myc), 3p(VHL, HoGG1), 5q(MCC, APC), 9p(CDKN2A), 10q(PTEN, MXI1), 17p(TP53), 17q(NME1), 18q(DCC), 21q(TFF1, PSEN2), and 22q(NF2).18–23 LOH mutations were categorized as high clonality (>75% of the DNA had LOH) or low clonality (50–75% of DNA had LOH) and assigned values of 1 and 0.5, respectively.17 The value of the first measured MSI mutations was assigned 0.75, and each additional MSI present at a genomic locus was assigned a value of 0.5.17 The ML score for each microdissected target was determined by adding together values based on the presence of LOH and MSI mutations and clonality of LOH mutations in each microdissected target. The maximum ML score among all microdissected targets at baseline was defined as the total ML (range: 0–10) for each subject.
Statistical analysis
Descriptive statistics were used to examine characteristics of progressors and nonprogressors, and bivariate analyses were performed to determine the relationship between the two groups and each independent variable. Conditional logistic regression was used to estimate the odds of predicting progression at varying ML cut-points after adjusting for sex, age, duration of surveillance, BE segment length, CT value, and number of surveillance endoscopies. Receiver operator characteristic (ROC) curves were created to assess the diagnostic utility of various cut-offs of ML for progression. All analyses were performed using Stata 13 (College Station, TX).
RESULTS
Of 343 subjects identified as progressors or nonprogressors, 51 were excluded due to lack of NDBE histology in recut tissue slides and 80 were excluded due to insufficient levels of amplifiable DNA in crude lysates. Of the 212 subjects who had amplifiable DNA, 159 were successfully matched and included in the final sample. Of these 159 subjects, there were 58 progressors and 101 nonprogressors. There were no differences between the groups in terms of demographics, baseline BE length, and duration of surveillance (Table 1). Progressors had a higher number of surveillance endoscopies compared to nonprogressors (2.1 ± 1.6 vs. 1.2 ± 2.0, P < 0.01). Most progressor FFPE tissue blocks (73%) were stored in archives for ≥10 years, compared to only 20% of nonprogressor FFPE tissue blocks. Mean CT value was significantly higher in progressors 31.4 ± 1.2 compared to nonprogressors 30.7 ± 1.8, P < 0.01.
Table 1.
Participant characteristics
| Total (n = 159) | Nonprogressor (n = 101) | Progressor (n = 58) | P value | |
|---|---|---|---|---|
| Age, mean ± SD | 58.9 ± 8.1 | 59.1 ± 8.2 | 58.4 ± 7.9 | 0.60 |
| Male, n(%) | 142 (89) | 91 (90) | 51 (88) | 0.67 |
| White, n(%) | 121 (95) | 80 (92) | 41 (100) | 0.32 |
| Barrett segment length, mean ± SD | 4.9 ± 2.7 | 4.8 ± 2.9 | 5.2 ± 2.4 | 0.35 |
| Number of surveillance EGDs, mean ± SD | 1.5 ± 1.9 | 1.2 ± 2.0 | 2.1 ± 1.6 | < 0.01 |
| Duration of surveillance, years, mean ± SD | 4.0 ± 1.6 | 3.9 ± 1.7 | 4.2 ± 1.4 | 0.41 |
Both progressors and nonprogressors had similar mean ML in preprogression tissue lysates: 0.73 ± 0.69 versus 0.74 ± 0.61, P = 0.93 (Table 2). A mean of 2 NDBE histological targets was evaluated in both groups, concordant with the previous study that showed that the test performance characteristics were optimal when ≥2 targets were used to calculate ML. In both groups, 33% had borderline levels of amplifiable DNA (CT 32–34). Approximately 50% of progressors and nonprogressors had either 1 NDBE histological target or borderline levels of amplifiable DNA.
Table 2.
Mutational load characteristics
| Total (n = 159) | Nonprogressor (n = 101) | Progressor (n = 58) | P value | |
|---|---|---|---|---|
| Mutational load, mean ± SD | 0.73 ± 0.64 | 0.74 ± 0.61 | 0.73 ± 0.69 | 0.93 |
| Number of targets, mean ± SD | 2.4 ± 1.4 | 2.3 ± 1.3 | 2.7 ± 1.5 | 0.09 |
| Count of MSI†, n(%) | 0.49 | |||
| 90 (57) | 57 (56) | 33 (57) | ||
| 1 | 55 (35) | 37 (37) | 18 (31) | |
| 2 | 14 (9) | 7 (7) | 7 (12) | |
| Count of low LOH‡, n(%) | 0.57 | |||
| 0 | 102 (64) | 64 (63) | 38 (66) | |
| 1 | 49 (31) | 32 (32) | 17 (29) | |
| 2 | 7 (4) | 5 (5) | 2 (3) | |
| 3 | 1 (1) | 0 (0) | 1 (2) | |
| Count of high LOH, n(%) | 0.73 | |||
| 0 | 135 (85) | 85 (84) | 50 (86) | |
| 1 | 23 (14) | 15 (15) | 8 (14) | |
| 2 | 1 (1) | 1 (1) | 0 (0) | |
| CT§ value, mean ± SD | 30.9 ± 1.6 | 30.7 ± 1.8 | 31.4 ± 1.2 | < 0.01 |
†microsatellite instability; ‡loss of heterozygosity;
§CT < 32 = sufficient amount, 32–34 = borderline amount of amplifiable DNA.
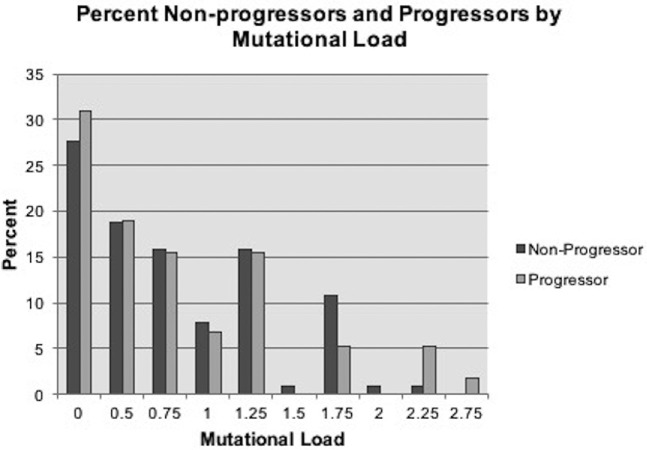
The percent of progressors and nonprogressors at each ML cut-point was assessed (Fig. 1). There were comparable percentages of progressors (31%) and nonprogressors (28%) with a ML = 0 in baseline tissue. A higher number of progressors had ML of 2.25 or higher. However, a higher number (11%) of nonprogressors had ML of ≥1.75 than progressors (5%). Overall, there was no clear trend of higher baseline ML in progressors.
Fig. 1.
Percent of nonprogressors and progressors in each mutational load category.
In multivariable analysis, the odds of predicting progression based on baseline ML was not significantly different between the cases and controls when tested at varying ML cut-points ranging from 0.5 to 1.75 (Table 3). ROC curves showed poor discrimination of the ML test in predicting progression to HGD or EAC in BE with AUC of 0.50 at ML ≥ 1, and AUC did not significantly change at varying ML cut-points.
Table 3.
Odds† of predicting progressor status at varying mutational load (ML) cutpoints and corresponding AUC from ROC curves
| ML | OR | 95%CI | P-value | AUC |
|---|---|---|---|---|
| ≥0.5 | 0.81 | 0.33–1.99 | 0.65 | 0.56 |
| ≥1 | 0.77 | 0.33–1.83 | 0.56 | 0.50 |
| ≥ 1.5 | 0.89 | 0.20–2.90 | 0.70 | 0.58 |
| ≥ 1.75 | 0.83 | 0.23–3.02 | 0.77 | 0.59 |
†Adjusted for sex, age, duration of surveillance, Barrett's segment length, ct value and number of surveillance EGDs.
CI, confidence interval; ML, mutational load; OR, odds ratio.
Post hoc analyses
Several post hoc analyses were performed to better understand the source of discrepancies between the current negative study and the prior study.15 Because we were concerned that the age of the specimen might influence the analysis, we assessed only more recently enrolled patients. However, ML analysis in a subset of subjects whose biopsies were obtained <4 years prior to progression did not improve signal detection in progressors compared to nonprogressors. Similarly, subset analyses in only those with high quality DNA (excluded subjects with borderline levels of PCR amplifiable DNA (CT 32–34) and/or only 1 assessable NDBE target) also did not improve signal detection. Finally, reweighting of LOH and MSI variables used in the ML score did not improve signal compared to noise due to the relatively few mutations detected in progressors.
As mentioned previously, all samples positive for LOH were originally run in triplicate with confirmation of mutations required in all three runs. In post hoc analyses, we applied less stringent criteria to see if it would impact signal strength. Therefore, when we analyzed for LOH signal that was positive in at least one run, the average ML signal in progressors improved to 1.9 but was still not statistically higher than that of nonprogressors who had an average ML of 1.6 (P = 0.14). Unfortunately, re-runs for mutations at all genomic loci in which results were originally negative could not be done due to crude lysate sample volume limitations.
While previous work successfully used purified DNA for ML assessment, this study used crude lysate given use of archival specimens. We hypothesized that ML signal could be relatively ‘muted’ in crude lysates, making the signal in progressors indistinguishable from background noise in nonprogressors at baseline. To better understand the magnitude of ML signal expected at baseline, we examined ML in crude lysates of 11 subjects with HGD or cancer histology from similarly archived tissue. Seven of the 11 subjects had sufficient amounts of PCR amplifiable DNA in crude lysates of FFPE tissue that was archived for an average of 6.2 years, and had an average ML of 2.4. In contrast, prior data showed an average ML in purified DNA of subjects with cancer and HGD histology was 4.1 and 3.3, respectively,17,18 suggesting that the ML in crude lysate samples may be muted compared to purified DNA from microdissection. An ML of <1.5 occurred in 43% of crude lysate DNA of microdissected targets with HGD, markedly less than the <1.5% of such targets based on past studies using purified DNA.17,18
DISCUSSION
This population-based nested case-control study comparing BE progressors and matched nonprogressors demonstrated that a previously identified biomarker panel did not predict progression to early neoplasia in the preprogression samples. Although previous work in a smaller study15 showed that the mean ML in preprogression tissue was significantly higher in BE subjects who progressed to HGD or EAC compared to nonprogressors, these results were not reproducible. Mean ML, the count of MSI, low clonality, and high clonality were not significantly different in progressors versus nonprogressors. Therefore, the utility of the ML score based on LOH and MSI at 10 genomic loci to stratify BE patients for risk of progression to EAC was not confirmed in these data.
A possible source of the inability to detect significant mutations in progressors could be variability in fixation and storage parameters of archived FFPE tissue biopsies. Of progressor biopsies, 73% were archived for ≥10 years, compared to only 20% of nonprogressor samples. Longer storage can decrease the amount of intact, amplifiable DNA.24 However, post hoc analysis of subjects whose biopsies were obtained <4 years prior to progression, to overcome the limitation of samples with longer storage times, did not improve signal detection. Another potential explanation could be the longer duration between baseline biopsy and progression in the current study versus prior study (4.1 years with a minimum requirement of 2 years versus 3.9 years with minimum requirement of 1 year). It may be that the baseline biopsies must be temporally closer to the date of progression in order to have acquired the genetic abnormalities assessed by this panel. Although we cannot exclude this as a potential factor, analysis of subjects with a surveillance duration of less than 4 years did not improve ML signal in progressors relative to nonprogressors.
Based on our post hoc analyses, by far the most notable difference was the use of purified DNA in the prior study and the use of crude lysate in this study for PCR-based ML assessment. While use of purified DNA would be preferable, in this investigation, given the use of archival specimens, crude lysate DNA was required to obtain sufficient levels of amplifiable DNA. The presence of PCR inhibitors in crude lysates and low amounts of DNA available for amplification methods are well-recognized sources of error in detecting genetic abnormalities.25 Although NDBE crude lysates were examined for PCR-based DNA amplifiability prior to the study, we did not do the same to assess the magnitude of ML signal expected in HGD or cancer tissue.17,18 In post hoc analysis, we found that the ML signal in crude lysates of HGD and EAC tissue was lower than the average ML signal observed in purified DNA of the same histology noted in previous studies. Furthermore, the average ML signal observed in crude lysate DNA of such tissue (2.4 ML) was similar to that observed in purified DNA at 3.9 years prior to the onset of HGD or EAC in our past study (2.2 ML). Therefore, it is not surprising that such ‘muted’ ML signal observed at progression in crude lysates was not discernible from noise at 4 years prior to progression. This seems the most likely explanation for the inability to reproduce the previous results.
Biomarkers are essential to accurately risk stratify BE patients who are at the highest risk of progression to guide surveillance strategies. Current risk stratification techniques and detection of dysplasia are based on endoscopy with biopsies, which are limited by sampling error, poor adherence to biopsy collection protocols,26 and variability in histologic assessment of the presence of dysplasia.27 Although the biomarker panel such as the one tested in this study is based on histologic specimens and cannot overcome sampling bias, it offers the advantage of predicting progression to HGD or EAC years prior to it occurring. Because the annual risk of BE progressing to HGD/EAC is <1%,28 testing biomarkers in a prospective fashion would require a large number of subjects and lengthy follow-up times, making this approach costly and time consuming. Therefore, a nested case-control study overcomes these issues and is a good study design when investigating biomarkers in conditions like BE with rare progression outcomes. Another strength of this study is that we conducted rigorous post hoc analyses to understand the reasons for the variation of results of this study compared to the prior.15 The lessons learned from these analyses can be utilized to inform future biomarker studies.
One of the limitations, which is inherent to our study design, is problems with the quantity of amplifiable DNA that could be extracted from the archived tissue specimens. In addition, this study only included NDBE baseline samples, while the previous study included both baseline LGD and NDBE, even though there was an insignificant difference in the proportion of cases with LGD at baseline between progressors and nonprogressors (30.4% cases, 17.4% controls, P = 0.23).15 Finally, in our previous study, subjects were matched by age, sex, and duration of surveillance but not by BE segment length as in this study. BE length is a well-known independent predictor for progression to HGD or EAC,29–31 and it is possible that this discrepancy could be contributing to the difference in results between the two studies.
With respect to next steps, given the conflicting nature of the outcomes of this study and its precursor, potential options include testing this biomarker panel in a prospective case-control trial using purified DNA samples. While time consuming and costly, this approach would overcome the limitations of archived specimens such as poor DNA quality. It would also more reliably confirm a threshold for risk of progression given potential differences in the magnitude of signal observed in archived specimens compared to those undergoing current pathology fixation methods. Based on our post hoc analysis, purified DNA rather than crude lysate should likely be used for ML analysis. Another approach is to re-evaluate the genomic loci used to constitute the ML, and investigate biomarkers that show a higher degree of signal difference from controls at a longer time point prior to progression. While this study was not able to reproduce the results of our prior work in using ML as a predictive marker of progression, it helps elucidate issues that arise with tissue-based, predictive, biomarker development in BE that are important to address as we work towards more optimal risk stratification approaches in BE.
Acknowledgments
This study was funded in part by NIH award number T32 DK07634 (SE), K24DK100548 (NJS). Interpace Diagnostics partly funded efforts to obtain archived pathology specimens, and performed the ML analysis. The authors have no potential competing interests.
Notes
Specific author contributions: (all authors approved the final draft submitted): Study design: Swathi Eluri, Esther Klaver, Lucas C. Duits, Sara A. Jackson, Jacques J. Bergman, Nicholas J. Shaheen; Data interpretation: Swathi Eluri, Esther Klaver, Lucas C. Duits, Jacques J. Bergman, Nicholas J. Shaheen; Manuscript drafting: Swathi Eluri, Esther Klaver, Sara A. Jackson, Nicholas J. Shaheen; Critical revision: Swathi Eluri, Esther Klaver, Lucas C. Duits, Sara A. Jackson, Jacques J. Bergman, Nicholas J. Shaheen; Project conception: Sara A. Jackson, Jacques J. Bergman, Nicholas J. Shaheen.
References
- 1. Incarbone R, Bonavina L, Saino G, Bona D, Peracchia A. Outcome of esophageal adenocarcinoma detected during endoscopic biopsy surveillance for Barrett's esophagus. Surg Endosc 2002; 16: 263–6. [DOI] [PubMed] [Google Scholar]
- 2. van Sandick J W, van Lanschot J J, Kuiken B W, Tytgat G N, Offerhaus G J, Obertop H. Impact of endoscopic biopsy surveillance of Barrett's oesophagus on pathological stage and clinical outcome of Barrett's carcinoma. Gut 1998; 43: 216–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shaheen N J, Falk G W, Iyer P G, Gerson L B. ACG clinical guideline: diagnosis and management of Barrett's esophagus. Am J Gastroenterol 2016; 111: 30–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Corley D A, Mehtani K, Quesenberry C, Zhao W, de Boer J, Weiss N S. Impact of endoscopic surveillance on mortality from Barrett's esophagus-associated esophageal adenocarcinomas. Gastroenterology 2013; 145: 312–319.e1.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shaheen N J, Richter J E. Barrett's oesophagus. Lancet North Am Ed 2009; 373: 850–61. [DOI] [PubMed] [Google Scholar]
- 6. Gatenby P, Soon Y. Barrett's oesophagus: evidence from the current meta-analyses. World J Gastrointest Pathophysiol 2014; 5: 178–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Downs-Kelly E, Mendelin J E, Bennett A E et al. . Poor interobserver agreement in the distinction of high-grade dysplasia and adenocarcinoma in pretreatment Barrett's esophagus biopsies. Am J Gastroenterol 2008; 103: 2333–40. [DOI] [PubMed] [Google Scholar]
- 8. Ormsby A, Petras R, Henricks W et al. . Observer variation in the diagnosis of superficial oesophageal adenocarcinoma. Gut 2002; 51: 671–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Galipeau P C, Li X, Blount P L et al. . NSAIDs modulate CDKN2A, TP53, and DNA content risk for progression to esophageal adenocarcinoma. PLoS Med 2007; 4: e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jin Z, Cheng Y, Gu W et al. . A multicenter, double-blinded validation study of methylation biomarkers for progression prediction in Barrett's esophagus. Cancer Res 2009; 69: 4112–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rabinovitch P S, Longton G, Blount P L, Levine D S, Reid B J. Predictors of progression in Barrett's esophagus III: baseline flow cytometric variables. Am J Gastroenterol 2001; 96: 3071–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu W, Bhagat T D, Yang X et al. . Hypomethylation of noncoding DNA regions and overexpression of the long noncoding RNA, AFAP1-AS1, in Barrett's esophagus and esophageal adenocarcinoma. Gastroenterology 2013; 144: 956–966.e4.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fels Elliott D R, Fitzgerald R C. Molecular markers for Barrett's esophagus and its progression to cancer. Curr Opin Gastroenterol 2013; 29: 437–45. [DOI] [PubMed] [Google Scholar]
- 14. Kastelein F, Biermann K, Steyerberg E W et al. . Aberrant p53 protein expression is associated with an increased risk of neoplastic progression in patients with Barrett's oesophagus. Gut 2013; 62: 1676–83. [DOI] [PubMed] [Google Scholar]
- 15. Eluri S, Brugge W R, Daglilar E S et al. . The presence of genetic mutations at key loci predicts progression to esophageal adenocarcinoma in Barrett's esophagus. Am J Gastroenterol 2015; 110: 828–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Casparie M, Tiebosch A T, Burger G et al. . Pathology databanking and biobanking in The Netherlands, a central role for PALGA, the nationwide histopathology and cytopathology data network and archive. Cell Oncol 2007; 29: 19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Khara H S, Jackson S A, Nair S et al. . Assessment of mutational load in biopsy tissue provides additional information about genomic instability to histological classifications of Barrett's esophagus. J Gastrointest Canc 2014; 45: 137–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ellsworth E, Jackson S A, Thakkar S J, Smith D M Jr, Finkelstein S. Correlation of the presence and extent of loss of heterozygosity mutations with histological classifications of Barrett's esophagus. BMC Gastroenterol 2012; 12: 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dolan K, Garde J, Walker S J, Sutton R, Gosney J, Field J K. LOH at the sites of the DCC, APC, and TP53 tumor suppressor genes occurs in Barrett's metaplasia and dysplasia adjacent to adenocarcinoma of the esophagus. Hum Pathol 1999; 30: 1508–14. [DOI] [PubMed] [Google Scholar]
- 20. Raja S, Finkelstein S D, Baksh F K et al. . Correlation between dysplasia and mutations of six tumor suppressor genes in Barrett's esophagus. Ann Thorac Surg 2001; 72: 1130–5. [DOI] [PubMed] [Google Scholar]
- 21. Lin X, Finkelstein S D, Zhu B, Ujevich B J, Silverman J F. Loss of heterozygosities in Barrett esophagus, dysplasia, and adenocarcinoma detected by esophageal brushing cytology and gastroesophageal biopsy. Cancer 2009; 117: 57–66. [DOI] [PubMed] [Google Scholar]
- 22. Barrett M T, Galipeau P C, Sanchez C A, Emond M J, Reid B J. Determination of the frequency of loss of heterozygosity in esophageal adenocarcinoma by cell sorting, whole genome amplification and microsatellite polymorphisms. Oncogene 1996; 12: 1873–8. [PubMed] [Google Scholar]
- 23. Sanz-Ortega J, Hernandez S, Saez M C et al. . 3p21, 5q21, 9p21 and 17p13.1 allelic deletions are potential markers of individuals with a high risk of developing adenocarcinoma in Barrett's epithelium without dysplasia. Hepatogastroenterology 2003; 50: 404–7. [PubMed] [Google Scholar]
- 24. Srinivasan M, Sedmak D, Jewell S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am J Pathol 2002; 161: 1961–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Budowle B, Eisenberg A J, van Daal A. Validity of low copy number typing and applications to forensic science. Croat Med J 2009; 50: 207–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Curvers W L, Peters F P, Elzer B et al. . Quality of Barrett's surveillance in The Netherlands: a standardized review of endoscopy and pathology reports. Eur J Gastroenterol Hepatol 2008; 20: 601–7. [DOI] [PubMed] [Google Scholar]
- 27. Kerkhof M, van Dekken H, Steyerberg E W et al. . Grading of dysplasia in Barrett's oesophagus: substantial interobserver variation between general and gastrointestinal pathologists. Histopathology 2007; 50: 920–7. [DOI] [PubMed] [Google Scholar]
- 28. Sikkema M, de Jonge P J, Steyerberg E W, Kuipers E J. Risk of esophageal adenocarcinoma and mortality in patients with Barrett's esophagus: a systematic review and meta-analysis. Clin Gastroenterol Hepatol 2010; 8: 235–44; quiz e32. [DOI] [PubMed] [Google Scholar]
- 29. Gatenby P A, Caygill C P, Ramus J R, Charlett A, Fitzgerald R C, Watson A. Short segment columnar-lined oesophagus: an underestimated cancer risk? A large cohort study of the relationship between Barrett's columnar-lined oesophagus segment length and adenocarcinoma risk. Eur J Gastroenterol Hepatol 2007; 19: 969–75. [DOI] [PubMed] [Google Scholar]
- 30. Sikkema M, Looman C W, Steyerberg E W et al. . Predictors for neoplastic progression in patients with Barrett's esophagus: a prospective cohort study. Am J Gastroenterol 2011; 106: 1231–8. [DOI] [PubMed] [Google Scholar]
- 31. Wong T, Tian J, Nagar A B. Barrett's surveillance identifies patients with early esophageal adenocarcinoma. Am J Med 2010; 123: 462–7. [DOI] [PubMed] [Google Scholar]