Abstract
The presence of Fusobacterium nucleatum in colorectal carcinoma tissue has been associated with microsatellite instability (MSI), lower-level T-cell infiltrates, and poor clinical outcomes. Considering differences in the tumor-immune microenvironment of carcinomas with high or low MSI, we hypothesized that the association of F. nucleatum with immune response might differ by tumor MSI status. Using samples from 1,041 rectal and colon cancer patients within the Nurses’ Health Study and Health Professionals Follow-up Study, we measured F. nucleatum DNA in tumor tissue by a quantitative polymerase chain reaction assay. Multivariable logistic regression models were used to examine the association between F. nucleatum status and histopathologic lymphocytic reactions or density of CD3+ cells, CD8+ cells, CD45RO (PTPRC)+ cells, or FOXP3+ cells in strata of tumor MSI status. We adjusted for potential confounders including CpG island methylator phenotype, LINE-1 methylation, and KRAS, BRAF, and PIK3CA mutations. The association of F. nucleatum with tumor-infiltrating lymphocytes (TIL) and intratumoral periglandular reaction differed by tumor MSI status (Pinteraction = 0.002). The presence of F. nucleatum was negatively associated with TIL in MSI-high tumors (multivariable odds ratio, 0.45; 95% confidence interval, 0.22–0.92), but positively associated with TIL in non-MSI-high tumors (multivariable odds ratio, 1.91; 95% confidence interval, 1.12–3.25). No significant differential association was observed for peritumoral lymphocytic reaction, Crohn’s-like lymphoid reaction, or T-cell densities. In conclusion, the association of F. nucleatum with immune response to colorectal carcinoma differs by tumor MSI status, suggesting that F. nucleatum and MSI status interact to affect anti-tumor immune reactions.
Keywords: colorectal neoplasms, immunology, microbiota, molecular pathological epidemiology, tumor microenvironment
Introduction
Accumulating evidence continues to illustrate the importance of the host immune system in regulating the evolution and progression of neoplastic diseases (1, 2). The colorectum is rich in microbes, and the gut microbiome plays a role in regulating local and systemic immune status (3). Fusobacterium nucleatum (F. nucleatum) has emerged as a potentially influential bacterium implicated in colorectal carcinogenesis (4–7). Metagenomic and clinical studies have highlighted the enrichment of F. nucleatum in colorectal cancer tissue relative to adjacent normal epithelium (5–9), and its persistence in metastatic lesions (10). Experimental evidence indicates that F. nucleatum can elicit inflammatory reactions, impair T cell-mediated anti-tumor immune response, and promote tumor cell proliferation (11–15). Studies have also demonstrated the association of F. nucleatum in colorectal cancer tissue with proximal tumor location, microsatellite instability (MSI), CpG island methylator phenotype (CIMP), lower levels of tumor-infiltrating CD3+ cells, and poor patient prognosis (8, 16–19).
Colorectal cancer consists of a heterogeneous collection of neoplasms that evolves through stepwise accumulation of genetic and epigenetic aberrations, a process influenced by environmental exposures, the microbiota, and host factors (20–22). MSI-high colorectal carcinomas typically contain numerous frameshift mutations and generate immunogenic peptides (“neoantigens”) due to mismatch repair deficiency, which encourage the anti-tumor immune response and lead to more favorable prognosis and better response to immunotherapies (23–28). In fact, the U.S. Food and Drug Administration (FDA) approved the use of anti-PDCD1 (programmed cell death 1, PD-1) antibodies, pembrolizumab and nivolumab, for colorectal cancer with high-level MSI or mismatch repair deficiency. Considering the differences in immune microenvironment between MSI-high and non-MSI-high tumors, we hypothesized that the association of F. nucleatum in colorectal carcinoma tissue with immune response might differ by tumor MSI status.
To test this hypothesis, we utilized a molecular pathological epidemiology (MPE) database of over 1,000 colorectal carcinoma cases in two U.S. prospective cohort studies, with data on clinical, pathological, and tumor molecular characteristics. This comprehensive dataset enabled us to assess whether the association of F. nucleatum in colorectal cancer tissue with immune response to tumor might differ by MSI status while adjusting for a number of potential confounders.
Materials and Methods
Study population
We utilized two prospective cohort studies in the U.S., the Nurses’ Health Study (NHS, 121,701 women aged 30–55 years followed since 1976) and the Health Professionals Follow-up Study (HPFS, 51,529 men aged 40–75 years followed since 1986). Every two years, participants have been sent follow-up questionnaires to report lifestyle factors and newly-diagnosed diseases including colorectal cancer. The follow-up rate was > 90% for each follow-up questionnaire in both cohort studies. In both cohorts, the National Death Index was used to ascertain deaths of participants and identify unreported lethal colorectal cancer cases. Study physicians, blinded to exposure data, reviewed medical records of identified colorectal cancer cases to confirm the disease diagnosis and to collect data on clinical characteristics (e.g., tumor location, disease stage).
Among participants diagnosed with colorectal cancer up to 2012, we analyzed 1,041 cases with available data on F. nucleatum and immune response in tumor tissue samples. We included both colon and rectal carcinomas, based on the colorectal continuum model (29, 30). We excluded patients who had been preoperatively treated. We collected formalin-fixed paraffin-embedded (FFPE) tumor tissue blocks from hospitals throughout the U.S. where colorectal cancer patients had undergone surgical resection. A single pathologist (S. Ogino), blinded to other data, reviewed hematoxylin and eosin-stained tissue sections from all collected blocks and recorded pathological features. Tumor differentiation was categorized into well/moderate vs. poor (> 50% vs. ≤ 50% gland formation, respectively).
Written informed consent was obtained from all participants at enrollment in the NHS and HPFS. This study was conducted in accordance with the Declaration of Helsinki, and after approval by the institutional review boards at Harvard T.H. Chan School of Public Health and Brigham and Women’s Hospital (Boston, MA).
Quantitative polymerase chain reaction (PCR)
DNA was extracted from colorectal cancer tissue in archival FFPE tissue sections using QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany). As previously described and validated (8), we performed a quantitative polymerase chain reaction (PCR) assay to measure the amount of F. nucleatum DNA in the tumor using custom TaqMan primer/probe sets (Applied Biosystems, Foster City, CA) for the nusG gene of F. nucleatum and the reference human gene SLCO2A1. Amplification and detection of DNA were performed using the StepOnePlus Real-Time PCR Systems (Applied Biosystems). Amount of intratumoral F. nucleatum DNA in each specimen were calculated as a relative value normalized to amount of SLCO2A1 using the 2−ΔCt method [where ΔCt (cycle threshold) = ‘the mean Ct value of F. nucleatum’ - ‘the mean Ct value of SLCO2A1’] (8). Each specimen was analyzed in duplicate for each target in a single batch, and the mean of the two Ct values was used for each target. In our previous validation study, the Ct values linearly decreased with the amount of input F. nucleatum DNA (r2 > 0.99), and the interassay coefficient of variation of the Ct values in five different batches was < 1% for all targets (8). Tumors with any detectable F. nucleatum DNA were classified as F. nucleatum-positive, whereas all other tumors were classified as F. nucleatum-negative.
We performed a quantitative PCR assay to measure the amount of Bifidobacterium genus DNA in the tumor using custom TaqMan primer/probe sets (Applied Biosystems) for the 16S ribosomal RNA gene of Bifidobacterium genus and for the reference gene 16S (31). The primer and probe sequences for each TaqMan Gene Expression Assay were as follows: Bifidobacterium forward primer, 5′-CGGGTGAGTAATGCGTGACC-3′; Bifidobacterium reverse primer, 5′-TGATAGGACGCGACCCCA-3′; Bifidobacterium FAM probe, 5′-CTCCTGGAAACGGGTG-3′; universal 16S forward primer, 5′-CGGTGAATACGTTCCCGG-3′; universal 16S reverse primer, 5′-TACGGCTACCTTGTTACGACTT-3′; and universal 16S FAM probe, 5′-CTTGTACACACCGCCCGTC-3′. We used 80 ng DNA in each reaction under the same reaction conditions as for F. nucleatum (8). Amounts of intratumoral Bifidobacterium genus DNA in each specimen were calculated as a relative value normalized to amounts of 16S using the 2−ΔCt method (8). Each specimen was analyzed in duplicate for each target in a single batch, and the mean of the two Ct values was used for each target. Tumors with any detectable Bifidobacterium genus DNA were classified as bifidobacteria-positive, whereas all other tumors were classified as bifidobacteria-negative.
Microsatellite instability (MSI) status and other molecular characteristics
MSI status was analyzed using 10 microsatellite markers (D2S123, D5S346, D17S250, BAT25, BAT26, BAT40, D18S55, D18S56, D18S67, and D18S487), as previously described (29). MSI-high was defined as presence of instability in ≥ 30% of the markers and non-MSI-high as instability in < 30% of the markers. Using bisulfite-treated DNA, we determined methylation status of eight CIMP-specific promoters (CACNA1G, CDKN2A, CRABP1, IGF2, MLH1, NEUROG1, RUNX3, and SOCS1) and long interspersed nucleotide element-1 (LINE-1) as previously described (29). CIMP-high was defined as ≥ 6 methylated promoters of eight promoters, and CIMP-low/negative as 0–5 methylated promoters. PCR and pyrosequencing were performed for KRAS (codons 12, 13, 61, and 146), BRAF (codon 600), and PIK3CA (exons 9 and 20), as previously described (29). We constructed tissue microarrays of colorectal cancer cases with sufficient tissue materials, including up to four tumor cores from each case in one tissue microarray block (32). As previously described (33, 34), immunohistochemistry for CD274 (PD-L1) and PDCD1LG2 (PDCD1 ligand 2, PD-L2) was performed using an anti-CD274 (clone MIH1; dilution, 1:50; eBioscience, San Diego, CA) and anti-PDCD1LG2 (clone 366C.9E5; dilution, 1:6,000; provided by the laboratory of G.J. Freeman), respectively. We assessed overall tumor CD274 expression as an ordinal scale of 0–4 by summing cytoplasmic expression score [absent (0), weak (1), moderate (2), or strong (3)] and membrane expression score [absent (0) or present (1)], and categorized CD274 as low (overall score of 0 to 1) or high (2 to 4) (33). We calculated the percentage of tumor cells expressing PDCD1LG2 in the cytoplasm or membrane, and grouped the data into bins of 0–20%, 21–50%, 51–80%, or 81–100% (34).
Lymphocytic reaction to colorectal cancer
Lymphocytic reaction to tumor was histopathologically evaluated on whole tissue sections, as previously described (23). Four components of lymphocytic reaction were examined, including tumor-infiltrating lymphocytes (TIL), intratumoral periglandular reaction, peritumoral lymphocytic reaction, and Crohn’s-like lymphoid reaction (Fig. 1). TIL was defined as lymphocytes on top of cancer cells in a tissue section. Intratumoral periglandular reaction was defined as lymphocytic reaction in tumor stroma within a tumor mass. Peritumoral lymphocytic reaction was defined as discrete lymphoid reaction surrounding a tumor mass.Crohn’s-like lymphoid reaction was defined as transmural lymphoid reaction.Each of the four components was graded as negative/low, intermediate, or high.A subset of cases (n = 398) were independently reviewed by a second pathologist (J.N. Glickman) with a good inter-observer correlation as previously described (23). We constructed tissue microarrays, and measured densities (cells/mm2) of CD3+ cells, CD8+ cells, CD45RO (an isoform of the PTPRC protein)+ cells, and FOXP3+ cells in colorectal cancer tissue based on immunohistochemistry and image analysis using an automated scanning microscope and the Ariol image analysis system (Genetix, San Jose, CA) as previously described (24).
Figure 1.
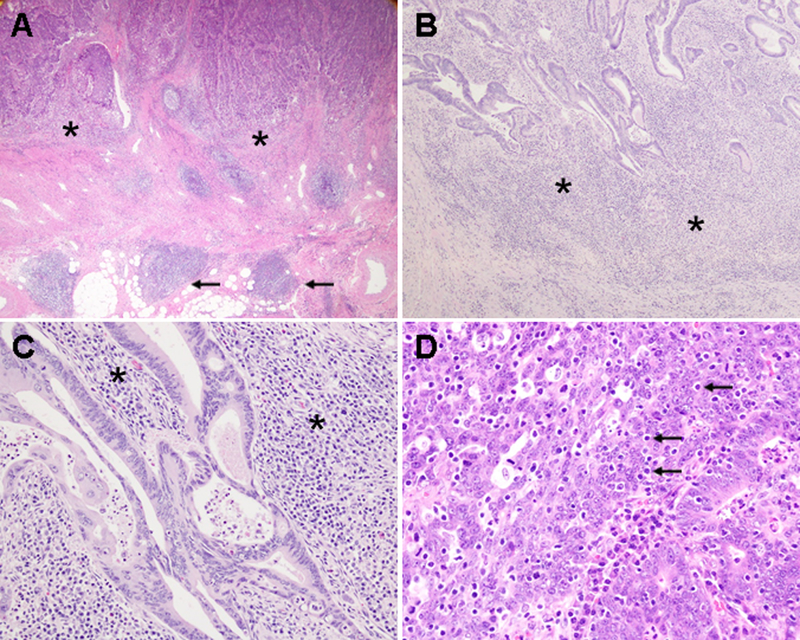
Lymphocytic reaction to colorectal cancer. A, Crohn-like lymphoid reaction (arrows) and peritumoral lymphocytic reaction (asterisks; original magnification, 20x). B, peritumoral lymphocytic reaction (asterisks; original magnification, 40x). C, intratumoral periglandular reaction (asterisks; original magnification, 200x). D, tumor-infiltrating lymphocytes (arrows; original magnification, 400x).
Statistical analysis
Our primary hypothesis test assessed a statistical interaction between F. nucleatum status in colorectal cancer tissue (positive vs. negative) and MSI status (high vs. non-high) in relation to immune response status. We assessed statistical interactions using the Wald test for the cross-product in multivariable-adjusted logistic regression models. In our primary hypothesis testing on new discoveries, we used the α level of 0.005. We conducted all other assessments including evaluations of individual odds ratios (ORs) as secondary analyses, and interpreted the results cautiously in addition to use of the α level of 0.005. Multivariable logistic regression models initially included age (continuous), sex, year of diagnosis (continuous), family history of colorectal cancer (absent vs. present), tumor location (proximal colon vs. distal colon vs. rectum), CIMP status (high vs. low/negative), LINE-1 methylation level (continuous), KRAS (mutant vs. wild-type), BRAF (mutant vs. wild-type), and PIK3CA (mutant vs. wild-type). A backward elimination with a threshold P of 0.05 was used to select variables for the final models. Cases with missing data [family history of colorectal cancer (0.8%), tumor location (0.3%), CIMP status (5.4%), KRAS (7.7%), BRAF (1.1%), and PIK3CA (6.6%)] were included in the majority category of a given categorical covariate to limit the degrees of freedom of the models. For cases with missing data on LINE-1 methylation level (2.6%), we assigned a separate indicator variable. We confirmed that the exclusion of cases with missing data in any of the variables did not substantially alter our results. The proportional odds assumption was assessed using the ordinal logistic regression model. We observed evidence of violation of this assumption in three of the four lymphocytic reaction patterns (TIL, intratumoral periglandular reaction, and peritumoral lymphocytic reaction) and T-cell quartile variables. We therefore used binary histological lymphocytic reaction variables and T-cell variables dichotomized at the median value as outcome variables in logistic regression analyses.
In secondary analyses, we assessed a statistical interaction between F. nucleatum status in colorectal cancer tissue (positive vs. negative) and MSI status (high vs. non-high) in relation to colorectal cancer-specific or overall mortality. We assessed statistical interactions using the Wald test for the cross-product in multivariable-adjusted Cox proportional hazards regression models. Multivariable Cox regression models initially included tumor differentiation (well-moderate vs. poor) and disease stage (I/II vs. III/IV) in addition to the same set of covariates as in the multivariable logistic regression models. A backward elimination with a threshold P of 0.05 was used to select variables for the final models. Cases with missing data on tumor differentiation (0.1%) were included in the majority category, and the other covariates were dealt as in the multivariable logistic regression models. We confirmed that the exclusion of cases with missing data in any of the variables did not substantially alter our results. The assumption of proportionality of hazards was generally satisfied using the assessment of a time-varying covariate; i.e., the cross-product of F. nucleatum status and survival time in strata of MSI status (P > 0.2). Cumulative survival probabilities were estimated using the Kaplan-Meier method and compared using the log-rank test. For colorectal cancer-specific mortality, deaths from other causes were treated as censored events. Using a single regression model with reparameterization of an interaction term (32), we calculated ORs for immune response status and hazard ratios for colorectal cancer mortality comparing F. nucleatum-positive to F. nucleatum-negative tumors in the two strata of MSI status.
All statistical analyses were performed using SAS software (version 9.4; SAS Institute, Cary, NC), and all P values were two-sided.
Results
Among the 1,041 colorectal carcinoma cases from the two prospective cohort studies, F. nucleatum DNA was detected using the quantitative PCR assay in 135 (13%) cases. Table 1 shows clinical, pathological and molecular features of colorectal cancer cases according to combined tumor tissue F. nucleatum and MSI status. As previously reported (8), MSI-high tumors were associated with detectable F. nucleatum DNA. There was a trend toward a higher rate of detectable intratumor F. nucleatum in more recent cases, particularly for MSI-high tumors. High persistence of F. nucleatum has been observed in metastatic liver lesions from F. nucleatum-positive colorectal cancer (10). We examined the correlations between F. nucleatum and disease stage in strata of tumor MSI status. We observed a significant positive correlation between F. nucleatum and disease stage in non-MSI-high tumors (P = 0.004, with the α level of 0.005), but not in MSI-high tumors (P = 0.34). However, statistical power was limited in the analysis of MSI-high tumors.
Table 1.
Clinical, pathological, and molecular characteristics of patients with colorectal cancer and their samples, according to Fusobacterium nucleatum (F. nucleatum) status in tumor tissue, stratified by microsatellite instability (MSI) status
| Non-MSI-high |
MSI-high |
|||||
|---|---|---|---|---|---|---|
|
F. nucleatum in tumor |
F. nucleatum in tumor |
|||||
| All casesa | Negativea | Positivea | Negativea | Positivea | ||
| Characteristic | (n = 1,041) | (n = 787) | (n = 77) | (n = 119) | (n = 58) | |
| Sex | Female (NHS) | 609 (59%) | 437 (56%) | 42 (55%) | 85 (71%) | 45 (78%) |
| Male (HPFS) | 432 (41%) | 350 (44%) | 35 (45%) | 34 (29%) | 13 (22%) | |
| Mean age ± SD (years) | 69.5 ± 8.9 | 68.9 ± 9.0 | 69.5 ± 9.6 | 72.1 ± 7.6 | 72.5 ± 8.4 | |
| Race/ethnicity | White | 995 (97%) | 751 (97%) | 74 (99%) | 114 (97%) | 56 (98%) |
| Black | 16 (1.6%) | 13 (1.7%) | 0 | 2 (1.7%) | 1 (1.8%) | |
| Other | 11 (1.1%) | 8 (1.0%) | 1 (1.3%) | 2 (1.7%) | 0 | |
| Year of diagnosis | 1995 or before | 343 (33%) | 285 (36%) | 21 (27%) | 29 (24%) | 8 (14%) |
| 1996-2000 | 286 (27%) | 216 (27%) | 25 (32%) | 33 (28%) | 12 (21%) | |
| 2001-2010 | 412 (40%) | 286 (36%) | 31 (40%) | 57 (48%) | 38 (66%) | |
| Family history of colorectal cancer in first-degree relative(s) | Absent | 829 (80%) | 632 (81%) | 66 (88%) | 84 (71%) | 47 (81%) |
| Present | 204 (20%) | 150 (19%) | 9 (12%) | 34 (29%) | 11 (19%) | |
| Prediagnosis body mass indexb | < 25 kg/m2 | 417 (40%) | 311 (40%) | 30 (39%) | 49 (42%) | 27 (47%) |
| 25-29.9 kg/m2 | 424 (41%) | 330 (42%) | 35 (46%) | 43 (36%) | 16 (28%) | |
| ≥ 30 kg/m2 | 194 (19%) | 142 (18%) | 11 (14%) | 26 (22%) | 15 (26%) | |
| Prudent dietary patternc | Quartile 1 (lowest) | 258 (25%) | 187 (24%) | 25 (33%) | 30 (25%) | 16 (28%) |
| Quartile 2 | 258 (25%) | 206 (26%) | 19 (25%) | 21 (18%) | 12 (21%) | |
| Quartile 3 | 257 (25%) | 187 (24%) | 21 (28%) | 32 (27%) | 17 (29%) | |
| Quartile 4 (highest) | 258 (25%) | 199 (26%) | 11 (14%) | 35 (30%) | 13 (22%) | |
| Western dietary patternc | Quartile 1 (lowest) | 258 (25%) | 198 (25%) | 10 (13%) | 31 (26%) | 19 (33%) |
| Quartile 2 | 258 (25%) | 192 (25%) | 21 (28%) | 32 (27%) | 13 (22%) | |
| Quartile 3 | 258 (25%) | 200 (26%) | 20 (26%) | 21 (18%) | 17 (29%) | |
| Quartile 4 (highest) | 258 (25%) | 190 (24%) | 25 (33%) | 34 (29%) | 9 (16%) | |
| Tumor location | Proximal colon | 522 (50%) | 332 (42%) | 33 (43%) | 104 (87%) | 53 (91%) |
| Distal colon | 297 (29%) | 264 (34%) | 20 (26%) | 10 (8.4%) | 3 (5.2%) | |
| Rectum | 219 (21%) | 189 (24%) | 23 (30%) | 5 (4.2%) | 2 (3.5%) | |
| Tumor differentiation | Well to moderate | 936 (90%) | 745 (95%) | 67 (88%) | 86 (72%) | 38 (66%) |
| Poor | 104 (10%) | 42 (5.3%) | 9 (12%) | 33 (28%) | 20 (34%) | |
| AJCC disease stage | I | 239 (25%) | 197 (27%) | 8 (11%) | 25 (22%) | 9 (17%) |
| II | 314 (33%) | 199 (28%) | 21 (30%) | 64 (56%) | 30 (56%) | |
| III | 280 (29%) | 222 (31%) | 28 (39%) | 18 (16%) | 12 (22%) | |
| IV | 127 (13%) | 103 (14%) | 14 (20%) | 7 (6.1%) | 3 (5.6%) | |
| CIMP status | CIMP-low/negative | 794 (81%) | 688 (92%) | 68 (94%) | 24 (21%) | 14 (25%) |
| CIMP-high | 191 (19%) | 56 (7.5%) | 4 (5.6%) | 89 (79%) | 42 (75%) | |
| Mean LINE-1 methylation level ± SD (%) | 63.6 ± 10.4 | 62.6 ± 10.2 | 62.2 ± 9.4 | 68.4 ± 10.3 | 70.5 ± 9.5 | |
| KRAS mutation | Wild-type | 541 (56%) | 390 (52%) | 30 (41%) | 88 (85%) | 33 (82%) |
| Mutant | 420 (44%) | 353 (48%) | 44 (59%) | 16 (15%) | 7 (18%) | |
| BRAF mutation | Wild-type | 858 (83%) | 709 (91%) | 73 (95%) | 55 (47%) | 21 (37%) |
| Mutant | 172 (17%) | 70 (9.0%) | 4 (5.2%) | 62 (53%) | 36 (63%) | |
| PIK3CA mutation | Wild-type | 812 (84%) | 617 (84%) | 59 (81%) | 91 (84%) | 45 (85%) |
| Mutant | 160 (16%) | 121 (16%) | 14 (19%) | 17 (16%) | 8 (15%) | |
| Tumor CD274 (PD-L1) expression | Low | 263 (41%) | 176 (37%) | 29 (52%) | 37 (46%) | 21 (64%) |
| High | 377 (59%) | 294 (63%) | 27 (48%) | 44 (54%) | 12 (36%) | |
| Tumor PDCD1LG2 (PD-L2) expression | 0-20% | 160 (25%) | 111 (24%) | 19 (37%) | 22 (28%) | 8 (25%) |
| 21-50% | 148 (23%) | 110 (23%) | 11 (21%) | 17 (21%) | 10 (31%) | |
| 51-80% | 197 (31%) | 150 (32%) | 12 (23%) | 25 (31%) | 10 (31%) | |
| 81-100% | 129 (20%) | 99 (21%) | 10 (19%) | 16 (20%) | 4 (13%) | |
| Bifidobacterium genus DNA | Negative | 748 (73%) | 576 (75%) | 54 (71%) | 82 (69%) | 36 (63%) |
| Positive | 276 (27%) | 197 (25%) | 22 (29%) | 36 (31%) | 21 (37%) | |
Abbreviations: AJCC, American Joint Committee on Cancer; CIMP, CpG island methylator phenotype; HPFS, Health Professionals Follow-up Study; LINE-1, long interspersed nucleotide element-1; MSI, microsatellite instability; NHS, Nurses’ Health Study; SD, standard deviation.
Percentage indicates the proportion of cases with a specific clinical, pathological, or molecular characteristic in all cases or in strata of F. nucleatum and MSI status. Total percentages may not equal 100% due to rounding.
Calculated by dividing weight (kilogram) by squared height (meter).
The prudent dietary pattern is characterized by high intake of vegetables, fruits, fish, poultry, and whole grains, and the western dietary pattern is characterized by high intake of red and processed meats, added sugar, and refined grains (42).
Table 2 shows the distribution of colorectal carcinoma cases according to F. nucleatum status and histological lymphocytic reaction patterns in strata of MSI status. The presence of F. nucleatum in tumor tissue was negatively correlated with intratumoral periglandular reaction in MSI-high tumors (P = 0.002, with the α level of 0.005), and was positively correlated with Crohn’s-like lymphoid reaction in non-MSI-high tumors (P = 0.001).
Table 2.
Distribution of colorectal cancer samples according to Fusobacterium nucleatum (F. nucleatum) status in tumor tissue and lymphocytic reaction status, stratified by microsatellite instability (MSI) status
|
F. nucleatum in colorectal cancer tissue |
|||||
|---|---|---|---|---|---|
| All casesa | Negativea | Positivea | Pb | ||
| Non-MSI-high | Tumor-infiltrating lymphocytes (TIL) (n = 862) | 0.013 | |||
| Negative/low | 699 (81%) | 644 (82%) | 55 (71%) | ||
| Intermediate | 119 (14%) | 107 (14%) | 12 (16%) | ||
| High | 44 (5.1%) | 34 (4.3%) | 10 (13%) | ||
| Intratumoral periglandular reaction (n = 863) | 0.57 | ||||
| Negative/low | 124 (14%) | 106 (13%) | 18 (23%) | ||
| Intermediate | 660 (76%) | 613 (78%) | 47 (61%) | ||
| High | 79 (9.2%) | 67 (8.5%) | 12 (16%) | ||
| Peritumoral lymphocytic reaction (n = 859) | 0.89 | ||||
| Negative/low | 138 (16%) | 123 (16%) | 15 (20%) | ||
| Intermediate | 615 (72%) | 567 (72%) | 48 (63%) | ||
| High | 106 (12%) | 93 (12%) | 13 (17%) | ||
| Crohn’s-like lymphoid reaction (n = 723) | 0.001 | ||||
| Negative/low | 586 (81%) | 544 (83%) | 42 (66%) | ||
| Intermediate | 111 (15%) | 92 (14%) | 19 (30%) | ||
| High | 26 (3.6%) | 23 (3.5%) | 3 (4.7%) | ||
| MSI-high | Tumor-infiltrating lymphocytes (TIL) (n = 177) | 0.055 | |||
| Negative/low | 46 (26%) | 25 (21%) | 21 (36%) | ||
| Intermediate | 64 (36%) | 45 (38%) | 19 (33%) | ||
| High | 67 (38%) | 49 (41%) | 18 (31%) | ||
| Intratumoral periglandular reaction (n = 177) | 0.002 | ||||
| Negative/low | 12 (6.8%) | 1 (0.8%) | 11 (19%) | ||
| Intermediate | 100 (56%) | 68 (57%) | 32 (55%) | ||
| High | 65 (37%) | 50 (42%) | 15 (26%) | ||
| Peritumoral lymphocytic reaction (n = 177) | 0.062 | ||||
| Negative/low | 13 (7.3%) | 4 (3.4%) | 9 (16%) | ||
| Intermediate | 91 (51%) | 62 (52%) | 29 (50%) | ||
| High | 73 (41%) | 53 (45%) | 20 (34%) | ||
| Crohn’s-like lymphoid reaction (n = 151) | 0.13 | ||||
| Negative/low | 64 (42%) | 41 (41%) | 23 (44%) | ||
| Intermediate | 48 (32%) | 26 (26%) | 22 (42%) | ||
| High | 39 (26%) | 32 (32%) | 7 (13%) | ||
Abbreviation: MSI, microsatellite instability.
Percentage indicates the proportion of cases with a specific level of lymphocytic reaction in strata of MSI status or in strata of F. nucleatum and MSI status. Total percentages may not equal 100% due to rounding.
In our primary hypothesis test using logistic regression models (Table 3 and Supplementary Table S1), we observed a statistically significant interaction between F. nucleatum and MSI status in relation to TIL or intratumoral periglandular reaction (Pinteraction = 0.002, with the α level of 0.005). The presence of F. nucleatum in tumor tissue was negatively associated with degrees of TIL in MSI-high colorectal cancer [multivariable OR, 0.45; 95% confidence interval (CI), 0.22–0.92], but positively associated with TIL in non-MSI-high cancer (multivariable OR, 1.91; 95% CI, 1.12–3.25). Similarly, the presence of F. nucleatum in tumor tissue was negatively associated with intratumoral periglandular reaction in MSI-high tumors (multivariable OR, 0.43; 95% CI, 0.21–0.87), but positively associated with intratumoral periglandular reaction (multivariable OR, 1.97, 95% CI, 1.00–3.86) in non-MSI-high tumors. The statistical interaction of F. nucleatum and MSI status in relation to peritumoral lymphocytic reaction or Crohn’s-like lymphoid reaction was not significant with the α level of 0.005 (Pinteraction > 0.01).
Table 3.
Logistic regression analyses to assess the association of Fusobacterium nucleatum (F. nucleatum) status in colorectal cancer tissue with lymphocytic reaction status, in strata of microsatellite instability (MSI) status
| Univariable OR (95% CI) |
Multivariable OR (95% CI)a |
||
|---|---|---|---|
| Model for tumor-infiltrating lymphocytes (TIL)b | |||
| (as an outcome variable) (n = 1,039) | |||
| Non-MSI-high | F. nucleatum-negative | 1 (referent) | 1 (referent) |
| F. nucleatum-positive | 1.83 (1.08-3.09) | 1.91 (1.12-3.25) | |
| MSI-high | F. nucleatum-negative | 1 (referent) | 1 (referent) |
| F. nucleatum-positive | 0.47 (0.23-0.94) | 0.45 (0.22-0.92) | |
| Pinteractionc | 0.002 | 0.002 | |
| Model for intratumoral periglandular reactionb | |||
| (as an outcome variable) (n = 1,040) | |||
| Non-MSI-high | F. nucleatum-negative | 1 (referent) | 1 (referent) |
| F. nucleatum-positive | 1.98 (1.02-3.85) | 1.97 (1.00-3.86) | |
| MSI-high | F. nucleatum-negative | 1 (referent) | 1 (referent) |
| F. nucleatum-positive | 0.48 (0.24-0.96) | 0.43 (0.21-0.87) | |
| Pinteractionc | 0.004 | 0.002 | |
| Model for peritumoral lymphocytic reactionb | |||
| (as an outcome variable) (n = 1,036) | |||
| Non-MSI-high | F. nucleatum-negative | 1 (referent) | 1 (referent) |
| F. nucleatum-positive | 1.53 (0.81-2.89) | 1.44 (0.76-2.76) | |
| MSI-high | F. nucleatum-negative | 1 (referent) | 1 (referent) |
| F. nucleatum-positive | 0.66 (0.34-1.26) | 0.54 (0.27-1.05) | |
| Pinteractionc | 0.068 | 0.038 | |
| Model for Crohn’s-like lymphoid reactionb | |||
| (as an outcome variable) (n = 874) | |||
| Non-MSI-high | F. nucleatum-negative | 1 (referent) | 1 (referent) |
| F. nucleatum-positive | 2.48 (1.42-4.31) | 2.86 (1.62-5.06) | |
| MSI-high | F. nucleatum-negative | 1 (referent) | 1 (referent) |
| F. nucleatum-positive | 0.89 (0.45-1.76) | 0.99 (0.49-1.98) | |
| Pinteractionc | 0.022 | 0.020 | |
Abbreviations: CI, confidence interval; MSI, microsatellite instability; OR, odds ratio.
The multivariable logistic regression model initially included sex, age at diagnosis, year of diagnosis, family history of colorectal cancer, tumor location, CpG island methylator phenotype-specific promoter status, long interspersed nucleotide element-1 methylation level, and KRAS, BRAF, and PIK3CA mutations. A backward elimination with a threshold P of 0.05 was used to select variables for the final models. The variables which remained in the final models for tumor-infiltrating lymphocytes and intratumoral periglandular reaction were shown in Supplementary Table S1.
To avoid violation of the proportional odds assumption, the most common score or less (i.e., 0 for tumor-infiltrating lymphocytes and Crohn’s-like lymphoid reaction; and 0-1 for intratumoral periglandular reaction and peritumoral lymphocytic reaction) was categorized as low, and higher score was categorized as high.
Pinteraction (two-sided) was calculated using the Wald test for the cross-product of F. nucleatum status (positive vs. negative) and MSI status (high vs. non-high) in the logistic regression model.
As secondary analyses using a subset of cases with available tissue microarray data (up to 580 cases), we examined the association of F. nucleatum with densities of T-cell subsets (CD3+ cells, CD8+ cells, CD45RO+ cells, or FOXP3+ cells) in strata of MSI status. Although we did not observe a significant interaction between F. nucleatum and MSI status in relation to the density of CD3+ cells, CD8+ cells, CD45RO+ cells, or FOXP3+ cells (Pinteraction > 0.1, with the α level of 0.005; Supplementary Table S2), statistical power was limited in these analyses.
We did not observe a statistically significant interaction between Bifidobacterium genus and MSI status in relation to lymphocytic reaction patterns or densities of T-cell subsets (Pinteraction > 0.1, with the α level of 0.005).
As secondary analyses to assess the prognostic association of F. nucleatum in strata of MSI status, we conducted Kaplan-Meier analyses and Cox regression analyses, and did not observe a significant interaction between F. nucleatum and MSI status in relation to colorectal cancer-specific or overall mortality (Supplementary Fig. S1 and Supplementary Table S3).
Discussion
Using two large prospective cohort studies in the U.S., we tested the hypothesis that the association of intratumoral F. nucleatum status with local immune response to colorectal cancer might differ by tumor MSI status. We found negative associations of F. nucleatum status with degrees of TIL and intratumoral periglandular reaction in MSI-high colorectal carcinomas and positive associations with those reaction patterns in non-MSI-high carcinomas. Our findings suggest interplay between F. nucleatum, MSI status, and immune cells in colorectal tumor.
High-level infiltration of lymphocytes in colorectal cancer has been associated with better clinical outcomes (23, 24, 35, 36), and therapeutic activation of anti-tumor immune response in the tumor microenvironment has become an attractive strategy for cancer immunotherapy (37–39). In particular, immune checkpoint inhibitors that target the PDCD1 (PD-1) or CD274 (PDCD1 ligand 1, PD-L1) protein have shown promise in treating various cancer types (37–39), including colorectal cancer with high MSI (26–28). However, despite the accompanying intense immune response, some MSI-high colorectal cancers can evade the immune eradication, and not all MSI-high tumors respond to the immune checkpoint blockade. Therefore, immunosuppressive factors in MSI-high colorectal cancer need to be elucidated to improve the efficacy of immunotherapies in colorectal cancer. In the tumor microenvironment, host cells, neoplastic cells, and microbiota form complex interactive networks. A better understanding of these interactions would have relevance to existing avenues of treatment and may also reveal additional strategies for combating cancer (40, 41).
Alterations in the gut microbial composition are considered to play a role in colorectal cancer development (3, 42, 43). Fiber-rich diets are associated with a lower risk of colorectal cancer containing abundant F. nucleatum (42). Our findings of a possible temporal increase in detectable intratumor F. nucleatum might be due to growing popularity of colonoscopy screening, changes in dietary and lifestyle patterns, and/or aging of the cohort participants. Such interactions might become evident with a longitudinal epidemiological investigation. Mechanistic studies indicate that F. nucleatum can exert carcinogenic effects through binding of its adhesin FadA to CDH1 (E-cadherin) and resultant activation of the CTNNB1 (beta-catenin) / WNT signaling pathway in colorectal carcinoma cells (44) as well as through suppression of the adaptive immune response (12). WNT signaling activation has been linked to immunosuppression in colorectal carcinoma (45). Experimental evidence suggests that binding of the virulence factor FAP2 to a host factor Gal-GalNAc may lead to interaction with the inhibitory immune cell receptor TIGIT, thereby inhibiting T-cell activity and NK cell cytotoxicity (11, 13, 46). In line with these studies, our previous population-based study has shown inverse associations of the enrichment of F. nucleatum in colorectal cancer tissue with CD3+ pan-T-cell density and patient survival (8, 18). F. nucleatum also increases numbers of myeloid-derived suppressor cells and tumor-associated macrophages in the colorectal tumor microenvironment (12, 47), contributing to proinflammatory reactions and impairment of tumor-specific immune responses.
Previous studies have linked F. nucleatum to MSI-high colorectal cancer (8, 16, 17), which is characterized by a vigorous anti-tumor immune response in the tumor microenvironment resulting from frameshift mutations and high expression of immunogenic peptides (23–27). However, there is a paradoxical finding that the presence of detectable F. nucleatum in tumor tissue is inversely associated with CD3+ T-cell density (8). Our current findings suggest that F. nucleatum may exert suppressive effects on adaptive anti-tumor immune response in MSI-high colorectal cancer with abundant immunogenic neoantigens (Fig. 2). During the development of colorectal carcinomas, MSI-high tumors need to acquire the means to resist anti-tumor immune activity. Our study provides evidence suggesting that infection with F. nucleatum may promote immune evasion in the immune cell-rich microenvironment of MSI-high colorectal carcinomas. We did not observe differential associations of F. nucleatum with T-cell subsets (CD3+ cells, CD8+ cells, CD45RO+ cells, and FOXP3+ cells) by tumor MSI status. These seemingly conflicting findings might be due to the roles of other cell types in immune reactions to tumor, measurement errors in T-cell densities, and/or a smaller overall sample size in analyses of T cells.
Figure 2.
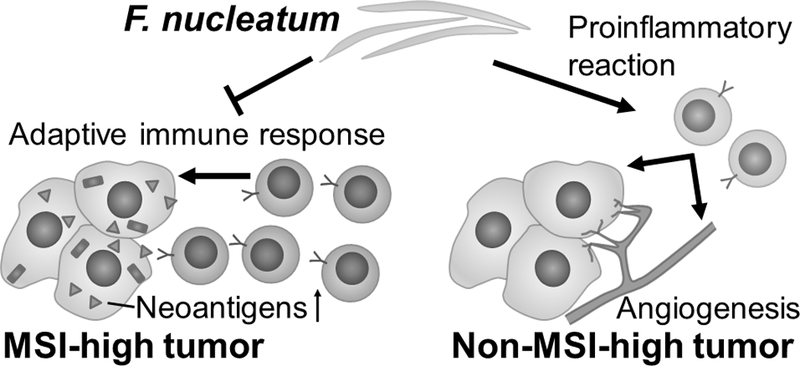
Fusobacterium nucleatum (F. nucleatum) and lymphocytic reaction to colorectal carcinoma according to tumor microsatellite instability (MSI) status. The current study suggests that F. nucleatum may attenuate adaptive immune response to MSI-high colorectal carcinomas which exhibit abundant immunogenic neoantigens. In contrast, the proinflammatory properties of F. nucleatum may lead to lymphocytic reaction to non-MSI-high carcinomas that contain lower amounts of neoantigens and tumor-infiltrating lymphocytes than MSI-high carcinomas. MSI, microsatellite instability.
The trend toward more lymphocytic reaction associated with F. nucleatum in non-MSI-high tumors warrants further discussion. In addition to suppression of the anti-tumor immune response, F. nucleatum can exert proinflammatory effects in the colorectal mucosa (12, 14, 15), and has been implicated in pathogenesis of inflammatory bowel diseases (14). Studies have shown that F. nucleatum may increase expression of proinflammatory cytokines and up-regulate the NF-κB pathway (12, 48) in colorectal tumors. Therefore, we speculate that the proinflammatory effects of F. nucleatum may outweigh its immunosuppressive effects in non-MSI-high tumors, which contain fewer neoantigens and fewer tumor-infiltrating immune cells on average relative to MSI-high tumors (Fig. 2). These divergent associations of tumor tissue F. nucleatum with lymphocytic reactions between MSI-high and non-MSI-high tumors underscore the importance of interactive effects of environmental and tumor molecular features on host immune response in the tumor microenvironment (40).
We acknowledge limitations of our study. Our study focused on limited numbers of microbial species and immune reaction components. Accumulating evidence points to the roles of a variety of microbes and immune cells in anti-tumor immune responses and carcinogenesis (2, 3, 49). Therefore, more comprehensive analyses are warranted to investigate the interactions between microbes other than F. nucleatum (e.g., enterotoxigenic Bacteroides fragilis and pks-positive Escherichia coli) (4) and other immune parameters (e.g., myeloid-derived suppressor cells, tumor-associated macrophages, and TIGIT-expressing cells). High throughput sequencing-based analyses including 16S rRNA gene sequencing, metagenomic analyses, gene expression profiling, and T-cell receptor sequencing would also provide insights into the interrelationship of tumor molecular markers and microbiota in relation to anti-tumor immune response. Detailed data on antibiotics and cancer treatments including immune checkpoint inhibitors were not available in our study. There might be confounding factors that were unaccounted for; nonetheless, we adjusted for a variety of demographic, clinical, and tumor characteristics, as we utilized the MPE database. Another limitation is the cross-sectional study design, which dictates that we cannot rule out reverse causation. It is possible that immune responses may alter the amount of F. nucleatum in the tumor microenvironment. However, our hypothesis was based on accumulating evidence for the suppressive effects of F. nucleatum on T cell-mediated anti-tumor immunity (8, 11–13). We used the quantitative PCR assay for F. nucleatum in FFPE tissue specimens; therefore, tissue processing procedures and storage conditions might have affected the detection rate. Nonetheless, our previous validation study using quantitative PCR showed concordance in detecting F. nucleatum in paired FFPE and frozen tissue specimens and linearity and reproducibility of F. nucleatum measurements in FFPE tissue specimens (8). Finally, our findings need to be validated in independent cohorts.
The current study has strengths, including the use of the MPE (40, 41) database derived from two U.S. prospective cohort studies, and this study represents the integration of immunology into the MPE paradigm (i.e., immunology-MPE) (50). The data on the amount of F. nucleatum DNA in the tumor, tumor molecular characteristics, and pathological findings in over 1,000 cases allowed us to comprehensively examine possible differential roles of F. nucleatum in MSI-high and non-MSI-high colorectal cancers. Our study population was derived from a large number of incident colorectal cancer cases that occurred in the well-defined populations of the prospective cohorts, and cases were also derived from hospitals in 48 states in the U.S.; both features increase the generalizability of our findings. Nevertheless, our findings need to be validated in independent studies.
In conclusion, the current study provides evidence for divergent effects of F. nucleatum in the tumor microenvironment according to tumor MSI status. There may be dominant suppressive effects of F. nucleatum on adaptive anti-tumor immune responses in MSI-high colorectal cancer and apparent proinflammatory effects in non-MSI-high cancer. Our findings, if validated, would inform future mechanistic studies examining the interplay of F. nucleatum and tumor characteristics in colorectal tumor evolution.
Supplementary Material
Acknowledgements
We would like to thank the participants and staff of the Nurses’ Health Study and the Health Professionals Follow-up Study for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data.
Grant Support: This work was supported by U.S. National Institutes of Health (NIH) grants (P01 CA87969 to M.J. Stampfer; UM1 CA186107 to M.J. Stampfer; P01 CA55075 to W.C. Willett; UM1 CA167552 to W.C. Willett; U01 CA167552 to W.C. Willett and L.A. Mucci; P50 CA127003 to C.S. Fuchs; R01 CA118553 to C.S. Fuchs; R01 CA169141 to C.S. Fuchs; R01 CA137178 to A.T. Chan; K24 DK098311 to A.T. Chan; R35 CA197735 to S. Ogino; R01 CA151993 to S. Ogino; K07 CA190673 to R. Nishihara; and K07 CA188126 to X. Zhang); by Nodal Award from the Dana-Farber Harvard Cancer Center (to S. Ogino); and by grants from the Project P Fund, The Friends of the Dana-Farber Cancer Institute, Bennett Family Fund, and the Entertainment Industry Foundation through National Colorectal Cancer Research Alliance. Research supported by a Stand Up To Cancer Colorectal Cancer Dream Team Translational Research Grant (Grant Number: SU2C-AACR-DT22–17) was awarded to C.S. Fuchs and M. Giannakis. Stand Up To Cancer is a division of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the Scientific Partner of SU2C. T. Hamada was supported by a fellowship grant from the Mitsukoshi Health and Welfare Foundation. K. Mima was supported by a grant from Program for Advancing Strategic International Networks to Accelerate the Circulation of Talented Researchers from Japan Society for the Promotion of Science. K. Kosumi was supported by grants from Overseas Research Fellowship (JP2017–775) and Program for Advancing Strategic International Networks to Accelerate the Circulation of Talented Researchers, from Japan Society for the Promotion of Science. M. Song was supported by the 2017 AACR-AstraZeneca Fellowship in Immuno-oncology Research (grant number 17–40-12-SONG). L. Liu was supported by a scholarship grant from Chinese Scholarship Council and a fellowship grant from Huazhong University of Science and Technology. M. Giannakis was supported by a KL2/Catalyst Medical Research Investigator Training award (NIH award, KL2 TR001100). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The funding source had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, and approval of the manuscript; and decision to submit the manuscript for publication.
Abbreviations:
- CI
confidence interval
- CIMP
CpG island methylator phenotype
- FFPE
formalin-fixed paraffin-embedded
- HPFS
Health Professionals Follow-up Study
- LINE-1
long interspersed nucleotide element-1
- MPE
molecular pathological epidemiology
- MSI
microsatellite instability
- NHS
Nurses’ Health Study
- OR
odds ratio
- PCR
polymerase chain reaction
- TIL
tumor-infiltrating lymphocytes
Footnotes
Disclosure of Potential Conflicts of Interest: A.T. Chan previously served as a consultant for Bayer Healthcare, Pfizer Inc., and Aralez Pharmaceuticals. This study was not funded by Bayer Healthcare, Pfizer Inc., or Aralez Pharmaceuticals. No other conflicts of interest exist. The other authors declare that they have no conflicts of interest
References
- 1.Zitvogel L, Pietrocola F, Kroemer G. Nutrition, inflammation and cancer. Nat Immunol 2017;18:843–50. [DOI] [PubMed] [Google Scholar]
- 2.Fridman WH, Zitvogel L, Sautes-Fridman C, Kroemer G. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol 2017;14:717–34. [DOI] [PubMed] [Google Scholar]
- 3.Chen J, Domingue JC, Sears CL. Microbiota dysbiosis in select human cancers: Evidence of association and causality. Semin Immunol 2017;32:25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tilg H, Adolph TE, Gerner RR, Moschen AR. The Intestinal Microbiota in Colorectal Cancer. Cancer Cell 2018. in press [DOI] [PubMed] [Google Scholar]
- 5.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res 2012;22:292–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res 2012;22:299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li YY, Ge QX, Cao J, Zhou YJ, Du YL, Shen B, et al. Association of Fusobacterium nucleatum infection with colorectal cancer in Chinese patients. World J Gastroenterol 2016;22:3227–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mima K, Sukawa Y, Nishihara R, Qian ZR, Yamauchi M, Inamura K, et al. Fusobacterium nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncol 2015;1:653–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Repass J, Iorns E, Denis A, Williams SR, Perfito N, Errington TM. Replication Study: Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Elife 2018;7:e25801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 2017;358:1443–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaplan CW, Ma X, Paranjpe A, Jewett A, Lux R, Kinder-Haake S, et al. Fusobacterium nucleatum outer membrane proteins Fap2 and RadD induce cell death in human lymphocytes. Infect Immun 2010;78:4773–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013;14:207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity 2015;42:344–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bashir A, Miskeen AY, Hazari YM, Asrafuzzaman S, Fazili KM. Fusobacterium nucleatum, inflammation, and immunity: the fire within human gut. Tumour Biol 2016;37:2805–10. [DOI] [PubMed] [Google Scholar]
- 15.Ye X, Wang R, Bhattacharya R, Boulbes DR, Fan F, Xia L, et al. Fusobacterium Nucleatum Subspecies Animalis Influences Proinflammatory Cytokine Expression and Monocyte Activation in Human Colorectal Tumors. Cancer Prev Res (Phila) 2017;10:398–409. [DOI] [PubMed] [Google Scholar]
- 16.Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V, et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis 2014;33:1381–90. [DOI] [PubMed] [Google Scholar]
- 17.Nosho K, Sukawa Y, Adachi Y, Ito M, Mitsuhashi K, Kurihara H, et al. Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer. World J Gastroenterol 2016;22:557–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut 2016;65:1973–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell 2017;170:548–63 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogino S, Chan AT, Fuchs CS, Giovannucci E. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut 2011;60:397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rajpoot M, Sharma AK, Sharma A, Gupta GK. Understanding the microbiome: Emerging biomarkers for exploiting the microbiota for personalized medicine against cancer. Semin Cancer Biol 2018. in press [DOI] [PubMed] [Google Scholar]
- 22.Morgillo F, Dallio M, Della Corte CM, Gravina AG, Viscardi G, Loguercio C, et al. Carcinogenesis as a Result of Multiple Inflammatory and Oxidative Hits: a Comprehensive Review from Tumor Microenvironment to Gut Microbiota. Neoplasia 2018;20:721–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogino S, Nosho K, Irahara N, Meyerhardt JA, Baba Y, Shima K, et al. Lymphocytic reaction to colorectal cancer is associated with longer survival, independent of lymph node count, microsatellite instability, and CpG island methylator phenotype. Clin Cancer Res 2009;15:6412–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nosho K, Baba Y, Tanaka N, Shima K, Hayashi M, Meyerhardt JA, et al. Tumour-infiltrating T-cell subsets, molecular changes in colorectal cancer, and prognosis: cohort study and literature review. J Pathol 2010;222:350–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giannakis M, Mu XJ, Shukla SA, Qian ZR, Cohen O, Nishihara R, et al. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell Rep 2016;15:857–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li SK, Martin A. Mismatch Repair and Colon Cancer: Mechanisms and Therapies Explored. Trends Mol Med 2016;22:274–89. [DOI] [PubMed] [Google Scholar]
- 27.Llosa NJ, Cruise M, Tam A, Wicks EC, Hechenbleikner EM, Taube JM, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov 2015;5:43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamauchi M, Morikawa T, Kuchiba A, Imamura Y, Qian ZR, Nishihara R, et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut 2012;61:847–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mima K, Cao Y, Chan AT, Qian ZR, Nowak JA, Masugi Y, et al. Fusobacterium nucleatum in Colorectal Carcinoma Tissue According to Tumor Location. Clin Transl Gastroenterol 2016;7:e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015;350:1084–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nosho K, Irahara N, Shima K, Kure S, Kirkner GJ, Schernhammer ES, et al. Comprehensive biostatistical analysis of CpG island methylator phenotype in colorectal cancer using a large population-based sample. PLoS One 2008;3:e3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamada T, Cao Y, Qian ZR, Masugi Y, Nowak JA, Yang J, et al. Aspirin Use and Colorectal Cancer Survival According to Tumor CD274 (Programmed Cell Death 1 Ligand 1) Expression Status. J Clin Oncol 2017;35:1836–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masugi Y, Nishihara R, Hamada T, Song M, da Silva A, Kosumi K, et al. Tumor PDCD1LG2 (PD-L2) Expression and the Lymphocytic Reaction to Colorectal Cancer. Cancer Immunol Res 2017;5:1046–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pages F, Mlecnik B, Marliot F, Bindea G, Ou FS, Bifulco C, et al. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet 2018;391:2128–39. [DOI] [PubMed] [Google Scholar]
- 36.Rozek LS, Schmit SL, Greenson JK, Tomsho LP, Rennert HS, Rennert G, et al. Tumor-Infiltrating Lymphocytes, Crohn’s-Like Lymphoid Reaction, and Survival From Colorectal Cancer. J Natl Cancer Inst 2016;108:djw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Basile D, Garattini SK, Bonotto M, Ongaro E, Casagrande M, Cattaneo M, et al. Immunotherapy for colorectal cancer: where are we heading? Expert Opin Biol Ther 2017;17:709–21. [DOI] [PubMed] [Google Scholar]
- 38.Grizzi F, Basso G, Borroni EM, Cavalleri T, Bianchi P, Stifter S, et al. Evolving notions on immune response in colorectal cancer and their implications for biomarker development. Inflamm Res 2018;67:375–89. [DOI] [PubMed] [Google Scholar]
- 39.Gang W, Wang JJ, Guan R, Yan S, Shi F, Zhang JY, et al. Strategy to targeting the immune resistance and novel therapy in colorectal cancer. Cancer Med 2018;7:1578–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogino S, Nowak JA, Hamada T, Phipps AI, Peters U, Milner DA, Jr., et al. Integrative analysis of exogenous, endogenous, tumour and immune factors for precision medicine. Gut 2018;67:1168–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamada T, Keum N, Nishihara R, Ogino S. Molecular pathological epidemiology: new developing frontiers of big data science to study etiologies and pathogenesis. J Gastroenterol 2017;52:265–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mehta RS, Nishihara R, Cao Y, Song M, Mima K, Qian ZR, et al. Association of Dietary Patterns With Risk of Colorectal Cancer Subtypes Classified by Fusobacterium nucleatum in Tumor Tissue. JAMA Oncol 2017;3:921–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park CH, Han DS, Oh YH, Lee AR, Lee YR, Eun CS. Role of Fusobacteria in the serrated pathway of colorectal carcinogenesis. Sci Rep 2016;6:25271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe 2013;14:195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grasso CS, Giannakis M, Wells DK, Hamada T, Mu XJ, Quist M, et al. Genetic Mechanisms of Immune Evasion in Colorectal Cancer. Cancer Discov 2018;8:730–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abed J, Emgard JE, Zamir G, Faroja M, Almogy G, Grenov A, et al. Fap2 Mediates Fusobacterium nucleatum Colorectal Adenocarcinoma Enrichment by Binding to Tumor-Expressed Gal-GalNAc. Cell Host Microbe 2016;20:215–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park HE, Kim JH, Cho NY, Lee HS, Kang GH. Intratumoral Fusobacterium nucleatum abundance correlates with macrophage infiltration and CDKN2A methylation in microsatellite-unstable colorectal carcinoma. Virchows Arch 2017;471:329–36. [DOI] [PubMed] [Google Scholar]
- 48.Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, et al. Fusobacterium nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor-kappaB, and Up-regulating Expression of MicroRNA-21. Gastroenterology 2017;152:851–66 e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, et al. The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov 2018;8:403–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ogino S, Giannakis M. Immunoscore for (colorectal) cancer precision medicine. Lancet 2018;391:2084–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


