Abstract
Drug naïve animals given a single dose of ethanol show changed responses to subsequent doses, including the development of ethanol tolerance and ethanol preference. These simple forms of behavioral plasticity are due in part to changes in gene expression and neuronal properties. Surprisingly little is known about how ethanol initiates changes in gene expression or what the changes do. Here we demonstrate a role in ethanol plasticity for Hr38, the sole Drosophila homolog of the mammalian Nr4a1/2/3 class of immediate early response transcription factors. Acute ethanol exposure induces transient expression of Hr38 and other immediate early neuronal activity genes. Ethanol activates the Mef2 transcriptional activator to induce Hr38, and the Sirt1 histone/protein deacetylase is required to terminate Hr38 induction. Loss of Hr38 decreases ethanol tolerance and causes precocious but short‐lasting ethanol preference. Similarly, reduced Mef2 activity in all neurons or specifically in the mushroom body α/β neurons decreases ethanol tolerance; Sirt1 promotes ethanol tolerance in these same neurons. Genetically decreasing Hr38 expression levels in Sirt1 null mutants restores ethanol tolerance, demonstrating that both induction and termination of Hr38 expression are important for behavioral plasticity to proceed. These data demonstrate that Hr38 functions as an immediate early transcription factor that promotes ethanol behavioral plasticity.
Keywords: behavioral plasticity, Drosophila, ethanol tolerance, Hr38, immediate early gene, Mef2, mushroom bodies, Sirt1
1. INTRODUCTION
Ethanol, one of the most widely used and frequently abused addictive drugs, is a small molecule that diffuses rapidly throughout the body and that binds to an as yet incompletely defined spectrum of molecules. Its effects vary based on dose, exposure time and pattern, an individuals' history of intake, and their genetic makeup. This complexity of action has hampered progress in reducing the prevalence of alcohol use disorders through rational interventions.1
One approach forward is to define, in detail, the stimulus‐response relationship for ethanol in ethanol naïve animals. While the first ethanol exposure rarely leads directly to alcoholism, it does cause changes in behavior that reflect changes in brain function; these changes provide an altered substrate for subsequent intake and they promote addiction risk. Furthermore, many genes are oppositely regulated by acute ethanol exposure and in ethanol withdrawal.2 Practically, this suggests that detailed mechanistic understanding of acute ethanol exposure action, especially when coupled to measures of behavioral plasticity, will provide insight into the more complex mechanisms underlying addiction. One form of ethanol‐induced behavioral plasticity is tolerance, the acquired resistance to the inebriating and sedating properties of ethanol.3 Ethanol tolerance facilitates increased intake, a risk factor for later developing alcohol use disorders.
Drugs of abuse, including ethanol, cause changes in gene expression in the brain that can alter the properties of the brain. Drosophila melanogaster is a useful organism for defining how acute ethanol alters behavior through gene regulation.4 In Drosophila, as in mammals, acute ethanol exposure progressively stimulates locomotion, motor incoordination, and sedation. Ethanol exposure also induces ethanol tolerance, ethanol preference, ethanol reward, and signs of ethanol withdrawal. Acute ethanol exposure causes marked changes in gene expression, and some ethanol‐regulated genes have been shown to be critical for ethanol‐induced behavioral plasticity.5, 6, 7, 8, 9, 10
Neural activity and drugs of abuse induce immediate early genes in the nervous system, including transcription factors like Fos that are important for driving programs of gene expression.11, 12 Neural activity in Drosophila also induces immediate early genes.13, 14, 15 As yet, miRNAs are the only class of immediate early genes that have been studied in the context of ethanol behaviors in Drosophila.14 Understanding how ethanol regulates immediate early response gene expression, and the consequences of their regulation, can help define how ethanol alters neural function and behavior.
Hr38 is a Drosophila immediate early response gene that is the sole homolog of the mammalian Nr4a1/Nr4a2/Nr4a3 gene family. Hr38 is strongly and consistently induced by artificial neural activation.13, 16 It functions in various processes in both development and adulthood, including ecdysis, carbohydrate storage and circadian rhythms.17, 18, 19 Here, we find that Hr38 and other immediate early genes are induced by ethanol, we define the mechanisms of Hr38 induction, and we demonstrate that Hr38 and its regulators function in the fly brain to promote the development of ethanol tolerance. These findings define early steps in gene regulation by ethanol that are important for the expression of ethanol‐induced behavioral plasticity.
2. MATERIALS AND METHODS
2.1. Drosophila culturing and strains
All strains were outcrossed for at least five generations to the Berlin genetic background carrying the w 1118 genetic marker mutation. The genetic background strain was used as an experimental control. Flies were cultured on standard cornmeal/molasses/yeast medium at 25°C and 70% relative humidity with a 12/12 hours light/dark schedule. Nicotinamide (70 mM, Sigma‐Aldrich, St Louis, MO USA) was fed to flies dissolved in 5% sucrose/2% yeast extract on Whatman filter paper for 48 hours, exchanged at 24 hours. Strains used in this study were UAS‐Mef2.EnR from Justin Blau,20 Hr38 y214 from Carl Thummel,21 MB‐Gal80 from Scott Waddell, University of Oxford, Oxford, UK, UAS‐Mef2.IR (Vienna Drosophila Resource Center, v15549, v15550), Hr38.GFP.FLAG (R. Spokony and K. White, Bloomington Drosophila Stock Center (BDSC) #38651), 17d‐Gal4 (BDSC #51631), elav‐Gal4 (c155, BDSC #458), elav‐Gal4 (3E1, BDSC #8760), and Sirt1 2A‐7‐11 (BDSC #8838).
2.2. RNA measurement
RNA was extracted from male heads, DNase treated, and reverse‐transcribed using MultiScribe (Applied Biosystems, Foster City, CA USA). Quantitative PCR reactions were performed using the SYBR Green method and custom designed primers on a StepOnePlus machine (Applied Biosystems). Ct values were normalized to RpL32, expression was calculated using the ΔΔCt method, and the mean of multiple independent biological replicates was calculated. Oligonucleotide primers used in this study were Cdc7: AATGGAGCTGCAGTCATGG (F), GGATTCGTGTGAGGAGATCATT (R); CG14186: GGCCAGCTAATCTCCAAGTT (F), GTTGTAGATCTCCTCGCCATC (R); CG17778: GCTGCGCTGACTTACTACTTAC (F), TGCATTGGCCACCGATTT (R); Hr38: GAGTGGCTCAACGACATCAT (F), CGTTCTGTGATCAGGGTTAGG (R); Jra: GTTCCCACCCACTGATTGA (F), GCTTGTTCTTGGCACTCTTG (R); Kayak: CCGATACTTCAAGTGCCCATAC (F), CCAGGACATTGGAGAAGTTGTT (R); Sirt1: GACTGCCGGATGAGTACC (F), ACGATCAGTAGATCGCAC (R); Stripe: CCGAGTATGCCGCTCAATTA (F), GGCGTATGGTGGTGATAAGG (R).
2.3. Whole mount immunohistochemistry
Brains were dissected in PBS and 0.05% Triton‐X 100 (0.05% PBT), fixed (2% paraformaldehyde in 0.05% PBT) overnight at 4°C or 1 hour at room temperature. They were washed 5× 10 minutes in 0.1% PBT, blocked 1 hour in 0.1% PBT with 0.5% wt/vol BSA and 5% normal goat serum and then incubated with primary antibodies overnight at 4°C. Brains were washed, blocked, and incubated with secondary antibodies overnight at 4°C, followed by further washes and then mounted on glass slides with Vectashield (Vector Laboratories, Burlingame, CA USA). Antibodies used were rabbit anti‐GFP (1:1000, Invitrogen A6455), mouse anti‐Elav (1:50, Developmental Studies Hybridoma Bank 9F8A9), goat anti‐rabbit Alexa 488, and goat anti‐mouse Alexa 594 (1:350, Cell Signaling Technologies Danvers, MA USA). Mushroom body kenyon cell nuclei were counted by drawing a 50 μm arc at the border between the mushroom body calyx and nuclei at the location with the greatest number of GFP‐positive nuclei, and counting positive nuclei within 25 μm of the border.
2.4. Ethanol behaviors
Ethanol sensitivity and tolerance were measured as previously described.5 Briefly, groups of 20 genetically identical flies (n = 1) were exposed to 55% ethanol vapor or 100% humidified air, and the number of flies that lost the righting reflex were counted at 6 minutes intervals. The time to 50% sedation (ST50) was calculated for each group, and the experiment was repeated across different days and from different parental crosses. Flies were allowed to rest for 3.5 hours and then re‐exposed to an identical concentration of ethanol vapor, and tolerance was calculated as the difference in ST50 between the two exposures. The capillary feeding assay (CAFE) was used to determine ethanol preference, as previously described.22 Groups of eight adult males were collected 3 to 4 days after eclosion and allowed to recover from CO2 for 1 day. They were pre‐exposed to either 55% ethanol vapor/air mixture or 100% humidified air alone for 30 minutes. After 16 hours recovery, flies were placed into the CAFE chamber, which consists of empty vials with capillary tubes containing liquid food with or without 15% ethanol, embedded in the vial plug. The preference index was the volume of food consumed from the ethanol capillaries minus that consumed from the no‐ethanol capillaries over the total volume consumed, corrected for evaporation by measuring the volume lost in vials with no flies. Bitter taste avoidance was measured by presenting flies with a choice of 1.25% agarose containing either 50 mM sucrose (S) or 100 mM sucrose and 1 mM quinine (SQ). Groups of approximately 20 male flies were food deprived on water for 14 hours, placed in a 40 × 90 × 10 mm clear acrylic arena, and 150 μL S and SQ dots were then placed in apposition at the center of the arena. The number of flies on each dot was counted at 30 minutes. Avoidance was calculated as (SQ − S)/(SQ + S) such that complete avoidance of bitter gives a value of −1.
2.5. Ethanol absorption and metabolism
Flies were frozen in liquid nitrogen and homogenized in 50 mM Tris‐HCl, pH 7.5. Ethanol concentrations were measured in fly homogenates using the Ethanol Assay Kit from Diagnostic Chemicals Ltd, Nova Scotia, Canada. (catalog #229‐29). To calculate the ethanol concentration in flies, the volume of one fly was estimated to be 1 μL.
2.6. Statistical analysis
GraphPad Prism 7.0c was used for unpaired t‐test, one sample t‐test, one‐way ANOVA with Tukey's post hoc test for normally distributed data, and Kruskal‐Wallis test with Dunn's post hoc test for non‐parametric data. Significance indicators on the figures indicate the results of t‐tests or post hoc tests for significant effects by ANOVA. Error bars represent the SEM.
3. RESULTS
3.1. Hr38 is induced by acute ethanol exposure
We surveyed a subset of immediate early genes—those that are broadly induced by neuronal activity—to ask if drug naïve Drosophila respond to ethanol transcriptionally through similar pathways.13, 15, 16 Of these genes, the Nr4a nuclear hormone receptor homolog Hr38 was the only transcription factor whose expression was induced to statistical significance (Figure 1A). The Jun‐related antigen Jra gene showed a strong trend towards induction, with a significant induction vs air exposure but not vs the no treatment control. Retrospective analysis of a gene expression time course following acute ethanol exposure revealed that Hr38 levels peaked 60 minutes after ethanol exposure termination and then decreased to baseline within 3 hours, kinetics that are typical for immediate early response genes (Figure 1B). Thus, ethanol induces immediate early genes in a pattern that partially overlaps that of neuronal activation.
Figure 1.
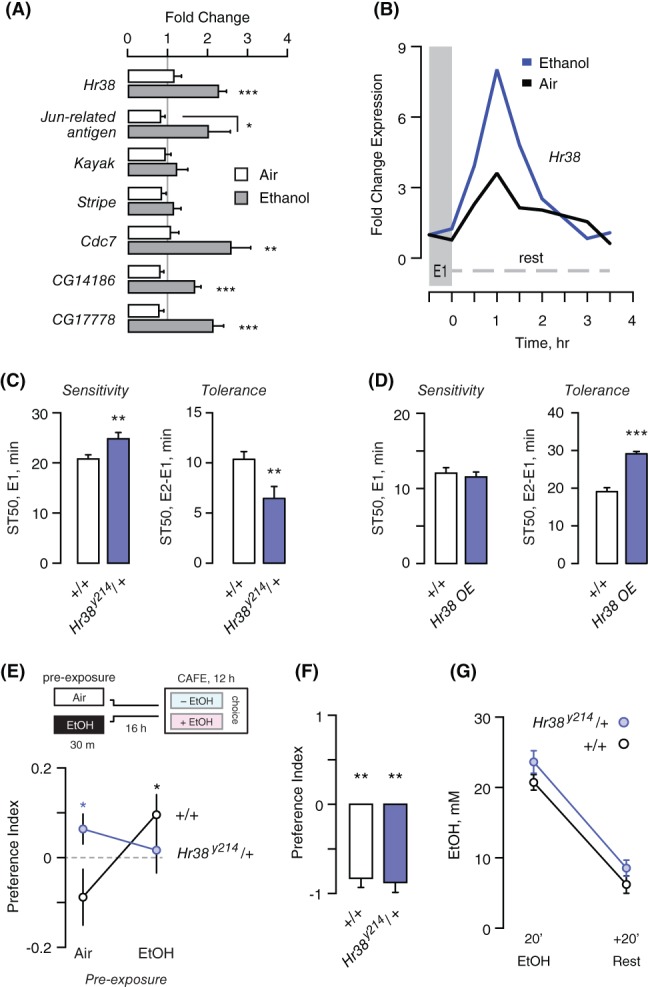
Hr38 is an ethanol immediate early response gene that bidirectionally regulates the development of ethanol tolerance. (A) Steady state transcript levels of genes induced by neuronal activity, 60 minutes after termination of 30 minutes ethanol or mock air exposure, presented as fold change vs no treatment (gray line) in the wild‐type control strain. One way ANOVA/Tukeys per gene, n = 5 biological replicates/condition. (B) Time course of Hr38 expression following air (black) or ethanol (blue). Data extracted from a published microarray experiment.8 (C) Time to 50% sedation (ST50) for Hr38 null mutant heterozygotes vs wild‐type controls (+/+) exposed to ethanol once (E1) for sensitivity or twice (E2‐E1) for tolerance, t‐test, n = 30 groups. (D) Ethanol sensitivity and tolerance in flies with three copies of the Hr38 genomic region (HR38.GFP) t‐test, n = 12 groups. (E) Ethanol preference in Hr38 null heterozygotes. Flies are pre‐exposed to either air or ethanol and after 16 hours placed into the 2 choice CAFÉ assay. A positive index indicates preference for ethanol intake. One‐sample t‐test compared to zero preference, n = 20 groups. (F) Hr38 null heterozygotes show avoidance of a bitter but sweeter food source in a two‐choice seeking assay. One sample t‐test compared to zero preference, n = 5 groups. (G) Ethanol accumulation immediately following a 20 minutes exposure, and after a 20 minutes rest, allowing ethanol to be metabolized; t‐test, n = 8 groups. *P < .05, **P < .01, ***P < .001
3.2. Hr38 promotes ethanol tolerance and ethanol preference
Induction of the transcription factor Hr38 suggested that it may regulate gene expression in the nervous system to promote ethanol behavioral plasticity. To ask if Hr38 functions in ethanol behaviors, we tested flies underexpressing or overexpressing the gene for ethanol sensitivity, ethanol rapid tolerance, and ethanol preference. Ethanol sensitivity was measured as the time to 50% sedation for groups of genetically identical flies. Ethanol tolerance was measured by giving these flies a second, identical ethanol exposure 3.5 hours after the first exposure, and calculating the difference in sedation time between exposures: flies acquire resistance to the sedative effects of ethanol.23 Hr38 y214 null mutants are pupal lethal, so we tested heterozygotes with 50% normal Hr38 levels, which we confirmed (53% ±12.5% SEM compared to the genetic background control, n = 8).21 Hr38 heterozygotes showed increased ethanol resistance and decreased ethanol tolerance (Figure 1C). A BAC insertion of the Hr38 genomic region, when heterozygous in wild‐type flies, increased Hr38 genomic copy number from two to three and expression 1.78‐fold (P = .0154 two‐tailed t‐test, n = 6 biological replicates). Hr38 overexpression did not affect ethanol sensitivity, but it markedly increased ethanol tolerance (Figure 1D). This suggests that Hr38 levels induced by ethanol are critical for setting the magnitude of ethanol tolerance, and that the role of Hr38 in ethanol sensitivity and tolerance may be separable.
Drosophila develops a preference for ethanol intake.24 Ethanol preference was measured in the two choice CAFÉ assay, where flies can drink from capillaries containing sucrose and yeast either with or without 15% ethanol.22, 25 Ethanol preference in wild‐type was induced by pre‐exposure to ethanol vapor (Figure 1E).26 In contrast, Hr38 loss‐of‐function mutants showed precocious ethanol preference, and this preference dissipated with ethanol pre‐exposure (Figure 1E). Ethanol exposure did not affect Hr38 mutant viability measured over a week, arguing against non‐specific tissue damage (not shown). A similar phenotype was previously observed in flies lacking Sirt1, where precocious preference and lack of induced preference were shown to be separable.5 Bitter taste avoidance was unaffected in the Hr38 mutants, suggesting that their ethanol taste reactivity is intact (Figure 1F). Moreover, ethanol absorption and metabolism were unaffected in Hr38 mutants (Figure 1G). Taken together, these results suggest that the levels of Hr38 expression are important for two forms of ethanol behavioral plasticity, tolerance and preference.
3.3. Hr38 is expressed in neurons where it is induced by ethanol
Hr38 on the genomic BAC is C‐terminal tagged with GFP (Hr38.GFP), which allowed us to determine its expression pattern in the fly brain (Figure 2). Hr38.GFP was localized to the cell nucleus but was also present in the cytoplasm in some brain regions, most notably in the axons of the mushroom body lobes (Figure 2A, Movie S1, Supporting Information). Co‐labeling indicated that Hr38‐positive cells were neurons (Figure 2A). Hr38.GFP expression was found sporadically throughout the adult brain, and the mushroom body kenyon cell nuclei were prominently labeled (Figure 2B). To ask if ethanol exposure recruited additional neurons to express Hr38, we counted GFP‐positive Kenyon cell nuclei. The number of Hr38‐labeled nuclei increased about 2‐fold in the mushroom body 1.5 hours after termination of ethanol treatment (Figure 2C).
Figure 2.
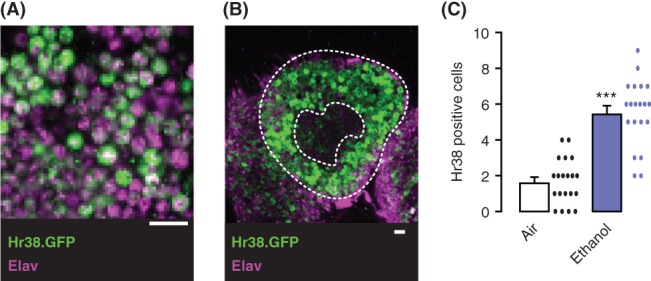
Hr38 protein expression in the adult brain is induced by ethanol in the mushroom bodies. (A) Cortex region of the adult fly brain expressing the Hr38.GFP fusion protein (green), showing co‐localization with the pan‐neuronal Elav nuclear protein (magenta). (B) High levels of Hr38.GFP expression in the Kenyon cell neurons (between dashed enclosures) of the mushroom body that surround the mushroom body calyx (inner dashed area) in the brain of an ethanol exposed fly. (C) Number of Hr38.GFP‐positive Kenyon cell nuclei 1.5 hours after termination of ethanol or mock air exposure. ***P < .0001, t‐test, n = 20 brains/treatment. Scale bars: 10 μm
3.4. Mef2 acts upstream of Hr38 for ethanol tolerance in the mushroom bodies
Mammalian homologs of Hr38 are transcriptionally induced by the Mef2 transcription factor.27, 28 We asked if ethanol upregulates Hr38 through Mef2 to promote tolerance development. Mef2 consensus DNA binding sites (CTAWWWWTAG) are overrepresented in the Hr38 genomic region, two of which are located less than 2 kb upstream of the Hr38 transcription start site (Figure 3A).29, 30 Two of the Mef2 consensus sites at the Hr38 locus bind Mef2 in chromatin immunoprecipitation from fly heads, and they are conserved in Drosophila simulans.30 Consensus sites also exist in the Apis melifera Hr38 enhancer region (not shown). We used a transgenic dominant negative Mef2 (Mef2.EnR) and Mef2 RNAi (Mef2.IR) to inhibit transcriptional activity at Mef2 enhancers in neurons.20 In Mef2.EnR, the Mef2 activation domain is replaced with the Engrailed repressor domain. Expression of Mef2.EnR in all neurons with elav‐Gal4 reduced Hr38 expression (Figure 3B). Further, Hr38 induction by ethanol was lost (Figure 3C). However, ethanol induction of Hr38 was also lost in the + >Mef2.EnR controls. Quantitative PCR confirmed the presence of Mef2.EnR fusion transcripts in these controls (not shown), which is consistent with previously observed phenotypic effects of the uninduced transgene.20 Ectopic leaky expression of dominant negative Mef2 in + >Mef2.EnR may more broadly interfere with ethanol induction of Hr38 in non‐neuronal tissues in the sample, or it may act indirectly through constitutive binding at other Mef2 enhancers in the genome. In contrast, pan‐neuronal expression of Mef2.IR specifically blocked Hr38 induction by ethanol (Figure 3D). These data suggest that Mef2 is an immediate early activator of Hr38 gene transcription, and that acute ethanol exposure acts at or upstream of Mef2 to change gene expression in neurons.
Figure 3.
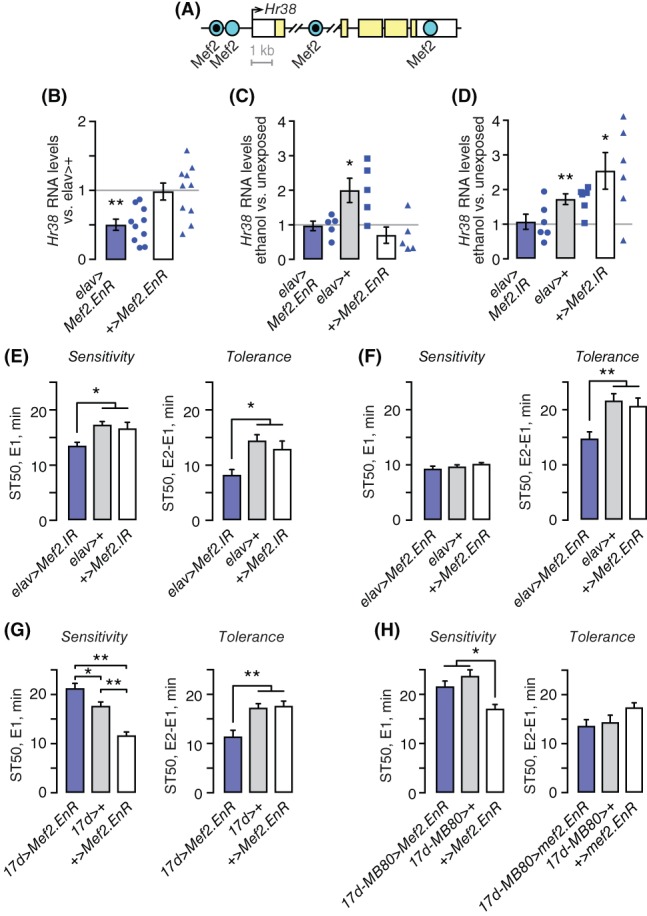
Mef2 promotes ethanol induction of Hr38, and ethanol tolerance in the mushroom bodies. (A) Diagram of the Hr38 genomic locus, indicating the positions of Mef2 consensus binding sites. Consensus sites conserved in other Drosophila species are indicated by a black dot. (B) Hr38 transcript levels in untreated flies expressing dominant negative Mef2.EnR pan‐neuronally, normalized to elav‐3E1 > + (gray line). Kruskal‐Wallis/Dunn's, n = 10 biological replicates. (C) Ethanol induction of Hr38 60 minutes after ethanol treatment is blocked by Mef2.EnR. Expressed as the difference between ethanol and untreated per biological replicate. No difference was detected between genotypes for the unexposed of mock air exposures. One sample t‐test compared to 1 (no induction), n = 5. (D) Ethanol induction of Hr38 is blocked by pan‐neuronal Mef2 RNAi (Mef2.IR). One sample t‐test compared to 1, n = 6. No difference in Hr38 expression was detected between the untreated genotypes. (E) Ethanol sensitivity and tolerance with pan‐neuronal expression (elav‐c155) of Mef2 RNAi. One way ANOVA/Tukey's, n = 30 groups. (F) Ethanol sensitivity and tolerance with pan‐neuronal expression of Mef2.EnR. One way ANOVA/Tukey's, n = 14 groups. (G) Ethanol sensitivity and tolerance with expression of Mef2.EnR restricted to the mushroom body α/β neurons (17d‐Gal4). One way ANOVA/Tukey's, n = 15 to 16 groups. *P < .05, **P < .01. (H) Ethanol sensitivity and tolerance in 17d > Mef2.Enr in the presence of the mushroom body‐specific MB‐Gal80 (MB80). One way ANOVA/Tukey's, n = 10 to 12 groups. *P < .05, **P < .01
These findings predicted that decreased Mef2 activity in neurons would decrease ethanol tolerance. Mef2 RNAi in all neurons increased ethanol sensitivity and decreased ethanol tolerance (Figure 3E). A second Mef2 RNAi appeared to be weaker, specifically reducing ethanol tolerance (not shown). Dominant negative Mef2 in all neurons also decreased ethanol tolerance, but it had no effect on ethanol sensitivity (Figure 3F).
Neuronal activity in the mushroom body α/β neurons promotes ethanol tolerance, where Mef2 and Hr38 expression are enriched.5, 31 Expression of dominant negative Mef2 in these neurons using the 17d‐Gal4 mushroom body driver reduced ethanol tolerance (Figure 3G). Co‐expression of the Gal4 repressor Gal80 specifically in the mushroom bodies blocked the effect of Mef2.EnR on ethanol tolerance, indicating that Mef2 promotes tolerance in the mushroom bodies (Figure 3H).
3.5. Sirt1 terminates ethanol‐induction of Hr38 to promote ethanol tolerance
Sirt1 (also known as Sir2) is a histone/protein deacetylase that regulates responses to drugs of abuse in Drosophila and mammals.5, 32 Mushroom body α/β neuron promotion of ethanol tolerance, preference, and reward requires Sirt1. Further, Sirt1 broadly allows gene expression regulation by acute ethanol exposure. We therefore asked if absence of Sirt1 affected ethanol induction of Hr38. Hr38 was induced normally at 1 hour after acute ethanol exposure in Sirt1 null mutants (Figure 4A). We also assessed Hr38 expression 3 hours after ethanol termination, when Hr38 levels have returned to pre‐exposure levels. Hr38 expression was markedly higher in Sirt1 null mutants at 3 hours (Figure 4A). The failure to terminate Hr38 induction in Sirt1 nulls may be a consequence of lacking Sirt1 throughout development and adulthood, or it may reflect loss of a more temporally direct action of Sirt1 in repressing Hr38. To help distinguish between these possibilities, we fed adult flies nicotinamide, a potent end‐product inhibitor of Sirt1 deacetylase activity that is active against Sirt1 in vivo, and that phenocopies Sirt1 nulls by decreasing ethanol tolerance.5 As in Sirt1 null mutants, adult wild‐type control flies treated with nicotinamide showed prolonged ethanol induction of Hr38 (Figure 4B). Thus, Sirt1 is required specifically for termination of Hr38 gene expression induction by acute ethanol exposure. Finally, we detected no change in Sirt1 levels with neuronally expressed dominant negative Mef2, indicating that Mef2 and Sirt1 act independently to regulate Hr38 expression (Figure 4C).
Figure 4.
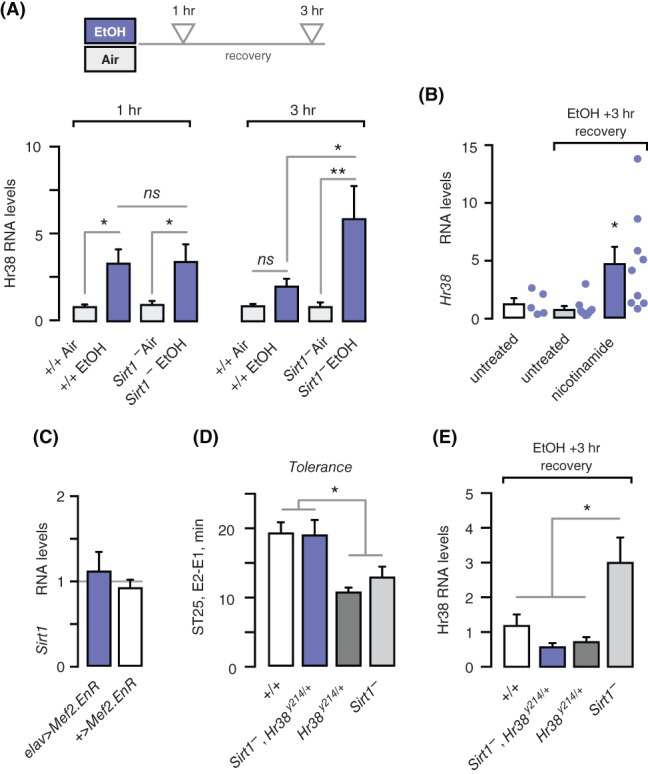
Sirt1 terminates Hr38 induction to promote ethanol tolerance. (A) Hr38 transcript expression levels in wild‐type controls (+/+) and Sirt1 null mutants 1 and 3 hours after treatment, normalized to untreated controls. One way ANOVA/Tukey's, n = 14 biological replicates. (B) Hr38 transcript expression levels in wild‐type control flies treated with the sirtuin end‐product inhibitor nicotinamide (70 mM). One‐sample t‐test compared to 1 (no induction), n = 5 to 10 biological replicates. (C) Sirt1 transcript levels in flies expressing dominant negative Mef2 in all neurons (elav‐3E1), normalized to elav>+. Kruskal‐Wallis/Dunn's, n = 4 to 5 biological replicates. (D) Ethanol tolerance in flies null for Sirt1 and heterozygous for Hr38. Because of the high resistance in exposure 2 in the double mutants, the time to 25% sedation (ST25) was measured. One way ANOVA/Tukey's, n = 8 to 21. (E) Hr38 transcript levels in flies mutant for Sirt1 and heterozygous for Hr38, normalized to untreated controls. One way ANOVA/Tukey's, n = 6 biological replicates. *P < .05, **P < .01
Our findings suggested that ethanol induction of Hr38 may need to be terminated rapidly in order for behavioral plasticity to proceed. We performed a test of this by making double mutants with Sirt1 and Hr38, predicting that genetically decreasing Hr38 expression by half may moderate the prolonged Hr38 induction in Sirt1 mutants and allow ethanol tolerance to develop. The double mutants showed restored ethanol tolerance, compared to either mutant alone (Figure 4D). Consistent with our hypothesis, 3 hours after ethanol exposure termination Hr38 transcript levels in the double mutants were reduced compared to Sirt1 mutants alone (Figure 4E). These findings suggest that termination of Hr38 expression is critical for the development of ethanol tolerance.
4. DISCUSSION
How ethanol changes gene expression in the nervous system is a key to understanding how ethanol causes maladaptive changes in brain function. Here we show that ethanol exposure in drug naïve Drosophila causes a Mef2‐dependent increase in Hr38 expression, that Sirt1 is required for Hr38 induction termination, and that Hr38 controls the extent of ethanol tolerance development. These data suggest that ethanol acts upstream of or directly on the Mef2 transcription factor to cause gene expression changes in the nervous system. Our studies also reveal a requirement for temporally precise termination of the immediate early transcriptional response. Genes regulated by the Hr38 transcription factor are candidate effectors for ethanol‐induced behavioral plasticity.
In flies and mammals, acute ethanol exposure causes changes in both gene expression and chromatin structure in the brain. In Drosophila, transcriptomic experiments that used varying ethanol exposures and recovery times on whole head samples discovered over 200 common ethanol responsive genes.8, 9, 10 Similarly, ethanol exposure increases histones H3 and H4 acetylation marks that generally indicate the opening of chromatin.5, 7 Like gene expression, histone H4 acetylation was modified at a large number of genomic loci 6 and 24 hours after ethanol exposure.33 Further, the increase in histone acetylation can be quite rapid, starting during inebriation.5 Therefore, acute ethanol induces genomic changes rapidly, broadly, and lastingly. Hr38 and the other ethanol‐induced immediate early response genes we identified are candidates for controlling major aspects of ethanol neuroadaptation. In particular, Hr38 as a transcription factor may control the expression of downstream effector genes. The reciprocal effects on ethanol tolerance of lowering and raising Hr38 levels, plus the consequences of prolonging Hr38 expression, all argue that Hr38 is a key regulator of the genomic program for ethanol neuroadaptation.
Hr38, the sole homolog of the mammalian Nr4a1/2/3 gene family, may carry out some of the functions ascribed to distinct mammalian family members. Mammalian Nr4a transcription factors are induced by neuronal activity, stress and drugs of abuse.34 In particular, Nr4a1, also known as Nur77 and NGFIB, is upregulated by cocaine and morphine in brain regions implicated in addiction, and deletion of Nr4a2, also known as Nurr1, decreases ethanol preference.35, 36, 37 Nr4a1 and Nr4a2 are also implicated in forms of long‐term memory.38 Nr4a1, induced by neuronal activity, regulates the density and distribution of dendritic spines, suggesting a possible link between acute ethanol activation of Hr38 and changes in the functional connectivity of the brain.39
Hr38 in Drosophila controls cuticle development and glycogen storage, and its expression is regulated by neuronal activity, social cues, light and extreme drops in temperature.16, 19, 21, 40, 41 Hr38’s role in ethanol responses is likely distinct from most of these roles, as manipulating its levels specifically in the nervous system affects the development of tolerance, and as we detected no change in ethanol absorption as would be expected for altered cuticle integrity.42 Further, we showed that blocking Mef2 activity in the mushroom bodies, where Hr38 expression is increased by ethanol, decreases ethanol tolerance. Mushroom body induction of Hr38 also occurs in males when presented with females, suggesting a possible molecular and anatomical link between ethanol and sexual behaviors.16
How is Hr38 expression controlled? We show that Mef2 sets Hr38 levels under steady state conditions and promotes Hr38 induction by ethanol. In mammals, Mef2A and Mef2D both increase Nr4a1 expression to regulate synapse number and dendrite differentiation.27, 28 Further, Mef2C and NR4a1 are coordinately increased in the striatum by cocaine, and Mef2 promotes cocaine sensitization in the nucleus accumbens, a brain region critical for drug reward.43, 44 Mef2 expression and activity can be regulated by many different signaling pathways, including those associated with neural activity like intracellular calcium levels, and also those that are known to be regulated by ethanol in mammals.27, 45, 46 The concomitant upregulation by ethanol of numerous other immediate early genes suggests that acute ethanol may act in part through pathways related to those engaged by neuronal activity.13
Transcript instability coupled with fast shutdown of transcription work concomitantly to keep immediate early gene expression transient.47 We showed that the rapid termination of Hr38 expression is critical for the development of ethanol tolerance, and that Hr38 induction termination depends on Sirt1. Whether Sirt1 is permissive or instructive for Hr38 expression termination requires additional mechanistic studies. However, an adult role for Sirt1 is suggested by pharmacological inhibition phenocopy of the Sirt1 null tolerance and Hr38 expression phenotypes.5 A possible model for a direct role for Sirt1 is as follows. As a histone deacetylase, Sirt1 may normally decrease Hr38 locus acetylation marks associated with open chromatin: complete lack of Sirt1 may prolong the opening of Hr38 chromatin and its availability for transcription. Sirt1 also deacetylates various transcription factors that may control Hr38 expression termination.48, 49 Two additional findings suggest a more complex mechanism. First, in wild‐type flies, acute ethanol exposure rapidly reduces Sirt1 protein levels by half, and this occurs at the same time as the termination of Hr38 expression.5 Accordingly, the 50% Sirt1 protein remaining after acute ethanol exposure in wild‐type flies may be sufficient to promote Hr38 expression termination. Second, whereas Sirt1 is broadly required for ethanol induction of gene expression, it is not required for ethanol induction of Hr38.5 Immediate early genes that are poised for transcriptional activation have distinct chromatin structure and may not need Sirt1 for activation to occur.50 This may imply that the chromatin structure at the Hr38 locus in Sirt1 mutants is by and in large intact. However, induction and termination of immediate early gene expression may be controlled by distinct chromatin‐based mechanisms operating on the same gene locus.
Timely termination of Hr38 may be important for setting the levels or the timing of downstream gene expression changes that influence tolerance development. Moreover, because reducing the amount of Hr38 in Sirt1 mutants restores ethanol tolerance, Hr38 dysregulation appears to be a central mechanism for Sirt1 ethanol behavioral phenotypes. Which Sirt1‐dependent ethanol response genes are regulated by Hr38 may help define the transcriptional pathway that ethanol uses to control behavioral plasticity. In particular, the regulation of presynaptic and postsynaptic properties by ethanol may proceed through an evolutionarily conserved Mef2/Sirt1/Hr38 pathway.5, 39
Supporting information
Movie S1. Hr38 expression pattern in the brain of an ethanol exposed Drosophila, showing expression in the axons of the mushroom body neurons and in nuclei scattered throughout the brain cortex.
ACKNOWLEDGMENTS
We thank the Wolf and Michael Cleary lab members and Ramendra Saha for discussions, and the Bloomington Drosophila Stock Center for strains. This work was supported by grants to F.W.W. from the NIH/NIAAA R03AA023262 and R21AA025560.
Conflict of interest
The authors state no conflict of interest.
Adhikari P, Orozco D, Randhawa H, Wolf FW. Mef2 induction of the immediate early gene Hr38/Nr4a is terminated by Sirt1 to promote ethanol tolerance. Genes, Brain and Behavior. 2019;18:e12486. 10.1111/gbb.12486
Funding information National Institute on Alcohol Abuse and Alcoholism, Grant/Award Numbers: R03AA023262, R21AA025560
REFERENCES
- 1. Edenberg HJ, Foroud T. Genetics and alcoholism. Nat Rev Gastroenterol Hepatol. 2013;10:487‐494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Palmisano M, Pandey SC. Epigenetic mechanisms of alcoholism and stress‐related disorders. Alcohol Fayettev N. 2017;60:7‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fadda F, Rossetti ZL. Chronic ethanol consumption: from neuroadaptation to neurodegeneration. Prog Neurobiol. 1998;56:385‐431. [DOI] [PubMed] [Google Scholar]
- 4. Kaun KR, Devineni AV, Heberlein U. Drosophila melanogaster as a model to study drug addiction. Hum Genet. 2012;131:959‐975. 10.1007/s00439-012-1146-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Engel GL, Marella S, Kaun KR, et al. Sir2/Sirt1 links acute inebriation to presynaptic changes and the development of alcohol tolerance, preference, and reward. J Neurosci. 2016;36:5241‐5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ghezzi A, Pohl JB, Wang Y, Atkinson NS. BK channels play a counter‐adaptive role in drug tolerance and dependence. Proc Natl Acad Sci. 2010;107:16360‐16365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ghezzi A, Krishnan HR, Lew L, Prado FJ, Ong DS, Atkinson NS. Alcohol‐induced histone acetylation reveals a gene network involved in alcohol tolerance. PLoS Genet. 2013;9:e1003986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kong EC, Allouche L, Chapot PA, et al. Ethanol‐regulated genes that contribute to ethanol sensitivity and rapid tolerance in Drosophila. Alcohol Clin Exp Res. 2010;34:302‐316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Morozova TV, Anholt RR, Mackay TF. Phenotypic and transcriptional response to selection for alcohol sensitivity in Drosophila melanogaster. Genome Biol. 2007;8:R231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Urizar NL, Yang Z, Edenberg HJ, Davis RL. Drosophila homer is required in a small set of neurons including the ellipsoid body for normal ethanol sensitivity and tolerance. J Neurosci. 2007;27:4541‐4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nat Rev Neurosci. 2011;12:623‐637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sheng M, Greenberg ME. The regulation and function of c‐fos and other immediate early genes in the nervous system. Neuron. 1990;4:477‐485. [DOI] [PubMed] [Google Scholar]
- 13. Chen X, Rahman R, Guo F, Rosbash M. Genome‐wide identification of neuronal activity‐regulated genes in Drosophila. eLife. 2016.5:e19942. [DOI] [PMC free article] [PubMed]
- 14. Ghezzi A, Zomeno M, Pietrzykowski AZ, Atkinson NS. Immediate‐early alcohol‐responsive miRNA expression in Drosophila. J Neurogenet. 2016;30:195‐204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guan Z, Saraswati S, Adolfsen B, Littleton JT. Genome‐wide transcriptional changes associated with enhanced activity in the Drosophila nervous system. Neuron. 2005;48:91‐107. [DOI] [PubMed] [Google Scholar]
- 16. Fujita N, Nagata Y, Nishiuchi T, Sato M, Iwami M, Kiya T. Visualization of neural activity in insect brains using a conserved immediate early gene, Hr38. Curr Biol. 2013;23:2063‐2070. [DOI] [PubMed] [Google Scholar]
- 17. Baker KD, Shewchuk LM, Kozlova T, et al. The Drosophila orphan nuclear receptor DHR38 mediates an atypical ecdysteroid signaling pathway. Cell. 2003;113:731‐742. [DOI] [PubMed] [Google Scholar]
- 18. Mezan S, Feuz JD, Deplancke B, Kadener S. PDF signaling is an integral part of the Drosophila circadian molecular oscillator. Cell Rep. 2016;17:708‐719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ruaud AF, Lam G, Thummel CS. The Drosophila NR4A nuclear receptor DHR38 regulates carbohydrate metabolism and glycogen storage. Mol Endocrinol. 2011;25:83‐91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blanchard FJ, Collins B, Cyran SA, Hancock DH, Taylor MV, Blau J. The transcription factor Mef2 is required for normal circadian behavior in Drosophila. J Neurosci. 2010;30:5855‐5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kozlova T, Lam G, Thummel CS. Drosophila DHR38 nuclear receptor is required for adult cuticle integrity at eclosion. Dev Dyn. 2009;238:701‐707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Devineni AV, Heberlein U. Preferential ethanol consumption in Drosophila models features of addiction. Curr Biol. 2009;19:2126‐2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Scholz H, Ramond J, Singh CM, Heberlein U. Functional ethanol tolerance in Drosophila. Neuron. 2000;28:261‐271. [DOI] [PubMed] [Google Scholar]
- 24. Devineni AV, McClure K, Guarnieri D, et al. The genetic relationships between ethanol preference, acute ethanol sensitivity, and ethanol tolerance in Drosophila melanogaster. Fly Austin. 2011;5:191‐199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ja WW, Carvalho GB, Mak EM, et al. Prandiology of Drosophila and the CAFE assay. Proc Natl Acad Sci USA. 2007;104:8253‐8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peru Y Colón de Portugal RL, Ojelade SA, Penninti PS, et al. Long‐lasting, experience‐dependent alcohol preference in Drosophila. Addict Biol. 2014;19:392‐401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Flavell SW, Cowan CW, Kim TK, et al. Activity‐dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science. 2006;311:1008‐1012. [DOI] [PubMed] [Google Scholar]
- 28. Shalizi A, Gaudillière B, Yuan Z, et al. A calcium‐regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311:1012‐1017. [DOI] [PubMed] [Google Scholar]
- 29. Andrés V, Cervera M, Mahdavi V. Determination of the consensus binding site for MEF2 expressed in muscle and brain reveals tissue‐specific sequence constraints. J Biol Chem. 1995;270:23246‐23249. [DOI] [PubMed] [Google Scholar]
- 30. Sivachenko A, Li Y, Abruzzi KC, Rosbash M. The transcription factor Mef2 links the Drosophila core clock to Fas2, neuronal morphology, and circadian behavior. Neuron. 2013;79:281‐292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schulz RA, Chromey C, Lu MF, Zhao B, Olson EN. Expression of the D‐MEF2 transcription in the Drosophila brain suggests a role in neuronal cell differentiation. Oncogene. 1996;12:1827‐1831. [PubMed] [Google Scholar]
- 32. Renthal W, Kumar A, Xiao G, et al. Genome‐wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron. 2009;62:335‐348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Park A, Ghezzi A, Wijesekera TP, Atkinson NS. Genetics and genomics of alcohol responses in Drosophila. Neuropharmacology. 2017;122:22‐35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Campos‐Melo D, Galleguillos D, Sánchez N, Gysling K, Andrés ME. Nur transcription factors in stress and addiction. Front Mol Neurosci. 2013;6:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chandra R, Francis TC, Konkalmatt P, et al. Opposing role for Egr3 in nucleus accumbens cell subtypes in cocaine action. J Neurosci Off J Soc Neurosci. 2015;35:7927‐7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Werme M, Olson L, Brené S. NGFI‐B and nor1 mRNAs are upregulated in brain reward pathways by drugs of abuse: different effects in Fischer and Lewis rats. Brain Res Mol Brain Res. 2000;76:18‐24. [DOI] [PubMed] [Google Scholar]
- 37. Werme M, Hermanson E, Carmine A, et al. Decreased ethanol preference and wheel running in Nurr1‐deficient mice. Eur J Neurosci. 2003;17:2418‐2424. [DOI] [PubMed] [Google Scholar]
- 38. McNulty SE, Barrett RM, Vogel‐Ciernia A, et al. Differential roles for Nr4a1 and Nr4a2 in object location vs. object recognition long‐term memory. Learn Mem. 2012;19:588‐592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen Y, Wang Y, Ertürk A, et al. Activity‐induced Nr4a1 regulates spine density and distribution pattern of excitatory synapses in pyramidal neurons. Neuron. 2014;83:431‐443. [DOI] [PubMed] [Google Scholar]
- 40. Adewoye AB, Kyriacou CP, Tauber E. Identification and functional analysis of early gene expression induced by circadian light‐resetting in Drosophila. BMC Genomics. 2015;16:570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. von Heckel K, Stephan W, Hutter S. Canalization of gene expression is a major signature of regulatory cold adaptation in temperate Drosophila melanogaster. BMC Genomics. 2016;17:574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Singh CM, Heberlein U. Genetic control of acute ethanol‐induced behaviors in Drosophila. Alcohol Clin Exp Res. 2000;24:1127‐1136. [PubMed] [Google Scholar]
- 43. Dietrich JB, Takemori H, Grosch‐Dirrig S, Bertorello A, Zwiller J. Cocaine induces the expression of MEF2C transcription factor in rat striatum through activation of SIK1 and phosphorylation of the histone deacetylase HDAC5. Synapse. 2012;66:61‐70. [DOI] [PubMed] [Google Scholar]
- 44. Pulipparacharuvil S, Renthal W, Hale CF, et al. Cocaine regulates MEF2 to control synaptic and behavioral plasticity. Neuron. 2008;59:621‐633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hawk JD, Abel T. The role of NR4A transcription factors in memory formation. Brain Res Bull. 2011;85:21‐29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ron D, Barak S. Molecular mechanisms underlying alcohol‐drinking behaviours. Nat Rev Neurosci. 2016;17:576‐591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Greenberg ME, Belasco JG. Control of the decay of labile proto‐oncogene and cytokine mRNAs Control of Messenger RNA Stability. Academic Press, San Diego, CA USA; 1993:199‐218. 10.1016/B978-0-08-091652-1.50013-X. [DOI] [Google Scholar]
- 48. Gomes AR, Yong JS, Kiew KC, et al. Sirtuin1 (SIRT1) in the acetylation of downstream target proteins. Methods Mol Biol. 2016;1436:169‐188. [DOI] [PubMed] [Google Scholar]
- 49. Yang Y, Yamada T, Hill KK, et al. Chromatin remodeling inactivates activity genes and regulates neural coding. Science. 2016;353:300‐305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen L‐F, Zhou AS, West AE. Transcribing the connectome: roles for transcription factors and chromatin regulators in activity‐dependent synapse development. J Neurophysiol. 2017;118:755‐770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Hr38 expression pattern in the brain of an ethanol exposed Drosophila, showing expression in the axons of the mushroom body neurons and in nuclei scattered throughout the brain cortex.


