Abstract
Gene regulatory networks underlie biological function and cellular physiology. Alternative splicing (AS) is a fundamental step in gene regulatory networks and plays a key role in development and disease. In addition to the identification of aberrant AS events, an increasing number of studies are focusing on molecular determinants of AS, including genetic and epigenetic regulators. We review here recent efforts to identify various deregulated AS events as well as their molecular determinants that alter biological functions, and discuss clinical features of AS and their druggable potential.
Keywords: alternative splicing, genetic and epigenetic, molecular regulators, computational and experimental methods
Widespread Alternative Splicing in Gene Regulation
The advent of high-throughput sequencing technologies has paved the way for genome-wide analyses. Recent analysis from Encyclopedia of DNA Elements (ENCODE) discovered more than 60,000 genes [1, 2], including protein-coding genes, long and small noncoding RNAs. However, the number of protein-coding genes (approximately 20,000) is surprisingly low given the complexity of the proteome. Alternative splicing (AS) provides a versatile mechanism to generate widespread structural transcript variation and proteome diversity [3]. Strikingly, recent data suggests that transcripts from up to 95% of multi-exon genes undergo some degree of alternative splicing [4, 5]. In addition to contributing to the regulation of gene expression, alternative splicing also alters protein sequences with subsequent effects on protein function.
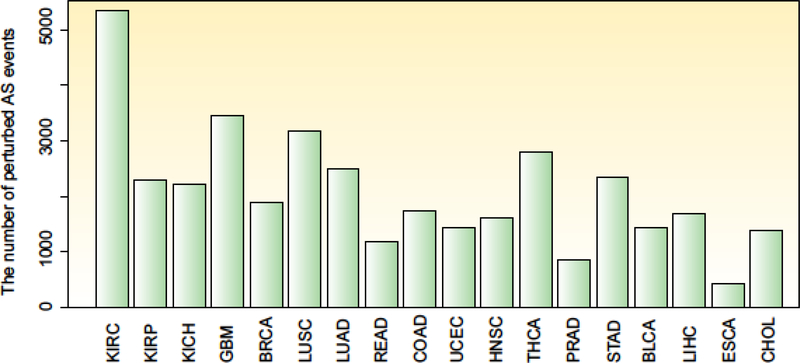
Given the importance of the splicing process in gene regulation, it is not surprising that RNA splicing is often disrupted in cancer [6–8]. An increasing number of studies have investigated the critical roles of alternative splicing in cancer, resulting in the demonstration of thousands of perturbed splicing events (Figure 1). Cancer-specific alternative splicing events in candidate genes or splicing factors had been shown to contribute to disease progression [9–11]; however, dysregulation of splicing patterns on a transcriptome-wide scale had been less well-studied until recently. Analogous to other biological processes, such as transcription and translation, alternative splicing is also regulated by both cis- and trans-acting elements [12]. This process forms an intricate splicing regulatory network that consists of a number of RNA regulatory sequences, RNA–protein complexes, splicing factors, as well as epigenetic regulators (including DNA methylation, histone modification and long noncoding RNAs). Integration of systems genetics with splicing regulatory networks will be crucial to uncover the underlying role of RNA splicing in the initiation and progression of cancer and to elucidate the potential of RNA splicing as a target for personalized medicine. Thus, we first briefly address the alternative splicing process in various types of cancer pathways, describe the potential regulators of alternative splicing, and review computational and experimental methods available to identify global changes in RNA splicing in cancer as well as the methods that aim to discover modulators of alternative splicing. Finally, we discuss the clinical utility of alternative splicing and the potential for exploiting alternative splicing as therapeutic target.
Figure-1. Widespread alternative splicing events across cancer types.
The number of perturbed alternative splicing events in 18 cancer types from the TCGA project. The Percentage Splicing Index (PSI) values for each alternative splicing event were compared between normal and cancer samples by Wilcox’s rank sum test. Alternative splicing events with FDR less than 0.01 were identified as perturbed. BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; STAD, stomach adenocarcinoma; THCA, thyroid carcinoma; UCEC, uterine corpus endometrial carcinoma.
Aberrant Splice-ome Networks with Functional Consequences
Splice-ome networks and signal transduction in the cell
Signal transduction in the cell is a highly orchestrated and regulated process. In the last few years it has become more clear that the efficacy and specificity of signal transduction in a cell is, at heart, a problem of molecular recognition and interaction in complex networks [13, 14]. Splicing is an important post-transcriptional component of gene regulatory networks (splice-ome). Many gene products mediate their function only when interacting with other genes in the context of networks and not in isolation [14, 15]. Indeed, widespread protein interaction rewiring has been observed in splice-ome networks [16, 17], revealing the plastic nature of alternative splicing and the necessity to precisely control it inside the cell [14]. In distinct cell types (with varying genotypes), precise signal transduction including the splicing process controls cell decision for adaptive phenotypic output. When this process goes awry, it would give rise to various disease types, such as cancer.
Alternative splicing and human cancer
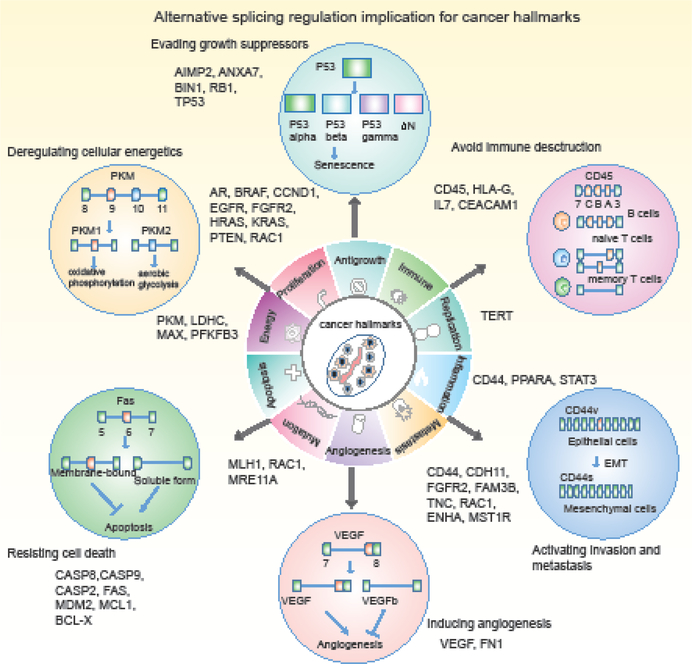
Many signaling pathways are modulated, at least in part, by alternative splicing in the cell [18, 19]. In cancer, it is likely that alternative splicing contributes to various hallmarks implicated in cancer development and progression (Figure 2). In particular, apoptosis, proliferation and metastasis are affected by alternative splicing in a large number of genes. For instance, alternative splicing in several genes (such as CD44 [20, 21], RAC1 [22] and FGFR2 [23]) are implicated in the regulation of more than one cancer hallmark. Apoptosis (programmed cell death) is critical to cellular homeostasis, however, cancer cells become insensitive to apoptotic signals through a number of mechanisms. Splicing-induced isoforms of several proteins can have opposing functions in regulating apoptosis [24]. In the case of the death receptor gene FAS, an isoform including exon 6 produces membrane-bound Fas and promotes apoptosis. In contrast, skipping of exon 6 produces a soluble protein that inhibits apoptosis [25, 26]. CASP9 serves as another example for which alternative splicing regulates cell apoptosis. An isoform (CASP9a) that includes a four-exon cassette results in the production of a pro-apoptotic protein while exclusion of the four-exon cassette (CASP9b) generates an anti-apoptotic protein [27]. Many other apoptosis-associated genes are subjected to alternative splicing regulation, such as CASP8 [28], MDM2 [29] and BCL2L1 [30]. These observations emphasize the prevalence of splicing dysregulation in cell apoptosis. Thus, manipulation of splicing favoring apoptotic isoforms of these genes could provide a novel therapeutic strategy.
Figure-2. Alternative splicing regulation implicated in cancer hallmarks.
Alternative splicing events related to ten cancer hallmarks are shown. Several examples are highlighted in circles.
In addition to resistance to cell death, alternative splicing plays an indispensable role in invasion and metastasis. An important example of this is CD44 (Figure 2), which undergoes extensive aberrant splicing. CD44 variants (CD44v) that contain one or more of variable exons have been detected in multiple cancer types. Exclusion of the variable exons produces the standard CD44 isoform (CD44s). Splicing of CD44 is dynamically regulated during epithelial–mesenchymal transition (EMT) [31]. The CD44v is predominant in epithelial cells while there is a gradual loss of this isoform and gain of the expression of CD44s isoform in cells that have undergone EMT [31]. CD44 isoform switching is required for cells to undergo EMT in breast cancer [32]. Moreover, CD44v promotes cell growth by activating Ras/MAPK signaling [33]. By contrast, CD44s which is devoid of variable exons mediates cell contact inhibition [34].
Moreover, recent efforts have shed new light on RNA splicing linking emerging cancer hallmarks, such as inflammation and avoiding immune detection. A compelling body of evidence suggests that inflammation and immune system dysregulation play critical roles in tumor progression [35, 36]. Different types of infiltrating immune cells can either support or inhibit tumor progression. Importantly, different splice isoforms are expressed in distinct immune cell types. For instance, the balance between alternative splice variants varies during virus-induced lymphomagenesis. Telomerase upregulation, critical for cell immortalization and oncogenesis, depends at least partly upon alternative splicing and requires an increase in active constitutively spliced isoform levels in lymphoma-derived T-cells [37]. The CD19 antigen, often expressed on many B-cell acute lymphoblastic leukemias (B-ALL), can be recognized by chimeric antigen receptor-armed T cells (CART). However, exon 2 skipping of CD19, which allows expression of the N-terminally truncated variant, fails to trigger CART-mediated killing of cancer cells [38].
Functional consequence of aberrant splicing
Alternative splicing often accompanies disease progression, and seems to play a functional role in causing disease. Could aberrant splicing exhibit a functional consequence and result in disease, or is it merely a byproduct during the disease process? Accumulating evidence has demonstrated a seemly causal role of aberrant splicing in cellular malignancy. For instance, knock-down of the exon 6-containing isoform of the gene DBF4B, which encodes a kinase with a role in DNA replication and proliferation, dramatically reduces tumorigenesis in colon both in vitro and in vivo [39]. In another study, upregulation of alternatively spliced isoform in the gene PACE4, encoding a member of the proprotein convertase family, leads to a dramatic increase in the progrowth differentiation factor GDF15 in prostate cancer [40]. Likewise, it was demonstrated that alternative splicing of NUMB, encoding an endocytic adaptor protein, could control cell proliferation in lung cancer using xenograft tumor models [41]. Furthermore, alternative spliced isoforms can modulate cellular functions by serving as a competitor with its common splice isoform. For instance, an aberrant splice isoform of PRMT1, encoding a protein arginine methyltransferase, modulates its kinase activity by competing for substrates, resulting in cellular malignancy [42]. Collectively, these data reflect the functional roles of aberrant splicing in eliciting disease progression.
Alternative splicing of non-coding genes and regulatory elements
While most studies have focused on AS of protein-coding genes, AS can also occur in noncoding RNAs. Long noncoding RNAs (lncRNAs) are a newly identified type of noncoding RNAs longer than 200nt that lack protein coding capacity [43]. It has been shown that >25% of lncRNA genes in human show evidence of AS with different functional outcomes [44]. AS events in lncRNAs are also observed in several cancer types. For example, AS of WT1-AS1 has been found in acute myeloid leukemia [45]. XIST encodes a lncRNA which plays a role in X-chromosome inactivation. It has two major splicing variants and evidence has shown that the short splicing isoform is sufficient to induce X-chromosome inactivation [46]. Indeed, a large fraction of the human genome including repetitive elements (RE) [47] is transcribed [48], and undergoes AS [49]. It has been shown that the AS of RE could generate functional isoforms that might have implications for genome instability in cancer [49]. In fact, at the global level, Deveson et al. have shown that noncoding exons are universally alternatively spliced by targeted single-molecule and short-read RNA-Seq [50].
Taken together, the studies discussed in this section demonstrate functional consequences of aberrant RNA splicing in regulating the phenotypic output at the cellular or organismal level. Understanding the mechanisms of alternative splicing will undoubtedly advance our knowledge of the processes underlying signal transduction rewiring and disease progression, and potentially facilitate biomarker discovery for clinical prevention or therapeutics.
Genetic and Epigenetic Regulation of Splice-ome Neworks
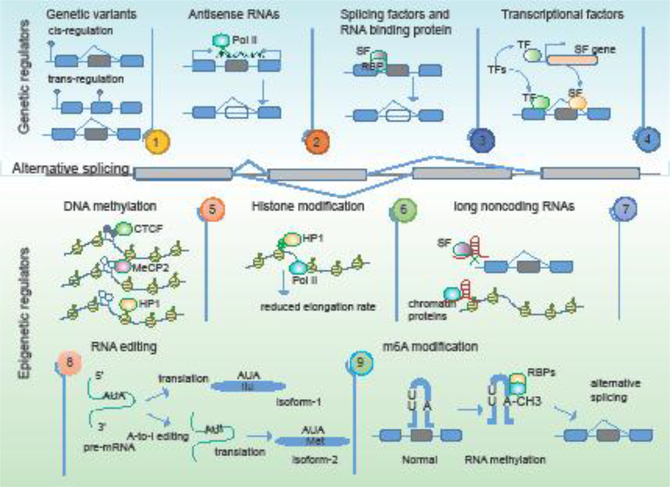
Given the complexity of gene expression regulation, it is not surprising that splicing is intricately modulated by multiple cis- and trans-regulatory elements. With the development of high-throughput sequencing technologies, both genetic and epigenetic regulators of alternative splicing have been identified (Figure 3).
Figure-3. Genetic and epigenetic determinants of alternative splicing.
The upper panels show the genetic regulators of alternative splicing events, including genetic variants, antisense RNAs, splicing factors and RNA binding proteins, and transcription factors. The bottom panels show the epigenetic regulators including DNA methylation, histone modification, long noncoding RNAs, RNA editing and RNA methylation.
Genetic variants
Whole-genome and whole-exome sequencing projects have identified thousands of somatic variants in patients stricken by various diseases including cancer [51]. Deregulation of AS during tumorigenesis is likely subject to regulation by somatic mutations that influence cancer risks by affecting the function of coding or noncoding genes [52, 53]. To determine the mutational impact, studies have been performed to assess their association with expression levels of specific genomic loci (expression quantitative trait loci, or eQTLs) [54], as well as the extent of alternative splicing (splicing QTLs) [55]. Mutations could modulate RNA splicing in cis or in trans across cancer types (Figure 3), and multiple ways in which mutations affect splicing fidelity have been observed [56, 57]. For instance, certain mutations in splicing factors such as SRSF2 and the hnRNP complexes may lead to genome-wide alterations in splicing [17, 58]. These mutations either enhance or impair the binding of splicing factors or mediators, thus resulting in the inclusion or skipping of exons in multiple genes [56, 59]. Moreover, mutations could introduce stop codons that target the transcript for degradation. Previous work on many cancer-related genes, including BRCA1 [60], CFTR, GH1 and ATM, demonstrate that mutations can induce aberrant splicing in complex diseases. The tumor suppressor gene BRCA1 is involved in DNA damage repair, and individuals with BRCA1 mutations exhibit high risk of breast cancer and ovarian cancer. For instance, a G/T mutation in exon 18 of BRCA1 results in exon skipping, by eliminating the first domain at the C-terminus, rendering the protein non-functional [61]. This example illustrates that genetic variants can alter RNA splicing patterns and affect the function of genes. However, genome-wide elucidation of genetic variants that mediate splicing perturbations in disease remains a challenging task.
Antisense RNAs
Recent reports have demonstrated aberrant splicing patterns could correlate with the expression of endogenous antisense mRNAs. Antisense RNAs are potential regulators of alternative splicing, transcriptional initiation and termination [62]. For example, antisense-correlated splicing is shown for the MSH6 gene, which is involved in DNA-mismatch repair. The splicing of two exons of MSH6 is significantly correlated with the expression of the antisense gene, FBXO11, at the same genomic locus [62]. These results demonstrate the ability of antisense RNAs to alter splice site selection and illustrate a powerful means to regulate cell fate in cancer. Moreover, these splicing events occur in a cancer subtype-specific manner [63], indicating antisense RNAs are likely an important component in the regulation of subtype-specific alternative splicing and tumorigenesis. A variety of mechanisms have been uncovered to explain the effects of antisense RNAs, such as direct interference of RNA Polymerase II binding, sterically blocking splice sites and thus redirecting the spliceosome to unhindered splice sites [64].
Splicing Factors and RNA Binding Proteins
In addition to cis-acting elements, trans-acting regulators can also affect RNA splicing in cancer. Splicing factors and RNA binding proteins are key trans-regulators of alternative splicing (Figure 3). For example, an exon skipping event in the target RNA MST1R is mediated by the splicing factors SRSF1 [65] or HNRNPA2B1 [66] in cancer. These trans-regulators can be altered by genomic mutations, which generally impair RNA-binding regulatory sites, thus affecting the splicing of multiple target oncogenes or tumor suppressors. For instance, mutations in trans-regulators SF3B1 and U2AF35 likely result in RNA splicing deregulation in various cancer types [67, 68]. trans-regulators could also be subject to transcriptional and post-transcriptional regulation, as well as epigenetic changes in cancer. Recently, systematic analysis has revealed widespread alternations in thousands of RNA binding proteins in cancer, including mutations, copy number variation and gene expression changes [69]. Together, these results collectively suggest that aberrant splicing changes contributing to the pathophysiology of cancer can be triggered by a complex network of cis- and trans-regulatory elements.
Transcription Factors
Comprehensive knowledge of the factors that regulate alternative splicing networks is critical for understanding splicing deregulation in disease. Transcription factors (TFs) emerge in recent years as a new class of alternative splicing regulators [70]. TBX5 has been established as a TF involved in development and disease [71]. TBX5 plays functional roles in regulating the AS process by forming a complex with the splicing factor SC35 [72]. Another TF named ZNF638 has also been shown to interact with splicing regulators and impact alternative splicing [73]. Recent studies have revealed on a global scale the mechanism of TFs in regulating AS. SPAR-seq (Systematic Parallel Analysis of endogenous RNA regulation coupled to barcode Sequencing) is a flexible screening platform able to probe more than 1,000 trans-acting factors for their impact on endogenous splicing events [74]. Based on this system, hundreds of TFs have been found to positively or negatively control AS events, and they regulate AS networks by directly binding RNA sequences which are adjacent to target exons. Furthermore, these TFs can modulate the expression of other splicing factors that regulate the same target exons. Last but not least, the coupling nature of transcription and splicing indicates that the changes in TF functions may influence splicing regulation. Together, these findings highlight that transcription factors represent a new layer of mediators that influence alternative splicing events with key functional roles in cancer.
DNA Methylation
DNA methylation is an epigenetic modification that plays an important role in regulating transcription [75]. However, epigenetic modifications also appear to regulate alternative splicing directly [76]. Alternatively spliced exons show lower levels of DNA methylation than constitutively spliced exons [77]. Three known factors (including CTCF, MeCP2 and HP1) can transmit information from DNA methylation levels to regulators of alternative splicing (Figure 3). CTCF and MeCP2 affect alternative splicing by modulating RNA Polymerase II (Pol II) elongation whereas HP1 recruits splicing factors from methylated DNAs to the mRNAs [77, 78]. As an example, alternative splicing of CD45 is regulated by DNA methylation [79]. CTCF can bind to exon 5 of CD45, which serves to slow elongation by Pol II. This binding increases the inclusion of exon 5. However, DNA methylation can inhibit CTCF binding, enabling Pol II to traverse pre-mRNA more rapidly. Thus, DNA methylation can result in exon 5 exclusion.
Histone Modification
Histone modification represents another key epigenetic modification that serves as an important regulator of gene transcription. A number of studies have shown that local histone modifications (such as H3K36me3, H3K9me3 and acetylation), can specifically influence the inclusion or exclusion of alternative exons [80]. It was demonstrated that the chromosomal regions of CD44 variable exons are highly enriched in H3K9me3 signals [81]. The enriched histone modification signals are recognized by HP1, which reduces Pol II-mediated elongation rates and facilitates inclusion of variable exons. Thus, the presence of H3K9me3 histone marks correlates with increased inclusion of variable exons. In addition, H3K9me2 was found to be associated with inclusion of alternative exons in Fibronectin (FN1), which plays an important role in cell adhesion, migration and differentiation [82]. HP1 can also recognize the H3K9me2 marks and slow down Pol II elongation, resulting in inclusion of exons. These data suggest that local histone modification marks can perturb the rate of Pol II progression leading to alternative splicing that could potentially contribute to the etiology of cancer [83].
Long Noncoding RNA
Recent studies have also suggested the critical roles of lncRNAs in the control of alternative splicing (Figure 3). In the past decade, applications of next-generation sequencing technologies to a number of cancer transcriptomes have revealed thousands of lncRNAs. Although only a small fraction of lncRNAs have been functionally characterized, several have been shown to have key roles in proliferation, migration or genomic stability [84, 85]. For instance, the expression levels of the lncRNA MALAT1 are often decreased in non-small cell lung cancer. In normal conditions, MALAT1 is associated with splicing factors and blocks the recruitment of these proteins to pre-mRNA. However, when MALAT1 is depleted, the levels of free splicing factor proteins are then increased, resulting in perturbed splicing pattern [86]. Furthermore, lncRNAs can modulate local histone modifications by directing chromatin complexes to DNA. Thus, it is valuable to determine whether lncRNAs can also influence alternative splicing by modulating chromatin modifications. Indeed, another evolutionarily conserved lncRNA, generated from within the FGFR2 locus [87], promotes epithelial-specific alternative splicing of FGFR2. This lncRNA mediates recruitment of Polycomb-group protein and histone demethylase KDM2a to impair binding of a repressive chromatin complex. Together, these examples uncover a new function of lncRNAs in splicing modulation in cancer.
RNA editing
RNA editing, which alters the sequences of RNAs, also contributes to an additional regulatory layer of alternative splicing [88]. A-to-I RNA editing (deamination of adenosine to inosine) is the main form of RNA editing in mammals, which occurs in regions of double-stranded RNA (dsRNA). Adenosine deaminases acting on RNAs (ADARs) are the RNA-editing enzymes involved in the hydrolytic deamination of A-to-I editing [89]. RNA editing in intronic sequences has been shown to influence alternative splicing of flanking exons. For example, RNA editing can result in elevated expression of signal transducer and activator of transcription 3 (STAT3), specifically the alternatively spliced STAT3β. Silencing ADAR1 causes a decrease in STAT3 editing and the decreased expression of STAT3β [90]. In addition to protein-coding genes, RNA editing can occur in noncoding genes, such as lncRNAs and microRNAs (miRNAs) [91, 92]. However, how RNA editing events regulate alternative splicing across cancer types still remains largely unknown.
RNA modification
In addition to DNA methylation, extensive modifications of mRNAs have been identified, such as N6-methyladenosine (m6A) [93]. RNA modifications play a role in regulating alternative splicing and gene expression, and perturbations of RNA modifications are associated with various types of cancer [94]. It has been shown that m6A influences the RNA-structure-dependent accessibility of RNA binding motifs to affect RNA-protein interactions. For instance, m6A can alter the local structure in mRNAs and lncRNAs to facilitate binding of the ribonucleoprotein HNRNPC, thus affecting the abundance as well as alternative splicing of target RNAs [95]. Moreover, it was shown that the m6A demethylase FTO binds preferentially to pre-mRNAs in intronic regions, in the proximity of alternatively spliced exons [96]. The function of m6A modification is also mediated by proteins capable of recognizing m6A, such as YTHDC1. These proteins have been reported to affect alternative splicing patterns but the underlying mechanisms remain unclear [97]. These representative examples illustrate how alternative splicing is regulated through m6A dependent RNA structural remodeling, providing a new direction for investigating the regulatory roles of alternative splicing in cancer.
Computational Identification of Alternative Splicing and Regulators in Splice-ome
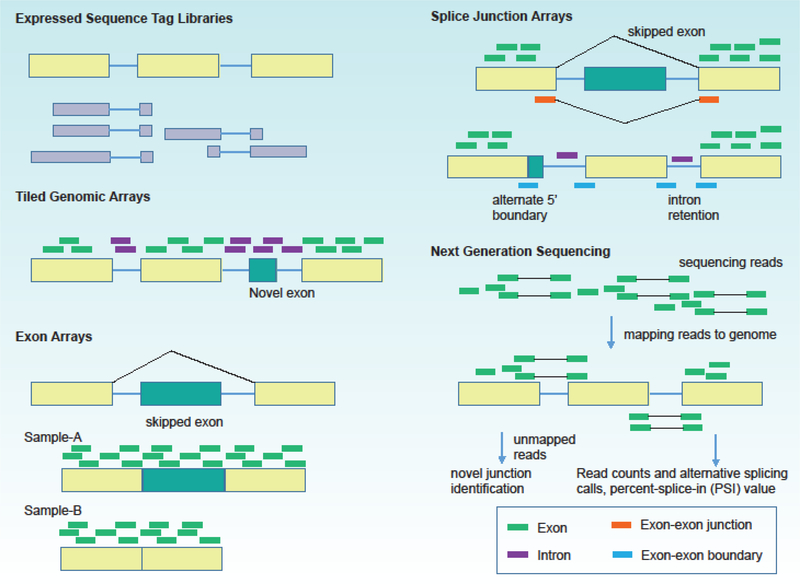
Although aberrant splicing is thought to play critical roles in disease signaling networks, a comprehensive overview in disease is not currently available. To address the challenge for genome-wide study of alternative splicing, several splice-sensitive platforms have been developed (Box 1 and Figure-4). The development of RNA sequencing (RNA-seq) has facilitated the discovery of splicing isoforms, allowing researchers to directly study the spectrum and expression of isoforms and their relative changes in cancer. Recently, the third-generation DNA sequencers, such as Oxford Nanopore Technologies [98], have led to long-read mRNA sequencing. These technologies allow to detect isoform structures at a high resolution, and therefore have successfully identified many novel AS events [99].
Box 1. Experimental Technologies for Studying Alternative Splicing.
Expressed Sequence Tag Libraries
Expressed sequence tags or ESTs are short sub-sequences of cDNA sequences, which are derived from fully processed mRNA. ESTs provide a broad sample of mRNA diversity and can be analyzed computationally [1]. ESTs or cDNA sequences are aligned to genomic sequences using bioinformatics programs that explore conserved splice site consensus sequences flanking the gaps formed by intron sequences between the aligned exons.
Microarrays
Tiled Genomic Arrays.
Genome tiling arrays use a set of overlapping probes to map transcribed regions to a very high resolution [2]. Previously non-annotated genes, exons and splicing events can be incorporated and interrogated, providing a comprehensive coverage. This method does not need for prior knowledge of exon coordinates, thus allowing de novo discovery of alternative splicing. The disadvantage of this platform is that it requires a significant expense and possess a challenge for computational analysis. Exon Arrays. Exon arrays can detect gene expression and alternative exon usage simultaneously. The resolution is increased with multiple exon centric probes within each annotated and predicted exon. This technology permits a comprehensive and unbiased transcriptome coverage [3, 4]. This platform does not contain splice-junction probes and permit discovery of novel alternative splicing variations.
Splice Junction Arrays.
This platform contains annotated exon-exon junction probes exclusive to individual isoforms and have been used to measure predetermined set of alternative splicing events [5, 6]. However, junction arrays cannot establish if two alternative splicing events are in the same or different transcripts and cross-hybridization may also generate false positives.
Next Generation Sequencing Technology
The recent development of the deep sequencing technologies has opened a new frontier for analyses of entire transcriptomes. RNA-sequencing (RNA-Seq) provides a more accurate measurement of transcript levels and their isoforms without a priori knowledge of genome [7, 8]. In this technology, cDNA fragment libraries with adaptors attached to one or both ends are obtained from a pool of RNA. High-throughput sequencing is performed to obtain short sequences from one end (single-end) or both ends (pair-end) from each RNA molecule. The acquired sequencing reads are aligned to a reference genome or gathered de novo without the reference genome sequence to get a genome-wide transcription map. This technology allows the detection of novel splice variants and genome-wide splice junctions.
Outstanding Questions Box
What are the mechanisms by which splice-ome networks are regulated and exploited by cancer cells?
Can aberrant splicing cause cancer? Or is splicing merely a secondary effect caused by defects in gene expression, epigenetic factors or chromatin structures?
How to integrate multi-omics datasets to determine molecular regulators of alternative splicing events in cancer?
To what extent and how do splice-ome networks contribute to phenotypic heterogeneity across cancer patient populations?
Figure-4.
Experimental platforms for characterizing alternative splicing events.
However, the advantages of these sequencing technologies cannot be realized without efficient and reliable bioinformatics processing. A number of computational tools have been proposed to identify alternative splicing events as well as genetic and epigenetic regulators of these processes (Table 1, Key Table). First, to map RNA sequencing data to reference human genomes, many bioinformatics packages are available for this task, including BWA [100], Bowtie [101] and SOAP [102]. Moreover, some methods have been developed for mapping sequencing reads to isoforms, such as TopHat [103] and SpliceMap [104]. After assignment of the reads to their possible gene of origin, the next step is quantification of alternative splicing events and isoforms. These methods either use annotation or de novo reconstruction of transcripts, such as Cufflinks [105] and iReckon [106]. Most of these methods can estimate the majority of alternative splicing types, while some methods focus on specific alternative splicing events, such as EBchangePoint [107] focusing on alternative 3’ and 5’ splicing sites.
Table 1.
Computational methods to identification of alternative splicing events or differential events in multiple conditions.
| Methods | Description | Link | ref |
|---|---|---|---|
| Cufflinks | Assembles transcripts, estimates abundances, and tests for differential expression. | http://cole-trapnell-lab.github.io/cufflinks/ | [1] |
| iReckon | A method for simultaneous determination of the isoforms and estimation of their abundances. | http://compbio.cs.toronto.edu/ireckon | [2] |
| NMFP | A non-negative matrix factorization based method for identifying isoforms | http://www.stat.ucla.edu/~jingyi.li/packages/NMFP.zip | [3] |
| SplicePie | Classification of RNA-Seq reads based on three major stages of splicing | https://github.com/pulyakhina/splicing_analysis_pipeline | [4] |
| EBChangePoint | An empirical Bayes change-point model to identify alternative 3’ and 5’ splicing sites | http://ebchangepoint.sourceforge.net/ | [5] |
| MISO | A probabilistic framework that quantitates the expression of alternatively spliced genes and identifies differentially regulated isoforms. | http://genes.mit.edu/burgelab/miso | [6] |
| DEXSeq | Uses generalized linear models to test for differential usage of exons and hence of isoforms. | http://www-huber.embl.de/pub/DEXSeq/ | [7] |
| MATS | A Bayesian framework for flexible detection of differential alternative splicing. | http://rnaseq-mats.sourceforge.net/ | [8] |
| SpliceSeq | A resource for analysis and visualization of RNA- Seq data on alternative splicing. | http://bioinformatics.mdanderson.org/main/SpliceSeq:Overview | [9] |
| ALEXA-Seq | Analyze massively parallel RNA sequence data to catalog transcripts and assess differential and alternative expression of known and predicted mRNA isoforms. | http://www.alexaplatform.org/alexa_seq/ | [10] |
| JuncBASE | A series of scripts that calculate exon exclusion and inclusion counts to splicing events and to identify statistically significant affected splicing events. | http://compbio.berkeley.edu/proj/juncbase/ | [11] |
| DiffSplice | An ab initio method for the detection and visualization of differential alternative transcription. | http://www.netlab.uky.edu/p/bioinfo/DiffSplice | [12] |
| rDiff | A parametric test using known isoform annotations to detect isoform changes and a nonparametric test to detect differential read coverages. | http://bioweb.me/rdiff | [13] |
| SplAdder | Identification, quantification and testing of alternative splicing events. | http://bioweb.me/spladder | [14] |
| jSplice | De novo extraction of alternative splicing events from RNAsequencing data with high accuracy, reliability, and speed | http://www.mhs.biol.ethz.ch/research/krek/jsplice | [15] |
| diffsplicing | Model the expression levels in three different settings: overall gene expression level, absolute transcript expression level and relative transcript expression level | https://github.com/PROBIC/diffsplicing | [16] |
| JunctionSeq | Detection and visualization of differential splicing | http://hartleys.github.io/JunctionSeq/ | [17] |
| SigFuge | single gene clustering of RNA- seq reveals differential isoform usage | http://www.bioconductor.org/packages/devel/bioc/html/SigFuge.html | [18] |
| rMATS | Robust and flexible detection of differential alternative splicing from replicate RNA- Seq data | http://rnaseq-mats.sourceforge.net/ | [19] |
| SNPlice | A computational approach for identifying cis-acting, splice-modulating variants from RNAseq data. | https://code.google.com/p/snplice/ | [20] |
| Jung et al. | Identified somatic exonic SNVs that disrupt splicing | Not provided the scripts. | [21] |
| SPANR | A machine-learning technique that scores how strongly genetic variants affect RNA splicing | http://tools.genes.toronto.edu/ | [22] |
| sQTLseekeR | A distance-based approach to identify the splicing QTLs | https://github.com/jmonlong/sQTLseekeR | [23] |
| GLiMMPS | A robust statistical method for detecting splicing QTLs from RNA-seq data | http://www.mimg.ucla.edu/faculty/xing/glimmps | [24] |
| SPAR-seq | A multiplexed and quantitative platform coupled to a sequencing output that is capable of linking trans-acting factors to endogenous gene regulation events of interest | https://github.com/vastgroup/vast-tools | [25] |
| DrAS-Net | A network-based method to identify the mutations that mediated the alternative splicing dysregulation in cancer | http://www.bio-bigdata.com/dras_net/index.jsp | [26] |
For the study of disease transcriptomes, the basic task is to find genes or isoforms that are differentially expressed in disease. The importance of studying alternative splicing alterations has been demonstrated by the observation that in many cancers changes in splicing isoforms predominate while the total expression levels of the parental genes do not vary. Thus, an increasing number of methods are being proposed to provide an isoform or event view of differential splicing or expression in cancer (Table 1). For instance, MISO takes an exon-centric strategy to detect differentially regulated exons or isoforms by comparison of estimated splicing levels [108]. In addition, some models are specifically designed for case-control matched pairs in clinical RNA-seq datasets, such as rMATS [109]. Alternative transcription start site is a major mechanism for diversity of human transcriptome. Qin et al. have developed a computational pipeline SEASTAR to identify first exons from RNA-seq data alone [110]. Alternative first exon usage is then quantified and compared across multiple samples. In addition, SUPPA2 was proposed to identify AS by taking biological variability into account [111]. All of these computational methods have shown promising performance, and have provided powerful approaches for quantitatively studying differential alternative splicing in human disease.
However, the functions of most AS events in disease are still not known. It remains challenging to address the question of which splicing events may be causal to disease, and which are secondary. In cancer, for example, it could be informative to perform analysis of cancer stem cells versus tumor mass cells derived from these stem cells. Identifying molecular determinants and mechanisms that perturb alternative splicing is fundamental for the development of disease-specific biomarkers for prognosis and therapy. In contrast to identification of perturbed alternative splicing events in disease, a number of computational methods have been proposed to identify the determinants of alternative splicing events. SNPlice is proposed for identifying cis-acting, splicemodulating variants from RNA-seq data [112]. In addition, Jung et al. used paired DNA sequencing and RNA-seq data across a number of cases to show the roles of somatic mutations in disrupting splicing in cancer [113]. Moreover, SPANR is developed to identify genetic splicing determinants in diseases. Several methods have also been developed to identify the splicing QTLs, such as sQTLseekeR [114] and GLiMMPS [115]. To systematically integrate different types of omics data, recent computational algorithms [69, 116] are devised for genome-wide identification of molecular determinants of AS. For instance, DrAS-Net employs a network-based approach to identify mutations at large scale that likely influence AS across cancer types [116]. SPAR-seq (Systematic Parallel Analysis of endogenous RNA regulation coupled to barcode Sequencing) is proposed to reveal global AS regulatory patterns by barcoded next-generation sequencing [74].
Systematic analyses have been performed to reveal the association between gene expression, DNA methylation, copy number variation and microRNA expression with cancer patient clinical features, such as survival [117]. In contrast, despite the importance of alternative splicing, limited efforts have been made to associate aberrant splicing to clinical features. A significant decrease in the overall survival of patients is observed in hyper-spliced tumors in medulloblastoma [63]. Prognostic alternative mRNA splicing signatures have been identified in non-small cell lung cancer [118]. In addition, a statistical method named SURVIV (Survival analysis of mRNA Isoform Variation) [119], is designed for identifying isoform variations that are associated with survival. By analysis of multiple cancer datasets, it is demonstrated that alternative splicing-based predictors consistently outperform gene expression-based survival predictors. A recent genomewide analysis of AS across cancer types further supports that AS network perturbations correlate with patient survival in general [116]. Integration of clinical and gene expression information with alternative splicing analysis leads to optimal predictions. These results collectively demonstrate potential functional significance of alternative isoform variation in disease.
Concluding Remarks
It is clear that functions of gene products are dynamically regulated by alternative splicing. Deregulation of alternative splicing influences many aspects of pathophysiology in human disease. Identification of alternative splicing deregulation in disease may generate signatures that could guide diagnostics and lead to the discovery of novel therapeutic targets. However, our current knowledge on the function and regulation of splice-ome network alterations in disease is only at its infancy.
Our understanding of alterative splicing alterations is limited to a handful of genes that are well studied. In cancer, for instance, systematic approaches aided by high-throughput sequencing are needed to map the AS landscape across various types of cancer. With increasing availability of high-quality cancer transcriptomes, it is anticipated that a large number of AS alterations would be identified and quantified. However, despite accumulating evidence of cancer associated AS events, the molecular determinants contributing to cancer-related splicing changes remain unknown in most cases. To systematically understand the determinants that lead to splicing deregulation, it will be important to integrate multiple omics datasets, such as (a) whole-genome sequencing data to identify somatic mutations that contribute to splicing alterations; (b) assess the copy number variation, and gene expression profiles to explore the alterations of splicing regulators or RNA binding proteins; and (c) combine large-scale epigenetic datasets to identify epigenetic regulators of splicing.
In the cell, there is often a mixture of protein isoforms generated from alterative splicing of the same gene, producing both “canonical” and “non-canonical”/”aberrant” splice products. Therefore, the ratio among this isoforms can vary, depending on if AS events are full (constitutive) or partial (minor). For instance, genomic mutations in RNA-binding proteins (RBPs) or in splice sites of the RNAs can result in alternative splicing, removing the canonical products [116]; while the expression levels of RBPs or RNA targets may have a relatively minor effect on alternative splicing. Different isoforms can exhibit distinct functional differences, and especially for genes that encode oligomeric complexes, the balance of splicing isoforms can play an important regulatory role. For instance, PRMT1, encoding a protein arginine methyltransferase, could produce an isoform PRMT1v2 in breast cancer. Overexpression of this isoform could promote cell survival and invasion [120]. Another isoform of PRMT1 that lacks exons 8 and 9 can not form catalytically active oligomers with the other endogenous PRMT1 isoforms, leading to oncogenesis [42].
Moreover, genes do not act in isolation but rather interact with each other in the context of splice-ome networks (such as RBP-RNA interactions) during cell signaling. Molecular networks provide an informative platform to investigate properties of cellular systems [14, 121]. Network-based approaches have been successfully applied to identifying cancer genes [122, 123]. Combining large-scale molecular interaction networks (protein-DNA, protein-RNA, and protein-protein) may provide novel insights into the splice-ome deregulation in disease. To accommodate integrative analyses, the development of robust computational methods and modeling approaches in combination with the stateof-art ‘omics’ techniques is key to understanding the nature of alternative splicing and unveiling their contribution to disease development, progression, and therapeutic resistance.
Taken together, different genetic or epigenetic regulators may contribute to the deregulation of alternative splicing in human disease. With a better understanding of splicing associated networks in disease, we may be able to gain insight into complex genotype-phenotype relationships at different molecular regulatory levels, and exploit personalized isoform-based therapeutics and biomarkers.
Highlights.
Alternative splicing has contributed to widespread structural transcript variation and proteome diversity.
Studies in various diseases showed that alternative splicing plays critical roles in different aspects of etiology, suggesting that alternative splice isoforms could potentially serve as therapeutic targets.
Various types of genetic and epigenetic regulators of alternative splicing have been revealed.
Combination of computational and experimental approaches is instrumental in identifying alternative splicing events as well as potential regulators.
Acknowledgments
This work was supported by the Cancer Prevention and Research Institute of Texas (CPRIT) New Investigator Grant RR160021 (N.S.), the University of Texas System Rising STARs award (N.S.), the AASLD Foundation Pinnacle Research Award in Liver Disease (N.S.), the University Center Foundation via the Institutional Research Grant program at the University of Texas MD Anderson Cancer Center (N.S.), and NIH/NCI Transition Career Development Award Grant 1K22CA214765 (S.Y.). Y.L. is supported by a postdoctoral fellowship from the Harold C. and Mary L. Daily Endowment Fund. Additional funding was provided by Susan G. Komen PDF17483544 to D.J.M. We would like to apologize to all our colleagues whose important work could not be cited here due to space restrictions. We also thank anonymous reviewers for their constructive feedback to improve the paper.
Glossary
- Alternative splicing
a process by which exons or portions of exons or noncoding regions within an mRNA transcript are differentially joined or skipped, resulting in multiple protein isoforms encoded by a gene.
- DNA methylation
a process by which methyl groups are added to the DNA molecule, which changes the activity of a DNA segment without changing the sequence
- Histone modification
a covalent post-translational modification to histone proteins which includes methylation, phosphorylation, acetylation, ubiquitylation, and sumoylation
- Long noncoding RNAs
RNAs of >200bp nucleotides in length that lack protein coding potential
- Cancer hallmarks
A set of biological processes acquired during the multistep development of human tumors
- Genome wide association studies
an examination of a genome-wide set of genetic variants in different individuals to explore whether any variant is associated with a trait
- eQTLs
DNA sequence variants that influence the expression level of one or more genes
- Splicing QTLs
DNA sequence variants that influence the alternative splicing level of one or more genes
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kellis M et al. (2014) Defining functional DNA elements in the human genome. Proc Natl Acad Sci U S A 111 (17), 6131–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Consortium EP (2011) A user’s guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol 9 (4), e1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nilsen TW and Graveley BR (2010) Expansion of the eukaryotic proteome by alternative splicing. Nature 463 (7280), 457–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerstein MB et al. (2014) Comparative analysis of the transcriptome across distant species. Nature 512 (7515), 445–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang ET et al. (2008) Alternative isoform regulation in human tissue transcriptomes. Nature 456 (7221), 470–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh RK and Cooper TA (2012) Pre-mRNA splicing in disease and therapeutics. Trends Mol Med 18 (8), 472–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scotti MM and Swanson MS (2016) RNA mis-splicing in disease. Nat Rev Genet 17 (1), 19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oltean S and Bates DO (2014) Hallmarks of alternative splicing in cancer. Oncogene 33 (46), 5311–8. [DOI] [PubMed] [Google Scholar]
- 9.Sebestyen E et al. (2015) Detection of recurrent alternative splicing switches in tumor samples reveals novel signatures of cancer. Nucleic Acids Res 43 (3), 1345–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karni R et al. (2007) The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol 14 (3), 185–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anczukow O and Krainer AR (2016) Splicing-factor alterations in cancers. RNA 22 (9), 1285–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chabot B and Shkreta L (2016) Defective control of pre-messenger RNA splicing in human disease. J Cell Biol 212 (1), 13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yi S et al. (2017) Functional variomics and network perturbation: connecting genotype to phenotype in cancer. Nat Rev Genet 18 (7), 395–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vidal M et al. (2011) Interactome networks and human disease. Cell 144 (6), 986–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sahni N et al. (2013) Edgotype: a fundamental link between genotype and phenotype. Curr Opin Genet Dev 23 (6), 649–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang X et al. (2016) Widespread Expansion of Protein Interaction Capabilities by Alternative Splicing. Cell 164 (4), 805–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gueroussov S et al. (2017) Regulatory Expansion in Mammals of Multivalent hnRNP Assemblies that Globally Control Alternative Splicing. Cell 170 (2), 324–339 e23. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J and Manley JL (2013) Misregulation of pre-mRNA alternative splicing in cancer. Cancer Discov 3 (11), 1228–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sveen A et al. (2016) Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene 35 (19), 2413–27. [DOI] [PubMed] [Google Scholar]
- 20.Birzele F et al. (2015) CD44 Isoform Status Predicts Response to Treatment with Anti-CD44 Antibody in Cancer Patients. Clin Cancer Res 21 (12), 2753–62. [DOI] [PubMed] [Google Scholar]
- 21.Cancer Genome Atlas Research, N. et al. (2015) Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl J Med 372 (26), 2481–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou C et al. (2013) The Rac1 splice form Rac1b promotes K-ras-induced lung tumorigenesis. Oncogene 32 (7), 903–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bai A et al. (2010) GP369, an FGFR2-IIIb-specific antibody, exhibits potent antitumor activity against human cancers driven by activated FGFR2 signaling. Cancer Res 70 (19), 7630–9. [DOI] [PubMed] [Google Scholar]
- 24.Schwerk C and Schulze-Osthoff K (2005) Regulation of apoptosis by alternative pre-mRNA splicing. Mol Cell 19 (1), 1–13. [DOI] [PubMed] [Google Scholar]
- 25.Cheng J et al. (1994) Protection from Fas-mediated apoptosis by a soluble form of the Fas molecule. Science 263 (5154), 1759–62. [DOI] [PubMed] [Google Scholar]
- 26.Cascino I et al. (1995) Three functional soluble forms of the human apoptosisinducing Fas molecule are produced by alternative splicing. J Immunol 154 (6), 2706–13. [PubMed] [Google Scholar]
- 27.Liu S and Cheng C (2013) Alternative RNA splicing and cancer. Wiley Interdiscip Rev RNA 4 (5), 547–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eckhart L et al. (2001) Alternative splicing of caspase-8 mRNA during differentiation of human leukocytes. Biochem Biophys Res Commun 289 (4), 777–81. [DOI] [PubMed] [Google Scholar]
- 29.Fan C and Wang X (2017) Mdm2 Splice isoforms regulate the p53/Mdm2/Mdm4 regulatory circuit via RING domain-mediated ubiquitination of p53 and Mdm4. Cell Cycle, 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li CY et al. (2004) Regulation of alternative splicing of Bcl-x by IL-6, GM-CSF and TPA. Cell Res 14 (6), 473–9. [DOI] [PubMed] [Google Scholar]
- 31.Brown RL et al. (2011) CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J Clin Invest 121 (3), 1064–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shapiro IM et al. (2011) An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet 7 (8), e1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orian-Rousseau V et al. (2002) CD44 is required for two consecutive steps in HGF/c-Met signaling. Genes Dev 16 (23), 3074–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morrison H et al. (2001) The NF2 tumor suppressor gene product, merlin, mediates contact inhibition of growth through interactions with CD44. Genes Dev 15 (8), 968–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grivennikov SI et al. (2010) Immunity, inflammation, and cancer. Cell 140 (6), 883–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Antonioli L et al. (2013) Immunity, inflammation and cancer: a leading role for adenosine. Nat Rev Cancer 13 (12), 842–57. [DOI] [PubMed] [Google Scholar]
- 37.Amor S et al. (2010) Alternative splicing and nonsense-mediated decay regulate telomerase reverse transcriptase (TERT) expression during virus-induced lymphomagenesis in vivo. BMC Cancer 10, 571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sotillo E et al. (2015) Convergence of Acquired Mutations and Alternative Splicing of CD19 Enables Resistance to CART-19 Immunotherapy. Cancer Discov 5 (12), 1282–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen L et al. (2017) SRSF1 Prevents DNA Damage and Promotes Tumorigenesis through Regulation of DBF4B Pre-mRNA Splicing. Cell Rep 21 (12), 3406–3413. [DOI] [PubMed] [Google Scholar]
- 40.Couture F et al. (2017) PACE4 Undergoes an Oncogenic Alternative Splicing Switch in Cancer. Cancer Res 77 (24), 6863–6879. [DOI] [PubMed] [Google Scholar]
- 41.Bechara EG et al. (2013) RBM5, 6, and 10 differentially regulate NUMB alternative splicing to control cancer cell proliferation. Mol Cell 52 (5), 720–33. [DOI] [PubMed] [Google Scholar]
- 42.Patounas O et al. (2018) A novel splicing isoform of protein arginine methyltransferase 1 (PRMT1) that lacks the dimerization arm and correlates with cellular malignancy. J Cell Biochem 119 (2), 2110–2123. [DOI] [PubMed] [Google Scholar]
- 43.Quinn JJ and Chang HY (2016) Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 17 (1), 47–62. [DOI] [PubMed] [Google Scholar]
- 44.Derrien T et al. (2012) The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22 (9), 1775–89. 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garzon R et al. (2014) Expression and prognostic impact of lncRNAs in acute myeloid leukemia. Proc Natl Acad Sci U S A 111 (52), 18679–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yue M and Ogawa Y (2018) CRISPR/Cas9-mediated modulation of splicing efficiency reveals short splicing isoform of Xist RNA is sufficient to induce X-chromosome inactivation. Nucleic Acids Res 46 (5), e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Koning AP et al. (2011) Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet 7 (12), e1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hall LL et al. (2014) Stable C0T-1 repeat RNA is abundant and is associated with euchromatic interphase chromosomes. Cell 156 (5), 907–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Darby MM et al. (2016) Widespread splicing of repetitive element loci into coding regions of gene transcripts. Hum Mol Genet 25 (22), 4962–4982. [DOI] [PubMed] [Google Scholar]
- 50.Deveson IW et al. (2018) Universal Alternative Splicing of Noncoding Exons. Cell Syst 6 (2), 245–255 e5. [DOI] [PubMed] [Google Scholar]
- 51.Welter D et al. (2014) The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res 42 (Database issue), D1001–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khurana E et al. (2016) Role of non-coding sequence variants in cancer. Nat Rev Genet 17 (2), 93–108. [DOI] [PubMed] [Google Scholar]
- 53.Albert FW and Kruglyak L (2015) The role of regulatory variation in complex traits and disease. Nat Rev Genet 16 (4), 197–212. [DOI] [PubMed] [Google Scholar]
- 54.Cookson W et al. (2009) Mapping complex disease traits with global gene expression. Nat Rev Genet 10 (3), 184–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Han Y et al. (2016) Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by downregulation of MMP2 and inactivation of ERK/MAPK signaling. Cell Death Dis 7, e2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jayasinghe RG et al. (2018) Systematic Analysis of Splice-Site-Creating Mutations in Cancer. Cell Rep 23 (1), 270–281 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seiler M et al. (2018) Somatic Mutational Landscape of Splicing Factor Genes and Their Functional Consequences across 33 Cancer Types. Cell Rep 23 (1), 282–296 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim E et al. (2015) SRSF2 Mutations Contribute to Myelodysplasia by Mutant-Specific Effects on Exon Recognition. Cancer Cell 27 (5), 617–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Faustino NA and Cooper TA (2003) Pre-mRNA splicing and human disease. Genes Dev 17 (4), 419–37. [DOI] [PubMed] [Google Scholar]
- 60.Orban TI and Olah E (2003) Emerging roles of BRCA1 alternative splicing. Mol Pathol 56 (4), 191–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu HX et al. (2001) A mechanism for exon skipping caused by nonsense or missense mutations in BRCA1 and other genes. Nat Genet 27 (1), 55–8. [DOI] [PubMed] [Google Scholar]
- 62.Morrissy AS et al. (2011) Extensive relationship between antisense transcription and alternative splicing in the human genome. Genome Res 21 (8), 1203–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dubuc AM et al. (2012) Subgroup-specific alternative splicing in medulloblastoma. Acta Neuropathol 123 (4), 485–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shearwin KE et al. (2005) Transcriptional interference--a crash course. Trends Genet 21 (6), 339–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ghigna C et al. (2005) Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell 20 (6), 881–90. [DOI] [PubMed] [Google Scholar]
- 66.Golan-Gerstl R et al. (2011) Splicing factor hnRNP A2/B1 regulates tumor suppressor gene splicing and is an oncogenic driver in glioblastoma. Cancer Res 71 (13), 4464–72. [DOI] [PubMed] [Google Scholar]
- 67.Wang L et al. (2011) SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med 365 (26), 2497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoshida K et al. (2011) Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478 (7367), 64–9. [DOI] [PubMed] [Google Scholar]
- 69.Sebestyen E et al. (2016) Large-scale analysis of genome and transcriptome alterations in multiple tumors unveils novel cancer-relevant splicing networks. Genome Res 26 (6), 732–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Licatalosi DD and Darnell RB (2010) RNA processing and its regulation: global insights into biological networks. Nat Rev Genet 11 (1), 75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bruneau BG et al. (2001) A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell 106 (6), 709–21. [DOI] [PubMed] [Google Scholar]
- 72.Fan C et al. (2009) Functional role of transcriptional factor TBX5 in pre-Mrna splicing and Holt-Oram syndrome via association with SC35. J Biol Chem 284 (38), 25653–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Du C et al. (2014) The adipogenic transcriptional cofactor ZNF638 interacts with splicing regulators and influences alternative splicing. J Lipid Res 55 (9), 1886–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Han H et al. (2017) Multilayered Control of Alternative Splicing Regulatory Networks by Transcription Factors. Mol Cell 65 (3), 539–553 e7. [DOI] [PubMed] [Google Scholar]
- 75.Jones PA et al. (2016) Targeting the cancer epigenome for therapy. Nat Rev Genet 17 (10), 630–41. [DOI] [PubMed] [Google Scholar]
- 76.Luco RF et al. (2011) Epigenetics in alternative pre-mRNA splicing. Cell 144 (1),16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lev Maor G et al. (2015) The alternative role of DNA methylation in splicing regulation. Trends Genet 31 (5), 274–80. [DOI] [PubMed] [Google Scholar]
- 78.Ong CT and Corces VG (2014) CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet 15 (4), 234–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shukla S et al. (2011) CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 479 (7371), 74–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhou HL et al. (2014) Regulation of alternative splicing by local histone modifications: potential roles for RNA-guided mechanisms. Nucleic Acids Res 42 (2), 701–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Saint-Andre V et al. (2011) Histone H3 lysine 9 trimethylation and HP1gamma favor inclusion of alternative exons. Nat Struct Mol Biol 18 (3), 337–44. [DOI] [PubMed] [Google Scholar]
- 82.White ES et al. (2008) New insights into form and function of fibronectin splice variants. J Pathol 216 (1), 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hammond CM et al. (2017) Histone chaperone networks shaping chromatin function. Nat Rev Mol Cell Biol 18 (3), 141–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Koch L (2017) Functional genomics: Screening for lncRNA function. Nat Rev Genet 18 (2), 70. [DOI] [PubMed] [Google Scholar]
- 85.Wahlestedt C (2013) Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov 12 (6), 433–46. [DOI] [PubMed] [Google Scholar]
- 86.Tripathi V et al. (2010) The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39 (6), 925–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gonzalez I et al. (2015) A lncRNA regulates alternative splicing via establishment of a splicing-specific chromatin signature. Nat Struct Mol Biol 22 (5), 370–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rueter SM et al. (1999) Regulation of alternative splicing by RNA editing. Nature 399 (6731), 75–80. [DOI] [PubMed] [Google Scholar]
- 89.Bass BL et al. (1997) A standardized nomenclature for adenosine deaminases that act on RNA. RNA 3 (9), 947–9. [PMC free article] [PubMed] [Google Scholar]
- 90.Goldberg L et al. (2017) Alternative Splicing of STAT3 Is Affected by RNA Editing. DNA Cell Biol 36 (5), 367–376. [DOI] [PubMed] [Google Scholar]
- 91.Wang Y et al. (2017) Systematic characterization of A-to-I RNA editing hotspots in microRNAs across human cancers. Genome Res 27 (7), 1112–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gong J et al. (2017) LNC editing: a database for functional effects of RNA editing in lncRNAs. Nucleic Acids Res 45 (D1), D79–D84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Meyer KD et al. (2012) Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 149 (7), 1635–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hsu PJ et al. (2017) Epitranscriptomic influences on development and disease. Genome Biol 18 (1), 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu N et al. (2015) N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518 (7540), 560–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bartosovic M et al. (2017) N6-methyladenosine demethylase FTO targets premRNAs and regulates alternative splicing and 3’-end processing. Nucleic Acids Res 45 (19), 11356–11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hartmann AM et al. (1999) The interaction and colocalization of Sam68 with the splicing-associated factor YT521-B in nuclear dots is regulated by the Src family kinase p59(fyn). Mol Biol Cell 10 (11), 3909–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Feng Y et al. (2015) Nanopore-based fourth-generation DNA sequencing technology. Genomics Proteomics Bioinformatics 13 (1), 4–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sharon D et al. (2013) A single-molecule long-read survey of the human transcriptome. Nat Biotechnol 31 (11), 1009–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li H and Durbin R (2009) Fast and accurate short read alignment with Burrows Wheeler transform. Bioinformatics 25 (14), 1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Langmead B et al. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10 (3), R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li R et al. (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25 (15), 1966–7. [DOI] [PubMed] [Google Scholar]
- 103.Trapnell C et al. (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25 (9), 1105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Au KF et al. (2010) Detection of splice junctions from paired-end RNA-seq data by SpliceMap. Nucleic Acids Res 38 (14), 4570–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Trapnell C et al. (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7 (3), 562–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mezlini AM et al. (2013) iReckon: simultaneous isoform discovery and abundance estimation from RNA-seq data. Genome Res 23 (3), 519–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang J and Wei Z (2016) An empirical Bayes change-point model for identifying 3’ and 5’ alternative splicing by next-generation RNA sequencing. Bioinformatics 32 (12), 1823–31. [DOI] [PubMed] [Google Scholar]
- 108.Katz Y et al. (2010) Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods 7 (12), 1009–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shen S et al. (2014) rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci U S A 111 (51), E5593–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Qin Z et al. (2018) SEASTAR: systematic evaluation of alternative transcription start sites in RNA. Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Trincado JL et al. (2018) SUPPA2: fast, accurate, and uncertainty-aware differential splicing analysis across multiple conditions. Genome Biol 19 (1), 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mudvari P et al. (2015) SNPlice: variants that modulate Intron retention from RNA-sequencing data. Bioinformatics 31 (8), 1191–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jung H et al. (2015) Intron retention is a widespread mechanism of tumor-suppressor inactivation. Nat Genet 47 (11), 1242–8. [DOI] [PubMed] [Google Scholar]
- 114.Monlong J et al. (2014) Identification of genetic variants associated with alternative splicing using sQTLseekeR. Nat Commun 5, 4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhao K et al. (2013) GLiMMPS: robust statistical model for regulatory variation of alternative splicing using RNA-seq data. Genome Biol 14 (7), R74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li Y et al. (2017) Revealing the Determinants of Widespread Alternative Splicing Perturbation in Cancer. Cell Rep 21 (3), 798–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hoadley KA et al. (2014) Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell 158 (4), 929–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li Y et al. (2017) Prognostic alternative mRNA splicing signature in non-small cell lung cancer. Cancer Lett. [DOI] [PubMed] [Google Scholar]
- 119.Shen S et al. (2016) SURVIV for survival analysis of mRNA isoform variation. Nat Commun 7, 11548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Baldwin RM et al. (2012) Alternatively spliced protein arginine methyltransferase 1 isoform PRMT1v2 promotes the survival and invasiveness of breast cancer cells. Cell Cycle 11 (24), 4597–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Barabasi AL and Oltvai ZN (2004) Network biology: understanding the cell’s functional organization. Nat Rev Genet 5 (2), 101–13. [DOI] [PubMed] [Google Scholar]
- 122.Sonachalam M et al. (2012) Systems biology approach to identify gene network signatures for colorectal cancer. Front Genet 3, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Barabasi AL et al. (2011) Network medicine: a network-based approach to human disease. Nat Rev Genet 12 (1), 56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.He C et al. (2009) A global view of cancer-specific transcript variants by subtractive transcriptome-wide analysis. PLoS One 4 (3), e4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Denoeud F et al. (2007) Prominent use of distal 5’ transcription start sites and discovery of a large number of additional exons in ENCODE regions. Genome Res 17 (6), 746–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Okoniewski MJ et al. (2007) An annotation infrastructure for the analysis and interpretation of Affymetrix exon array data. Genome Biol 8 (5), R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Clark TA et al. (2007) Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biol 8 (4), R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Clark TA et al. (2002) Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science 296 (5569), 907–10. [DOI] [PubMed] [Google Scholar]
- 129.Johnson JM et al. (2003) Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302 (5653), 2141–4. [DOI] [PubMed] [Google Scholar]
- 130.Bryant DW Jr. et al. (2012) Detection and quantification of alternative splicing variants using RNA-seq. Methods Mol Biol 883, 97–110. [DOI] [PubMed] [Google Scholar]
- 131.Conesa A et al. (2016) A survey of best practices for RNA-seq data analysis. Genome Biol 17, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pulyakhina I et al. (2015) SplicePie: a novel analytical approach for the detection of alternative, non-sequential and recursive splicing. Nucleic Acids Res 43 (12), e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Anders S et al. (2012) Detecting differential usage of exons from RNA-seq data. Genome Res 22 (10), 2008–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Park JW et al. (2013) Identifying differential alternative splicing events from RNA sequencing data using RNASeq-MATS. Methods Mol Biol 1038, 171–9. [DOI] [PubMed] [Google Scholar]
- 135.Ryan MC et al. (2012) SpliceSeq: a resource for analysis and visualization of RNA-Seq data on alternative splicing and its functional impacts. Bioinformatics 28 (18), 2385–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Griffith M et al. (2010) Alternative expression analysis by RNA sequencing. Nat Methods 7 (10), 843–7. [DOI] [PubMed] [Google Scholar]
- 137.Brooks AN et al. (2011) Conservation of an RNA regulatory map between Drosophila and mammals. Genome Res 21 (2), 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hu Y et al. (2013) DiffSplice: the genome-wide detection of differential splicing events with RNA-seq. Nucleic Acids Res 41 (2), e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Drewe P et al. (2013) Accurate detection of differential RNA processing. Nucleic Acids Res 41 (10), 5189–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kahles A et al. (2016) SplAdder: identification, quantification and testing of alternative splicing events from RNA-Seq data. Bioinformatics 32 (12), 1840–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Christinat Y et al. (2016) jSplice: a high-performance method for accurate prediction of alternative splicing events and its application to large-scale renal cancer transcriptome data. Bioinformatics 32 (14), 2111–9. [DOI] [PubMed] [Google Scholar]
- 142.Topa H and Honkela A (2016) Analysis of differential splicing suggests different modes of short-term splicing regulation. Bioinformatics 32 (12), i147–i155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hartley SW and Mullikin JC (2016) Detection and visualization of differential splicing in RNA-Seq data with JunctionSeq. Nucleic Acids Res 44 (15), e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kimes PK et al. (2014) SigFuge: single gene clustering of RNA-seq reveals differential isoform usage among cancer samples. Nucleic Acids Res 42 (14), e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Xiong HY et al. (2015) RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science 347 (6218), 1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]