Abstract
Background
Galectin-3 as a β-galactoside-binding protein, has been found to be involved in tumor cell growth, anti-apoptosis, adhesion, angiogenesis, invasion, and distant metastases, indicating that it may play a pivotal role in cancer development and progression. However, their results remain debatable and inconclusive. Hence, this meta-analysis was performed to clarify the precise predictive value of galectin-3 in various cancers.
Methods
PubMed, Web of Science, Embase, Cochrane Library, CNKI and Wanfang databases were searched comprehensively for eligible studies up to July 15, 2018. Pooled hazard ratios (HRs) with 95% confidence intervals (CIs) of OS or DFS/PFS/RFS were calculated to demonstrate their associations.
Results
A total of 36 relevant studies were ultimately enrolled in this meta-analysis. Our results shed light on the significant association of elevated galectin-3 expression with reduced OS or DFS/RFS/PFS in overall cancer patients (pooled HR = 1.79, 95% CI 1.42–2.27, I2= 67.3%, p < 0.01; pooled HR = 1.57, 95% CI 1.04–2.37, I2= 67.1%, p = 0.001). In tumor type subgroup analysis, we found high expression of galectin-3 was correlated with shorter OS or DFS/RFS/PFS in colorectal cancer (pooled HR = 3.05, 95% CI 2.13–4.35, I2= 0.0%, p = 0.734; pooled HR = 2.49, 95% CI 1.82–3.41, I2 = 0.0%, p = 0.738; respectively) and meanwhile it merely associated with reduced OS in ovarian cancer or non-small cell lung cancer (pooled HR = 2.24, 95% CI 1.38–3.64, I2= 0.0%, p = 0.910; pooled HR = 2.07, 95% CI 1.48–2.88, I2= 0.0%, p = 0.563; separately).
Conclusions
Taken together, our results suggested that galectin-3 played an oncogenic role in colorectal cancer, ovarian cancer and non-small cell lung cancer, indicating it could be a promising biomarker and a novel therapeutic target for them. Further studies were warranted to validate our findings.
Keywords: Prognostic role, Galectin-3, Cancer, Meta-analysis
Background
Galectins are a large family of widely distributed carbohydrate-binding proteins, characterized by their binding affinity for β-galactosides and conserved sequences in the binding site [1]. Meanwhile, galectins are often exhibited a high level of expression in cancer cells or cancer-associated stromal cells with the aggressiveness of tumors and the acquisition of the metastatic phenotype [2]. Because of their significant involvement in various biological functions and pathology, the role of galectins seems to be of importance [3]. Therein, galectin-3 also knew as LGALS3, L31, GAL3, MAC2, CBP35, GALBP and GALIG, belongs to the family of galectins [4]. In both extracellular and intracellular manners, galectin-3 exhibits its pleiotropic biological and molecular functions. Extracellularly, it has the ability to adjust microenvironment by means of interacting with the cell surface and extracellular matrix glycoproteins or glycolipids. Intracellularly, it was capable of modulating signaling pathways via interacting with cytoplasmic and nuclear proteins [5]. Up to now, a growing number of researches have suggested the involvement of galectin-3 in tumor progression and disease outcome [6–8].
Galectin-3 has been found to be differently expressed in various normal and malignant tissues. Previous studies indicated that down-regulation of galectin-3 was associated with loss of the transformed phenotypes in thyroid papillary carcinoma cells, but up-regulation of it could induce the transformed phenotype in normal thyroid follicular cell lines [9]. Accumulating data have demonstrated that different galectin-3 expression in tumor tissues was associated with unfavorable survival in cancer patients [10–14]. These studies concentrated on colorectal carcinoma, cervical carcinoma, breast cancers, gastric carcinoma, laryngeal squamous-cell carcinoma and so on. However, their results remained inconsistent. The discrepancies among these studies highlighted the importance of evaluating the prognostic significance of galectin-3 in multiple human malignant neoplasms. Hence, this meta-analysis was conducted to clarify the relationship between galectin-3 expression and the prognosis of patients with carcinoma. Last but not least, it is the first time for us to shed light on their relationship and galectin-3 is anticipated to be a prognostic marker in clinical applications.
Materials and methods
Literature search strategy
We conducted a comprehensive search of online databases PubMed, EMBASE and Web of Science, Cochrane Library, Chinese National Knowledge Infrastructure (CNKI) and Wanfang database (Chinese) to identify relevant literature published before July 15, 2018. The search strategy was mainly consisted of the following keywords in combination with Medical Subject Headings (MeSH) terms and text words: (“cancer” or “carcinoma” or “neoplasm” or “tumor” or “tumour”) and (“galectin-3” or “GAL3” or “LGALS3” or “L31” or “MAC2” or “CBP35” or “GALBP” or “GALIG”). In addition, potentially eligible articles were identified via meticulously searching from the reference lists of relevant reviews and original literature.
Inclusion and exclusion criteria
The eligible studies needed to meet the following four inclusion criteria: (1) English or Chinese publications; (2) patients with carcinoma; (3) a relationship of galectin-3 expression with cancer prognosis; (4) sufficient data could be extracted. Additionally, the exclusion criteria included the following points: (1) non-English or non-Chinese research; (2) duplicates of the previous publication; (3) reviews or letters or case reports or comments or editorials; (4) unrelated to galectin-3 or human patients; (5) absence of key information.
Quality assessment
The following information should be extracted from included articles before being evaluated: (1) the study population and country; (2) the study design; (3) assay method to determine galectin-3 expression; (4) the prognosis or survival assessment; (5) the detected tumor and pathology information; (6) the cutoff point of galectin-3; and (7) the follow-up duration. In addition, Newcastle–Ottawa Scale (NOS), as one of the most useful scale to evaluate the quality of non-randomized studies (http://www.ohri.ca/programs/clinical_epidemiology/oxford.htm), was independently evaluated by two blind reviewers [15]. The criteria of quality assessment were as follows: (1) representativeness of the exposed cohort; (2) selection of the non-exposed cohort; (3) ascertainment of exposure; (4) outcome of interest not present at start of study; (5) control for important factor or additional factor; (6) assessment of outcome; (7) follow-up long enough for outcomes to occur; (8) adequacy of follow up of cohorts. Total quality score of NOS was ranged from 0 to 9, which was regarded as high quality with the final score > 6. Details were presented in Table 1.
Table 1.
Newcastle–Ottawa quality assessments scale
| Studies | Year | Quality indicators from Newcastle–Ottawa Scale | Scores | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |||
| Chou [20] | 2018 | ★ | – | ★ | ★ | ★ | ★ | ★ | ★ | 7 |
| Lu [10] | 2017 | ★ | ★ | ★ | ★ | ★★ | ★ | – | ★ | 8 |
| Huang [4] | 2017 | ★ | ★ | ★ | ★ | ★ | ★ | – | ★ | 7 |
| Li [11] | 2017 | ★ | ★ | ★ | ★ | ★★ | ★ | – | – | 7 |
| Shimura [41] | 2017 | ★ | ★ | ★ | ★ | ★★ | ★ | – | ★ | 8 |
| Wang [49] | 2017 | ★ | ★ | ★ | ★ | ★ | ★ | – | ★ | 7 |
| Liu [37] | 2017 | ★ | ★ | ★ | ★ | ★★ | ★ | – | – | 7 |
| Gopalan [21] | 2016 | ★ | ★ | ★ | ★ | ★★ | ★ | – | ★ | 8 |
| Ilmer [12] | 2016 | ★ | ★ | – | ★ | ★★ | ★ | – | ★ | 7 |
| Yang [48] | 2016 | ★ | ★ | ★ | ★ | ★★ | ★ | – | ★ | 8 |
| Tas [36] | 2016 | ★ | – | ★ | ★★ | ★ | – | ★ | 7 | |
| Cheng [40] | 2015 | ★ | ★ | ★ | ★ | ★★ | ★ | ★ | – | 7 |
| Lu [47] | 2015 | ★ | ★ | ★ | ★ | ★★ | ★ | ★ | – | 8 |
| Jiang [22] | 2014 | ★ | ★ | ★ | ★ | ★ | ★ | ★ | ★ | 8 |
| Gomes [23] | 2014 | ★ | – | ★ | ★ | ★★ | ★ | ★ | ★ | 8 |
| Mu [44] | 2013 | ★ | ★ | ★ | ★ | ★ | ★ | – | ★ | 7 |
| Wu [45] | 2013 | ★ | ★ | ★ | ★ | ★★ | ★ | – | – | 7 |
| Liu [46] | 2013 | ★ | – | ★ | ★ | ★ | ★ | – | ★ | 6 |
| Yamaki [24] | 2012 | ★ | ★ | ★ | ★ | ★★ | ★ | ★ | – | 8 |
| Yang [25] | 2012 | ★ | – | ★ | ★ | ★ | – | – | ★ | 5 |
| Kim [26] | 2012 | ★ | ★ | ★ | ★ | ★★ | ★ | – | – | 7 |
| Kosacka [38] | 2011 | ★ | ★ | – | – | ★★ | ★ | ★ | – | 6 |
| Povegliano [27] | 2010 | ★ | ★ | ★ | ★ | ★ | ★ | – | ★ | 7 |
| Canesin [42] | 2010 | ★ | – | ★ | – | ★★ | ★ | – | ★ | 6 |
| Vereecken [43] | 2009 | ★ | ★ | ★ | – | ★ | ★ | – | ★ | 6 |
| Miranda [28] | 2009 | ★ | – | – | ★ | ★★ | ★ | ★ | – | 6 |
| Szoke [29] | 2007 | ★ | – | – | ★ | ★ | ★ | – | ★ | 5 |
| Kang [30] | 2007 | ★ | ★ | ★ | ★ | ★ | ★ | ★ | – | 7 |
| Moisa [39] | 2007 | ★ | ★ | – | ★ | ★ | – | ★ | ★ | 6 |
| Okada [13] | 2006 | ★ | ★ | ★ | ★ | ★ | ★ | – | ★ | 7 |
| Plzak [31] | 2004 | ★ | ★ | ★ | – | ★ | – | ★ | – | 5 |
| Piantelli [14] | 2002 | ★ | ★ | ★ | ★ | ★★ | ★ | ★ | – | 8 |
| Brule [32] | 2000 | ★ | – | ★ | ★ | ★ | ★ | ★ | – | 6 |
| Honjo [33] | 2001 | ★ | ★ | ★ | – | ★ | ★ | ★ | – | 6 |
| Nakamura [34] | 1999 | ★ | ★ | ★ | ★ | ★★ | ★ | – | ★ | 8 |
| Sanjuan [35] | 1997 | ★ | – | ★ | – | ★ | ★ | – | – | 4 |
1. Representativeness of the exposed cohort; 2. Selection of the non-exposed cohort; 3. Ascertainment of exposure; 4. Outcome of interest not present at start of study; 5. Control for important factor or additional factor; 6. Assessment of outcome; 7. Follow-up long enough for outcomes to occur; 8. Adequacy of follow up of cohorts
Data extraction
All available data from the identified studies were extracted respectively by two reviewers (Y.W and SW.L). If any disagreement achieved, a third reviewer (Y.T) would join in and reached a consensus. Extracted data were recorded in a standardized form including following items: first author’s surname, publication year, patients’ median or mean age, nationality, dominant ethnicity, number of patients, investigating method, cutoff value, follow-up time, and hazard ratios (HRs) for prognostic outcomes (overall survival [OS] and disease/recurrence/progression-free survival [DFS/RFS/PFS]) along with their 95% CI and p-values. Data were extracted from Kaplan–Meier curves to extrapolate HRs with 95% CIs by using previously described methods, when it could not be directly obtained from each article [16, 17]. Details of the aforementioned data were displayed in Tables 2 and 3.
Table 2.
Main characteristics of studies included in this meta-analysis
| First author | Publication year | Case nationality | Dominant ethnicity | Median or mean age | Study design | Malignant disease | Main type of pathology | Detected sample | Assay method | Survival analysis | Source of HR | Maximum months of follow-up |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chou [20] | 2018 | China | Asian | 50 | R | Glioblastoma multiforme | Glioma | Tissue | ihc | OS/PFS | Reported | 207 |
| Lu [10] | 2017 | China | Asian | 60 | R | Colorectal cancer | AdenoCa | Tissue | ihc | OS | SC | 40 |
| Huang [4] | 2017 | China | Asian | 60 | R | Colorectal cancer | AdenoCa | Tissue | ihc | OS | Reported | 50 |
| Li [11] | 2017 | China | Asian | 40 | R | Cervical carcinoma | SqCa | Tissue | ihc | OS | Reported | 78 |
| Shimuraa [41] | 2017 | Japan | Asian | 55 | R | Biliary cancer | AdenoCa | Serum | ELISA | OS | Reported | 69.9 |
| Shimurab [41] | 2017 | Japan | Asian | 55 | R | Pancreatic cancer | AdenoCa | Serum | ELISA | OS | Reported | 66 |
| Wang [49] | 2017 | China | Asian | NM | R | Ovarian cancer | SqCa | Tissue | ihc | OS | Reported | 72 |
| Liu [37] | 2017 | China | Asian | 65.1 | R | Colorectal cancer | AdenoCa | Tissue | ihc | DFS | Reported | 60 |
| Gopalan [21] | 2016 | Australia | Caucasian | 60 | R | Colorectal cancer | AdenoCa | Tissue | ihc | OS | SC | 110 |
| Ilmer [12] | 2016 | American | Caucasian | 47 | R | Breast cancer | AdenoCa | Tissue | ihc | OS | SC | 232 |
| Yang [48] | 2016 | China | Asian | 66.8 | R | Colorectal cancer | AdenoCa | tissue | ihc | DFS | Reported | 60 |
| Tas [36] | 2016 | Turkey | Caucasian | 59.5 | R | Gastric cancer | AdenoCa | Serum | ELISA | OS | SC | 97 |
| Cheng [40] | 2015 | China | Asian | 55.2 | R | Gastric cancer | AdenoCa | Serum | ELISA | OS | SC | 60 |
| Lu [47] | 2015 | China | Asian | 51 | R | Ovarian cancer | SqCa | Tissue | ihc | OS | Reported | 77 |
| Jiang [22] | 2014 | China | Asian | 50 | R | Hepatocellular carcinoma | AdenoCa | Tissue | ihc | OS | Reported | 87 |
| Gomes [23] | 2014 | Brazil | Caucasian | 50 | R | Gastric cancer | AdenoCa | Tissue | ihc | OS | SC | 55 |
| Mu [44] | 2013 | China | Asian | 66 | R | Gastric cancer | AdenoCa | Tissue | ihc | OS | Reported | NM |
| Wu [45] | 2013 | China | Asian | 59.6 | R | Non-small cell lung cancer | SqCa | Tissue | ihc | OS | Reported | 90 |
| Liu [46] | 2013 | China | Asian | 57.1 | R | Non-small cell lung cancer | SqCa | Tissue | ihc | OS | SC | 80 |
| Yamaki [24] | 2012 | Japan | Asian | 53 | R | Breast cancer | AdenoCa | Tissue | ihc | OS/PFS | SC | 13 |
| Yang [25] | 2012 | China | Asian | 45 | R | Gallbladder carcinoma | AdenoCa | Tissue | ihc | OS | SC | 18 |
| Kim [26] | 2012 | Korea | Asian | 60 | R | Gastric cancer | AdenoCa | Tissue | ihc | OS | Reported | 96 |
| Kosacka [38] | 2011 | Poland | Caucasian | 59.3 | R | Non-small cell lung cancer | SqCa | Tissue | ihc | OS | SC | 24 |
| Povegliano [27] | 2010 | Brazil | Caucasian | 50 | R | Colorectal cancer | AdenoCa | Tissue | ihc | OS | SC | 83 |
| Canesin [42] | 2010 | American | Caucasian | NM | R | Bladder cancer | SqCa | Tissue | ihc | OS | SC | 173 |
| Vereecken [43] | 2009 | American | Caucasian | 60 | R | Melanoma | NM | Serum | ELISA | OS | Reported | 60 |
| Miranda [28] | 2009 | Brazil | Caucasian | 59 | R | Laryngeal carcinoma | SqCa | Tissue | ihc | DFS | SC | 166 |
| Szoke [29] | 2007 | German | Caucasian | 58.8 | R | Non-small cell lung cancer | SqCa | Tissue | ihc | OS | SC | 127 |
| Kang [30] | 2007 | Korea | Asian | 63 | R | Esophageal cancer | SqCa | Tissue | ihc | OS | SC | 108 |
| Moisa [39] | 2007 | Germany | Caucasian | 56.8 | R | Breast cancer | AdenoCa | Tissue | ihc | OS/DFS | Reported | 185 |
| Okada [13] | 2006 | Japan | Asian | 63.9 | R | Gastric cancer | AdenoCa | tissue | ihc | OS | Reported | 72 |
| Plzak [31] | 2004 | Prague | Caucasian | 60 | R | Head and neck carcinoma | SqCa | Tissue | ihc | OS | SC | 60 |
| Piantelli [14] | 2002 | Rome | Caucasian | 60 | R | Laryngeal carcinoma | SqCa | Tissue | ihc | OS/RFS | SC | 90 |
| Brule [32] | 2000 | Belgium | Caucasian | 65 | R | Prostate carcinomas | AdenoCa | Tissue | ihc | PFS | SC | 86 |
| Honjo [33] | 2001 | Japan | Asian | 60 | R | Tongue carcinoma | SqCa | Tissue | ihc | OS/DFS | SC | 118 |
| Nakamura [34] | 1999 | Japan | Asian | NM | R | Colorectal cancer | AdenoCa | Tissue | ihc | OS/DFS | SC | 103 |
| Sanjuan [35] | 1997 | Spain | Caucasian | NM | R | Colorectal cancer | AdenoCa | Tissue | ihc | OS/RFS | SC | 96 |
R retrospective, AdenoCa adenocarcinoma, SqCa squamous carcinoma, IHC immunohistochemistry, OS overall survival, DFS disease-free survival, PFS progression-free survival, RFS recurrence-free survival, SC survival curve
a, bData extracted from one study due to different malignant disease (biliary cancer and pancreatic cancer)
Table 3.
HRs and 95% CIs of patient survival or cancer progression relating to galectin-3 expression in eligible studies
| First author | Year | Malignant disease | Main type of pathology | Survival analysis | Cut off point | Case number | OS | DFS/RFS/PFS | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| High expression | Low expression | HR (95% CI) | p-value | HR (95% CI) | p-value | ||||||
| Chou [20] | 2018 | Glioblastoma multiforme | Glioma | OS/PFS | IRS score ≥ 2 (range 0–2) | NM | NM | 1.34 (0.59–3.03) | 0.478 | 0.181 (0.025–1.299) | 0.089 |
| Lu [10] | 2017 | Colorectal cancer | AdenoCa | OS | IRS score ≥ 2 (range 0–3) | 43 | 14 | 1.88 (0.88–5.23) | 0.0086 | NM | NM |
| Huang [4] | 2017 | Colorectal cancer | AdenoCa | OS | IRS score ≥ 2 (range 0–4) | 51 | 66 | 2.39 (1.12–4.75) | 0.015 | NM | NM |
| Li [11] | 2017 | Cervical carcinoma | SqCa | OS | IRS score ≥ 7 (range 0–12) | 45 | 39 | 14.00 (1.75–112.31) | 0.013 | NM | NM |
| Shimuraa [41] | 2017 | Biliary cancer | AdenoCa | OS | ≥ 10.3 ng/ml | 22 | 2 | 6.19 (1.18–32.36) | 0.031 | NM | NM |
| Shimurab [41] | 2017 | Pancreatic cancer | AdenoCa | OS | ≥ 10.3 ng/ml | 18 | 3 | 4.59 (1.17–17.68) | 0.028 | NM | NM |
| Wang [49] | 2017 | Ovarian cancer | SqCa | OS | 30% of tumor cells stained | 75 | 23 | 2.19 (1.17–4.02) | 0.014 | NM | NM |
| Liu [37] | 2017 | Colorectal cancer | AdenoCa | DFS | 50% of tumor cells stained | 38 | 23 | NM | NM | 2.10 (1.05–4.17) | < 0.05 |
| Gopalan [21] | 2016 | Colorectal cancer | AdenoCa | OS | IRS score ≥ 3 (range 0–4) | 69 | 4 | 4.00 (0.90–20.00) | 0.052 | NM | NM |
| Ilmer [12] | 2016 | Breast cancer | AdenoCa | OS | Hscore ≥ 150 | 23 | 64 | 0.69 (0.17–2.86) | 0.019 | NM | NM |
| Yang [48] | 2016 | Colorectal cancer | AdenoCa | DFS | IRS score ≥ 4 | 40 | 24 | NM | NM | 2.09 (1.09–3.79) | < 0.05 |
| Tas [36] | 2016 | Gastric cancer | AdenoCa | OS | NM | 29 | 29 | 0.79 (0.37–1.67) | 0.54 | NM | NM |
| Cheng [40] | 2015 | Gastric cancer | AdenoCa | OS | ≥ 16.4 ng/ml | 43 | 43 | 1.63 (0.72–3.66) | 0.099 | NM | NM |
| Lu [47] | 2015 | Ovarian cancer | SqCa | OS | IRS score ≥ 5 | 23 | 54 | 2.32 (1.05–5.10) | 0.036 | NM | NM |
| Jiang [22] | 2014 | Hepatocellular carcinoma | AdenoCa | OS | IRS score ≥ 4 | 135 | 30 | 7.51 (3.00–18.78) | < 0.01 | NM | NM |
| Gomes [23] | 2014 | Gastric cancer | AdenoCa | OS | 50% of tumor cells stained | 31 | 26 | 0.73 (0.27–1.98) | 0.798 | NM | NM |
| Mu [44] | 2013 | Gastric cancer | AdenoCa | OS | ≥ 10.0 ng/ml | NM | NM | 1.58 (1.11–2.86) | 0.013 | NM | NM |
| Wu [45] | 2013 | Non-small cell lung cancer | SqCa | OS | IRS score ≥ 2 (range 0–2) | 102 | 58 | 2.05 (1.15–3.67) | 0.015 | NM | NM |
| Liu [46] | 2013 | Non-small cell lung cancer | SqCa | OS | 10% of tumor cells stained | 52 | 10 | 3.09 (1.23–5.26) | 0.045 | NM | NM |
| Yamaki [24] | 2012 | Breast cancer | AdenoCa | OS/PFS | 30% of tumor cells stained | 67 | 49 | 0.90 (0.15–5.35) | 0.041 | 0.46 (0.18–1.22) | 0.018 |
| Yang [25] | 2012 | Gallbladder carcinoma | AdenoCa | OS | 25% of tumor cells stained | 67 | 41 | 1.68 (1.05–2.69) | 0.028 | NM | NM |
| Kim [26] | 2012 | Gastric cancer | AdenoCa | OS | 10% of tumor cells stained | 397 | 74 | 0.80 (0.51–1.26) | 0.331 | NM | NM |
| Kosacka [38] | 2011 | Non-small cell lung cancer | SqCa | OS | 10% of tumor cells stained | 18 | 29 | 1.24 (0.38–4.05) | 0.84 | NM | NM |
| Povegliano [27] | 2010 | Colorectal cancer | AdenoCa | OS | 50% of tumor cells stained | 32 | 43 | 1.28 (0.01–138.79) | 0.056 | NM | NM |
| Canesin [42] | 2010 | Bladder cancer | SqCa | OS | 20% of tumor cells stained | 194 | 194 | 2.34 (1.81–3.02) | < 0.001 | NM | NM |
| Vereecken [43] | 2009 | Melanoma | NM | OS | ≥ 10.0 ng/ml | NM | NM | 4.64 (2.17–9.91) | 0.0001 | NM | NM |
| Miranda [28] | 2009 | Laryngeal carcinoma | SqCa | DFS | NM | 47 | 18 | NM | NM | 1.06 (0.44–2.60) | 0.5284 |
| Szoke [29] | 2007 | Non-small cell lung cancer | SqCa | OS | NM | 51 | 41 | 1.86 (1.09–3.15) | 0.003 | NM | NM |
| Kang [30] | 2007 | Esophageal cancer | SqCa | OS | IRS score ≥ 2 (range 0–4) | 18 | 44 | 0.98 (0.56–1.70) | 0.227 | NM | NM |
| Moisa [39] | 2007 | Breast cancer | AdenoCa | OS/DFS | IRS score ≥ 2 (range 0–3) | 52 | 146 | 1.41 (1.16–3.89) | 0.013 | 1.65 (0.91–2.87) | 0.09 |
| Okada [13] | 2006 | Gastric cancer | AdenoCa | OS | 60% of tumor cells stained | 60 | 55 | 0.26 (0.11–0.64) | 0.0031 | NM | NM |
| Plzak [31] | 2004 | Head and neck carcinoma | SqCa | OS | 50% of tumor cells stained | 23 | 30 | 0.30 (0.06–1.64) | 0.0024 | NM | NM |
| Piantelli [14] | 2002 | Laryngeal carcinoma | SqCa | OS/RFS | 5% of tumor cells stained | 42 | 31 | 0.54 (0.13–2.23) | 0.0001 | 0.49 (0.20–1.21) | 0.0013 |
| Brule [32] | 2000 | Prostate carcinomas | AdenoCa | PFS | IRS score ≥ 1.5 (range 0–2) | 25 | 102 | NM | NM | 3.45 (1.49–7.95) | 0.044 |
| Honjo [33] | 2001 | Tongue carcinoma | SqCa | OS/DFS | 85% of tumor cells stained | 31 | 23 | 3.51 (1.32–9.37) | 0.012 | 2.30 (0.83–6.33) | 0.021 |
| Nakamura [34] | 1999 | Colorectal cancer | AdenoCa | OS/DFS | 66.7% of tumor cells stained | 36 | 71 | 3.63 (1.88–7.01) | 0.014 | 2.65 (1.54–4.58) | 0.0224 |
| Sanjuan [35] | 1997 | Colorectal cancer | AdenoCa | OS/RFS | 25% of tumor cells stained | 83 | 68 | 4.15 (2.01–8.55) | 0.0086 | 3.32 (1.67–6.60) | 0.01 |
AdenoCa adenocarcinoma, SqCa squamous carcinoma, OS overall survival, DFS disease-free survival, PFS progression-free survival, RFS recurrence-free survival, NM not mentioned, IRS immunoreactivity score, Hscore the intensity and respective percentage cells that stain at each intensity were multiplied to reach a Hscore that ranged from 0 to 300, OS overall survival, HR hazard ratio, CI confidence interval
a, bData extracted from one study due to different malignant disease (biliary cancer and pancreatic cancer)
Statistical analysis
Based on available data, the relationship between galectin-3 and multiple human malignant neoplasms was conducted by OS or DFS/RFS/PFS and the pooled hazard ratios (HRs) with 95% confidence intervals (CIs) were utilized to evaluate their efficacy. The effect of heterogeneity was quantified via I2= 100% × (Q − df)/Q. If significant heterogeneity (p < 0.1 or I2> 50%) existed, the random-effects model (DerSimonian–Laird method) would be applied; otherwise, a fixed-effects model (Mantel–Haenszel method) would be utilized [18]. Moreover, in the case of significant heterogeneity, subgroup analysis was carried out by the type of malignant disease and dominant ethnicity to further minimize the influence. Sensitivity analysis was conducted to access the stability of results by deleting one single study each time to reflect the impact of the individual to overall. Publication bias was evaluated by the Begg’s funnel plot and Egger linear regression test with a funnel plot [19]. If p < 0.05, it indicated the existence of publication bias. All p-values were calculated using a two-sided test and p < 0.05 was considered statistically significant. Besides, all statistical data were conducted by Stata software (version 12.0; StataCorp LP, College Station, TX) and Microsoft Excel (V.2007, Microsoft Corporation, Redmond, WA, USA).
Results
Summary of enrolled studies
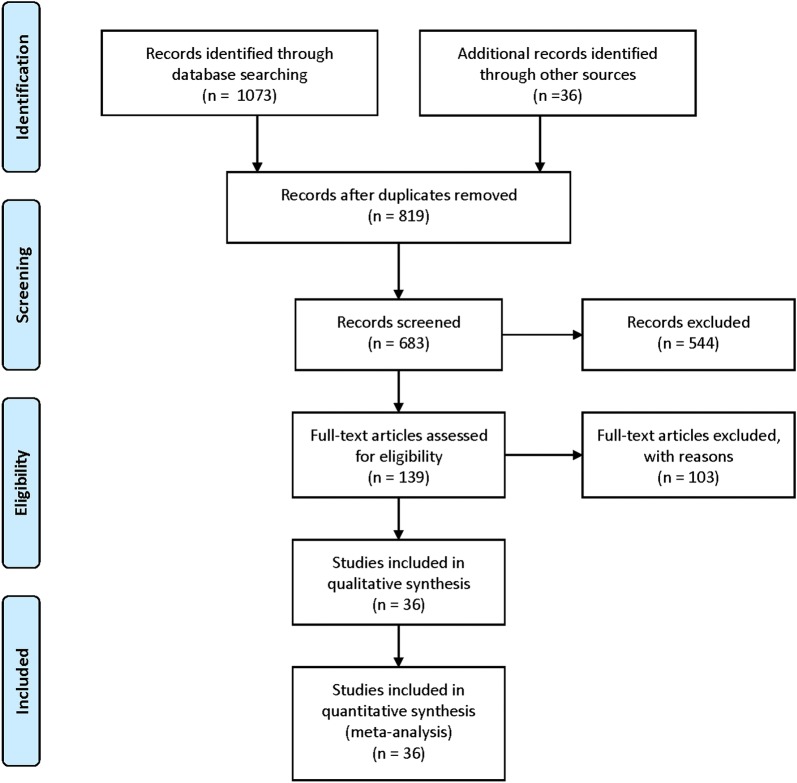
The literature search yielded 1109 citations through online databases by means of previous search strategy. Amongst them, 970 records were excluded because of reviews, letters, case-reports, duplicates and so on, after screening the tittles and abstracts. The full texts of the remaining 139 articles were evaluated by the reviewers. Among them, 103 potentially suitable studies were excluded because of lacking sufficient survival data (HRs and 95% CIs), not related to OS or DFS/RFS/PFS, absence of key information. Ultimately, 36 studies were considered to be eligible for this meta-analysis (Fig. 1) [4, 10–14, 20–49].
Fig. 1.
Flow diagram of the literature selection process
Detailed quality assessments of each eligible article were presented in Table 1 and the main characteristics of these 36 enrolled studies were summarized in Tables 2 and 3. Amongst them, 33 studies focused on OS and 11 articles investigated DFS or PFS or RFS. 15 of these records focused on Caucasian populations, which mainly came from European countries, and 22 focused on Asian populations. As for cancer type, malignant neoplasms assessed in this article included colorectal carcinoma, gastric carcinoma, breast cancer, laryngeal squamous cell carcinoma (LSCC), esophageal squamous cell carcinoma (ESCC), glioblastoma multiforme, cervical carcinoma, hepatocellular carcinoma, gallbladder carcinoma, non-small cell lung cancer, head and neck carcinoma, prostate carcinomas, tongue carcinoma, biliary cancer, pancreatic cancer, ovarian cancer, bladder cancer and melanoma. Besides, all these aforementioned studies were retrospective.
OS associated with galectin-3 expression
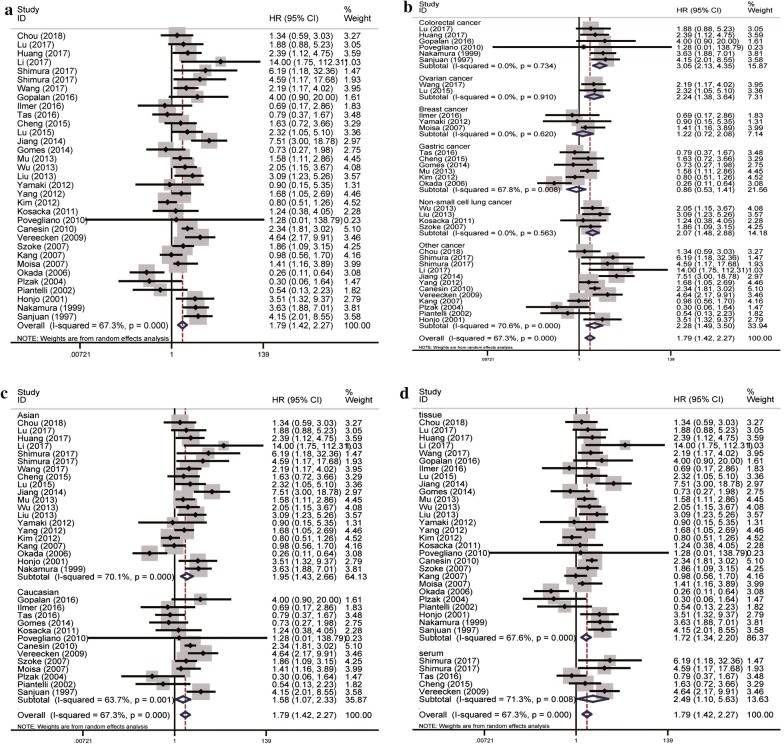
A total of 33 eligible studies were enrolled to evaluate the role of elevated galectin-3 expression in multiple human malignant neoplasms by OS, within a random-effects model. Our results did indicate that high galectin-3 expression was significantly associated with unfavorable OS in overall cancer patients (pooled HR = 1.79, 95% CI 1.42–2.27, I2= 67.3%, p < 0.01; Fig. 2a). In the subgroup analysis of specific cancer type, we found high expression of galectin-3 correlated with reduced OS in colorectal cancer, ovarian cancer and non-small cell lung cancer (pooled HR = 3.05, 95% CI 2.13–4.35, I2= 0.0%, p = 0.734; pooled HR = 2.24, 95% CI 1.38–3.64, I2= 0.0%, p = 0.910; pooled HR = 2.07, 95% CI 1.48–2.88, I2= 0.0%, p = 0.563; respectively) (Fig. 2b). Furthermore, in terms of dominant ethnicity subgroup analysis, both the Asian and Caucasian ethnicity were statistically significant (pooled HR = 1.95, 95% CI 1.43–2.66, I2= 70.1%, p < 0.01; pooled HR = 1.58, 95% CI 1.07–2.33, I2= 63.7%, p = 0.001; separately) (Fig. 2c). Besides, no matter galectin-3 in the tissue or in the plasma, its elevated expression was associated with reduced OS (pooled HR = 1.72, 95% CI 1.34–2.20, I2= 67.6%, p < 0.01; pooled HR = 2.49, 95% CI 1.10–5.63, I2= 71.3%, p = 0.008; respectively) (Fig. 2d).
Fig. 2.
Forest plots of OS in association with galectin-3 in various cancers. a The overall group; b the subgroup analysis of cancer types; c the subgroup analysis of dominant ethnicity; d the subgroup analysis of detected samples
DFS/RFS/PFS associated with galectin-3 expression
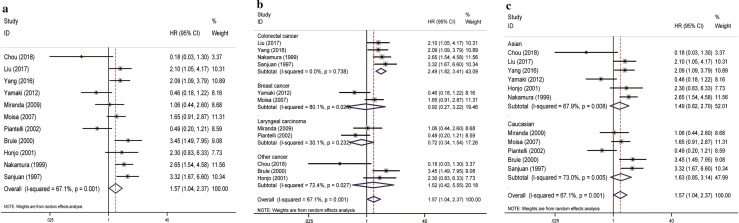
A total of 11 original studies were included to evaluate the role of elevated galectin-3 expression in patients with various solid tumors by DFS/RFS/PFS, within a random-effects model. Our results successfully identified the significant association of high galectin-3 expression with reduced DFS/RFS/PFS in overall cancer patients (pooled HR = 1.57, 95% CI 1.04–2.37, I2= 67.1%, p = 0.001; Fig. 3a). In the subgroup analysis of specific cancer type, we found that high expression of galectin-3 was correlated with shorter DFS/RFS/PFS in colorectal cancer (pooled HR = 2.49, 95% CI 1.82–3.41, I2 = 0.0%, p = 0.738; Fig. 3b). However, In terms of dominant ethnicity subgroup analysis, both the Asian and Caucasian ethnicity were not statistically significant (Fig. 3c).
Fig. 3.
Forest plots of DFS/RFS/PFS in association with galectin-3 in various cancers. a The overall group; b the subgroup analysis of cancer types; c the subgroup analysis of dominant ethnicity
Sensitivity analyses
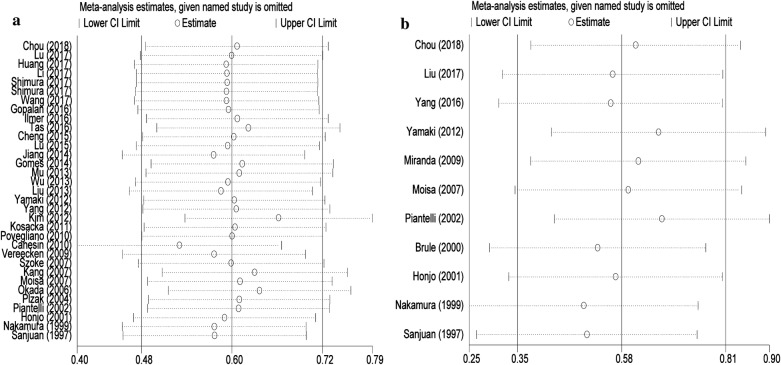
In order to determine the robustness and the stability of our results, sensitivity analysis was conducted to access the stability of results by deleting one single study each time, to reflect the impact of the individual to overall. Our results indicated that no single study significantly influenced the pooled OR and 95% CIs. Namely, our results are comparatively reliable and stable (Fig. 4).
Fig. 4.
Sensitivity analysis of each included study. a OS for individual studies. b DFS/RFS/PFS for individual studies
Publication bias
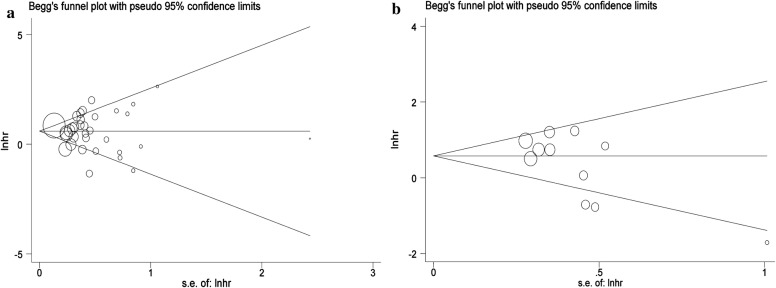
The combined application of Begg’s and Egger’s test was utilized to evaluate the publication bias and meanwhile the funnel plots were displayed in Fig. 5. In the pooled analysis of OS or DFS/RFS/PFS, the p values of Begg’s test and the p values of Egger’s test were all above 0.05, indicating no publication bias in this study.
Fig. 5.
Begg’s funnel plots of the publication bias. a OS for individual studies. b DFS/RFS/PFS for individual studies
Discussion
Up to now, elaborate efforts have been made to establish reliable and convincing evidence to detect promising biomarkers for patients with solid tumors. Galectins, as a family of animal carbohydrate-binding proteins, which had the ability to agglutinate cells, were considered to be potential biomarkers of cancer prognosis given their unique structure and functions into consideration [50, 51]. Over the past years, galectins have been implicated in the development of cancer, the pathogenesis of heart failure and ventricular remodeling, infectious processes, and inflammatory processes [52]. Amongst them, due to its differential expression between cancer and normal tissues, galectin-3 was regarded as one important member of galectins family. However, the definite role of galectin-3 in various human malignant neoplasms remained inconsistent. Hence, this meta-analysis was conducted to clarify this question.
It was the first time for us to shed light on the association between elevated galectin-3 expression and the prognosis of patients with solid tumors. Meanwhile, our results were the systematic evaluation of the prognostic outcomes (OS or DFS/RFS/PFS) in a larger population. Our results did suggest that galectin-3 play an oncogenic role in overall cancer patients. Moreover, we found that high expression of galectin-3 was correlated with shorter OS or DFS/RFS/PFS in colorectal cancer and meanwhile it merely associated with reduced OS in ovarian cancer or non-small cell lung cancer, indicating that it could be a promising biomarker and a novel therapeutic target for them. Furthermore, in subgroup analyses of dominant ethnicity, we observed that both the Asian and Caucasian ethnicity were statistically significant for OS, suggesting that the detection of high galectin-3 expression in these patients might be useful for prognosis prediction. Besides, the outcomes of us shed light on that no matter galectin-3 in the tissue or in the plasma, its role remained stable, indicating it could be a promising biomarker and a novel therapeutic target. Meanwhile, according to the results of sensitivity analyses and publication bias, no single study significantly influenced the pooled OR and 95% CIs and no obvious publication bias was detected in this meta-analysis, indicating the robustness and the stability of our results.
Previous researches indicated that increased expression of galectin-3 often predicted unfavorable outcomes and the level of galectin-3 was positively correlated with invasion of depth, vessel invasion, lymph node metastasis, distant metastasis, and TNM stages of various cancers [26, 53]. Tao et al. [37] demonstrated that the positive expression of galectin-3 was associated with more malignant biological behavior of colorectal cancer and it could be used as a predictor of poor prognosis for patients. As for tongue carcinomas, Honjo showed that cytoplasmic galectin-3 expression increased during the progression from normal to cancerous states, whereas nuclear galectin-3 expression decreased during the progression from normal to cancerous states, indicating that enhanced expression of cytoplasmic galectin-3 could serve as a predictor of disease recurrence in these patients [33].
As for its relevant mechanisms, several studies found that galectin-3 was expressed in both cytosol and nucleus [10, 54]. Therein as an important regulator of the Wnt/β-catenin signaling pathway, galectin-3 could activate the epithelial–mesenchymal transition (EMT) in tumor cells to promote the invasion and metastasis of cancer [55, 56]. Furthermore, it could subsequently activate the Ras-mediated Akt signaling pathway to inhibit cell apoptosis by interacting with the activated GTP-bound K-Ras [57]. Besides, it could also modulate VEGF- and bFGF-mediated angiogenesis by binding its carbohydrate recognition domains (CRDs) to integrate αvβ3, and then promote the growth of new blood vessels [58].
As for the effects on heterogeneity, subgroup analysis was a way to discover their potential sources and even decrease the huge heterogeneity. As presented by our results, we could easily find that there might be the existence of significant heterogeneity of elevated galectin-3 expression in the overall cancer patients. So we conducted a subgroup analysis based on the specific cancer types and found that most of their heterogeneity decreased significantly, even with no heterogeneity. However, subgroup analysis of dominant ethnicity was not associated with significant reduction of heterogeneity, indicating that the dominating source of heterogeneity might be the different cancer types.
Sometimes, galectin-3 combined with another biomarker was often utilized simultaneously in prognostic outcome analyses, showing it might not be an independent factor affecting the prognosis of cancer patients. As indicated by Li et al. [11] the expressions of ezrin and galectin-3 were correlated with the development of cervical cancer, and over-expressions of those proteins were indicative of poor prognosis in patients with cervical cancer. Galectin-3 associated with cyclin D1 expression was also studied in non-small cell lung cancer. As a result, no important correlations with clinicopathological findings and no prognostic values were revealed between them. However, higher cyclin D1 expression was found in galectin-3 negative tumor tissues and the differences in correlations between their expressions in two main histopathological types of non-small cell lung cancer were also discovered [38].
The strength of this study was our broad search strategy with few restrictions to minimize any potential publication bias. Moreover, this was the first meta-analysis reporting the prognostic value of galectin-3 for cancers in the medical literature, which could provide some references for clinical work. Although this meta-analysis was performed with rigorous statistics, our conclusion still had several limitations for the following reasons. Firstly, different studies had their own varied expression cut-off values, which brought many difficulties for us to define the standard cutoff value, resulting in bias in the results of the effectiveness of galectin-3 as a prognostic factor in cancer patients. Secondly, heterogeneity existed in the total OS and DFS/RFS/PFS group and it was likely due to the different characteristics of the patients, such as the age, cancer type, different method in detecting samples and the varied cut-off values of galectin-3 expression. Thirdly, due to the insufficient studies, correlation between galectin-3 and OS or DFS/RFS/PFS in other tumor types has not been further analyzed. Fourthly, some essays studied galectin-3 combined with another biomarker in prognostic outcome analyses, showing galectin-3 was not an independent factor affecting the prognosis of cancer patients. Last but not least, all of these enrolled studies were derived from retrospective or observational data, which could not have a clear impact on group baseline features as RCTs. Upcoming prospective RCTs were required to provide more available data. Taking these aforementioned limitations into consideration, our results could be interpreted rigorously and meanwhile more well-designed studies were required to verify our findings.
Conclusions
In summary, it was the first time for us to shed light on the prognostic role of elevated galectin-3 expression in various cancers. Our results did suggest that galectin-3 played an oncogenic role in colorectal cancer, ovarian cancer and non-small cell lung cancer, indicating that it could be a promising biomarker for predicting the prognosis of patients with malignant neoplasms, and the biological functions of galectin-3 were of great research value of the subject. Due to the aforementioned limitations, larger samples of more strictly designed studies were required to provide more high-quality data to elaborate their associations.
Authors’ contributions
NHS, XHM: protocol/project development; QJZ, XZ: data collection or management; YMW, YT: data analysis; YW, SWL: manuscript writing/editing. All authors read and approved the final manuscript.
Acknowledgements
We would like to thank the researchers and study participants for their contributions.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All the data (pooled hazard ratios with 95% confidence intervals of OS or DFS/PFS/RFS) used to support the findings of this study are included within the article. Please contact author for data requests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This article was funded by Medical key talent of Jiangsu Province: ZDRCA2016009 and the National Natural Science Foundation of China (Grant Number: 81871151; 81801438).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yi Wang, Email: wangyi_urology@163.com.
Shiwei Liu, Email: 495995677@qq.com.
Ye Tian, Email: urologytianye@163.com.
Yamin Wang, Email: wangyamin231@163.com.
Qijie Zhang, Email: 15950537490@163.com.
Xiang Zhou, Email: zhouxiang199302@163.com.
Xianghu Meng, Phone: +08615951945520, Email: xhmeng888@163.com.
Ninghong Song, Phone: +08613851490672, Email: songninghong_urol@163.com.
References
- 1.Dings RPM, Miller MC, Griffin RJ, Mayo KH. Galectins as molecular targets for therapeutic intervention. Int J Mol Sci. 2018;19(3):905. doi: 10.3390/ijms19030905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chung LY, Tang SJ, Wu YC, Sun GH, Liu HY, Sun KH. Galectin-3 augments tumor initiating property and tumorigenicity of lung cancer through interaction with β-catenin. Oncotarget. 2015;6(7):4936–4952. doi: 10.18632/oncotarget.3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barondes SH, Castronovo V, Cooper DN, Cummings RD, Drickamer K, Feizi T, Gitt MA, Hirabayashi J, Hughes C, Kasai K, et al. Galectins: a family of animal beta-galactoside-binding lectins. Cell. 1994;76(4):597–598. doi: 10.1016/0092-8674(94)90498-7. [DOI] [PubMed] [Google Scholar]
- 4.Huang Z, Ai Z, Li N, Xi H, Gao X, Wang F, Tan X, Liu H. Over expression of galectin-3 associates with short-term poor prognosis in stage II colon cancer. Cancer Biomark. 2016;17(4):445–455. doi: 10.3233/CBM-160661. [DOI] [PubMed] [Google Scholar]
- 5.Liu FT, Rabinovich GA. Galectins as modulators of tumour progression. Nat Rev Cancer. 2005;5(1):29–41. doi: 10.1038/nrc1527. [DOI] [PubMed] [Google Scholar]
- 6.Knapp JS, Lokeshwar SD, Vogel U, Hennenlotter J, Schwentner C, Kramer MW, Stenzl A, Merseburger AS. Galectin-3 expression in prostate cancer and benign prostate tissues: correlation with biochemical recurrence. World J Urol. 2013;31(2):351–358. doi: 10.1007/s00345-012-0925-y. [DOI] [PubMed] [Google Scholar]
- 7.Cheng CL, Hou HA, Lee MC, Liu CY, Jhuang JY, Lai YJ, Lin CW, Chen HY, Liu FT, Chou WC, et al. Higher bone marrow LGALS3 expression is an independent unfavorable prognostic factor for overall survival in patients with acute myeloid leukemia. Blood. 2013;121(16):3172–3180. doi: 10.1182/blood-2012-07-443762. [DOI] [PubMed] [Google Scholar]
- 8.Kramer MW, Kuczyk MA, Hennenlotter J, Serth J, Schilling D, Stenzl A, Merseburger AS. Decreased expression of galectin-3 predicts tumour recurrence in pTa bladder cancer. Oncol Rep. 2008;20(6):1403–1408. [PubMed] [Google Scholar]
- 9.Yoshii T, Inohara H, Takenaka Y, Honjo Y, Akahani S, Nomura T, Raz A, Kubo T. Galectin-3 maintains the transformed phenotype of thyroid papillary carcinoma cells. Int J Oncol. 2001;18(4):787–792. doi: 10.3892/ijo.18.4.787. [DOI] [PubMed] [Google Scholar]
- 10.Lu W, Wang J, Yang G, Yu N, Huang Z, Xu H, Li J, Qiu J, Zeng X, Chen S, et al. Posttranscriptional regulation of galectin-3 by miR-128 contributes to colorectal cancer progression. Oncotarget. 2017;8(9):15242–15251. doi: 10.18632/oncotarget.14839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li M, Feng YM, Fang SQ. Overexpression of ezrin and galectin-3 as predictors of poor prognosis of cervical cancer. Braz J Med Biol Res. 2017;50(4):e5356. doi: 10.1590/1414-431X20165356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ilmer M, Mazurek N, Gilcrease MZ, Byrd JC, Woodward WA, Buchholz TA, Acklin K, Ramirez K, Hafley M, Alt E, et al. Low expression of galectin-3 is associated with poor survival in node-positive breast cancers and mesenchymal phenotype in breast cancer stem cells. Breast Cancer Res. 2016;18(1):97. doi: 10.1186/s13058-016-0757-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okada K, Shimura T, Suehiro T, Mochiki E, Kuwano H. Reduced galectin-3 expression is an indicator of unfavorable prognosis in gastric cancer. Anticancer Res. 2006;26(2b):1369–1376. [PubMed] [Google Scholar]
- 14.Piantelli M, Iacobelli S, Almadori G, Iezzi M, Tinari N, Natoli C, Cadoni G, Lauriola L, Ranelletti FO. Lack of expression of galectin-3 is associated with a poor outcome in node-negative patients with laryngeal squamous-cell carcinoma. J Clin Oncol. 2002;20(18):3850–3856. doi: 10.1200/JCO.2002.01.078. [DOI] [PubMed] [Google Scholar]
- 15.Stang A. Critical evaluation of the Newcastle–Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 16.Williamson PR, Smith CT, Hutton JL, Marson AG. Aggregate data meta-analysis with time-to-event outcomes. Stat Med. 2002;21(22):3337–3351. doi: 10.1002/sim.1303. [DOI] [PubMed] [Google Scholar]
- 17.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dersimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 19.Egger M, Davey SG, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7129):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chou SY, Yen SL, Huang CC, Huang EY. Galectin-1 is a poor prognostic factor in patients with glioblastoma multiforme after radiotherapy. BMC Cancer. 2018;18(1):105. doi: 10.1186/s12885-018-4025-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gopalan V, Saremi N, Sullivan E, Kabir S, Lu CT, Salajegheh A, Leung M, Smith RA, Lam AK. The expression profiles of the galectin gene family in colorectal adenocarcinomas. Hum Pathol. 2016;53:105–113. doi: 10.1016/j.humpath.2016.02.016. [DOI] [PubMed] [Google Scholar]
- 22.Jiang SS, Weng DS, Wang QJ, Pan K, Zhang YJ, Li YQ, Li JJ, Zhao JJ, He J, Lv L, et al. Galectin-3 is associated with a poor prognosis in primary hepatocellular carcinoma. J Transl Med. 2014;12:273. doi: 10.1186/s12967-014-0273-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gomes TS, Oshima CT, Forones NM, De Oliveira Lima F, Ribeiro DA. Expression of galectin-3 in gastric adenocarcinoma. Indian J Med Res. 2014;140(1):69–76. [PMC free article] [PubMed] [Google Scholar]
- 24.Yamaki S, Fujii T, Yajima R, Hirakata T, Yamaguchi S, Fujisawa T, Tsutsumi S, Asao T, Yanagita Y, Iijima M, et al. Clinicopathological significance of decreased galectin-3 expression and the long-term prognosis in patients with breast cancer. Surg Today. 2013;43(8):901–905. doi: 10.1007/s00595-012-0378-3. [DOI] [PubMed] [Google Scholar]
- 25.Yang LP, Jiang S, Liu JQ, Miao XY, Yang ZL. Up-regulation of galectin-3 and Sambucus nigra agglutinin binding site is associated with invasion, metastasis and poor-progression of the gallbladder adenocarcinoma. Hepatogastroenterology. 2012;59(119):2089–2094. doi: 10.5754/hge12129. [DOI] [PubMed] [Google Scholar]
- 26.Kim SJ, Kim DC, Kim MC, Jung GJ, Kim KH, Jang JS, Kwon HC, Kim YM, Jeong JS. Fascin expression is related to poor survival in gastric cancer. Pathol Int. 2012;62(12):777–784. doi: 10.1111/pin.12012. [DOI] [PubMed] [Google Scholar]
- 27.Povegliano LZ, Oshima CTF, de Oliveira Lima F, Scherholz PLA, Forones NM. Immunoexpression of galectin-3 in colorectal cancer and its relationship with survival. J Gastrointest Cancer. 2011;42(4):217–221. doi: 10.1007/s12029-010-9189-1. [DOI] [PubMed] [Google Scholar]
- 28.Miranda FA, Hassumi MK, Guimaraes MC, Simoes RT, Silva TG, Lira RC, Rocha AM, Mendes CT, Jr, Donadi EA, Soares CP, et al. Galectin-3 overexpression in invasive laryngeal carcinoma, assessed by computer-assisted analysis. J Histochem Cytochem. 2009;57(7):665–673. doi: 10.1369/jhc.2009.952960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szoke T, Kayser K, Trojan I, Kayser G, Furak J, Tiszlavicz L, Baumhakel JD, Gabius HJ. The role of microvascularization and growth/adhesion-regulatory lectins in the prognosis of non-small cell lung cancer in stage II. Eur J Cardiothorac Surg. 2007;31(5):783–787. doi: 10.1016/j.ejcts.2007.01.072. [DOI] [PubMed] [Google Scholar]
- 30.Kang SY, Han JH, Lee KJ, Choi JH, Park JI, Kim HI, Lee HW, Jang JH, Park JS, Kim HC, et al. Low expression of Bax predicts poor prognosis in patients with locally advanced esophageal cancer treated with definitive chemoradiotherapy. Clin Cancer Res. 2007;13(14):4146–4153. doi: 10.1158/1078-0432.CCR-06-3063. [DOI] [PubMed] [Google Scholar]
- 31.Plzak J, Betka J, Smetana K, Jr, Chovanec M, Kaltner H, Andre S, Kodet R, Gabius HJ. Galectin-3—an emerging prognostic indicator in advanced head and neck carcinoma. Eur J Cancer (Oxford, England: 1990) 2004;40(15):2324–2330. doi: 10.1016/j.ejca.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 32.van den Brule FA, Waltregny D, Liu FT, Castronovo V. Alteration of the cytoplasmic/nuclear expression pattern of galectin-3 correlates with prostate carcinoma progression. Int J Cancer. 2000;89(4):361–367. doi: 10.1002/1097-0215(20000720)89:4<361::AID-IJC8>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 33.Honjo Y, Inohara H, Akahani S, Yoshii T, Takenaka Y, Yoshida J, Hattori K, Tomiyama Y, Raz A, Kubo T. Expression of cytoplasmic galectin-3 as a prognostic marker in tongue carcinoma. Clin Cancer Res. 2000;6(12):4635–4640. [PubMed] [Google Scholar]
- 34.Nakamura M, Inufusa H, Adachi T, Aga M, Kurimoto M, Nakatani Y, Wakano T, Nakajima A, Hida JI, Miyake M, et al. Involvement of galectin-3 expression in colorectal cancer progression and metastasis. Int J Oncol. 1999;15(1):143–148. doi: 10.3892/ijo.15.1.143. [DOI] [PubMed] [Google Scholar]
- 35.Sanjuan X, Fernandez PL, Castells A, Castronovo V, van den Brule F, Liu FT, Cardesa A, Campo E. Differential expression of galectin 3 and galectin 1 in colorectal cancer progression. Gastroenterology. 1997;113(6):1906–1915. doi: 10.1016/S0016-5085(97)70010-6. [DOI] [PubMed] [Google Scholar]
- 36.Tas F, Bilgin E, Tastekin D, Erturk K, Duranyildiz D. Clinical significance of serum galectin-3 levels in gastric cancer patients. J Gastrointest Cancer. 2016;47(2):182–186. doi: 10.1007/s12029-016-9817-5. [DOI] [PubMed] [Google Scholar]
- 37.Tao L, Jin L, Dechun L, Hongqiang Y, Changhua K, Guijun L. Galectin-3 expression in colorectal cancer and its correlation with clinical pathological characteristics and prognosis. Open Med (Warsaw, Poland) 2017;12:226–230. doi: 10.1515/med-2017-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kosacka M, Piesiak P, Kowal A, Golecki M, Jankowska R. Galectin-3 and cyclin D1 expression in non-small cell lung cancer. J Exp Clin Cancer Res. 2011;30:101. doi: 10.1186/1756-9966-30-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moisa A, Fritz P, Eck A, Wehner HD, Murdter T, Simon W, Gabius HJ. Growth/adhesion-regulatory tissue lectin galectin-3: stromal presence but not cytoplasmic/nuclear expression in tumor cells as a negative prognostic factor in breast cancer. Anticancer Res. 2007;27(4b):2131–2139. [PubMed] [Google Scholar]
- 40.Cheng D, Liang B, Li Y. Serum galectin-3 as a potential marker for gastric cancer. Med Sci Monit. 2015;21:755–760. doi: 10.12659/MSM.892386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimura T, Shibata M, Gonda K, Kofunato Y, Okada R, Ishigame T, Kimura T, Kenjo A, Kono K, Marubashi S. Significance of circulating galectin-3 in patients with pancreatobiliary cancer. Anticancer Res. 2017;37(9):4979–4986. doi: 10.21873/anticanres.11909. [DOI] [PubMed] [Google Scholar]
- 42.Canesin G, Gonzalez-Peramato P, Palou J, Urrutia M, Cordon-Cardo C, Sanchez-Carbayo M. Galectin-3 expression is associated with bladder cancer progression and clinical outcome. Tumour Biol. 2010;31(4):277–285. doi: 10.1007/s13277-010-0033-9. [DOI] [PubMed] [Google Scholar]
- 43.Vereecken P, Awada A, Suciu S, Castro G, Morandini R, Litynska A, Lienard D, Ezzedine K, Ghanem G, Heenen M. Evaluation of the prognostic significance of serum galectin-3 in American joint committee on cancer stage III and stage IV melanoma patients. Melanoma Res. 2009;19(5):316–320. doi: 10.1097/CMR.0b013e32832ec001. [DOI] [PubMed] [Google Scholar]
- 44.Mu XS, Wang SQ, Long B, Mou YF, Wu Y, Liao KY, Xu Y, Mo SJ, Ma B, Tang XL. The clinical significance of galectin-3 and P27 involved in treatment of elderly early gastric cancer with endoscopic submucosal dissection. Chin J Gerontol. 2014;19:5423–5425. [Google Scholar]
- 45.Wu SW, Yu L, Zhou L, Cheng ZN, Tao YS. Expression of Gal-3 and CD82/KAI1 proteins in non-small cell lung cancer and their clinical significance. Chin J Oncol. 2013;35(2):124–128. doi: 10.3760/cma.j.issn.0253-3766.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 46.Liu MY, Du B, Li CH, Zhao YB, Meng QW, Cai L. Expression and related factors of galectin-3 in non-small cell lung cancer. Chin J Lung Cancer. 2013;16(8):417–421. doi: 10.3779/j.issn.1009-3419.2013.08.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu HW, Wang DY, Liu YY, Xu GC, Xie LL, Lin ZQ. Expression of galectin-3 and β-catenin in epithelial ovarian cancer and their clinical significance. J Sun Yat-sen Univ (Medical Sciences) 2015;36(6):883–888. [Google Scholar]
- 48.Yang XZ. Galectin-3 expression in colorectal cancer and its clinical significance. Chin J Integr Tradit Chin West Med. 2016;22(5):438–441. [Google Scholar]
- 49.Wang DY, Lin ZQ, Liu YY, Huang CX, Lu HW. Expression of galectin-3 and Bcl-2 in epithelial ovarian cancer and their clinical significance. J Sun Yat-sen Univ (Medical Sciences) 2017;38(3):379–385. [Google Scholar]
- 50.Barondes SH, Cooper DN, Gitt MA, Leffler H. Galectins. Structure and function of a large family of animal lectins. J Biol Chem. 1994;269(33):20807–20810. [PubMed] [Google Scholar]
- 51.Buttery R, Monaghan H, Salter DM, Sethi T. Galectin-3: differential expression between small-cell and non-small-cell lung cancer. Histopathology. 2004;44(4):339–344. doi: 10.1111/j.1365-2559.2004.01815.x. [DOI] [PubMed] [Google Scholar]
- 52.Ebrahim AH, Alalawi Z, Mirandola L, Rakhshanda R, Dahlbeck S, Nguyen D, Jenkins M, Grizzi F, Cobos E, Figueroa JA, et al. Galectins in cancer: carcinogenesis, diagnosis and therapy. Ann Transl Med. 2014;2(9):88. doi: 10.3978/j.issn.2305-5839.2014.09.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou X, Jing J, Peng J, Mao W, Zheng Y, Wang D, Wang X, Liu Z, Zhang X. Expression and clinical significance of galectin-3 in osteosarcoma. Gene. 2014;546(2):403–407. doi: 10.1016/j.gene.2014.04.066. [DOI] [PubMed] [Google Scholar]
- 54.Rubio D, Garcia S, De la Cueva T, Paz MF, Lloyd AC, Bernad A, Garcia-Castro J. Human mesenchymal stem cell transformation is associated with a mesenchymal–epithelial transition. Exp Cell Res. 2008;314(4):691–698. doi: 10.1016/j.yexcr.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 55.Thiery JP. Epithelial–mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2(6):442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 56.Steinestel K, Eder S, Schrader AJ, Steinestel J. Clinical significance of epithelial–mesenchymal transition. Clin Transl Med. 2014;3:17. doi: 10.1186/2001-1326-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Elad-Sfadia G, Haklai R, Balan E, Kloog Y. Galectin-3 augments K-Ras activation and triggers a Ras signal that attenuates ERK but not phosphoinositide 3-kinase activity. J Biol Chem. 2004;279(33):34922–34930. doi: 10.1074/jbc.M312697200. [DOI] [PubMed] [Google Scholar]
- 58.Markowska AI, Liu FT, Panjwani N. Galectin-3 is an important mediator of VEGF- and bFGF-mediated angiogenic response. J Exp Med. 2010;207(9):1981–1993. doi: 10.1084/jem.20090121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data (pooled hazard ratios with 95% confidence intervals of OS or DFS/PFS/RFS) used to support the findings of this study are included within the article. Please contact author for data requests.