Abstract
To systematically explore complex genetic interactions, we constructed ~200,000 yeast triple mutants and scored negative trigenic interactions. We selected double mutant query genes across a broad spectrum of biological processes, spanning a range of quantitative features of the global digenic interaction network and tested for a genetic interaction with a third mutation. Trigenic interactions often occurred among functionally related genes and essential genes were hubs on the trigenic network. Despite their functional enrichment, trigenic interactions tended to link genes in distant bioprocesses and display a weaker magnitude than digenic interactions. We estimate that the global trigenic interaction network is ~100-fold larger than the global digenic network, highlighting the potential for complex genetic interactions to impact the biology of inheritance, including the genotype to phenotype relationship.
One Sentence Summary:
Exploring the expanse and nature of the trigenic interaction landscape in yeast.
Main Text:
Genetic interactions occur when a combination of mutations in different genes leads to an unexpected phenotype that deviates from a model incorporating the combined effects of the corresponding single mutant phenotypes. In humans, each individual carries thousands of different variants, which means there is incredible potential for combinatorial genetic interactions to determine our personal phenotype (1, 2). Indeed, genetic interactions are thought to represent a significant component of the missing heritability associated with current genome wide association studies (GWAS) (3); however, the statistical limitations associated with GWAS datasets preclude the detection of specific genetic interactions and thus potential genetic networks underlying inherited traits, including diseases, remain elusive (3–5). To address the role of genetic interactions in the genotype to phenotype relationship, we have been exploring their general principles through systematic analysis of genetic networks underlying cellular fitness in a genetically-tractable model system, the budding yeast, Saccharomyces cerevisiae (6). Our previous studies focused predominantly on genetic interactions involving two genes (“digenic interactions”) (7). Here, we analyzed a series of single, double, and triple mutants by quantifying their colony size, as a proxy for fitness, to systematically explore complex genetic interactions.
There are two basic types of fitness-based genetic interactions. A negative genetic interaction refers to a combination of mutations that results in a more severe fitness defect than expected (8). Synthetic lethality is an extreme example of a negative genetic interaction and occurs when two mutations, neither of which is lethal on its own, combine and lead to an inviable double mutant phenotype (9, 10). Conversely, a positive genetic interaction occurs when a combination of genetic perturbations results in a fitness phenotype that is greater than expected, including genetic suppression, in which the fitness defect of a query mutant is alleviated by a mutation in a second gene (11). To map a global digenic interaction network for yeast, we constructed millions of double mutants and identified hundreds of thousands of negative and positive genetic interactions (7). To put these results in perspective, while only ~1000 of the ~6000 total yeast genes are individually essential and cause lethality when deleted (12), and an equivalent number of nonessential genes cause a slow growth defect under standard laboratory conditions (13), ~550,000 different yeast gene pairs display a combinatorial negative genetic interaction, including a subset of ~10,000 extreme synthetic lethal interactions involving nonessential gene pairs (7). Thus, there are numerous potential ways to generate extreme lethal phenotypes through negative digenic interactions of nonessential gene pairs.
The set of digenic interactions for a query gene, its genetic interaction profile, provides a quantitative measure of function, because genes with similar roles have overlapping profiles (14, 15). Indeed, genes belonging to the same biological pathway or protein complex display highly similar genetic interaction profiles. Moreover, a global network based on digenic interaction profile similarity reveals a hierarchy of functional modules, which includes detailed pathways and complexes, that in turn cluster into larger modules corresponding to bioprocesses, and those cluster together into modules corresponding to cellular compartments, to outline the functional architecture of a cell (7).
A complete understanding of the role of genetic interactions in the genotype to phenotype relationship requires that we also investigate complex, higher-order genetic interactions, involving more than two genes. Because there are ~2000-fold more yeast triple mutants than the ~18 million double mutants, it is possible that there is a substantially larger number of trigenic than digenic interactions and that higher-order interactions may be important for driving inherited traits. Here, we survey yeast trigenic interactions, sampling quantitative features of the digenic network, and explore the implications of the higher-order genetic interaction network.
Mapping trigenic interactions quantitatively and surveying the global trigenic landscape
To explore the trigenic interaction landscape, we designed query strains that sampled three key quantitative features of our global digenic interaction network (7). We designed query strains carrying mutations in two genes spanning a range of the following features: (1) digenic interaction strength; (2) number of digenic interactions (average digenic interaction degree); (3) digenic interaction profile similarity (Fig. 1A, Table S1). Gene pairs were selected to fill bins of varying digenic interaction attributes and to cover all major biological processes in the cell enabling a sample that would provide a diverse survey of the trigenic interaction landscape. We largely focused on unambiguous singletons since duplicated genes represent a relatively small subset of genes and thus can only represent a small fraction of the global trigenic interaction network. For this survey, we constructed 151 double mutant query strains and 302 single mutant strains, encompassing 47 temperature-sensitive alleles of different essential genes and 254 deletion alleles of unique nonessential genes. The query strains in this set were selected to span the different digenic attribute bins according to predefined thresholds (Table S1). An additional 31 double mutant queries fell outside of the defined thresholds but were included for validation and comparison purposes (Additional Data S1, S2, S3) (16). The fitness of the resulting query strains was measured using a quantitative growth assay, and the behavior of the single and double mutant query strains showed strong agreement with our previously published dataset (Fig. S1, S2, Additional Data S4) (7, 15).
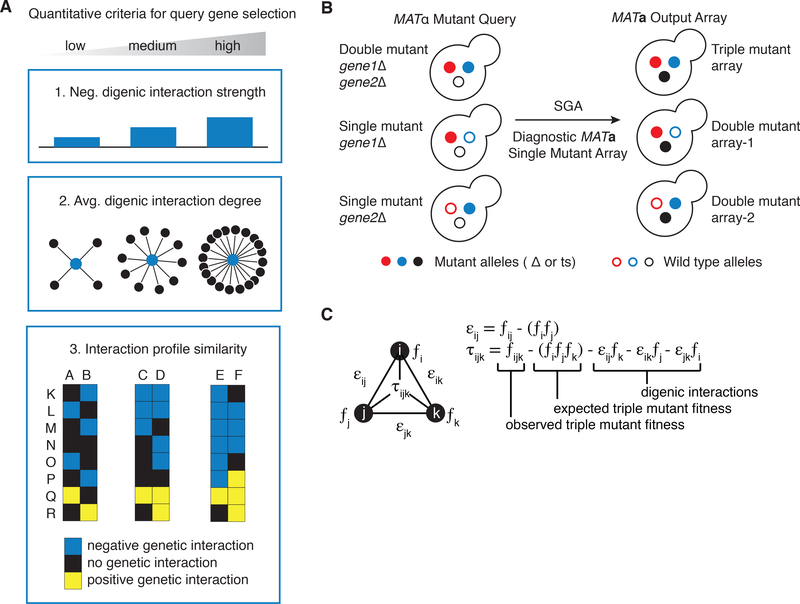
Fig. 1. Triple mutant Synthetic Genetic Array (SGA) analysis.
(A) Criteria for selecting query strains for sampling trigenic interaction landscape of singleton genes in yeast. The gene pairs were grouped into three general categories based on a range of feature as follows: 1) gene pairs directly connected by zero-very weak (0 to −0.08, n = 74), weak (−0.08 to −0.1, n = 32) or moderate (< −0.1, n = 45) negative digenic interaction; 2) low (10 to 45, n = 50), intermediate (46 to 70, n = 53), and high (> 71, n = 48) average digenic interaction degree (denoted by the number of black edges of each node); and 3) low (−0.02 to 0.03, n = 46, represented by genes A & B, which show a relatively low overlap of genetic interactions with genes K to R), intermediate (0.03 to 0.1, n = 59, represented by gene pair C & D, which display an intermediate overlap of genetic interactions) and high (> 0.1, n = 46, represented by gene pair E & F, which display a relatively high level of overlap of genetic interactions) functional similarity, as measured by their digenic interaction profile similarity and co-annotation to the same Gene Ontology term(s). Query mutant genes were either nonessential deletion mutant alleles (Δ) or conditional temperature sensitive (ts) alleles of essential genes. (B) Diagram illustrating the triple mutant SGA experimental strategy. To quantify a trigenic interaction, 3 types of screens are conducted in parallel. To estimate triple mutant fitness a double mutant query strain carrying two desired mutated genes of interest (red and blue filled circles) is crossed to a diagnostic array of single mutants (black filled circle). Meiosis is induced in heterozygous triple mutants and haploid triple mutant progeny is selected in sequential replica pinning steps. In parallel, single mutant control query strains are used to generate double mutants for fitness analysis. (C) Triple mutant SGA quantitative scoring strategy. The top equation shows the quantification of a digenic interaction, where ε is the digenic interaction score, ƒij is the observed double mutant fitness and the expected double mutant fitness is expressed as the product of single mutant fitness estimates ƒiƒj. The trigenic interaction score (τ) is derived from the digenic interaction score, where ƒijk is the observed triple mutant fitness, ƒiƒjƒk is triple mutant fitness expectation expressed as the product of three single mutant fitness estimates; the influence of digenic interactions is subtracted from the expectation, each digenic interaction is scaled by the fitness of the third mutation.
Trigenic interaction screening required development and implementation of three operational components. First, Synthetic Genetic Array (SGA) analysis, an automated form of yeast genetics that is often used to cross a query gene mutation into an array of single mutants to generate a defined set of haploid double mutants (6), was adapted such that a double-mutant query strain is crossed into an array of single mutants, to generate triple mutants for trigenic interaction analysis (Fig. 1B). Because the identification of a trigenic interaction requires comparison with the corresponding double mutants, we also conducted screens in which the individual mutants of the query gene pair were scored for digenic interactions (Fig. 1B). Second, for experimental feasibility, we assembled a diagnostic array of 1,182 strains, comprising 990 nonessential gene deletion mutants and 192 essential gene mutants carrying temperature-sensitive alleles, which combine to span ~20% of the yeast genome (Additional Data S5). Importantly, the diagnostic array was designed to be highly representative of the rest of the genome in terms of exhibited genetic interaction profiles (Fig. S3). Briefly, array strains are selected from a larger genetic interaction dataset for their ability to represent different regions of the global network in a minimally redundant way. This is accomplished by iteratively selecting strains to maximize the performance of profile similarities when predicting co-annotations to a functional gold-standard (17). Third, we developed a scoring method, the τ-SGA score, which combines double and triple mutant fitness estimates derived from colony size to identify trigenic interactions quantitatively (Fig. 1C). The τ-SGA score differs from the MinDC score reported previously (18), because it accounts for all cases where two of the genes are not independent, resulting in an expectation that contains digenic interaction effects that are scaled by the fitness of the non-interacting genes (Fig. S4) (16). The final trigenic τ-SGA interaction score then accounts for digenic effects, but also enables detection of trigenic interactions where digenic effects of insufficient explanatory power can be found.
We focused exclusively on the analysis of deleterious negative trigenic interactions for two reasons. First, quantitative scoring of negative genetic interactions is often more accurate than that for positive interactions because there is a greater signal to noise ratio for negative genetic interactions, which means they are associated with lower false positive and false negative rates (8), a feature that is important for the robust statistical analysis necessary to differentiate true trigenic interactions from the extensive background digenic network. Second, negative digenic interactions are generally more functionally informative than positive digenic interactions (8) and, thus, the large-scale mapping of a negative trigenic interaction network is expected to provide the most mechanistic insight into gene function and pathway wiring.
Trigenic interactions are enriched for functionally related genes
To obtain sufficient precision, we carried out each analysis, which involved screening the individual query genes for digenic interactions and the double mutant query for trigenic interactions, in at least two replicates (Fig. S5). In total, we tested 410,399 double and 195,666 triple mutants for fitness defects meeting a previously established intermediate magnitude cut-off (15) (Additional Data S2), which identified 9,363 digenic and 3,196 trigenic negative interactions. From detailed validation of trigenic interactions of our CLN1-CLN2 double mutant query, which was screened previously (19), we estimated a false negative rate of ~40%, a false positive rate of ~20%, and a true positive rate between ~60 and ~75% (Table S2, Fig. S6) (16), which is consistent with our previous global digenic network analysis (8).
The distribution of trigenic interaction degree for array strains shows that the majority of low degree genes (70%) account for ~88% of all trigenic interactions, whereas highly connected genes contribute the remaining ~12% of interactions. Thus, the trigenic interactions are not associated with a small set of highly connected genes, rather the interactions are distributed across many different genes (Fig. S7). On the other hand, with a smaller more biased set of double mutant query genes, the distribution of trigenic interaction degree shows that ~22% of them accounted for 51% of trigenic interactions, indicating that a particular subset of the digenic queries were enriched for trigenic interactions (Fig. S7). Approximately 1/3 of the newly mapped trigenic interactions identified connections that were not observed in our digenic control network; we refer to these as ‘novel’ trigenic interactions. The remaining ~2/3 of the trigenic interactions overlapped a digenic interaction, while still exhibiting a stronger than expected fitness defect in the triple mutant; these we refer to as ‘modified’ trigenic interactions (Fig. S8A). Thus, while a substantial fraction of trigenic interactions elucidate totally novel functional information, the majority of the trigenic interactions we mapped expand upon the digenic interaction network.
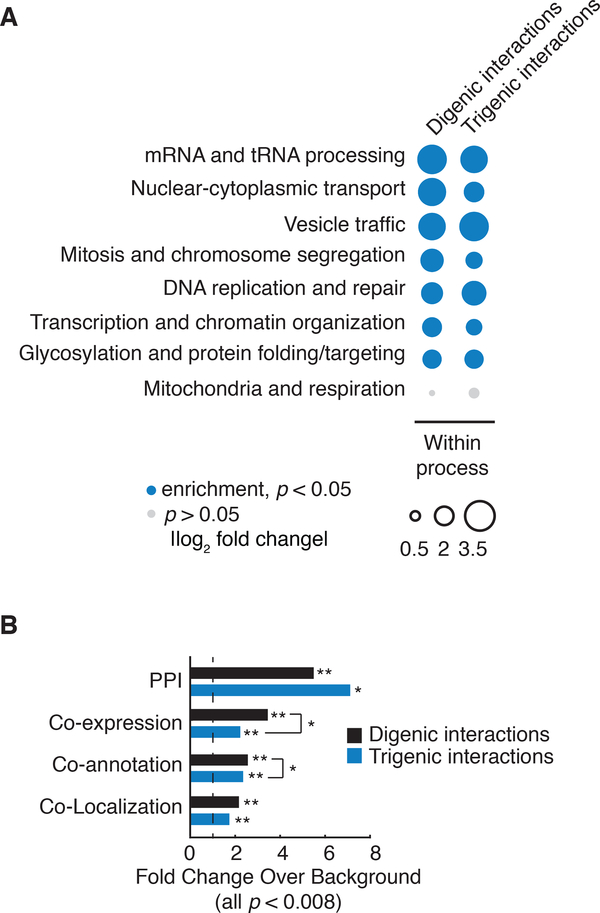
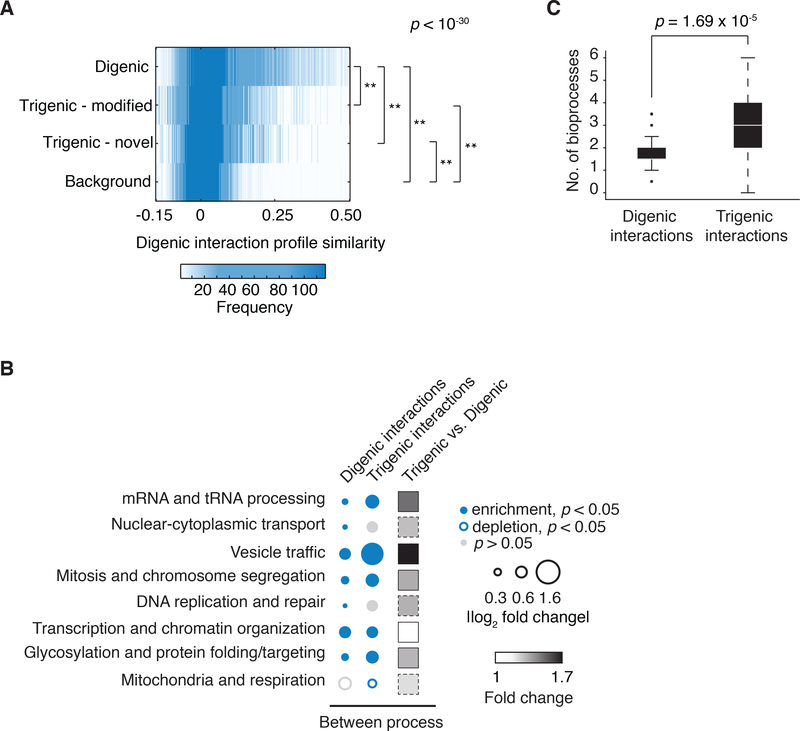
We first assessed the functional information embedded in the trigenic network by comparing the distributions of digenic and trigenic interactions across different biological processes. As observed previously (15), digenic interactions were enriched among genes annotated to the same biological process and, although the magnitude of trigenic interaction enrichment was somewhat lower, they were comparably enriched for genes within the same bioprocess (Fig. 2A). We also evaluated the enrichment of digenic and trigenic interactions across common functional standards, including annotation to the same Gene Ontology (GO) biological process, subcellular-localization pattern, protein-protein interaction, and gene coexpression (Fig. 2B). Like digenic interactions (7, 15), genes involved in trigenic interactions were significantly enriched for all of these standards with genes participating in the ‘modified’ class of trigenic interactions exhibiting stronger functional relationships (Fig. S8B) as well as a stronger magnitude of interactions (Fig. S8C). Thus, trigenic interactions resemble digenic interactions in that they are rich in functional information, which means that genes participating in many trigenic interactions can be predicted from alternative datasets and general knowledge of cellular function.
Fig. 2. Functional characterization of trigenic interactions.
(A) Frequency of negative genetic interactions within biological processes. The fraction of screened query-array combinations exhibiting negative interactions belonging to functional gene sets annotated by SAFE on the global genetic interaction network (55). Within process received a count for any combination in which both genes for digenic interactions or all three genes for trigenic interactions, were annotated to the same term. The size of the circle assigned to each process-process element reflects the fold-increase over the background fraction of interactions (digenic = 0.023, trigenic = 0.016). Significance was assessed using hypergeometric cumulative distribution test, p < 0.05. Significant enrichment is shown in filled blue circle and no significant change is in grey. (B) Enrichment of negative digenic (black) and trigenic (blue) interactions across four functional standards. Black dashed line marks no enrichment. The functional standards are the following: merged protein-protein interaction (PPI) standard (56–60), co-annotation is based on SAFE terms (7), co-expression (61), co-localization (62). Significance was assessed using hypergeometric cumulative distribution test, * represents 10−4 ≤ p < 0.01, ** represents p < 10-4.
Trigenic interactions expand functional connections mapped by the global digenic network
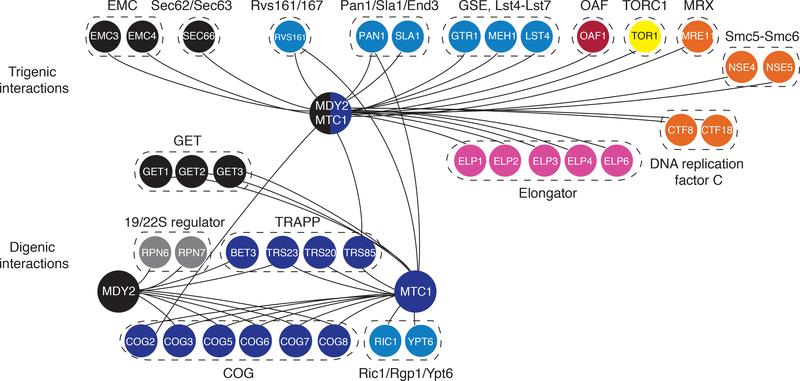
Our functional analysis revealed that trigenic interactions have some properties distinct from digenic interactions, suggesting that they may be useful for discovering new connections between genes and their corresponding pathways. As an illustrative example, we examined the MDY2-MTC1 double mutant query, which is a highly-connected hub within the trigenic network. MDY2 encodes a protein that interacts and functions with components of the GET (Guided Entry of Tail-anchor) pathway (20), which is important for Golgi to ER (Endoplasmic Reticulum) trafficking and inserting tail-anchored proteins into ER membranes. MTC1 encodes a protein of unknown function that localizes to the early Golgi. The MTC1 digenic interaction profile is similar to that of USO1, which is involved in vesicle-mediated ER to Golgi transport (21), and RUD3, which encodes a Golgi matrix protein important for the structural organization of the cisGolgi (22), suggesting MTC1 also has a role in the early secretory pathway.
As expected from these previous findings, the MDY2 and MTC1 digenic interactions identified in our screen were enriched for genes involved in cell polarity and the early secretory pathway (Fig. 3, S9A, Additional Data S6). However, MDY2-MTC1 double mutant query profile encompassed a much more functionally diverse set of genes (Fig. 3). For example, while we observed novel trigenic interactions with ER to Golgi transport genes, we also observed trigenic interactions with genes involved in other modes of vesicle trafficking, including endocytosis and peroxisome biology. Moreover, MDY2-MTC1 trigenic interactions identified connections to genes with more diverse functions, such as components of the elongator complex (Fig. 3), which controls the modification of wobble nucleosides in tRNAs, and several genes involved in DNA replication and repair. Notably, the MDY2-MTC1 query also showed a trigenic interaction with TOR1 (target of rapamycin), which encodes the key kinase subunit of the TORC1 complex that is required for growth in response to nutrients by regulating ribosome biogenesis, nutrient transport, and autophagy (23). Consistent with this observation, the MDY2-MTC1 trigenic interaction network captures a set of genes that have a dual role in TORC1 signaling and sorting of the general amino acid permease, including GTR1, MEH1, and LST4 (24–26).
Fig. 3. MDY2-MTC1 – a hub on the trigenic interaction network.
Representative digenic interactions are highlighted for MDY2 and MTC1 single mutant query genes, and representative trigenic interactions are shown for the MDY2-MTC1 double mutant query. The network was visualized using Cytoscape (63). Genes were chosen from representative protein complexes (8) in which ≥ 50% of members on the diagnostic array display genetic interactions. Negative genetic interactions, ε or τ < −0.08, p < 0.05, are depicted. All of the digenic and trigenic interactions displayed have been confirmed by tetrad analysis. Nodes are color coded based on their biological roles. Nodes are labeled with gene names and genes are grouped according to specific protein complexes.
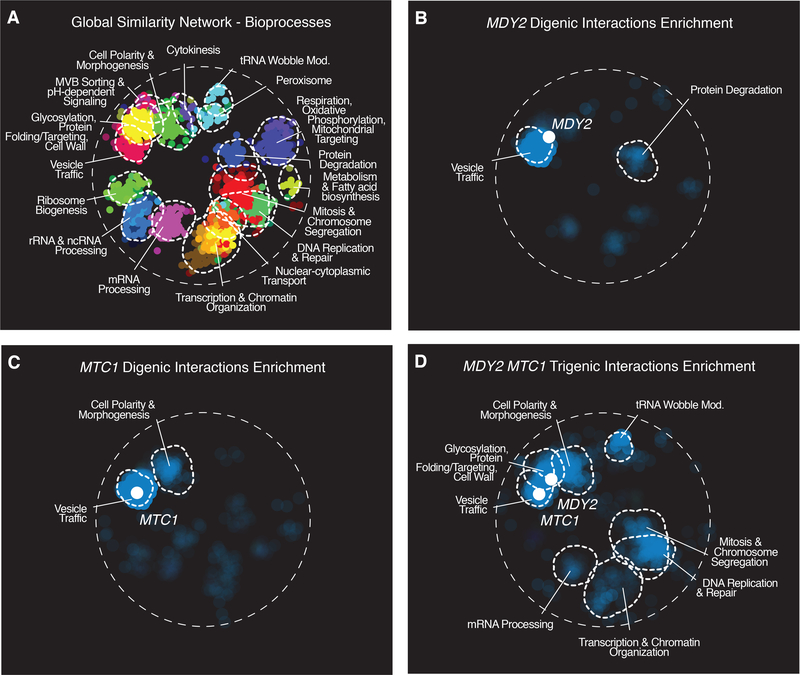
The spectrum of bioprocesses that are represented in genetic interaction profiles can be visualized by mapping functional enrichment within the context of the global yeast digenic interaction profile similarity network, which clusters genes into 17 distinct bioprocesses (7, 27) (Fig. 4A). In comparison to the MDY2 and MTC1 digenic interaction profiles (Fig. 4B and C), the MDY2-MTC1 trigenic interactions were enriched not only for vesicle trafficking and cell polarity bioprocess regions of the network, but also in regions encompassing genes annotated to the t-RNA wobble modification bioprocess and DNA replication and repair/mitosis and chromosome segregation (Fig. 4D). Thus, the MDY2-MTC1 trigenic interaction profile exhibited a more expanded and functionally diverse set of connections than either of the corresponding MDY2 or MTC1 digenic interaction profiles.
Fig. 4. Enrichment of genetic interactions within bioprocesses defined by a global network of digenic interaction profile similarities.
(A) The global digenic interaction profile similarity network (7) was annotated using SAFE (55), identifying network regions enriched for similar GO biological process terms as outlined in dashed white lines. (B) MDY2 digenic interactions showing bioprocess enrichments are highlighted. (C) MTC1 digenic interactions showing bioprocess enrichments are highlighted. (D) MDY2-MTC1 trigenic interactions showing bioprocess enrichment are highlighted.
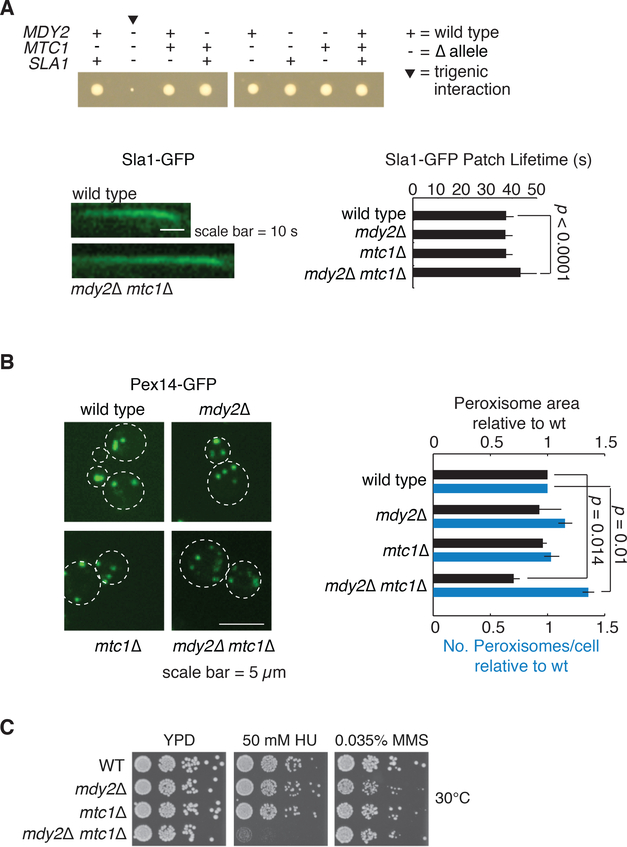
We utilized a variety of assays to test three functional connections revealed by the MDY2-MTC1 trigenic interaction profile. First, while the MDY2-MTC1 double mutant strain did not show an exaggerated cell biological phenotype associated with the early trafficking function (Fig. S9B), it displayed a dramatic synthetic sick phenotype when combined with deletion of SLA1, which is involved in cortical actin assembly and endocytic vesicle formation, which translates into an extended Sla1 patch lifetime, reflecting a defect in endocytosis (Fig. 5A) (28). Second, given a negative trigenic interaction with OAF1, which encodes an oleate-activated transcription factor involved in peroxisome organization and biogenesis (29), we used fluorescence microscopy to explore peroxisome morphology. The MDY2-MTC1 double mutant displayed an accumulation of relatively small peroxisomes (Fig. 5B), which may be indicative of a defect in ER-derived peroxisome membrane biogenesis (30). Third, the MDY2-MTC1 double mutant showed pronounced sensitivity to hydroxyurea (HU), but not methyl methanesulfonate (MMS), which is consistent with a specific defect in DNA replication, and reflects the negative genetic interactions we observed with a number of DNA replication and repair genes, including NSE4 and NSE5, which encode components of the Smc5-Smc6 complex that mediates resolution of DNA structures spanning sister chromatids (Fig. 5C, Fig. S10A-C) (31). We suspect that the MDY2-MTC1 double mutant may be primarily defective in trafficking functions that can modulate signaling or metabolic pathways and thereby influence DNA synthesis/repair pathways indirectly (Fig. S10D-H).
Fig. 5. Trigenic interactions reflect the physiology of MDY2-MTC1 double mutant query strain.
(A) Endocytic membrane trafficking is impaired in mdy2Δ mtc1Δ double mutant query strain. Example of tetrad analysis confirmations for the mdy2Δ mtc1Δ sla1Δ triple mutant strain. Endocytic uptake dynamics were examined with the Sla1-GFP reporter. Lifetime of Sla1-GFP endocytic vesicle formation was quantified across ~100 different patches in two independent experiments. Error bars represent s.d. Representative kymographs are displayed for wild-type and the mdy2Δ mtc1Δ double mutant. (B) Peroxisome biogenesis was monitored in wild-type, mdy2Δ, mtc1Δ, and mdy2Δ mtc1Δ mutants using Pex14p-GFP reporter. (C) Growth response to HU and MMS for wild-type, mdy2Δ, mtc1Δ, and mdy2Δ mtc1Δ mutants.
Trigenic interaction profiles are more functionally diverse than their corresponding digenic profiles.
To test the generality of whether query genes connect to more functionally divergent genes through trigenic interactions than digenic interactions, we compared digenic profile similarity of pairs of genes spanned by either digenic or trigenic interactions. Indeed, genes involved in trigenic interactions tend to show profiles that are less similar than those connected by digenic interactions, suggesting they are less functionally related than those connected by digenic interactions (Fig. 6A, S11A). We also found that trigenic interactions were more enriched than digenic interactions for connections that bridge several different biological processes, including mRNA and tRNA processing, vesicle trafficking, mitosis and chromosome segregation and glycosylation and protein folding/targeting (Fig. 6B). Moreover, as we showed for the MDY2-MTC1 double mutant query (Fig. 4), trigenic interaction profiles were generally enriched for genes spanning more diverse bioprocesses than the corresponding digenic interaction profiles (Fig. 6C, S11B-D). Genes involved in vesicle trafficking were particularly enriched for trigenic interactions occurring between bioprocesses (Fig 6B, S7, S11D). Interestingly, as we observed for MDY2-MTC1, other double mutant queries carrying mutations in genes implicated in membrane trafficking were enriched for trigenic interactions with genes involved in DNA replication and repair machinery, which may indicate a general connection between these two bioprocesses. For example, the digenic query strains MVP1-MRL1, which carry mutations in genes required for sorting proteins to the vacuole (32, 33), and SEC27-GET4, which carry mutations in genes involved in ER-to-Golgi transport (34) and the insertion of tailanchored proteins into ER membrane (20), both exhibited an enrichment of trigenic interactions with DNA replication and repair machinery (Fig. S11D). In general, our findings show that trigenic interaction profiles are composed of connections involving a set of more functionally diverse genes than their corresponding digenic interaction profiles (Fig. 6C). However, we note that, despite their higher tendency to connect diverse processes, a significant fraction of trigenic interactions occurs among genes within the same bioprocess (p < 1×10−16; hypergeometric test) (Fig. 2A).
Fig. 6. Trigenic interactions are more functionally distant than digenic interactions.
(A) Distribution of genetic interaction profile similarities of genes showing digenic and trigenic interactions. P-values are based on rank sum median test, all p < 10-34. (B) Frequency of negative genetic interactions between biological processes using SAFE annotations for digenic and trigenic interactions (55). The size of the circle assigned to each process-process element reflects the fold-increase over the background fraction of interactions (digenic = 0.023, trigenic = 0.016), p < 0.05 based on hypergeometric cumulative distribution test. The “between process” category received a count for any combinations that were not counted in the “within process” category shown in Fig. 2A. Significant enrichment is shown in filled blue circle, significant underenrichment in open blue circle, and no significant change is in grey. Trigenic vs. digenic fold change is the ratio of trigenic interaction enrichment to digenic interaction enrichment, and is shown as a filled square (black is maximal fold change, white is no fold change). In cases where the between-process enrichment was observed but is not significant at a p < 0.05, the square is outlined with a dashed line. (C) The number of SAFE bioprocess clusters enriched for digenic or trigenic interactions.
Gene features of trigenic interactions and the expanse of the global trigenic landscape
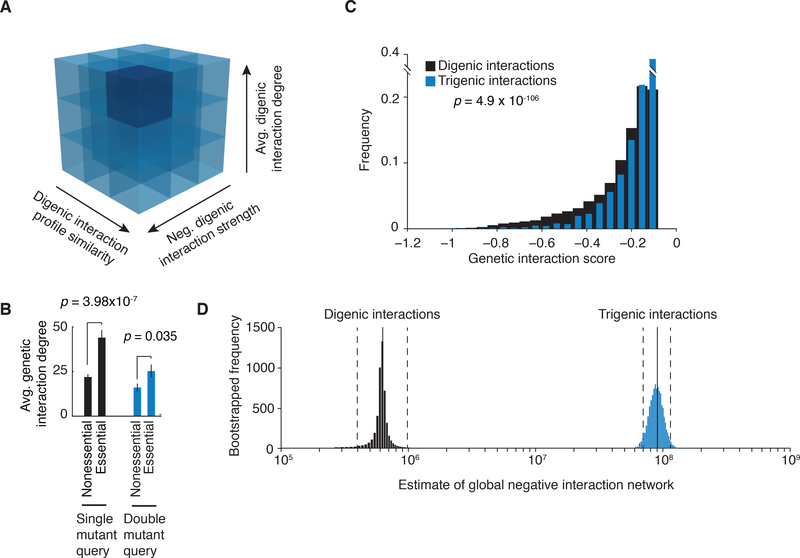
Having selected the query gene pairs based on the properties of the global digenic interaction network, we can assess how these properties relate to trigenic interaction frequency. The strongest correlation with the number of observed trigenic interactions for each gene pair was the digenic genetic interaction profile similarity (r = 0.41, p = 1.2×10−7), followed by the average number of digenic interactions of the query genes (r = 0.25, p = 1.9×10−3), and the strength of a direct negative genetic interaction between the query gene pair (r = 0.23, p = 5.4×10−3) (Fig. S12A-C). Thus, numerous trigenic interactions were observed for functionally related query genes, which display overlapping profiles on the digenic similarity network and often show a digenic interaction with each other (7) (Fig. 7A). As observed for digenic interactions (7), the frequency of trigenic interactions was highly correlated with the fitness defect of the double mutant query strain (Fig. S13). Consistent with this observation, essential genes exhibited high connectivity on the trigenic interaction network; a double mutant query that carries at least one temperature-sensitive allele of an essential gene, which is often associated with a fitness defect at the semi-permissive screening temperature, exhibited more genetic interactions than a query deleted for a pair of nonessential genes (p = 0.035) (Fig. 7B). More generally, query genes that are highly connected on the digenic network are also highly connected on the trigenic network (Fig. S12D).
Fig. 7. Relation of digenic and trigenic interaction networks.
(A) Trigenic interaction degree distribution correlated with three quantitative features of genes on the digenic interaction network: 1) Interaction profile similarity of the two genes in the double mutant query gene pair (bin thresholds: −0.02, 0.03, 0.1, +∞), which generates three bins for average digenic interaction profile similarity (r): −0.02 < r < 0.03; 0.03 ≤ r < 0.1; 0.1 ≤ r; 2) Negative digenic interaction strength associated with the double mutant query gene pair (bin thresholds: 0, −0.08, −0.1, −∞), which generates three bins for digenic interaction score (ε): ε < −0.1; −0.1 ≤ ε <−0.08; −0.08 ≤ ε < 0. Average digenic interaction degree, which represents the average number of negative genetic interactions associated with each of the genes of the double mutant query gene pair (bin thresholds: 10, 45, 70, +∞), which generates 3 bins for average digenic interaction degree: 10 ≤ degree < 45; 45 ≤ degree <70; 70 ≤ degree. The bin with the highest average negative trigenic interaction degree at the intermediate interaction score cut-off (τ < −0.08) of 63.5 is colored dark blue. (B) Essentiality determines trigenic interaction degree. No. single mutants: 254 nonessential genes, 47 essential genes. No. double mutants: 111 nonessential gene pairs, 40 essential or mixed essentiality gene pairs. Mean genetic interaction is shown, error bars: SEM, p values are based on a t-test. Negative genetic interactions, ε or τ < −0.08, p < 0.05, are depicted. (C) Cumulative distribution of negative digenic and trigenic interaction score magnitudes, pairwise significance was assessed using a Wilcoxon rank-sum test. (D) Estimates of the number of digenic and trigenic interactions at the intermediate score cut-off (ε or τ < −0.08, p < 0.05). Bootstrapping was used to generate the estimate by sampling 10,000 times with replacement, 95% C.I. is depicted by the dashed lines, the estimate of the extent of trigenic interaction landscape is denoted with a solid black line. This conservative estimate of the total number of trigenic interactions in the yeast genome covers approximately 26% of the interaction space, for the total genome-wide estimate see Fig. S15B, Table S3.
Interestingly, trigenic interactions tend to be ~25% weaker than digenic interactions (p < 1.71×10−98) (Fig. 7C), which means the average digenic interaction often has a more profound phenotype than the average trigenic interaction. However, to fully understand the potential for trigenic interactions to drive fitness defects, we also need to estimate the frequency at which they occur. Because we have mapped digenic interactions comprehensively and we know the false positive and false negative rates associated with this analysis, we can estimate the number of digenic interactions within the yeast genome, revealing a distribution that centers around ~6×105 total negative interactions (Fig. 7D) (16). Furthermore, because digenic interaction properties are predictive of trigenic interaction degree, we can also extrapolate our findings to estimate the number of negative trigenic interactions across the whole genome. As noted earlier, we selected gene pairs for trigenic analysis to fill bins of varying attributes, including double mutant queries with either weak or strong interactions, as well as those with either sparse or rich genetic interaction profiles, as depicted schematically in Fig. 1A and Fig. 7A (Table S1). For extrapolation to the whole genome, we used the mean trigenic interaction degree of double mutants in a given bin as the expected degree for any hypothetical pair from the genome with similar characteristics (Additional Data S7, Fig. S14). Integrating this value across the total number of gene pairs with the given characteristics, which preserves the double mutant distribution across different digenic interaction features, and summing over all bins yielded an estimate of the total number of trigenic interactions.
In the yeast genome, there are 2000-fold more possible triple gene combinations (36 billion) than possible gene pairs (18 million), but the density of interactions (as both observed and extrapolated) is similar, reduced by only ~3 fold for trigenic interactions (Table S3). We predict that approximately 108 trigenic combinations exhibit a negative genetic interaction, generating a conservative estimate of on the order of 100-fold more trigenic interactions than observed for the global digenic network (Fig. 7D, Table S3). To establish confidence intervals for the estimate, we repeated the extrapolation process with 10,000 boot-strapped samplings of the 151 double mutant query pairs, keeping their associated trigenic interactions degrees and the corresponding digenic interaction features constant. The bin with the lowest digenic interaction degree that encompasses a large fraction of the potential double mutants in the genome and is assigned a low trigenic interaction degree, which means the summarized estimate provided is likely a conservative underestimate. Moreover, because our binning scheme restricts our extrapolation to ~25% of the potential trigenic interaction space (e.g. by omitting potential double mutant queries that show a positive digenic interaction), we are underestimating its extent, and the true number of trigenic interactions is likely to be several fold higher (Fig. S15, Table S3) (16). The vast expanse of the global trigenic interaction network points to the potential for higher-order interactions to impact all aspects of the genetics of outbred populations, including the genotype to phenotype relationship.
Discussion
Systematic mapping of trigenic interactions revealed that their properties resemble digenic interactions because they often connect functionally related genes, which means they have the potential to contribute to our understanding of the functional wiring diagram of the cell. The global digenic network is predictive of trigenic interactions because query gene pairs showing properties associated with shared functionality, such as overlapping digenic interaction profiles, or a negative digenic genetic interaction, often map numerous trigenic interactions (Fig. 7A). Thus, if two query genes are in the same or similar bioprocess cluster on the global digenic profile similarity network (Fig. 4A), they will likely show a rich trigenic interaction profile, as we observed for the MDY2-MTC1 double mutant query (Fig. 3, 4D). Gene essentiality and the average digenic interaction degree of the query gene pair was also correlated with trigenic connectivity (Fig. 7B), indicating that highly connected hubs are consistent on both the digenic and trigenic interaction networks (Fig. S12C & D).
Many of the trigenic interactions we observed overlapped with at least one digenic interaction. In some cases, we chose query gene pairs displaying a negative genetic interaction and so all of the trigenic interactions in these profiles accentuated the query interaction (Fig. S8). Moreover, a substantial proportion of trigenic interactions measured for non-interacting query pairs exacerbated a digenic interaction that was previously seen between one or both of the query genes and the third gene (Fig. S8). Thus, our findings show that negative trigenic interactions often highlight the potential for a third mutation to amplify the phenotype of a digenic interaction. Analogously, in human genetics, the variation in an individual’s genetic background can have profound influence on the penetrance of the phenotype associated with a disease gene (35).
While we found that trigenic interactions tend to be slightly weaker than digenic interactions, they are ~100 times more numerous and are more functionally diverse than their digenic counterparts. The expanse of possible three gene combinations makes exhaustive trigenic interaction mapping intractable with our current methodology. However, the substantial overlap of the digenic and trigenic networks indicates that the genetic landscape of the cell expands with higher-order genetic interactions but does not change drastically in terms of its functional modularity. Thus, the global digenic network is highly informative of potential trigenic interactions and it can be used effectively to predict candidate query gene pairs for efficient trigenic interaction analysis.
Trigenic interaction data may inform a wide variety of subjects within biology. For example, the number, magnitude, and properties of digenic and trigenic interactions clarifies aspects of speciation theory in evolutionary biology (36, 37). Hybrids between species exhibit reduced fitness, which is usually attributed to negative (‘epistatic’) interactions among genes that diverged in isolated populations. Each population may evolve fixed variants that are neutral or adaptive in their own genetic backgrounds. When these variants are brought together for the first time in hybrid genomes, they may cause novel deleterious genetic interactions, also termed Dobzhansky-Muller incompatibilities. As populations diverge from one another, the number of potential digenic interactions increases as the square of the number of substitutions, the so-called snowball effect (36, 38, 39). That is, each subsequent substitution in a distinct population has the potential to interact with any substitution from the other population (and vice versa), and thus the probability of a speciation event grows with each step. Most speciation genetics research has focused on these digenic interactions. However, the number of trigenic combinations accumulates exponentially faster than digenic combinations. Both digenic and trigenic interactions have been implicated in speciation (40, 41), but the general extent to which digenic or complex negative genetic interactions drive speciation remains unknown. If digenic interactions do, in fact, play a major role in orchestrating speciation, then either the frequency and/or the strength of deleterious trigenic interactions must be relatively smaller than that of digenic interactions. Our systematic analysis shows that trigenic interactions are somewhat less likely (3-fold, ~3% vs ~1% for digenic vs trigenic, respectively) and generally weaker (~25% weaker) than digenic interactions. Nevertheless, modeling based upon our findings suggests that trigenic interactions are substantially common and often strong enough to play a key role in the evolution of hybrid inviability (Fig. S16, Table S4) (16). Because the connections associated with higher-order interactions may often overlap with those of simpler interactions, and because those simpler interactions require fewer substitutions and will often manifest first, our findings may also suggest that the evolution of even more highly complex interactions may be limited, even though their absolute numbers increase exponentially, a possibility that is consistent with evolutionary theory (38).
Our trigenic interaction study is also relevant to synthetic biology efforts aimed at efficient synthesis (42, 43) and design of minimal genomes (44, 45). Indeed, digenic synthetic lethal interactions were recently noted as a major constraint in the design of the minimal genome for the bacteria M. mycoides, for which a viable genome could only be constructed after resolving lethal interactions that arose between nonessential genes (44). For species in which systematic gene perturbation studies have been conducted, the proportion of essential genes is relatively small (e.g. ~20% in yeast, ~10% in human cells, which increases to 20% when only expressed genes are considered) (13, 46–48). However, we expect that digenic and trigenic interactions will dictate much larger minimal genomes than the essential gene set, even for growth under simple laboratory conditions. With the complete digenic network (7), we estimate the minimal genome would encompass greater than ~70% of genes after accounting for digenic interactions (Table S5) (16). With the inclusion of constraints imposed by trigenic interactions, we expect that a minimal viable genome, without a substantial fitness defect, may nearly approach the complete set of genes encoded in the genome. Thus, genetic interactions may help to explain the large gap between the number of genes with strong individual fitness defects and the total genome size, and the prevalence of yeast negative trigenic interactions suggests that many genomes may lack the potential for significant compression while maintaining normal fitness.
It is important to consider other types of genetic interactions, in addition to those associated with severe loss of function alleles due to entire ORF deletions of non-essential genes or temperature-sensitive alleles of essential genes. Our analysis revealed that double mutants with strong fitness defects often show rich trigenic interaction profiles (Fig. S13C). Similarly, single mutant fitness defect also correlates with digenic interaction degree (Fig S13A) (7). Presumably the weaker fitness effects associated with the variation found in natural populations may require higher-order combinations, involving more than two genes, to influence trait heritability through genetic interactions (49). In the yeast model system, genetic interactions were found to play a significant role in the heritability of a number of different quantitative traits, possibly with a greater contribution made by digenic interactions versus higher-order interactions (4, 5, 50). The genetic mechanism underlying conditional essentiality, in which a given yeast gene is nonessential in one genetic background but essential in another, often appears to be associated with a complex set of modifier loci (49), as do a number of other traits (51, 52). Thus, both digenic and higher-order interactions are established components of the genetic architecture of yeast complex traits and similar findings have been made in a number of other organisms (53). In part because model organism populations have allele distributions that are different than those in humans, it remains to be seen just how much higher-order genetic interactions will contribute to the genetics of complex human disease (50, 54). Nevertheless, the extensive landscape of trigenic interactions revealed here for yeast, as well as their capacity for generating functionally diverse phenotypes and driving speciation, suggests that higher-order genetic interactions may play a key role in the genotype to phenotype relationship.
Materials and Methods:
Materials and methods for construction of yeast single, double and triple mutant strains as well as quantification of genetic interactions and any associated analyses are described in detail in the Supplementary Materials. General methodological information and references to specific sections appear throughout the text.
Supplementary Material
Acknowledgments:
We thank Helena Friesen, Jing Hou, and Jason Moffat for insightful discussions. We also thank Myra Paz Masinas and Justin Nelson for logistical help.
Funding: This work was primarily supported by the National Institutes of Health (R01HG005853) (C.B., B.J.A., and C.L.M.), Canadian Institutes of Health Research (FDN-143264 and FDN-143265) (C.B. and B.J.A.), National Institutes of Health (R01HG005084 and R01GM104975) (C.L.M.) and the National Science Foundation (DBI\0953881) (C.L.M.). Computing resources and data storage services were partially provided by the Minnesota Supercomputing Institute and the UMN Office of Information Technology, respectively. Additional support was provided by Canadian Institutes of Health Research (MOP-79368) (G.W.B.), National Science Foundation (DEB-1456462) (D.B.), Swiss National Science Foundation (R.L.), Canton of Geneva and the European Research Council Consolidator Grant programme (R.L.), Natural Science and Engineering Research Council of Canada Postgraduate Scholarship-Doctoral PGS D2 (E.K.), University of Toronto Open Fellowship (E.K.), U of Minnesota Doctoral Dissertation Fellowship (B.V). C.L.M, B.J.A., and C.B are fellows of the Canadian Institute for Advanced Research (CIFAR).
Footnotes
Competing interests: All authors declare that they have no competing interests.
Data and materials availability: All data files (Additional Data S1 to S7) associated with this study are described in detail in the supplementary materials and can be downloaded from http://boonelab.ccbr.utoronto.ca/supplement/kuzmin2017/supplement.html. Data files S1 to S7 were also deposited in the DRYAD Digital Repository (http://datadryad.org/review?doi=doi:10.5061/dryad.tt367)
References and Notes:
- 1.Hartman J. L. t., Garvik B, Hartwell L, Principles for the buffering of genetic variation. Science 291, 1001–1004 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Genomes Project C et al. , A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zuk O, Hechter E, Sunyaev SR, Lander ES, The mystery of missing heritability: Genetic interactions create phantom heritability. Proceedings of the National Academy of Sciences 109, 1193–1198 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bloom JS, Ehrenreich IM, Loo WT, Lite TL, Kruglyak L, Finding the sources of missing heritability in a yeast cross. Nature 494, 234–237 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloom JS et al. , Genetic interactions contribute less than additive effects to quantitative trait variation in yeast. Nat Commun 6, 8712 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tong AH et al. , Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294, 2364–2368 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Costanzo M et al. , A global genetic interaction network maps a wiring diagram of cellular function. Science 353, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baryshnikova A et al. , Quantitative analysis of fitness and genetic interactions in yeast on a genome scale. Nat Methods 7, 1017–1024 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Novick P, Botstein D, Phenotypic analysis of temperature-sensitive yeast actin mutants. Cell 40, 405–416 (1985). [DOI] [PubMed] [Google Scholar]
- 10.Bender A, Pringle JR, Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol Cell Biol 11, 1295–1305 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Leeuwen J et al. , Exploring genetic suppression interactions on a global scale. Science 354, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winzeler EA et al. , Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906 (1999). [DOI] [PubMed] [Google Scholar]
- 13.Giaever G et al. , Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Tong AH et al. , Global mapping of the yeast genetic interaction network. Science 303, 808–813 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Costanzo M et al. , The genetic landscape of a cell. Science 327, 425–431 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Materials and methods are available as supporting materials on Science online.
- 17.Deshpande R et al. , Efficient strategies for screening large-scale genetic interaction networks. bioRxiv doi.org/10.1101/159632, (2017). [Google Scholar]
- 18.Haber JE et al. , Systematic triple-mutant analysis uncovers functional connectivity between pathways involved in chromosome regulation. Cell Rep 3, 2168–2178 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zou J et al. , Regulation of cell polarity through phosphorylation of Bni4 by Pho85 G1 cyclin-dependent kinases in Saccharomyces cerevisiae. Mol Biol Cell 20, 3239–3250 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jonikas MC et al. , Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science 323, 1693–1697 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noda Y, Yamagishi T, Yoda K, Specific membrane recruitment of Uso1 protein, the essential endoplasmic reticulum-to-Golgi tethering factor in yeast vesicular transport. J Cell Biochem 101, 686–694 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Gillingham AK, Tong AH, Boone C, Munro S, The GTPase Arf1p and the ER to Golgi cargo receptor Erv14p cooperate to recruit the golgin Rud3p to the cis-Golgi. J Cell Biol 167, 281–292 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzalez A, Hall MN, Nutrient sensing and TOR signaling in yeast and mammals. EMBO J 36, 397–408 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Zand A, Gent J, Braakman I, Tabak HF, Biochemically distinct vesicles from the endoplasmic reticulum fuse to form peroxisomes. Cell 149, 397–409 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Kira S et al. , Dynamic relocation of the TORC1-Gtr1/2-Ego1/2/3 complex is regulated by Gtr1 and Gtr2. Mol Biol Cell 27, 382–396 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peli-Gulli MP, Sardu A, Panchaud N, Raucci S, De Virgilio C, Amino Acids Stimulate TORC1 through Lst4-Lst7, a GTPase-Activating Protein Complex for the Rag Family GTPase Gtr2. Cell Rep 13, 1–7 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Usaj M et al. , TheCellMap.org: A Web-Accessible Database for Visualizing and Mining the Global Yeast Genetic Interaction Network. G3 (Bethesda) 7, 1539–1549 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun Y, Leong NT, Wong T, Drubin DG, A Pan1/End3/Sla1 complex links Arp2/3-mediated actin assembly to sites of clathrin-mediated endocytosis. Mol Biol Cell 26, 3841–3856 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karpichev IV, Small GM, Global regulatory functions of Oaf1p and Pip2p (Oaf2p), transcription factors that regulate genes encoding peroxisomal proteins in Saccharomyces cerevisiae. Mol Cell Biol 18, 6560–6570 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hettema EH, Erdmann R, van der Klei I, Veenhuis M, Evolving models for peroxisome biogenesis. Curr Opin Cell Biol 29, 25–30 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bustard DE et al. , During replication stress, non-SMC element 5 (NSE5) is required for Smc5/6 protein complex functionality at stalled forks. J Biol Chem 287, 11374–11383 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chi RJ et al. , Fission of SNX-BAR-coated endosomal retrograde transport carriers is promoted by the dynamin-related protein Vps1. J Cell Biol 204, 793–806 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whyte JR, Munro S, A yeast homolog of the mammalian mannose 6-phosphate receptors contributes to the sorting of vacuolar hydrolases. Curr Biol 11, 1074–1078 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Eugster A, Frigerio G, Dale M, Duden R, The alpha- and beta’-COP WD40 domains mediate cargoselective interactions with distinct di-lysine motifs. Mol Biol Cell 15, 1011–1023 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Argmann CA, Houten SM, Zhu J, Schadt EE, A Next Generation Multiscale View of Inborn Errors of Metabolism. Cell Metab 23, 13–26 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gavrilets S, Fitness Landscapes and the Origin of Species Levin SA, Horn Henry S., Ed., Monographs in Population Biology (Princeton University Press, Princeton and Oxford, 2004), pp. 149–194. [Google Scholar]
- 37.Foley BR, Rose CG, Rundle DE, Leong W, Edmands S, Postzygotic isolation involves strong mitochondrial and sex-specific effects in Tigriopus californicus, a species lacking heteromorphic sex chromosomes. Heredity (Edinb) 111, 391–401 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orr HA, The population genetics of speciation: the evolution of hybrid incompatibilities. Genetics 139, 1805–1813 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Welch JJ, Accumulating Dobzhansky-Muller incompatibilities: reconciling theory and data. Evolution 58, 1145–1156 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Moyle LC, Nakazato T, Hybrid incompatibility “snowballs” between Solanum species. Science 329, 1521–1523 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Tang S, Presgraves DC, Evolution of the Drosophila nuclear pore complex results in multiple hybrid incompatibilities. Science 323, 779–782 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richardson SM et al. , Design of a synthetic yeast genome. Science 355, 1040–1044 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Gibson DG et al. , One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. Proc Natl Acad Sci U S A 105, 20404–20409 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hutchison CA 3rd et al. , Design and synthesis of a minimal bacterial genome. Science 351, aad6253 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Gibson DG et al. , Creation of a bacterial cell controlled by a chemically synthesized genome. Science 329, 52–56 (2010). [DOI] [PubMed] [Google Scholar]
- 46.Wang T et al. , Identification and characterization of essential genes in the human genome. Science 350, 1096–1101 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hart T et al. , High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell 163, 1515–1526 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Blomen VA et al. , Gene essentiality and synthetic lethality in haploid human cells. Science 350, 1092–1096 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Dowell RD et al. , Genotype to phenotype: a complex problem. Science 328, 469 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Forsberg SK, Bloom JS, Sadhu MJ, Kruglyak L, Carlborg O, Accounting for genetic interactions improves modeling of individual quantitative trait phenotypes in yeast. Nat Genet 49, 497–503 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taylor MB, Ehrenreich IM, Genetic interactions involving five or more genes contribute to a complex trait in yeast. PLoS Genet 10, e1004324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taylor MB, Ehrenreich IM, Higher-order genetic interactions and their contribution to complex traits. Trends Genet 31, 34–40 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Phillips PC, Epistasis--the essential role of gene interactions in the structure and evolution of genetic systems. Nat Rev Genet 9, 855–867 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sackton TB, Hartl DL, Genotypic Context and Epistasis in Individuals and Populations. Cell 166, 279–287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baryshnikova A, Systematic Functional Annotation and Visualization of Biological Networks. Cell Syst, (2016). [DOI] [PubMed] [Google Scholar]
- 56.Gavin AC et al. , Proteome survey reveals modularity of the yeast cell machinery. Nature 440, 631–636 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Krogan NJ et al. , Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440, 637–643 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Tarassov K et al. , An in vivo map of the yeast protein interactome. Science 320, 1465–1470 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Yu H et al. , High-quality binary protein interaction map of the yeast interactome network. Science 322, 104–110 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Babu M et al. , Interaction landscape of membrane-protein complexes in Saccharomyces cerevisiae. Nature 489, 585–589 (2012). [DOI] [PubMed] [Google Scholar]
- 61.Huttenhower C, Hibbs M, Myers C, Troyanskaya OG, A scalable method for integration and functional analysis of multiple microarray datasets. Bioinformatics 22, 2890–2897 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Chong YT et al. , Yeast Proteome Dynamics from Single Cell Imaging and Automated Analysis. Cell 161, 1413–1424 (2015). [DOI] [PubMed] [Google Scholar]
- 63.Shannon P et al. , Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Z et al. , Systematic exploration of essential yeast gene function with temperature-sensitive mutants. Nat Biotechnol 29, 361–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C, A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res 21, 3329–3330 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gietz D, St Jean A, Woods RA, Schiestl RH, Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20, 1425 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kuzmin E, Costanzo M, Andrews B, Boone C, Synthetic Genetic Array Analysis. Cold Spring Harb Protoc 2016, pdb prot088807 (2016). [DOI] [PubMed] [Google Scholar]
- 68.Myers CL, Barrett DR, Hibbs MA, Huttenhower C, Troyanskaya OG, Finding function: evaluation methods for functional genomic data. BMC Genomics 7, 187 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kuzmin E et al. , Synthetic genetic array analysis for global mapping of genetic networks in yeast. Methods Mol Biol 1205, 143–168 (2014). [DOI] [PubMed] [Google Scholar]
- 70.Leung GP, Lee L, Schmidt TI, Shirahige K, Kobor MS, Rtt107 is required for recruitment of the SMC5/6 complex to DNA double strand breaks. J Biol Chem 286, 26250–26257 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bellay J et al. , Putting genetic interactions in context through a global modular decomposition. Genome Res 21, 1375–1387 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pellicioli A et al. , Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. The EMBO journal 18, 6561–6572 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weisman R, Cohen A, Gasser SM, TORC2-a new player in genome stability. EMBO Mol Med 6, 995–1002 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Winston F, Dollard C, Ricupero-Hovasse SL, Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11, 53–55 (1995). [DOI] [PubMed] [Google Scholar]
- 75.Addinall SG et al. , A genomewide suppressor and enhancer analysis of cdc13–1 reveals varied cellular processes influencing telomere capping in Saccharomyces cerevisiae. Genetics 180, 2251–2266 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Greenall A et al. , A genome wide analysis of the response to uncapped telomeres in budding yeast reveals a novel role for the NAD+ biosynthetic gene BNA2 in chromosome end protection. Genome Biol 9, R146 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.VanderSluis B et al. , Genetic interactions reveal the evolutionary trajectories of duplicate genes. Mol Syst Biol 6, 429 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bateson WRSE, Mendel’s Principles of Heredity. (Cambridge University Press, London, England, 1909). [Google Scholar]
- 79.Dobzhansky T, Genetic nature of species differences. American Naturalist 71, 404–420 (1937). [Google Scholar]
- 80.Muller HJ, Isolating mechanisms, evolution and temperature. Biological Symposia 6, 71–125 (1942). [Google Scholar]
- 81.Orr HA, Turelli M, The evolution of postzygotic isolation: accumulating Dobzhansky-Muller incompatibilities. Evolution 55, 1085–1094 (2001). [DOI] [PubMed] [Google Scholar]
- 82.Livingstone K et al. , A stochastic model for the development of Bateson-Dobzhansky-Muller incompatibilities that incorporates protein interaction networks. Math Biosci 238, 49–53 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.