Abstract
Pseudomonas aeruginosa can manipulate eukaryotic host cells using secreted effectors delivered by the type III or the type VI Secretion Systems (T3SS and T6SS). The T3SS allows the injection of bacterial effectors (Exo toxins) into eukaryotic cell. P. aeruginosa, encodes three T6SSs, H1-, H2- and H3-T6SS. The H1-T6SS is mainly involved in delivering toxins to kill bacterial competitors. Recently, two T6SS-secreted phospholipases D, PldA (H2-T6SS) and PldB (H3-T6SS), were identified as trans-kingdom virulence effectors, triggering both killing of bacterial competitors and internalization into non-phagocytic cells. We deciphered the prevalence of T3SS and T6SS effectors encoding genes in 185 clinical isolates responsible for infections (septicaemia, pulmonary infections, urinary tract infections, and chronic infections in CF patients), 47 environmental strains, and on 33 carbapenemase-producers. We included 107 complete genomes of P. aeruginosa available in public databases. The prevalence of pldA is increased in clinical isolates responsible for severe acute infection and particularly in multi-drug resistant strains. In contrast, the pldB prevalence was high (96.8%) in all isolates. Regarding T3SS effectors, exoT and exoY are present in nearly all isolates while exoS and exoU were found to be exclusive with a higher prevalence of exoU+ strains in severe acute infections. The hypervirulent exoU+ isolates are more prone to be pldA+, suggesting a role of PldA in virulence. Finally, we observed that extremely drug resistant isolates producing an IMP-type carbapenemase were all pldA+. Our results suggest that PldA might have a role during pulmonary infections and have been co-selected in multidrug resistant strains particularly IMP-producers.
Keywords: type III secretion system, type VI secretion system, PldA, PldB, Exo toxins, multidrug resistance, carbapenemase, IMP
Introduction
Pseudomonas aeruginosa is an extracellular Gram-negative bacterium responsible for more than 51,000 healthcare associated infections per year in the United States according to Centers for Disease Control and Prevention (2013). P. aeruginosa related-infections are mostly healthcare associated and cause a large subset of diseases including urinary tract infections (UTIs), acute pulmonary infections, wound infections, acute otitis and septicaemia (Stryjewski and Sexton, 2003). It is also a major cause of chronic infection in patients with cystic fibrosis (CF) (Lund-Palau et al., 2016). Indeed, this opportunistic pathogen is responsible of 85% chronic infections in the CF patient during adolescence and its propagation is involved in the progression of lung damage in CF patients (Davies, 2002).
Like many other Gram-negative bacteria, P. aeruginosa is able to manipulate eukaryotic host cells using its type III secretion system (T3SS) (Dortet et al., 2018) and more precisely secreted exoenzymes ExoS, ExoT, ExoY, and ExoU (Cornelis, 2006; Galle et al., 2012). More recently the type VI secretion system (T6SS) was shown to deliver bacterial toxins. The genome of P. aeruginosa encodes three T6SSs (H1-, H2-, and H3-T6SS) (Hachani et al., 2016). The H1-T6SS and its main effectors, Tse1-3, has been demonstrated to have a crucial role in bacterial warfare and thus possibly help to outcompete and manipulate the resident microbiota in the host (Cianfanelli et al., 2016). At the opposite, very little is known about H2- and H3-T6SS. Recently, a novel class of T6SS effectors that trigger both killing of bacterial competitors and internalization into non-phagocytic cells have been identified in P. aeruginosa (Russell et al., 2013). The T6SS-secreted phospholipases D (Pld), PldA and PldB, are thus the first example of trans-kingdom virulence effectors (Russell et al., 2013; Jiang et al., 2014). PldA and PldB have been proposed to be secreted via H2- and H3-T6SS, respectively (Russell et al., 2013; Jiang et al., 2014; Hachani et al., 2016). These two virulence factors are encoded in two distinct pld clusters that are not present in all P. aeruginosa isolates (Wilderman et al., 2001).
Pseudomonas aeruginosa is on the World Health Organization list of antibiotic resistant priority pathogens. Indeed, P. aeruginosa is intrinsically resistant to a number of antimicrobials due to the low permeability of its outer-membrane, the constitutive expression of various efflux pumps, and the production of β-lactamases (El Zowalaty et al., 2015). During the last decade, multidrug-resistant (MDR) isolates have been increasingly reported worldwide. The rise of multidrug resistance in P. aeruginosa is linked to the dissemination of extended-spectrum β-lactamases (ESBLs) and more recently of carbapenemase-producing isolates (Mesaros et al., 2007). Currently, the most widespread carbapenemases in Pseudomonas spp. are metallo-β-lactamases of VIM- (Verona imipenemase) and IMP- (Imipenemase) types (Gupta, 2008; Miriagou et al., 2010). Of note, VIM- and IMP-producers are usually resistant to all antimicrobial molecules except polymyxins and sometimes aztreonam, resulting in critical therapeutic issues in case of severe infections, such as septicaemia and pneumonia (Barbier et al., 2013). Thus, according to the CDC report, infections caused by these MDR P. aeruginosa represents 13% (6,700 / 51,000) of P. aeruginosa related infections and were responsible to 440 deaths per year in the United States in 2013 (Centers for Disease Control and Prevention, 2013).
In the context of the rapid emergence of pan-drug resistant P. aeruginosa isolates, the development of alternative ways to treat infected patients is crucial. Virulence inhibition strategies seem to interesting approaches. A prerequisite is, however, to establish among clinical isolates the prevalence of virulence factors encoding genes. Although the prevalence and the role of the T3SS effectors (ExoS, ExoT, ExoY, and ExoU) has been extensively described in clinical isolates, the prevalence of T6SS effectors, and particularly the trans-kingdom phospholipases PldA and PldB, remain less understood. Here, we determined the prevalence of genes encoding T3SS and T6SS effectors, including phospholipase PldA and PldB, in (i) P. aeruginosa isolates responsible for human infections (UTIs, acute pulmonary infection, septicaemia, CF-related chronic infections) compared to environmental strains, and (ii) in a collection of MDR P. aeruginosa isolates including the most relevant worldwide disseminated MDR clones [e.g., the VIM-2 producing clone of sequence type (ST) 11 and the IMP-13-producing clone of ST 621] (Santella et al., 2010).
Materials and Methods
Strain Collection
A collection of 185 P. aeruginosa isolates responsible for human infections were collected at the Bicêtre and Besançon hospital from 2016 to 2017. These infections included septicaemia (n = 46), UTIs (n = 32), pulmonary infections (n = 58) and chronic infections in CF patient (n = 49). The 58 pulmonary infections were acute infections occurring in patients deprived of any chronic pulmonary underlying diseases (e.g., CF patients, obstructive bronchopneumopathy). In addition, 47 environmental isolates from Besançon area were also tested. These environmental isolates were all recovered from natural setting without any link with hospital effluents. Finally, an additional collection of 33 well-characterized MDR P. aeruginosa isolates was also included in this study. These MDR isolates were all carbapenemase-producers including KPC-2 (n = 4); two GES-type (GES-2 and GES-5) producers; 10 VIM-producers (VIM-1, -2, and -4), 9 IMP-producers (IMP-1, -2, and -13), one NDM-1 producer; 3 GIM-1 producers; 3 AIM-1 producers and one SPM-1 producer.
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility was determined by disk diffusion on Mueller Hinton (MH) agar plates (Biorad, Marnes la Coquette, France) at 37°C. All results were interpreted using EUCAST breakpoint as updated in 20171.
Molecular Detection of T3SS and T6SS Effectors Encoding Genes
PCR assays were performed for detection of the exoU, exoS, exoT, and exoY (T3SS effectors encoding genes), tse1, tse2, and tse3 (H1-T6SS effectors encoding genes), pldA and pldB (H2- and H3-T6SS effectors encoding genes, respectively). PCR amplification of structural component encoding genes of T3SS (pscC) and H1-, H2-, H3-T6SS (tssB1, tssB2, and tssB3, respectively) were performed to confirm the presence or absence of these secretion machineries. DNA extracts were obtained by boiling method. Briefly, one bacterial colony was mixed in 100 μl of sterile water and boiled at 100°C for 10 min. Bacteria residues were pelleted by centrifugation at 13000 g for 5 min. The DNA extract was present in the supernatant. All primers used in this study are indicated in Table 1. The PCR mix was as follow: 2 μl of DNA extract obtained by boiling extract, 2.5 μl of reverse and forward primers, 5 μl of 10X PCR buffer, 1 μl of dNTP, 0.4 μl Taq DNA polymerase (5 U/μl; Roche) and 36.6 μl of water. The PCR protocol was as follow : 5 min initial denaturation at 95°C; 35 cycles including 30 s denaturation at 95°C, 1 min hybridization at 55°C and 1 min elongation at 72°C; and a final elongation step at 72°C for 10 min. Amplification PCR products were visualized using an UV transilluminator after electrophoresis in 2% agar gel for 40 min at 80 V.
Table 1.
List or primers used for the detection of T3SS and T6SS effectors.
| Secretion system | Gene | Primers |
|
|---|---|---|---|
| Name | Sequences | ||
| T3SS | pscC∗ | F pscC | 5′-ccaggcctgccttacgactat-3′ |
| R pscC | 5′-tcgtagtaccagcccgaggtt-3′ | ||
| exoS | R exoS | 5′-atgccggtgtagagaccaag-3′ | |
| F exoS | 5′-aggtcagcagagtatcggc-3′ | ||
| exoT | R exoT | 5′-tcgatcgatatctcgctgac-3′ | |
| F exoT | 5′-tgaacaggtcgtgaagacc-3′ | ||
| exoU | R exoU | 5′-acgtaacagagctacgttgg-3′ | |
| F exoU | 5′-ttgatcatcagcagttactcg-3′ | ||
| exoY | R exoY | 5′-tgccatagaatccgtcctcg-3′ | |
| F exoY | 5′-atgacctcttcctggtagcg-3′ | ||
| H1-T6SS | tssB1∗ | F tssB1 | 5′-gtgcagatcgagtacgacg-3′ |
| R tssB1 | 5′-tcttgccgtccatgtaggt-3′ | ||
| tse1 | R tse1 | 5′-agggagtagacgtagtag-3′ | |
| F tse1 | 5′-aagtacccgatgtgctggtg-3′ | ||
| tse2 | R tse2 | 5′-acgaagttgtccagttgg-3′ | |
| F tse2 | 5′-ttaaggtgaagcaggacagc-3′ | ||
| tse3 | R tse3 | 5′-ttgctgtacttggtgacgg-3′ | |
| F tse3 | 5′-atccagggtccgttcag-3′ | ||
| H2-T6SS | tssB2∗ | F tssB2 | 5′-atggccaaagaaggctcggt-3′ |
| R tssB2 | 5′-tggtgcttggccaggacctc-3′ | ||
| pldA | R pldA | 5′-tgtcgatcaccacggatttc-3′ | |
| F pldA | 5′-tatgacttcgaaaccatgctcg-3′ | ||
| H3-T6SS | tssB3∗ | F tssB3 | 5′-gtacgcagcacaagctggac-3′ |
| R tssB3 | 5′-ttgaccgggtcgaagtcctc-3′ | ||
| pldB | R pldb | 5′-ttcgatgttgttttcctggcc-3′ | |
| F pldb | 5′-aagaccacctacgacatgatg-3′ | ||
∗genes encoded a structural protein of the secretion system (T3SS or T6SS).
In silico Detection of T3SS and T6SS Effectors in Complete Genomes of P. aeruginosa
From the European Nucleotide Archive (ENA) 107 complete genome of P. aeruginosa were identified and analyzed. Among these 107 P. aeruginosa strains, 20 were carbapenemase-producers including 2 GES-20, 2 IMP-1, 2 IMP-34, 3 NDM-1, 1 DIM-1, 5 SPM-1, and 5 VIM-2. T3SS/T6SS effectors encoding genes were searched with the Basic local alignment search tool nucleotide (BLASTn) program on Pseudomonas genome website2 using P. aeruginosa PAO1 and UCBPP-PA14 as reference genomes.
Statistical Data Analysis
Statistical analysis was performed using GraphPad Prism v6 software. Data were compared using χ2 test. P-values < 0.05 were considered statistically different.
Results
Prevalence of Genes Encoding T3SS and T6SS Effectors in P. aeruginosa
All the 339 P. aeruginosa isolates (185 clinical isolates, 47 environmental isolates, and 107 complete genome of P. aeruginosa) were positive for genes encoding structural components of T3SS (pscC) and H1-, H2-, and H3-T6SS (tss1, tss2, tss3, respectively) except for two strains recovered from a CF patient, which gave negative results for tssB3 and one environmental strain that was negative for pscC gene.
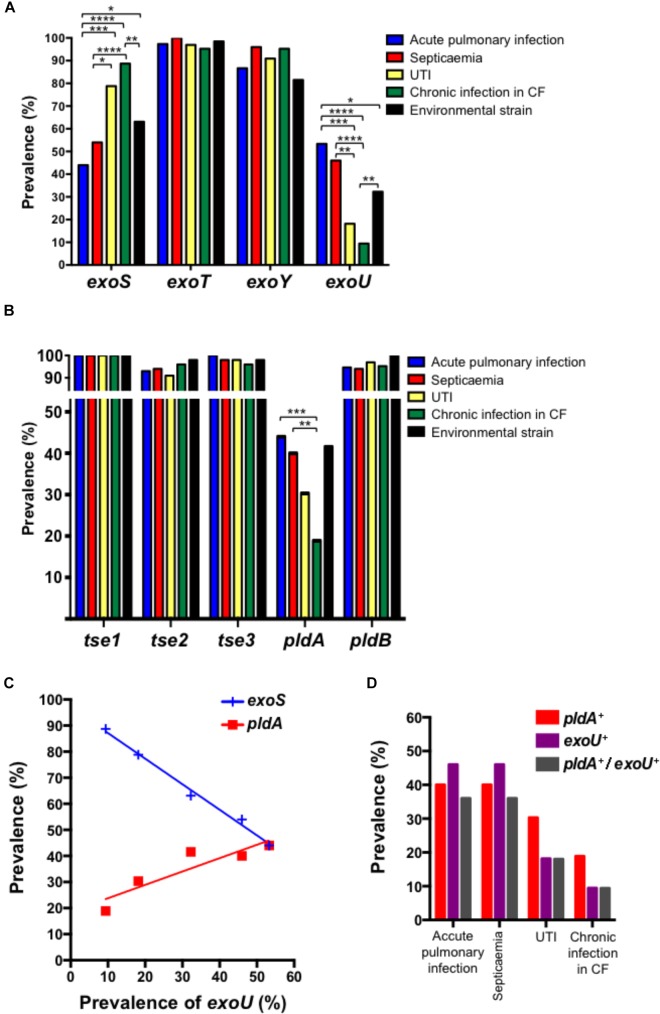
Among the 339 P. aeruginosa isolates the global prevalences of genes encoding T3SS-secreted exotoxins were 65.2% (221/339) for exoS, 98.5% (334/339) for exoT, 32.4% (110/339) for exoU, and 90.0% (305/339) for exoY. Regardless of the biological sample (urine, pulmonary sample, blood, and environmental samples) from which the strain was cultured, the prevalences of exoT and exoY were high, ranging from 95.2 to 100% for exoT and 86.2 to 95.9% for exoY (Figure 1A). As previously observed, we identified an inverse correlation between the presence of exoS and exoU in P. aeruginosa isolates (Figure 1C; Feltman et al., 2001). Of note, the prevalence of exoU was higher in invasive acute infections (53.3% in acute pulmonary, infection from non-CF patients and 46.0% in septicaemia) compared to environmental strain (32.3%), UTIs (18.2%) and chronic infection in CF patients (5.4%).
FIGURE 1.
Prevalence of T3SS effectors (A) and T6SS effectors (B) in Pseudomonas aeruginosa isolates recovered from acute pulmonary infections, septicaemia, UTIs, chronic infections in CF patients and environment. Correlation between the prevalence of exoS, exoU, and pldA is indicated in (C). Data were compared two-by-two using χ2 test (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; and ∗∗∗∗p < 0.0001). (D) Prevalence of pldA, exoU and pldA+ exoU in Pseudomonas aeruginosa isolates recovered from accute pulmonary infections, septicaemia, UTIs and chronic infections in CF patients.
Regarding T6SS effectors, H1-T6SS effectors were highly prevalent in all P. aeruginosa isolates. Indeed, we observed prevalences of 100%, 91–98%, and 96–100% for tse1, tse2, and tse3, respectively (Figure 1B). This prevalence did not depend on the sample from which the strain was isolated. The same observation was made for pldB (H3-TSS6 effector) with a global prevalence of 96.8% (328/339), ranging from 94% in P. aeruginosa isolates responsible for acute pulmonary infection in non-CF patient to 100% in environmental strains. The prevalence of pldA was higher in strains responsible for acute invasive infection (acute pulmonary infection and septicaemia) as compared to UTI, environmental strain and strains isolated from chronic infection in CF patients (Figure 1B). Accordingly, we observed a correlation between pldA and exoU and consequently an inverse correlation with exoS (Figure 1C). Despite pldA+ and exoU+ correlation was found, the mutual exclusivity which was observed between exoS+ and exoU+ do not exist between pldA+ and exoU+ (Figure 1D).
Increased Prevalence of pldA and exoU With Antimicrobial Resistance in Clinical Isolates
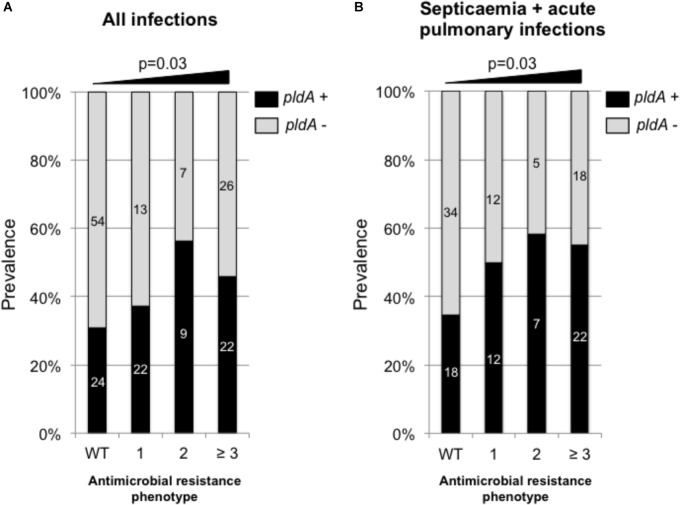
Using our collection of 185 P. aeruginosa isolates responsible for infections we found that the pldA negative isolates were significantly more susceptible to antimicrobial than pldA positive isolates with a percentage of wild-type phenotype of 49.3% for pldA negative strain versus 30.5% for pldA positive isolates (χ2 test, p = 0.03). Of note, the tendency of being more resistant for pldA+ positive isolates was significant (χ2 test, p = 0.03) for all type of infections including acute infections (UTIs, pulmonary infections and septicaemia) and chronic infections in CF patients (Figure 2A). However, this tendency is exacerbated in acute infection only (Figure 2B). Since exoU+ and pldA+ were found to be correlated (Figure 1C), the same type of correlation between exoU+ and antimicrobial resistance was observed. Indeed, the percentage of exoU+ increased from 24.6% in isolates with wild-type phenotype to 43.6% on isolates harboring resistance to at least 3 antimicrobial families (data not shown).
FIGURE 2.
Prevalence of pldA in P. aeruginosa responsible for all type of infections (acute and chronic) (A) and acute infections (B) depending on the antimicrobial resistance phenotype. WT, wild-type phenotype; 1, resistance to one antimicrobial family, 2, resistance to two antimicrobial families; ≥3, resistance to three and more than three antimicrobial families. The antimicrobial families considered in the analysis were: penicillins (ticarcillin and piperacillin), broad-spectrum cephalosporins (ceftazidime and cefepime), carbapenems (imipenem and meropenem), fluroroquinolones (ciprofloxacine) and aminoglycosides (amikacin, tobramycin, and gentamicin). Raw numbers are indicated in side the bars. Statistical analysis was performed using χ2 test for trend.
To validate this hypothesis of increased prevalence of pldA positive P. aeruginosa isolates with the antimicrobial resistance, we tested our collection of carbapenemase-producing strains. In this collection of 33 extremely drug resistant isolates, the prevalence of pldA positive strain was maximum as compared to the other phenotypes with a global prevalence at 56.8%. Again, the prevalence of exoU+ strains followed the same overall tendency with a global prevalence of 41.8% in this extremely drug resistant isolates, close to the 43.6% observed previously with isolates harboring resistance to at least 3 antimicrobial families.
IMP-Type Carbapenemase Production Is Associated With pldA Positivity
Of note, in our collection of 33 extremely drug resistant P. aeruginosa, all isolates that produce a carbapenemase of IMP-type (4 IMP-1, 1 IMP-2, and 4 IMP-13) were pldA positive whereas the prevalence of pldA was of 41.6% with other carbapenemase types (VIM, NDM, GES, SPM, and AIM). Among these IMP-producers, all IMP-13-producing isolates were of the same ST, the worldwide disseminated ST 621 (Santella et al., 2010). The other isolates were of ST 244-like and 304. More systematically, we looked for pldA gene in complete genomes of IMP-producing P. aeruginosa available in NCBI. Among the 107 complete genomes of P. aeruginosa, we identified four additional isolates producing IMP-1 (one ST-2613 and one ST-357) and two IMP-34 (both ST-2613). This analysis confirmed the systematic association between pldA presence and IMP-type carbapenemase production.
As opposed to pldA, exoU positivity was not associated with a specific carbapenemase type. Indeed, the prevalence of exoU+ was not significantly different in IMP-producers (30%) and in other carbapenemase types producers (29.4%).
Discussion
Regarding of P. aeruginosa virulence factors during infection, the implication of T3SS-effectors has been deeply demonstrated (Feltman et al., 2001; Garey et al., 2008; Hauser, 2009). Our results regarding the prevalence of Exo toxins encoding genes are in accordance with previous studies that found prevalence of 58–78% for exoS, 92% for exoT, 89% for exoY and 28–48% for exoY in P. aeruginosa clinical isolates (Hauser, 2009). An inverse correlation between the presence of exoS and exoU in P. aeruginosa isolates has been previously demonstrated (Feltman et al., 2001). Our results confirm this inverse correlation (Figure 1). Of note exoU+ isolates are also considered to be more virulent (Schulert et al., 2003) and more often associated with severe acute infections, such as septicaemia (Wareham and Curtis, 2007) and severe acute pulmonary infections (Sullivan et al., 2014). Our results confirm the higher prevalence of exoU+ P. aeruginosa isolates in acute pneumonia and septicaemia. In addition, we also identified a correlation between pldA and exoU expression (Figure 1). Together, our results confim those of a very recent study which reports that the presence of pldA and exoU in mucoid P. aeruginosa is associated with high risk of exacerbations in non-CF patients (Luo et al., 2018). Since it has been demonstrated that PldA is a trans-kingdom T6SS effector that allows P. aeruginosa to interact with epithelial cells (Jiang et al., 2014; Hachani et al., 2016), we might hypothesize that the worst outcome observed with exoU+ isolates might also depend on the expression of pldA, at least partially. Interestingly, it has been recently described that the expression of T6SS encoding genes is up-regulated in presence of respiratory mucus (Cattoir et al., 2013), suggesting a potential role of T6SS during the infectious process. Unfortunately, the studied P. aeruginosa isolate was pldA-, and only the H1-T6SS genes expression was reported. Since we showed that pldA is more prevalent in isolates responsible for acute pulmonary infection, septicaemia and in a lesser extend UTIs, further studies are necessary to unravel whether H2-T6SS genes including pldA are also overexpressed in presence of relevant biological fluids (respiratory mucus, blood, and urine). In addition, one of the limitations of our study is the lack of information regarding the clinical outcomes of patients. Indeed, these data might be useful to identify any relevant clinical impact of PldA during infectious process. However, the suggested role of pldA in pulmonary infection is in agreement with the results of Wilderman et al. (2001). Indeed, they first identified that not all strains of P. aeruginosa carry pldA thus suppose to be part of a ∼7 kb mobile genetic element. In this study, pldA was found to be most prevalent in non-CF patients than in CF patients. Unfortunately, no data were presented regarding the type of infections in non-CF patients. Then, using competition studies between PldA knock-out mutant and its PAO1 parental strain, they demonstrated the contribution of PldA during chronic pulmonary infection in a rat model.
Unlike pldA results, no difference in the prevalence of pldB, another trans-kingdom T6SS that was demonstrated to interact with the host cells (Russell et al., 2013). Indeed, we found that pldB is barely present in all P. aeruginosa isolates. This finding is not in favor of a specific role of this phospholipase during certain types of infection.
Finally, as it was previously observed for exoU (Garey et al., 2008; Mitov et al., 2010), we found that pldA+ isolates were significantly more resistant than pldA- isolates. We demonstated that pldA prevalence was maximum (∼60%) in extremely resistant P. aeruginosa isolates that produce a carbapenemase. Interestingly, among carbapenemase-producers, all IMP-type-producing strains were pldA+. Further studies are now needed to explore this close association between pldA and blaIMP genes, particularly in the context of the worldwide dissemination high-risk clone such as IMP-13-producing P. aeruginosa of ST621 that are responsible for outbreaks of UTIs (Pagani et al., 2005; Santella et al., 2010).
To our knowledge, this is the first study that evaluated the prevalence of genes encoding a large subset of T6SS effectors, including the more recently described pldA and pldB genes, in clinical isolates of P. aeruginosa. We found that pldA, but not pldB, prevalence is increased in clinical isolates responsible for severe acute infection and particularly in multi-drug resistant isolates. In addition the hypervirulent exoU+ isolates are more prone to be pldA+, thus suggesting PldA might also play a role in this increased virulence. Finally, we showed that extremely drug resistant P. aeruginosa isolates that produce an IMP-type carbapenemase were all pldA+, an unexpected relationship that needs to be further explored.
Ethics Statement
Written informed consent was not required as the samples were not recovered specifically for the study and no information regarding the patients were included in this study.
Author Contributions
LD and TB had full access to all of the data in the study, and took responsibility for the integrity of the data and the accuracy of the data analysis. LD contributed to the study of concept and design. LD, TB, and Y-MB acquired, analyzed, or interpreted the data. LD, TB, and TN drafted the manuscript. LD, PP, AF, and TN contributed to critical revision of the manuscript for important intellectual content.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by People Program (Marie Skłodowska-Curie Actions) of the European Union’s Horizon 2020 (REA Grant Agreement No. 654909) and from the University Paris-Sud, France. LD and TN are members of the Laboratory of Excellence in Research on Medication and Innovative Therapeutics (LERMIT) supported by a grant from the French National Research Agency (ANR-10-LABX-33).
References
- Barbier F., Andremont A., Wolff M., Bouadma L. (2013). Hospital-acquired pneumonia and ventilator-associated pneumonia: recent advances in epidemiology and management. Curr. Opin. Pulm. Med. 19 216–228. 10.1097/MCP.0b013e32835f27be [DOI] [PubMed] [Google Scholar]
- Cattoir V., Narasimhan G., Skurnik D., Aschard H., Roux D., Ramphal R., et al. (2013). Transcriptional response of mucoid Pseudomonas aeruginosa to human respiratory mucus. MBio 3:e00410-12. 10.1128/mBio.00410-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (2013). Antibiotic Resistance Threats in The United States, 2013. Available at: https://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf [Google Scholar]
- Cianfanelli F. R., Monlezun L., Coulthurst S. J. (2016). Aim, load, fire: the type VI secretion system, a bacterial nanoweapon. Trends Microbiol. 24 51–62. 10.1016/j.tim.2015.10.005 [DOI] [PubMed] [Google Scholar]
- Cornelis G. R. (2006). The type III secretion injectisome. Nat. Rev. Microbiol. 4 811–825. 10.1038/nrmicro1526 [DOI] [PubMed] [Google Scholar]
- Davies J. C. (2002). Pseudomonas aeruginosa in cystic fibrosis: pathogenesis and persistence. Paediatr. Respir. Rev. 3 128–134. 10.1016/S1526-0550(02)00003-3 [DOI] [PubMed] [Google Scholar]
- Dortet L., Lombardi C., Cretin F., Dessen A., Filloux A. (2018). Pore-forming activity of the Pseudomonas aeruginosa type III secretion system translocon alters the host epigenome. Nat. Microbiol. 3 378–386. 10.1038/s41564-018-0109-7 [DOI] [PubMed] [Google Scholar]
- El Zowalaty M. E., Al Thani A. A., Webster T. J., El Zowalaty A. E., Schweizer H. P., Nasrallah G. K., et al. (2015). Pseudomonas aeruginosa: arsenal of resistance mechanisms, decades of changing resistance profiles, and future antimicrobial therapies. Future Microbiol. 10 1683–1706. 10.2217/fmb.15.48 [DOI] [PubMed] [Google Scholar]
- Feltman H., Schulert G., Khan S., Jain M., Peterson L., Hauser A. R. (2001). Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Microbiology 147(Pt 10), 2659–2669. 10.1099/00221287-147-10-2659 [DOI] [PubMed] [Google Scholar]
- Galle M., Carpentier I., Beyaert R. (2012). Structure and function of the Type III secretion system of Pseudomonas aeruginosa. Curr. Protein Pept. Sci. 13 831–842. 10.2174/138920312804871210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garey K. W., Vo Q. P., Larocco M. T., Gentry L. O., Tam V. H. (2008). Prevalence of type III secretion protein exoenzymes and antimicrobial susceptibility patterns from bloodstream isolates of patients with Pseudomonas aeruginosa bacteremia. J. Chemother. 20 714–720. 10.1179/joc.2008.20.6.714 [DOI] [PubMed] [Google Scholar]
- Gupta V. (2008). Metallo-beta-lactamases in Pseudomonas aeruginosa and acinetobacter species. Expert Opin. Investig. Drugs 17 131–143. 10.1517/13543784.17.2.131 [DOI] [PubMed] [Google Scholar]
- Hachani A., Wood T. E., Filloux A. (2016). Type VI secretion and anti-host effectors. Curr. Opin. Microbiol. 29 81–93. 10.1016/j.mib.2015.11.006 [DOI] [PubMed] [Google Scholar]
- Hauser A. R. (2009). The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat. Rev. Microbiol. 7 654–665. 10.1038/nrmicro2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F., Waterfield N. R., Yang J., Yang G., Jin Q. (2014). A Pseudomonas aeruginosa type VI secretion phospholipase D effector targets both prokaryotic and eukaryotic cells. Cell Host Microbe 15 600–610. 10.1016/j.chom.2014.04.010 [DOI] [PubMed] [Google Scholar]
- Lund-Palau H., Turnbull A. R., Bush A., Bardin E., Cameron L., Soren O., et al. (2016). Pseudomonas aeruginosa infection in cystic fibrosis: pathophysiological mechanisms and therapeutic approaches. Expert Rev. Respir. Med. 10 685–697. 10.1080/17476348.2016.1177460 [DOI] [PubMed] [Google Scholar]
- Luo R. G., Miao X. Y., Luo L. L., Mao B., Yu F. Y., Xu J. F. (2018). Presence of pldA and exoU in mucoid Pseudomonas aeruginosa is associated with high risk of exacerbations in non-cystic fibrosis bronchiectasis patients. Clin. Microbiol. Infect. 10.1016/j.cmi.2018.07.008 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Mesaros N., Nordmann P., Plesiat P., Roussel-Delvallez M., Van Eldere J., Glupczynski Y., et al. (2007). Pseudomonas aeruginosa: resistance and therapeutic options at the turn of the new millennium. Clin. Microbiol. Infect. 13 560–578. 10.1111/j.1469-0691.2007.01681.x [DOI] [PubMed] [Google Scholar]
- Miriagou V., Cornaglia G., Edelstein M., Galani I., Giske C. G., Gniadkowski M., et al. (2010). Acquired carbapenemases in Gram-negative bacterial pathogens: detection and surveillance issues. Clin. Microbiol. Infect. 16 112–122. 10.1111/j.1469-0691.2009.03116.x [DOI] [PubMed] [Google Scholar]
- Mitov I., Strateva T., Markova B. (2010). Prevalence of virulence genes among bulgarian nosocomial and cystic fibrosis isolates of Pseudomonas aeruginosa. Braz. J. Microbiol. 41 588–595. 10.1590/S1517-83822010000300008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani L., Colinon C., Migliavacca R., Labonia M., Docquier J. D., Nucleo E., et al. (2005). Nosocomial outbreak caused by multidrug-resistant Pseudomonas aeruginosa producing IMP-13 metallo-beta-lactamase. J. Clin. Microbiol. 43 3824–3828. 10.1128/JCM.43.8.3824-3828.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell A. B., LeRoux M., Hathazi K., Agnello D. M., Ishikawa T., Wiggins P. A., et al. (2013). Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature 496 508–512. 10.1038/nature12074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santella G., Pollini S., Docquier J. D., Mereuta A. I., Gutkind G., Rossolini G. M., et al. (2010). Intercontinental dissemination of IMP-13-producing Pseudomonas aeruginosa belonging in sequence type 621. J. Clin. Microbiol. 48 4342–4343. 10.1128/JCM.00614-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulert G. S., Feltman H., Rabin S. D., Martin C. G., Battle S. E., Rello J., et al. (2003). Secretion of the toxin ExoU is a marker for highly virulent Pseudomonas aeruginosa isolates obtained from patients with hospital-acquired pneumonia. J. Infect. Dis. 188 1695–1706. 10.1086/379372 [DOI] [PubMed] [Google Scholar]
- Stryjewski M., Sexton D. (2003). Severe Infections Caused by Pseudomonas Aeruginosa Perspectives on Critical Care Infectious Diseases Berlin: Springer. [Google Scholar]
- Sullivan E., Bensman J., Lou M., Agnello M., Shriner K., Wong-Beringer A. (2014). Risk of developing pneumonia is enhanced by the combined traits of fluoroquinolone resistance and type III secretion virulence in respiratory isolates of Pseudomonas aeruginosa. Crit. Care Med. 42 48–56. 10.1097/CCM.0b013e318298a86f [DOI] [PubMed] [Google Scholar]
- Wareham D. W., Curtis M. A. (2007). A genotypic and phenotypic comparison of type III secretion profiles of Pseudomonas aeruginosa cystic fibrosis and bacteremia isolates. Int. J. Med. Microbiol. 297 227–234. 10.1016/j.ijmm.2007.02.004 [DOI] [PubMed] [Google Scholar]
- Wilderman P. J., Vasil A. I., Johnson Z., Vasil M. L. (2001). Genetic and biochemical analyses of a eukaryotic-like phospholipase D of Pseudomonas aeruginosa suggest horizontal acquisition and a role for persistence in a chronic pulmonary infection model. Mol. Microbiol. 39 291–303. 10.1046/j.1365-2958.2001.02282.x [DOI] [PubMed] [Google Scholar]