Abstract
The protozoan Entamoeba gingivalis resides in the oral cavity and is frequently observed in the periodontal pockets of humans and pets. This species of Entamoeba is closely related to the human pathogen Entamoeba histolytica, the agent of amoebiasis. Although E. gingivalis is highly enriched in people with periodontitis (a disease in which inflammation and bone loss correlate with changes in the microbial flora), the potential role of this protozoan in oral infectious diseases is not known. Periodontitis affects half the adult population in the world, eventually leads to edentulism, and has been linked to other pathologies, like diabetes and cardiovascular diseases. As aging is a risk factor for the disorder, it is considered an inevitable physiological process, even though it can be prevented and cured. However, the impact of periodontitis on the patient's health and quality of life, as well as its economic burden, are underestimated. Commonly accepted models explain the progression from health to gingivitis and then periodontitis by a gradual change in the identity and proportion of bacterial microorganisms in the gingival crevices. Though not pathognomonic, inflammation is always present in periodontitis. The recruitment of leukocytes to inflamed gums and their passage to the periodontal pocket lumen are speculated to fuel both tissue destruction and the development of the flora. The individual contribution to the disease of each bacterial species is difficult to establish and the eventual role of protozoa in the fate of this disease has been ignored. Following recent scientific findings, we discuss the relevance of these data and propose that the status of E. gingivalis be reconsidered as a potential pathogen contributing to periodontitis.
Keywords: Entamoeba gingivalis, periodontitis, gingivitis, inflammation, parasitic infection, infectious disease
Introduction: a disease with underestimated impact
Periodontitis is a disease leading to alveolar bone destruction and eventually tooth loss. The prevalence of periodontitis is constant among the defined World Health Organization (WHO) regions, with around one person out of two between 35 and 44 years old (Petersen and Ogawa, 2005). This prevalence increases with age (Demmer and Papapanou, 2010). In the USA, between 2009 and 2012, 46% of adults aged 30 years or more suffer from periodontitis (Eke et al., 2015).
Periodontitis is a handicapping disease, for which the WHO calculated the loss of 3,518,002 DALYs (Disability-adjusted life years, a measure of disease burden as the loss of healthy life years) in 2015 in the world, meaning 0.132% of the worldwide DALYs (Organization, 2016), though the associated disability weight is low (0.007), reflecting only “minor bleeding of the gums from time to time, with mild discomfort” (Evaluation, 2016). Though periodontitis is linked to systemic diseases like diabetes (Nascimento et al., 2018) and ischemic stroke (Leira et al., 2017), the etiological link is difficult to demonstrate and the possible impact of periodontitis on other ailments is ignored for the calculation of DALYs.
The etiology of periodontitis is still unclear and it is classified by the WHO as a non-communicable disease. Some human genetic factors linked to periodontitis were demonstrated (Vieira and Albandar, 2014) and are still investigated to explain its prevalence in the global population, in conjunction with age (Demmer and Papapanou, 2010). Modifiable risk factors for the disease were also sought and some parameters have been identified: smoking (Eke et al., 2016), alcohol consumption (Wang et al., 2016), and poor oral hygiene (Lertpimonchai et al., 2017). Beyond pain from wounds, eventual edentulism, and defects in occlusion, the patients experience halitosis (Silva et al., 2017) and esthetic issues (Nieri et al., 2013). Altogether, these factors may account for their psychological and social distress (Lopez et al., 2012; Hsu et al., 2015; Dumitrescu, 2016; Reynolds and Duane, 2018).
Evolution toward gum disease goes through three stages (i) formation of dental plaque; (ii) gingivitis, which is an inflammation of the gums due to the dental plaque, and (iii) periodontitis, in which alveolar bone and fibers that hold the teeth in place are irreversibly damaged. The pathophysiology of the disease is harshly debated, but a consensus was reached about some key points. First, inflammation is compulsory and prior to bone loss, evidenced by pocket formation [reviewed in Van Dyke (2017)]. Second, the microbial flora in periodontal pockets differs from that observed in healthy sulci (Marsh, 1994). Last, plaque and calculi worsen prognosis (Löe et al., 1965). Consensual treatment in clinics is thus based on the mechanical and/or surgical removal of dental plaque, calculi, and damaged/inflamed tissues (Smiley et al., 2015). These paths lead to an inefficient solution dealing with late symptoms without considering the evoked causes of the disease. The keystone for the improvement of periodontitis management worldwide is a better knowledge of its pathophysiology.
Due to the important correlation of periodontitis with the presence of Entamoeba gingivalis in the oral cavity, here, we searched for the facts that can shed light on the question of whether E. gingivalis plays a role in the occurrence of the periodontitis. In this review, we summarize existing data on the biology of the amoeba Entamoeba gingivalis and on its potential role as an infectious agent in periodontitis. We aim at highlighting perspectives for new research on the pathophysiology and prophylaxis of this neglected disease.
Microbiology of periodontitis: the bacterial paradigm
Though the saliva contains low nutrient concentrations and antimicrobial defense systems [reviewed in van 'T Hof et al. (2014)], the healthy oral cavity houses a commensal microbiota, composed of bacterial communities [about 1,000 species across humans, Consortium (2012)], whereas the contribution of viruses, parasites, archaea, and fungi is still to be characterized. Microorganisms and oral mucosae maintain a mutualistic, resilient symbiotic relationship (Rosier et al., 2018). The bacterial ecosystem of healthy sulci is intriguingly similar between individuals and it comprises immotile bacilli and cocci, as seen in microscopy (Listgarten, 1976), with bacterial species differing from those encountered on the tongue (Aas et al., 2005; Consortium, 2012; Rogers and Bruce, 2012). At the tooth surface, in particular in the dental sulcus, nutrients coming from food and cellular debris accumulate and support the survival of bacteria that adhere and colonize the dental enamel. Bacterial flagella, pili, and wall proteins can recruit other bacteria, by co-aggregation (Kolenbrander and Celesk, 1983; Gibbons et al., 1988). Furthermore, the secretion of polysaccharides initiates the formation and organization of a scaffold (Jakubovics, 2010), while intercellular signaling molecules regulate biofilm development, in particular through a quorum sensing mechanism mediated by different types of messengers, as cyclic di-guanosine monophosphate or LuxS [reviewed in Marsh et al. (2011)]. This intra- and inter-species communication leads to coordination of activities and increases the chances of genetic material transfer. The resulting dental plaque is an organized biofilm, whose formation is not pathologic (Gibbons and Van Houte, 1973), though it was thought to be responsible for gingivitis and periodontitis (Schultz-Haudt et al., 1954).
Some bacteria are associated with periodontitis and this led to the proposal of a specific plaque explanation for the disease (Loesche, 1979). These bacteria group in clusters associated with disease progression (Socransky et al., 1998), reflecting the sequential colonization of the periodontal sulcus and pocket (Li et al., 2004; Feres et al., 2016). The “periodontopathogenic” red complex is comprised of anaerobic bacteria (Aggregatibacter actinomycetemcomitans, Tannerella forsythia, and Porphyromonas gingivalis), supporting the hypothesis of a gradual modification of the environment prior to and necessary for colonization by other bacterium types (Marsh, 2003; Darveau, 2010). However, P. gingivalis is present in some healthy patients (Socransky et al., 1998) and is not abundant, even in periodontitis (Moore et al., 1982), while this “keystone pathogen” provokes environmental changes in the sulcus promoting inflammation (Hajishengallis et al., 2011). Thus, P. gingivalis cannot be considered an etiological agent for periodontitis by itself, at least with respect to Koch's postulates.
Koch's postulates are the extreme case of Hill's criteria for causation (Hill, 1965) in which infection by a single etiologic agent is the unique parameter influencing the occurrence of the disease (Inglis, 2007). Thus, the quest for a single pathogen explaining the etiology of periodontitis by itself, following Koch's postulates, may be in vain. Contrariwise, periodontitis, as a biofilm disease (Schaudinn et al., 2009), may result from the integration of various causative parameters. Bacteria are among these parameters and the composition of the microbial communities accurately correlates with clinical outcome (Feres et al., 2016; Hunter et al., 2016). Indeed, some species can be efficiently used as markers for diagnosis (Meuric et al., 2017), re-opening ways for considerations about the use of bacterial identification–though in a multi-variate fashion–for epidemiology or treatment follow-up.
Beside changes in its composition, the bacterial community can harbor changes in its functions, generating a new equilibrium (dysbiosis) that is possible in the new dental plaque environment (Hajishengallis and Lamont, 2012). Bacterial entities collaborate and functional changes, such as synergism, are evidenced at the transcriptional level (Kirst et al., 2015; Yost et al., 2015; Deng et al., 2018). Nevertheless, the abundance of some bacterium species is not synonymous for their activity (Mark Welch et al., 2016), a fact compatible with the “keystone pathogen” theory and the role of P. gingivalis in shaping the ecology of the periodontal pocket. This ecology is impacted by the dysbiotic communities, as evidenced in vitro with deregulated host inflammatory responses (Yost et al., 2017; Herrero et al., 2018). The consequences of dysbiosis were also evidenced in vivo at the systemic level, with metabolic changes linked to diabetes (Branchereau et al., 2016; Blasco-Baque et al., 2017).
Immuno-pathological processes TOWARD periodontitis
While the dental plaque stacks, mineralization leads to formation of tartar deposits, which can cause injury, as well as overhanging restorations or repetitive wounding. In parallel, the constant presence of bacterial components and the possible colonization by periodontopathogens can be sensed by the host and can cause chronic inflammation and an initial tissue lesion [reviewed in Kurgan and Kantarci (2018)]. The host takes part in fueling progression to disease by inflammation and active mediators of inflammation resolution improve the disease's outcome (Lee et al., 2016; Mizraji et al., 2018). Resident leukocytes and endothelial cells respond to bacterial biofilms: vascular permeability increases and interleukin 8 attracts neutrophils to the affected tissues (Tonetti et al., 1998). Neutrophils play a pivotal role in periodontitis (Ryder, 2010), producing reactive oxygen species (ROS), with probable impact at the systemic level [reviewed in Wang et al. (2017)]. While the early lesion progresses, other immune cells are recruited, including macrophages, lymphocytes, plasma cells, and mast cells. The inflammation can be visible at the microscopic level, where rete pegs–epithelial projections into the underlying connective tissue–form in the pocket epithelium and blood vessels proliferate [reviewed in Zoellner et al. (2002)], and at the macroscopic level, in particular by reddening and bleeding. Macrophages are predominant at this stage (Dennison and Van Dyke, 1997); they can derive from circulating monocytes produced in the spleen or be resident macrophages from embryonic origin, like Langerhans cells, with a possible different role during gum inflammation (Moughal et al., 1992). In established lesions, adaptive responses take place and lymphocytes are abundantly detected (Gemmell et al., 2007). Collagen fibers are increasingly altered, leading to a severe tissue remodeling and loosening of the pocket epithelium (Payne et al., 1975). Thus, greater amounts of dental plaque can accumulate in subgingival locations and aggravate gingivitis, which is considered reversible after elimination of the biofilm, in particular due to the absence of bone and periodontal ligament destruction (Ebersole et al., 2017).
Left untreated, gingivitis evolves to periodontitis, which is characterized by an inflammatory infiltrate composed of plasma cells, and by degradation of collagen fibers, loss of connective tissue, and bone destruction in an anaerobic environment (Mettraux et al., 1984). Periodontitis results in clinical attachment loss, i.e., the deeper positioning of the junction between the pocket epithelium and the cementum of the tooth root. The resulting volume below the gum forms the periodontal pocket, witnessing the breaking down of the initial epithelial attachment, the destruction of the connective tissues constituting the periodontal ligament, and the lysis of the alveolar bone. The sulcus depth–the distance between the free gingival margin and the epithelial attachment–is inferior to 3 mm in healthy or gingivitis sites. When superior to 3 mm, periodontitis is suspected and confirmed by inflammation (redness and swelling) and bleeding on probing.
During periodontitis, the balance between bone resorption and regeneration is displaced: Th17 cells induce osteoclastogenesis (Sato et al., 2006). Recently, it has been described that immunopathogenic Th17 lymphocytes [converted from Foxp3+ T cells; a recent review on T cell contribution to periodontitis in Kinane et al. (2017)] that cause bone damage in rheumatoid arthritis can also determine bone resorption and antimicrobial immunity in the oral cavity (Tsukasaki et al., 2018). In human periodontal lesions, Th17 lymphocytes are abundant (Hajishengallis, 2014) and the major source of IL-17. Foxp3+IL-17+ cells are found in the transition state. The generation of pathogenic exFoxp3+TH17 cells in the oral mucosa is dependent on IL-6, which is expressed by periodontal ligament fibroblasts in periodontitis and stimulated by bacterial PAMPs as LPS (Yamaji et al., 1995) through PRRs as TLR-2 and -4 (Sun et al., 2010; Makkawi et al., 2017). Furthermore, osteogenesis is impaired during periodontitis, while bone resorption by osteoclasts is promoted (Zhou et al., 2018), highlighting that not only immune mechanisms are involved in periodontitis pathophysiology. Beside the bacteria and human cells, some archaea, viruses, protozoa, and fungi are differentially present in healthy and diseased sites (Deng et al., 2017). The contributions of these different phenomena, as well as lysis by pathogens or other host immune cells, still need to be elucidated to solve the current paradigm of periodontitis physiopathology, in which only some of the players are visible in the game.
Entamoeba gingivalis and periodontal infections
Debate about the presence of Entamoeba gingivalis during periodontitis
Discovery of Entamoeba gingivalis
Though periodontitis was described since antiquity (Langsjoen, 1998), its association with parasites has been evidenced only a century ago. The first description of Entamoeba gingivalis–then named “Amoebea gengivalis”–was laconically performed in 1849 from dental plaque samples, mentioning amoebic movement and the presence of internal vesicles (Gros, 1849). Though free amoebae were described since 1755 (Rösel Von Rosenhof, 1755), E. gingivalis is the first amoeba which was found in humans. The pathogenic association of amoebae with humans was first documented in 1875, validating Koch's postulates in an animal model for the pathogen Entamoeba histolytica, then named “Ameba coli” (Lösch, 1875). The pathogenicity of Entamoeba gingivalis was questioned early (Kartulis, 1893) and the first systematic study associating it with periodontitis was preliminarily published in 1914 (Barrett, 1914): amoebae were detected in the totality of the 46 cases of pyorrhea (periodontitis) that were enrolled in the study. The authors later included 7 healthy individuals from the same group of patients in the “Insane Department of the Philadelphia Hospital” and claimed they could not detect amoebae in “the detritus collected around the neck of the teeth” (Smith and Barrett, 1915a). Furthermore, administration of emetine caused the withdrawal of amoebae and was followed by the cure of pyorrhea in 13 patients (Barrett, 1914). As emetine was thought to be a specific amoebicidal alkaloid with poor bactericidal effect, the etiological link between Entamoeba gingivalis and periodontitis was extrapolated and led Smith and Barrett to rename the disease “amoebic pyorrhea” (Barrett, 1914) or “oral endamebiasis” (Smith and Barrett, 1915b).
Rejection of amoebic etiology for periodontitis
It is of epistemological importance to underline that systematic studies, some with a low number of participants, about the involvement of Entamoeba gingivalis in periodontitis were countered by specialist opinions without formal experimental proofs, as reported by Craig (Craig, 1916). This controversy led to an almost total abandon of the etiology- and emetine-based therapy, discrediting at the same time its original scientific background. Further statements about the non-permanent cure were made but were unsupported by scientific data, and incriminating relapses (Howitt, 1925). However, the possibility of re-infections was completely ignored and casts doubts about the understanding of both pathophysiology and epidemiology of the disease at that time. In parallel, a method for the culture of E. gingivalis was described, which allowed the study of the effects of emetine hydrochloride on the parasite: “emetine hydrochloride has, apparently, no very marked amoebicidal action in vitro against either of the strains of E. gingivalis used” (Howitt, 1925). However, in a single experiment were evaluated the minimum lethal concentration (MLC, around 116.5 μM, 1:16,600 dilution) and the subjectively determined minimal inhibitory concentration (MIC, around 38.5 μM, 1:50,000) were evaluated. It is noteworthy that the MLC obtained for the emetine-sensitive HM1:IMSS E. histolytica strain in a more recent study is higher than 100 μM, the IC50 is 29.9 μM, and the MIC is lower than 1 μM (Bansal et al., 2004). This underlines that the experimental methodology to study the effect of emetine on E. gingivalis was not accurate. The conclusions of this paper should thus be considered with caution because this in vitro study did not corroborate the results reported during the treatment of patients, as previously cited.
Entamoeba gingivalis infections in the genetic Era
After this controversy, only a few studies based on the microscopic detection of E. gingivalis were published, but almost all of them revealed a prevalence of the parasite close to 100% in advanced periodontal pockets (Fisher, 1927; Hinshaw and Simonton, 1928; Wantland and Wantland, 1960; Wantland and Lauer, 1970; Gottlieb and Miller, 1971; Keyes and Rams, 1983; Lange, 1984; Linke et al., 1989). In the cited publications, the prevalence of E. gingivalis in healthy sulci–when studied–ranged from 0 to 26%, suggesting possible errors in the identification of the amoeba. Development of gene amplification by polymerase chain reaction (PCR) and the sequencing of a gene of E. gingivalis (Yamamoto et al., 1995) opened ways for the molecular identification of the parasite and accurate epidemiological studies. The first study–using a long amplicon (1.4 kb) and a sub-optimal DNA purification protocol–revealed a prevalence of 6.25% (2 sites out of 32, from 8 patients) in gingivitis or periodontitis sites, without precision of either their relative number or grade; no amplification was obtained from 20 healthy sites (Kikuta et al., 1996). In the second study, 69.2% of periodontitis sites were positive in real-time PCR for E. gingivalis, while none of the 12 healthy sites included in the study were (Trim et al., 2011). A third study showed a prevalence of 80.6% (58/72) in periodontitis sites and 33.3% (11/33) in healthy sites by conventional PCR, with controls of PCR inhibition and matrix degradation (Bonner et al., 2014). Recently, transcriptomics revealed that E. gingivalis 18s rRNA sequence was detected in all (4/4) periodontal pockets and was less abundant in 60% (6/10), or undetected in 40% (4/10) of healthy sites (Deng et al., 2017). Furthermore, genetic variants of E. gingivalis have been identified (Cembranelli et al., 2013; Garcia et al., 2018b) and different levels of virulence reflected at the transcriptomic levels in genetically identical parasites (Santi-Rocca et al., 2008) may account for discrepancies in their molecular detection as compared with microscopy or clinical diagnoses. The new clinical characterization of periodontitis (Tonetti et al., 2018) will avoid further confusion about the definition of health and various disease grades, that may also be responsible for variability between the studies.
Altogether, these data suggest that E. gingivalis may be asymptomatically present in some sulci and may be associated with the disease after environmental changes, reminiscent of the intestinal pathogen E. histolytica.
Life cycle of E. gingivalis
In most species of the genus Entamoeba, two cellular forms have been identified in nature: the cyst, which is the contaminant form found in the environment, and trophozoites, the vegetative cell able to divide, that initially derives from excystation of cysts ingested by the host. The survival of these Entamoeba species is ensured by their encystment in response to environmental changes (Mi-Ichi et al., 2016), permitting the survival in environments exposed to oxygen, like human stools, where identification of Entamoeba species is made by a simple morphological phenotyping that relies on the number of nuclei carried by the cyst. The sole E. gingivalis would not encyst, though cysts of E. gingivalis were reported in the literature at the beginning of the twentieth century (Chiavaro, 1914; Smith and Barrett, 1915b; Craig, 1916). However, it is now commonly accepted that E. gingivalis does not produce cysts, considering the absence of proof as a proof of absence. Nevertheless, the parasite E. gingivalis is essentially observed in periodontal pockets, suggesting that low oxygen levels are important for the survival of trophozoites, as in the case of E. histolytica and E. dispar [reviewed in Olivos-Garcia et al. (2016)]. Direct transmission of trophozoites to a new host would imply that they are resistant to oxygen, which raises questions about how E. gingivalis is transmitted in nature, and what ecological niche serves as a reservoir for this microorganism. Unfortunately, the complete life cycle of E. gingivalis is still missing and not addressed yet, hampering efficient prophylaxis.
In the closely-related specie E. histolytica, resistance to oxygen is modulated by interaction with bacteria (Varet et al., 2018), as well as virulence (Bracha and Mirelman, 1984; Galvan-Moroyoqui et al., 2008). The microbiota could be of major importance in switching from commensal to pathogenic forms and explain why only a minor part of E. histolytica intestinal infections are invasive and symptomatic. During periodontitis, bacterial virulence genes are strongly modulated (Deng et al., 2017) and the frequent and abundant detection of E. gingivalis in periodontitis pockets (Bonner et al., 2014) suggests and warns that interactions between constituents of the microbiota could be essential for their functions during the pathophysiology of the disease.
Ingestion of human cells by E. gingivalis
Entamoeba gingivalis resembles E. histolytica in several aspects: trophozoites measure about 30 μm, they are both endowed with mobility, and they ingest human cells and bacteria. The ability of E. histolytica to kill and phagocytose host cells correlates with parasite virulence and this amoeba is able to feed on human cells: erythrocytes, lymphocytes, and epithelial cells (Christy and Petri, 2011). Two mechanisms of cell killing and uptake have been discovered for E. histolytica: phagocytosis and trogocytosis (Ralston et al., 2014).
Phagocytosis is the phenomenon by which single cells ingest large volumes of material, like other cells or big particles; phagocytic cells include diverse unicellular entities as amoebae, but also macrophages and neutrophils that are cells from the immune system [a recent review in Niedergang and Grinstein (2018)]. In E. histolytica, phagocytosis is indispensable for its nutritional needs since this amoeba ingests bacteria in the intestinal lumen. Moreover, phagocytosis is correlated with virulence because E. histolytica kills human cells that are eventually phagocytosed. The current model is that E. histolytica first kills the host cells in a contact-dependent manner and then phagocytosis of dead cells takes place. However, live cells like bacteria and erythrocytes are also phagocytosed by E. histolytica.
In a recently-discovered second mechanism for cell damage, E. histolytica ingests fragments of live host cells in a nibbling-like process termed “trogocytosis” (Ralston et al., 2014). Though a specific AGC kinase1 was found exclusively involved in trogocytosis (Somlata and Nozaki, 2017), it seems premature to completely dissociate them from the phagocytic process. Furthermore, previous evidence has shown the existence of these structures during phagocytosis of epithelial and endothelial cells by E. histolytica [(Lejeune and Gicquaud, 1987; Nakada-Tsukui et al., 2009), Figure 2 in Faust et al. (2011)] and variability of phagocytic cup shape exist during phagocytosis (Tollis et al., 2010). It has been suggested that amoebic trogocytosis essentially concerns bits of live cells that are internalized, and phagocytosis is the process by which an entire cell is internalized (Ralston, 2015).
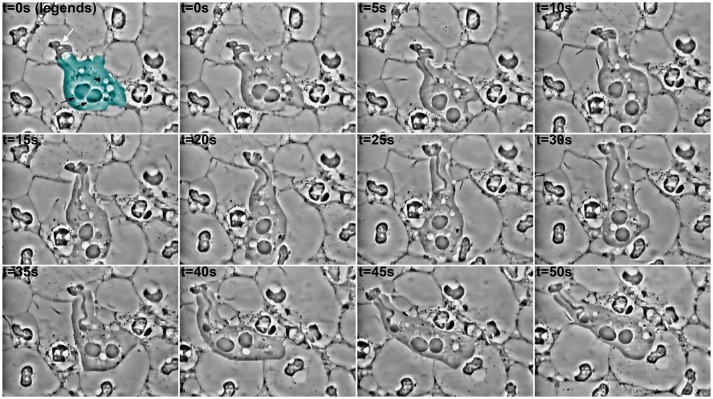
Phagocytosis of parts of human cells by E. gingivalis was reported almost a century ago (Child, 1926) but poorly studied since then. Some exceptions however existed and, thanks to video microscopy, the impressive process of cell ingestion by this amoeba has been highlighted (Lyons and Stanfield, 1989; Bonner et al., 2014). Entamoeba gingivalis is able to engulf one or more human cells at the time by a yet-undescribed mechanism (Figure 1). In the observed samples, cells around the amoebae present an altered cellular content, suggesting E. gingivalis can trigger signals leading to modification of human cells. As the only human cells observed in these samples were polymorphonuclear cells, and the literature mentions neutrophils are predominant in periodontal pockets, the target cells of amoebic phagocytosis may be the latter. The processes leading to the modifications in nuclear and cytoplasmic morphology in these cells remain to be defined and could be linked, for instance, to proteolytic activity of the amoebae and bacteria, or to a delayed/frustrated NETosis (Neutrophil extracellular traps). Whatever this process is, and whether the amoeba phagocytoses or trogocytosis, the cellular content of the neutrophils leads to one certain point: the first line of defense of cellular innate immunity against E. gingivalis and other organisms in the dysbiotic biofilm lacks its weapon (nuclei for NET formation and gene expression) and is thus unable to accomplish its functions.
Figure 1.
Ingestion of material by an amoeba in a periodontal pocket. Pictures extracted every 5 s from a video-microscopy of saliva-mounted plaque from the deepest part of the periodontal pocket, at 1,000 × magnification. In the first panel, the amoeba is pseudo colored in cyan; a black arrowhead indicates its nucleus with the typical peripheral chromatin, while the central karyosome is out of focus. The black arrows indicate food vacuoles. The white arrow designates the internal material (possibly a modified nucleus, perhaps with other subcellular structures) from a host cell (probably a leukocyte), whose ingestion has begun through a “channel,” as already observed for trogocytosis and erythrophagocytosis in E. histolytica. After 30 s, a food vacuole begins to form at the extremity of the channel. It is noteworthy that the amoeba continues to emit pseudopods and to move during the process, and that it is surrounded by cells with nuclei of different shapes, or even lacking.
Hot topics about E. gingivalis infections
The parasite E. gingivalis was identified for its amoeboid movement (Gros, 1849) and characteristics of its locomotion have not been studied since, unlike the closely-related species E. histolytica (Aguilar-Rojas et al., 2016). While observations in wet-mounted slides are possible, culture of the parasite is still a bottleneck for the study of E. gingivalis biology. Division events have not recently been documented and the whole life cycle is yet to be described.
The genetic variability in the species E. gingivalis seems important (Garcia et al., 2018b) and the genetic distance between the ST1 and ST2 variants may indicate great differences in their biology, accounting for their probable association with different pathologies (Garcia et al., 2018a). As the genotyping of E. gingivalis is based on the only gene that has been sequenced, a greater genetic variability may be expected, and further epidemiological studies will precise if some subtypes colonize specific sulcus/pocket environments and, thus, correlate with different clinical outcomes.
The role of the microbiota cannot be ignored and the strong modulation of bacterial genes during periodontitis supports a dysbiotic environment that may impact and be impacted by E. gingivalis parasites. Transcriptomic studies will provide clues about these interactions and will rely on the identification of amoebic genetic material. This will be rendered possible by sequencing the E. gingivalis genome, probably after the axenic culture of the parasite.
The parasite E. gingivalis is more prevalent and more abundant in periodontal pockets, suggesting that this ecological niche is either propitious for its survival, or that the parasite induces changes leading to this environment. Further studies will have to take into consideration the physicochemical and biological characteristics of the periodontal pockets to allow relevant studies of the biology of the parasites, either in vitro or in animal models.
Conclusion
The absence of reliable animal models for infection by E. gingivalis after its axenic culture impedes the ability to conclude about its etiological role in periodontitis, with respect to Koch's postulates. However, recent advances in the field of periodontitis have introduced moderation about this vision, considering that keystone pathogens can participate in changing the environment and consequently in causing dysbiosis, without being the exclusive etiological agent of the disease. Entamoeba gingivalis can be an important agent in the pathophysiology of periodontitis and, since its presence is documented and undoubted, it cannot be ignored.
Interestingly, the natural diversity in the human host has allowed the identification of various components of periodontal infections: some pathological traits are preferentially associated with human genetic variants (Offenbacher et al., 2016) and the P. gingivalis paradigm may be only one of the possibilities or one of the steps of periodontitis pathophysiology. Indeed, evidence about the kinetics of periodontitis setup is scarce and other possible agents must be considered. First, the ST2 “kamaktli” variant, with a high genetic divergence from E. gingivalis ST1 (Garcia et al., 2018b) reminds us that strains of parasites may not be equally virulent or may not have the same tropism (Garcia et al., 2018a). Second, some transcripts from archaea species and viruses are differently abundant between the healthy and the diseased (Deng et al., 2017). In particular, herpes viruses are associated with periodontitis (Slots, 2015; Zhu et al., 2015; Li et al., 2017). Finally, another protozoan is present in some cases of human periodontitis: Trichomonas tenax (Marty et al., 2017). Further studies are needed to decipher the ecosystem of the different stages of the periodontal pockets, assigning roles for all of the detected biological entities, which can be opportunistic, neutral, pathogenic, or mutualistic with the organisms within the pocket, and which can have the same type of interactions with the host tissues. Interestingly, the host directly participates in the pathogenic process: osteogenesis is impaired during periodontitis, while bone resorption by osteoclasts is promoted (Zhou et al., 2018). Furthermore, inflammation has an important role in the disease, since active mediators of inflammation resolution improve the disease's outcome (Lee et al., 2016; Mizraji et al., 2018). The contributions of these different phenomena, as well as lysis by pathogens and host immune cells, still need to be elucidated.
All species of Entamoeba are not necessarily pathogenic and some of them are commensals (e.g., E. dispar or E. coli). The study of E. gingivalis biology during infection, in particular its virulence factors and pathogenic processes, will allow us to better understand the whole interactions in the ecosystem of the periodontal pockets and to determine the potential participation of E. gingivalis in the pathophysiogenesis of periodontitis. Further research should determine if it is taking part in the pathophysiology of periodontitis or just a bona fide marker of the disease. In addition, as the picture is getting more complex and the genetic susceptibility of patients shapes different microbiota (Offenbacher et al., 2016), what we call periodontitis might be a group of related diseases with comparable outcomes. Fundamental research must consider this variability to elucidate the pathophysiology of periodontal diseases and to implement efficient public health measures.
Author contributions
All authors contributed equally to the redaction of the manuscript, conceived, and directed by JS-R.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Aas J. A., Paster B. J., Stokes L. N., Olsen I., Dewhirst F. E. (2005). Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43, 5721–5732. 10.1128/JCM.43.11.5721-5732.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Rojas A., Olivo-Marin J. C., Guillen N. (2016). The motility of Entamoeba histolytica: finding ways to understand intestinal amoebiasis. Curr. Opin. Microbiol. 34, 24–30. 10.1016/j.mib.2016.07.016 [DOI] [PubMed] [Google Scholar]
- Bansal D., Sehgal R., Chawla Y., Mahajan R. C., Malla N. (2004). In vitro activity of antiamoebic drugs against clinical isolates of Entamoeba histolytica and Entamoeba dispar. Ann. Clin. Microbiol. Antimicrob. 3:27. 10.1186/1476-0711-3-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett M. (1914). The protozoa of the mouth in relation to pyorrhea alveolaris. Dent. Cosm. 56, 948–953. [Google Scholar]
- Blasco-Baque V., Garidou L., Pomie C., Escoula Q., Loubieres P., Le Gall-David S., et al. (2017). Periodontitis induced by Porphyromonas gingivalis drives periodontal microbiota dysbiosis and insulin resistance via an impaired adaptive immune response. Gut 66, 872–885. 10.1136/gutjnl-2015-309897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner M., Amard V., Bar-Pinatel C., Charpentier F., Chatard J. M., Desmuyck Y., et al. (2014). Detection of the amoeba Entamoeba gingivalis in periodontal pockets. Parasite 21:30. 10.1051/parasite/2014029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracha R., Mirelman D. (1984). Virulence of Entamoeba histolytica trophozoites. Effects of bacteria, microaerobic conditions, and metronidazole. J. Exp. Med. 160, 353–368. 10.1084/jem.160.2.353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branchereau M., Reichardt F., Loubieres P., Marck P., Waget A., Azalbert V., et al. (2016). Periodontal dysbiosis linked to periodontitis is associated with cardiometabolic adaptation to high-fat diet in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 310, G1091–1101. 10.1152/ajpgi.00424.2015 [DOI] [PubMed] [Google Scholar]
- Cembranelli S. B., Souto F. O., Ferreira-Paim K., Richinho T. T., Nunes P. L., Nascentes G. A., et al. (2013). First evidence of genetic intraspecific variability and occurrence of Entamoeba gingivalis in HIV(+)/AIDS. PLoS ONE 8:e82864. 10.1371/journal.pone.0082864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiavaro A. (1914). Researches upon Entamoeba buccalis. Dent. Rev. 28, 1122–1135. [Google Scholar]
- Child H. J. (1926). Studies on the ingestion of leucocytes and on mitosis in Endamoeba gingivalis. Univ. Calif. Publ. Zool. 28, 251–284. [Google Scholar]
- Christy N. C., Petri W. A., Jr. (2011). Mechanisms of adherence, cytotoxicity and phagocytosis modulate the pathogenesis of Entamoeba histolytica. Future Microbiol. 6, 1501–1519. 10.2217/fmb.11.120 [DOI] [PubMed] [Google Scholar]
- Consortium, H. M. P. (2012). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig C. F. (1916). Observations upon the Endamebae of the Mouth. I. Endamoeba Gingivalis (Buccalis). J. Infect.Dis. 18, 220–238. 10.1093/infdis/18.2.220 [DOI] [Google Scholar]
- Darveau R. P. (2010). Periodontitis: a polymicrobial disruption of host homeostasis. Nat. Rev. Microbiol. 8, 481–490. 10.1038/nrmicro2337 [DOI] [PubMed] [Google Scholar]
- Demmer R. T., Papapanou P. N. (2010). Epidemiologic patterns of chronic and aggressive periodontitis. Periodontol. 2000 53, 28–44. 10.1111/j.1600-0757.2009.00326.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z. L., Szafranski S. P., Jarek M., Bhuju S., Wagner-Dobler I. (2017). Dysbiosis in chronic periodontitis: key microbial players and interactions with the human host. Sci. Rep. 7:3703. 10.1038/s41598-017-03804-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z. L., Sztajer H., Jarek M., Bhuju S., Wagner-Dobler I. (2018). Worlds apart–transcriptome profiles of key oral microbes in the periodontal pocket compared to single laboratory culture reflect synergistic interactions. Front. Microbiol. 9:124. 10.3389/fmicb.2018.00124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennison D. K., Van Dyke T. E. (1997). The acute inflammatory response and the role of phagocytic cells in periodontal health and disease. Periodontol. 2000 14, 54–78. 10.1111/j.1600-0757.1997.tb00192.x [DOI] [PubMed] [Google Scholar]
- Dumitrescu A. L. (2016). Depression and inflammatory periodontal disease considerations-an interdisciplinary approach. Front. Psychol. 7:347. 10.3389/fpsyg.2016.00347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersole J. L., Dawson D., III., Emecen-Huja P., Nagarajan R., Howard K., Grady M. E., et al. (2017). The periodontal war: microbes and immunity. Periodontol. 2000 75, 52–115. 10.1111/prd.12222 [DOI] [PubMed] [Google Scholar]
- Eke P. I., Dye B. A., Wei L., Slade G. D., Thornton-Evans G. O., Borgnakke W. S., et al. (2015). Update on prevalence of periodontitis in adults in the United States: NHANES 2009 to 2012. J. Periodontol. 86, 611–622. 10.1902/jop.2015.140520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eke P. I., Wei L., Thornton-Evans G. O., Borrell L. N., Borgnakke W. S., Dye B., et al. (2016). Risk indicators for periodontitis in US adults: NHANES 2009 to 2012. J. Periodontol. 87, 1174–1185. 10.1902/jop.2016.160013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evaluation, I. F. H. M. A. (2016). Global Burden of Disease Study 2015. Global Burden of Disease Study 2015 (GBD 2015) Disability Weights. Seattle, WA. Available online at: http://ghdx.healthdata.org/record/global-burden-disease-study-2015-gbd-2015-disability-weights
- Faust D. M., Marquay Markiewicz J., Santi-Rocca J., Guillen N. (2011). New insights into host-pathogen interactions during Entamoeba histolytica liver infection. Eur. J. Microbiol. Immunol. 1, 10–18. 10.1556/EuJMI.1.2011.1.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feres M., Teles F., Teles R., Figueiredo L. C., Faveri M. (2016). The subgingival periodontal microbiota of the aging mouth. Periodontol. 2000 72, 30–53. 10.1111/prd.12136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher J. H. (1927). Pyorrhea alveolaris: the rôle of certain microörganisms found in the lesions. Am. J. Pathol. 3:169. [PMC free article] [PubMed] [Google Scholar]
- Galvan-Moroyoqui J. M., Del Carmen Dominguez-Robles M., Franco E., Meza I. (2008). The interplay between Entamoeba and enteropathogenic bacteria modulates epithelial cell damage. PLoS Negl. Trop. Dis. 2:e266. 10.1371/journal.pntd.0000266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia G., Ramos F., Maldonado J., Fernandez A., Yanez J., Hernandez L., et al. (2018a). Prevalence of two Entamoeba gingivalis ST1 and ST2-kamaktli subtypes in the human oral cavity under various conditions. Parasitol. Res. 117, 2941–2948. 10.1007/s00436-018-5990-8 [DOI] [PubMed] [Google Scholar]
- Garcia G., Ramos F., Martinez-Hernandez F., Hernandez L., Yanez J., Gaytan P. (2018b). A new subtype of Entamoeba gingivalis: “E. gingivalis ST2, kamaktli variant”. Parasitol. Res. 117, 1277–1284. 10.1007/s00436-018-5798-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmell E., Yamazaki K., Seymour G. J. (2007). The role of T cells in periodontal disease: homeostasis and autoimmunity. Periodontol. 2000 43, 14–40. 10.1111/j.1600-0757.2006.00173.x [DOI] [PubMed] [Google Scholar]
- Gibbons R. J., Hay D. I., Cisar J. O., Clark W. B. (1988). Adsorbed salivary proline-rich protein 1 and statherin: receptors for type 1 fimbriae of Actinomyces viscosus T14V-J1 on apatitic surfaces. Infect. Immun. 56, 2990–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons R. J., Van Houte J. (1973). On the formation of dental plaques. J. Periodontol. 44, 347–360. 10.1902/jop.1973.44.6.347 [DOI] [PubMed] [Google Scholar]
- Gottlieb D. S., Miller L. H. (1971). Entamoeba gingivalis in periodontal disease. J. Periodontol. 42, 412–415. 10.1902/jop.1971.42.7.412 [DOI] [PubMed] [Google Scholar]
- Gros G. (1849). Fragments d'helminthologie et de physiologie microscopique. Bull. Soc. Impériale Nat. Moscou 22, 549–573. [Google Scholar]
- Hajishengallis G. (2014). Immunomicrobial pathogenesis of periodontitis: keystones, pathobionts, and host response. Trends Immunol. 35, 3–11. 10.1016/j.it.2013.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G., Lamont R. J. (2012). Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol. Oral Microbiol. 27, 409–419. 10.1111/j.2041-1014.2012.00663.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G., Liang S., Payne M. A., Hashim A., Jotwani R., Eskan M. A., et al. (2011). Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microb. 10, 497–506. 10.1016/j.chom.2011.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero E. R., Fernandes S., Verspecht T., Ugarte-Berzal E., Boon N., Proost P., et al. (2018). Dysbiotic biofilms deregulate the periodontal inflammatory response. J. Dent. Res. 97, 547–555. 10.1177/0022034517752675 [DOI] [PubMed] [Google Scholar]
- Hill A. B. (1965). The environment and disease: association or causation? Proc. R. Soc. Med. 58, 295–300. [PMC free article] [PubMed] [Google Scholar]
- Hinshaw H. C., Simonton F. (1928). The amoebas of the mouth in relation to incidence of pyorrhea, clinical symptoms, extent of lesions, and age of patients. J. Dent. Res. 8, 507–528. 10.1177/00220345280080040501 [DOI] [Google Scholar]
- Howitt B. F. (1925). The cultivation of Endamoeba gingivalis (Gros). Univ. Calif. Publ. Zool. 28, 65–126. [Google Scholar]
- Hsu C. C., Hsu Y. C., Chen H. J., Lin C. C., Chang K. H., Lee C. Y., et al. (2015). Association of periodontitis and subsequent depression: a nationwide population-based study. Medicine 94:e2347. 10.1097/MD.0000000000002347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter M. C., Pozhitkov A. E., Noble P. A. (2016). Microbial signatures of oral dysbiosis, periodontitis and edentulism revealed by Gene Meter methodology. J. Microbiol. Methods 131, 85–101. 10.1016/j.mimet.2016.09.019 [DOI] [PubMed] [Google Scholar]
- Inglis T. J. (2007). Principia aetiologica: taking causality beyond Koch's postulates. J. Med. Microbiol. 56, 1419–1422. 10.1099/jmm.0.47179-0 [DOI] [PubMed] [Google Scholar]
- Jakubovics N. S. (2010). Talk of the town: interspecies communication in oral biofilms. Mol. Oral Microbiol. 25, 4–14. 10.1111/j.2041-1014.2009.00563.x [DOI] [PubMed] [Google Scholar]
- Kartulis S. (1893). Muskelparasiten beim Menschen. Z. Hyg. Infektionskr. 13:2. [Google Scholar]
- Keyes P. H., Rams T. E. (1983). A rationale for management of periodontal diseases: rapid identification of microbial “therapeutic targets” with phase-contrast microscopy. J. Am. Dent. Assoc. 106, 803–812. [DOI] [PubMed] [Google Scholar]
- Kikuta N., Yamamoto A., Goto N. (1996). Detection and identification of Entamoeba gingivalis by specific amplification of rRNA gene. Can. J. Microbiol. 42, 1248–1251. 10.1139/m96-161 [DOI] [PubMed] [Google Scholar]
- Kinane D. F., Stathopoulou P. G., Papapanou P. N. (2017). Periodontal diseases. Nat. Rev. Dis. Primers 3:17038 10.1038/nrdp.2017.38 [DOI] [PubMed] [Google Scholar]
- Kirst M. E., Li E. C., Alfant B., Chi Y. Y., Walker C., Magnusson I., et al. (2015). Dysbiosis and alterations in predicted functions of the subgingival microbiome in chronic periodontitis. Appl. Environ. Microbiol. 81, 783–793. 10.1128/AEM.02712-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander P. E., Celesk R. A. (1983). Coaggregation of human oral cytophaga species and Actinomyces israelii. Infect. Immun. 40, 1178–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurgan S., Kantarci A. (2018). Molecular basis for immunohistochemical and inflammatory changes during progression of gingivitis to periodontitis. Periodontol. 2000 76, 51–67. 10.1111/prd.12146 [DOI] [PubMed] [Google Scholar]
- Lange D. E. (1984). Microbiological aspects of periodontal diseases. Zahnarztl. Mitt. 74, 2420, 2422–2423, 2426–2427. [PubMed] [Google Scholar]
- Langsjoen O. (1998). “Diseases of the dentition,” in The Cambridge Encyclopedia of Human Paleopathology, eds Aufderheide A. C., Rodríguez-Martín C. (Cambridge: University Press Cambridge: ), 393–412. [Google Scholar]
- Lee C. T., Teles R., Kantarci A., Chen T., Mccafferty J., Starr J. R., et al. (2016). Resolvin E1 reverses experimental periodontitis and dysbiosis. J. Immunol. 197, 2796–2806. 10.4049/jimmunol.1600859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leira Y., Seoane J., Blanco M., Rodriguez-Yanez M., Takkouche B., Blanco J., et al. (2017). Association between periodontitis and ischemic stroke: a systematic review and meta-analysis. Eur. J. Epidemiol. 32, 43–53. 10.1007/s10654-016-0170-6 [DOI] [PubMed] [Google Scholar]
- Lejeune A., Gicquaud C. (1987). Evidence for two mechanisms of human erythrocyte endocytosis by Entamoeba histolytica-like amoebae (Laredo strain). Biol. Cell 59, 239–245. 10.1111/j.1768-322X.1987.tb00536.x [DOI] [PubMed] [Google Scholar]
- Lertpimonchai A., Rattanasiri S., Arj-Ong Vallibhakara S., Attia J., Thakkinstian A. (2017). The association between oral hygiene and periodontitis: a systematic review and meta-analysis. Int. Dent. J. 67, 332–343. 10.1111/idj.12317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Zhu C., Deng F. Y., Wong M. C. M., Lu H. X., Feng X. P. (2017). Herpesviruses in etiopathogenesis of aggressive periodontitis: a meta-analysis based on case-control studies. PLoS ONE 12:e0186373. 10.1371/journal.pone.0186373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Helmerhorst E. J., Leone C. W., Troxler R. F., Yaskell T., Haffajee A. D., et al. (2004). Identification of early microbial colonizers in human dental biofilm. J. Appl. Microbiol. 97, 1311–1318. 10.1111/j.1365-2672.2004.02420.x [DOI] [PubMed] [Google Scholar]
- Linke H. A., Gannon J. T., Obin J. N. (1989). Clinical survey of Entamoeba gingivalis by multiple sampling in patients with advanced periodontal disease. Int. J. Parasitol. 19, 803–808. 10.1016/0020-7519(89)90069-6 [DOI] [PubMed] [Google Scholar]
- Listgarten M. A. (1976). Structure of the microbial flora associated with periodontal health and disease in man. A light and electron microscopic study. J. Periodontol. 47, 1–18. 10.1902/jop.1976.47.1.1 [DOI] [PubMed] [Google Scholar]
- Löe H., Theilade E., Jensen S. B. (1965). Experimental gingivitis in man. J. Periodontol. 36, 177–187. 10.1902/jop.1965.36.3.177 [DOI] [PubMed] [Google Scholar]
- Loesche W. J. (1979). Clinical and microbiological aspects of chemotherapeutic agents used according to the specific plaque hypothesis. J. Dent. Res. 58, 2404–2412. 10.1177/00220345790580120905 [DOI] [PubMed] [Google Scholar]
- Lopez R., Ramirez V., Marro P., Baelum V. (2012). Psychosocial distress and periodontitis in adolescents. Oral Health Prev. Dent. 10, 211–218. 10.3290/j.ohpd.a28516 [DOI] [PubMed] [Google Scholar]
- Lösch F. (1875). Massenhafte Entwicklung von Amöben im Dickdarm. Virchow. Arch. 65, 196–211. 10.1007/BF02028799 [DOI] [Google Scholar]
- Lyons T., Stanfield E. (1989). Introduction to Protozoa and Fungi in Periodontal Infections: A Manual of Microbiological Diagnosis and Non-Surgical Treatment. Ottawa, ON: Trevor Lyons. [Google Scholar]
- Makkawi H., Hoch S., Burns E., Hosur K., Hajishengallis G., Kirschning C. J., et al. (2017). Porphyromonas gingivalis stimulates TLR2-PI3K signaling to escape immune clearance and induce bone resorption independently of MyD88. Front. Cell. Infect. Microbiol. 7:359. 10.3389/fcimb.2017.00359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark Welch J. L., Rossetti B. J., Rieken C. W., Dewhirst F. E., Borisy G. G. (2016). Biogeography of a human oral microbiome at the micron scale. Proc. Natl. Acad. Sci. U.S.A. 113, E791–800. 10.1073/pnas.1522149113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh P. D. (1994). Microbial ecology of dental plaque and its significance in health and disease. Adv. Dent. Res. 8, 263–271. 10.1177/08959374940080022001 [DOI] [PubMed] [Google Scholar]
- Marsh P. D. (2003). Are dental diseases examples of ecological catastrophes? Microbiology 149, 279–294. 10.1099/mic.0.26082-0 [DOI] [PubMed] [Google Scholar]
- Marsh P. D., Moter A., Devine D. A. (2011). Dental plaque biofilms: communities, conflict and control. Periodontol. 2000 55, 16–35. 10.1111/j.1600-0757.2009.00339.x [DOI] [PubMed] [Google Scholar]
- Marty M., Lemaitre M., Kemoun P., Morrier J. J., Monsarrat P. (2017). Trichomonas tenax and periodontal diseases: a concise review. Parasitology 144, 1417–1425. 10.1017/S0031182017000701 [DOI] [PubMed] [Google Scholar]
- Mettraux G. R., Gusberti F. A., Graf H. (1984). Oxygen tension (pO2) in untreated human periodontal pockets. J. Periodontol. 55, 516–521. 10.1902/jop.1984.55.9.516 [DOI] [PubMed] [Google Scholar]
- Meuric V., Le Gall-David S., Boyer E., Acuna-Amador L., Martin B., Fong S. B., et al. (2017). Signature of microbial dysbiosis in periodontitis. Appl. Environ. Microbiol. 83:e00462-17. 10.1128/AEM.00462-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi-Ichi F., Yoshida H., Hamano S. (2016). Entamoeba Encystation: new targets to prevent the transmission of Amebiasis. PLoS Pathog. 12:e1005845. 10.1371/journal.ppat.1005845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizraji G., Heyman O., Van Dyke T. E., Wilensky A. (2018). Resolvin D2 Restrains Th1 immunity and prevents alveolar bone loss in murine periodontitis. Front. Immunol. 9:785. 10.3389/fimmu.2018.00785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore W. E., Holdeman L. V., Smibert R. M., Hash D. E., Burmeister J. A., Ranney R. R. (1982). Bacteriology of severe periodontitis in young adult humans. Infect. Immun. 38, 1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moughal N. A., Adonogianaki E., Kinane D. F. (1992). Langerhans cell dynamics in human gingiva during experimentally induced inflammation. J. Biol. Buccale 20, 163–167. [PubMed] [Google Scholar]
- Nakada-Tsukui K., Okada H., Mitra B. N., Nozaki T. (2009). Phosphatidylinositol-phosphates mediate cytoskeletal reorganization during phagocytosis via a unique modular protein consisting of RhoGEF/DH and FYVE domains in the parasitic protozoon Entamoeba histolytica. Cell. Microbiol. 11, 1471–1491. 10.1111/j.1462-5822.2009.01341.x [DOI] [PubMed] [Google Scholar]
- Nascimento G. G., Leite F. R. M., Vestergaard P., Scheutz F., Lopez R. (2018). Does diabetes increase the risk of periodontitis? A systematic review and meta-regression analysis of longitudinal prospective studies. Acta Diabetol. 55, 653–667. 10.1007/s00592-018-1120-4 [DOI] [PubMed] [Google Scholar]
- Niedergang F., Grinstein S. (2018). How to build a phagosome: new concepts for an old process. Curr. Opin. Cell Biol. 50, 57–63. 10.1016/j.ceb.2018.01.009 [DOI] [PubMed] [Google Scholar]
- Nieri M., Pini Prato G. P., Giani M., Magnani N., Pagliaro U., Rotundo R. (2013). Patient perceptions of buccal gingival recessions and requests for treatment. J. Clin. Periodontol. 40, 707–712. 10.1111/jcpe.12114 [DOI] [PubMed] [Google Scholar]
- Offenbacher S., Divaris K., Barros S. P., Moss K. L., Marchesan J. T., Morelli T., et al. (2016). Genome-wide association study of biologically informed periodontal complex traits offers novel insights into the genetic basis of periodontal disease. Hum. Mol. Genet. 25, 2113–2129. 10.1093/hmg/ddw069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivos-Garcia A., Saavedra E., Nequiz M., Santos F., Luis-Garcia E. R., Gudino M., et al. (2016). The oxygen reduction pathway and heat shock stress response are both required for Entamoeba histolytica pathogenicity. Curr. Genet. 62, 295–300. 10.1007/s00294-015-0543-5 [DOI] [PubMed] [Google Scholar]
- Organization, W. H. (2016). Global Health Estimates 2015: Deaths by Cause, Age, Sex, by Country and by Region, 2000–2015. Geneva. [Google Scholar]
- Payne W. A., Page R. C., Ogilvie A. L., Hall W. B. (1975). Histopathologic features of the initial and early stages of experimental gingivitis in man. J. Periodont. Res. 10, 51–64. 10.1111/j.1600-0765.1975.tb00008.x [DOI] [PubMed] [Google Scholar]
- Petersen P. E., Ogawa H. (2005). Strengthening the prevention of periodontal disease: the WHO approach. J. Periodontol. 76, 2187–2193. 10.1902/jop.2005.76.12.2187 [DOI] [PubMed] [Google Scholar]
- Ralston K. S. (2015). Chew on this: amoebic trogocytosis and host cell killing by Entamoeba histolytica. Trends Parasitol. 31, 442–452. 10.1016/j.pt.2015.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston K. S., Solga M. D., Mackey-Lawrence N. M., Somlata Bhattacharya A., Petri W. A., Jr. (2014). Trogocytosis by Entamoeba histolytica contributes to cell killing and tissue invasion. Nature 508, 526–530. 10.1038/nature13242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds I., Duane B. (2018). Periodontal disease has an impact on patients' quality of life. Evid. Based Dent. 19, 14–15. 10.1038/sj.ebd.6401287 [DOI] [PubMed] [Google Scholar]
- Rogers G. B., Bruce K. D. (2012). Exploring the parallel development of microbial systems in neonates with cystic fibrosis. MBio 3:e00408–00412. 10.1128/mBio.00408-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rösel Von Rosenhof A. J. (1755). Der Monatlich-Herausgegebenen Insecten-Belustigung. Nürnberg: Fleischmann JJ. [Google Scholar]
- Rosier B. T., Marsh P. D., Mira A. (2018). Resilience of the oral microbiota in health: mechanisms that prevent dysbiosis. J. Dent. Res. 97, 371–380. 10.1177/0022034517742139 [DOI] [PubMed] [Google Scholar]
- Ryder M. I. (2010). Comparison of neutrophil functions in aggressive and chronic periodontitis. Periodontol. 2000 53, 124–137. 10.1111/j.1600-0757.2009.00327.x [DOI] [PubMed] [Google Scholar]
- Santi-Rocca J., Weber C., Guigon G., Sismeiro O., Coppee J. Y., Guillen N. (2008). The lysine- and glutamic acid-rich protein KERP1 plays a role in Entamoeba histolytica liver abscess pathogenesis. Cell. Microbiol. 10, 202–217. 10.1111/j.1462-5822.2007.01030.x [DOI] [PubMed] [Google Scholar]
- Sato K., Suematsu A., Okamoto K., Yamaguchi A., Morishita Y., Kadono Y., et al. (2006). Th17 functions as an osteoclastogenic helper T cell subset that links T cell activation and bone destruction. J. Exp. Med. 203, 2673–2682. 10.1084/jem.20061775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaudinn C., Gorur A., Keller D., Sedghizadeh P. P., Costerton J. W. (2009). Periodontitis: an archetypical biofilm disease. J. Am. Dent. Assoc. 140, 978–986. 10.14219/jada.archive.2009.0307 [DOI] [PubMed] [Google Scholar]
- Schultz-Haudt S., Bruce M. A., Bibby B. G. (1954). Bacterial factors in nonspecific gingivitis. J. Dent. Res. 33, 454–458. 10.1177/00220345540330040301 [DOI] [PubMed] [Google Scholar]
- Silva M. F., Cademartori M. G., Leite F. R. M., Lopez R., Demarco F. F., Nascimento G. G. (2017). Is periodontitis associated with halitosis? A systematic review and meta-regression analysis. J. Clin. Periodontol. 44, 1003–1009. 10.1111/jcpe.12786 [DOI] [PubMed] [Google Scholar]
- Slots J. (2015). Periodontal herpesviruses: prevalence, pathogenicity, systemic risk. Periodontol. 2000 69, 28–45. 10.1111/prd.12085 [DOI] [PubMed] [Google Scholar]
- Smiley C. J., Tracy S. L., Abt E., Michalowicz B. S., John M. T., Gunsolley J., et al. (2015). Evidence-based clinical practice guideline on the nonsurgical treatment of chronic periodontitis by means of scaling and root planing with or without adjuncts. J. Am. Dent. Assoc. 146, 525–535. 10.1016/j.adaj.2015.01.026 [DOI] [PubMed] [Google Scholar]
- Smith A., Barrett M. (1915a). Emetin in the treatment of peridental sup-purations. Dent. Cosm. 57:1201. [Google Scholar]
- Smith A. J., Barrett M. (1915b). The parasite of oral Endamebiasis, Endameba gingivalis (Gros). J. Parasitol. 1, 159–174. [Google Scholar]
- Socransky S. S., Haffajee A. D., Cugini M. A., Smith C., Kent R. L., Jr. (1998). Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25, 134–144. 10.1111/j.1600-051X.1998.tb02419.x [DOI] [PubMed] [Google Scholar]
- Somlata Nakada-Tsukui, K., Nozaki T. (2017). AGC family kinase 1 participates in trogocytosis but not in phagocytosis in Entamoeba histolytica. Nat. Commun. 8:101 10.1038/s41467-017-00199-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Shu R., Li C. L., Zhang M. Z. (2010). Gram-negative periodontal bacteria induce the activation of Toll-like receptors 2 and 4, and cytokine production in human periodontal ligament cells. J. Periodontol. 81, 1488–1496. 10.1902/jop.2010.100004 [DOI] [PubMed] [Google Scholar]
- Tollis S., Dart A. E., Tzircotis G., Endres R. G. (2010). The zipper mechanism in phagocytosis: energetic requirements and variability in phagocytic cup shape. BMC Syst. Biol. 4:149. 10.1186/1752-0509-4-149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonetti M. S., Greenwell H., Kornman K. S. (2018). Staging and grading of periodontitis: framework and proposal of a new classification and case definition. J. Periodontol. 89(Suppl. 1), S159–S172. 10.1002/JPER.18-0006 [DOI] [PubMed] [Google Scholar]
- Tonetti M. S., Imboden M. A., Lang N. P. (1998). Neutrophil migration into the gingival sulcus is associated with transepithelial gradients of interleukin-8 and ICAM-1. J. Periodontol. 69, 1139–1147. 10.1902/jop.1998.69.10.1139 [DOI] [PubMed] [Google Scholar]
- Trim R. D., Skinner M. A., Farone M. B., Dubois J. D., Newsome A. L. (2011). Use of PCR to detect Entamoeba gingivalis in diseased gingival pockets and demonstrate its absence in healthy gingival sites. Parasitol. Res. 109, 857–864. 10.1007/s00436-011-2312-9 [DOI] [PubMed] [Google Scholar]
- Tsukasaki M., Komatsu N., Nagashima K., Nitta T., Pluemsakunthai W., Shukunami C., et al. (2018). Host defense against oral microbiota by bone-damaging T cells. Nat. Commun. 9:701. 10.1038/s41467-018-03147-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke T. E. (2017). Pro-resolving mediators in the regulation of periodontal disease. Mol. Aspects Med. 58, 21–36. 10.1016/j.mam.2017.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van 'T Hof W., Veerman E. C., Nieuw Amerongen A. V., Ligtenberg A. J. (2014). Antimicrobial defense systems in saliva. Monogr. Oral Sci. 24, 40–51. 10.1159/000358783 [DOI] [PubMed] [Google Scholar]
- Varet H., Shaulov Y., Sismeiro O., Trebicz-Geffen M., Legendre R., Coppée J.-Y., et al. (2018). Enteric bacteria boost defences against oxidative stress in Entamoeba histolytica. Sci. Rep. 8:9042. 10.1038/s41598-018-27086-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira A. R., Albandar J. M. (2014). Role of genetic factors in the pathogenesis of aggressive periodontitis. Periodontol. 2000 65, 92–106. 10.1111/prd.12021 [DOI] [PubMed] [Google Scholar]
- Wang J., Lv J., Wang W., Jiang X. (2016). Alcohol consumption and risk of periodontitis: a meta-analysis. J. Clin. Periodontol. 43, 572–583. 10.1111/jcpe.12556 [DOI] [PubMed] [Google Scholar]
- Wang Y., Andrukhov O., Rausch-Fan X. (2017). Oxidative stress and antioxidant system in periodontitis. Front. Physiol. 8:910. 10.3389/fphys.2017.00910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wantland W. W., Lauer D. (1970). Correlation of some oral hygiene variables with age, sex, and incidence of oral protozoa. J. Dent. Res. 49, 293–297. 10.1177/00220345700490021601 [DOI] [PubMed] [Google Scholar]
- Wantland W. W., Wantland E. M. (1960). Incidence, ecology, and reproduction of oral protozoa. J. Dent. Res. 39:863. 10.1177/00220345600390041801 [DOI] [PubMed] [Google Scholar]
- Yamaji Y., Kubota T., Sasaguri K., Sato S., Suzuki Y., Kumada H., et al. (1995). Inflammatory cytokine gene expression in human periodontal ligament fibroblasts stimulated with bacterial lipopolysaccharides. Infect. Immun. 63, 3576–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., Kikuta N., Hashimoto T., Oyaizu H., Goto N. (1995). Nucleotide sequence of the SrRNA gene of Entamoeba gingivalis: applications for construction of a species-specific DNA probe and phylogenetic analysis. Microbiol. Immunol. 39, 185–192. 10.1111/j.1348-0421.1995.tb02187.x [DOI] [PubMed] [Google Scholar]
- Yost S., Duran-Pinedo A. E., Krishnan K., Frias-Lopez J. (2017). Potassium is a key signal in host-microbiome dysbiosis in periodontitis. PLoS Pathog. 13:e1006457. 10.1371/journal.ppat.1006457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yost S., Duran-Pinedo A. E., Teles R., Krishnan K., Frias-Lopez J. (2015). Functional signatures of oral dysbiosis during periodontitis progression revealed by microbial metatranscriptome analysis. Genome Med. 7:27 10.1186/s13073-015-0153-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R., Shen L., Yang C., Wang L., Guo H., Yang P., et al. (2018). Periodontitis may restrain the mandibular bone healing via disturbing Osteogenic and Osteoclastic balance. Inflammation 41, 972–983. 10.1007/s10753-018-0751-5 [DOI] [PubMed] [Google Scholar]
- Zhu C., Li F., Wong M. C., Feng X. P., Lu H. X., Xu W. (2015). Association between Herpesviruses and Chronic Periodontitis: a meta-analysis based on case-control studies. PLoS ONE 10:e0144319. 10.1371/journal.pone.0144319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoellner H., Chapple C. C., Hunter N. (2002). Microvasculature in gingivitis and chronic periodontitis: disruption of vascular networks with protracted inflammation. Microsc. Res. Tech. 56, 15–31. 10.1002/jemt.10009 [DOI] [PubMed] [Google Scholar]