Abstract
Microsatellite Instability (MSI) status is an established predictive biomarker for the treatment of the anti‐programmed death 1 (PD‐1) antibody. The current approach to determine the MSI status in tumours requires matched normal DNA. Some mononucleotide microsatellite markers are known to have few variant alleles in both Caucasians and Asians. Therefore, the length of these microsatellite makers is almost confined within the quasi‐monomorphic variation range (QMVR). Considering the application of MSI testing for various types of cancers, a simple, sensitive and inexpensive method is desired. This study assessed the clinical utility of the QMVR for determining the MSI status in patients with unresectable metastatic colorectal cancer (mCRC). The study enrolled 435 patients with mCRC. The concordance of the MSI status in mCRC between the standard method using tumour DNA plus matched normal DNA and the testing method using only tumour DNA was evaluated. Eleven (2.5%) MSI‐high cases were detected by both the standard and testing methods. The sensitivity and specificity of the testing method were both 100%, indicating complete concordance between the methods. Among the mononucleotide markers, three and two patients showed discordance for NR‐21 and BAT‐25, respectively. Results from MSI testing with normal tissue indicated that four of five patients had rare germline variants outside the QMVR. For BAT‐26, NR‐24 and MONO‐27, all patients showed complete concordance. Using the QMVR, the MSI status of mCRC can be determined without matched normal DNA.
Keywords: metastatic colorectal cancer, microsatellite instability, mononucleotide microsatellite markers, Promega panel, quasi‐monomorphic variation range
1. INTRODUCTION
Microsatellite instability (MSI) is defined as alteration of the lengths of repetitive sequences in tumour DNA, which is not observed in corresponding germline DNA. MSI is recognised as a hallmark of mismatch repair (MMR) deficiency.1, 2
Recently, pembrolizumab and nivolumab, which are IgG4 monoclonal antagonist antibodies against programmed cell death‐1 (PD‐1), were shown to have durable antitumor activity in patients with MMR‐deficient (dMMR) metastatic colorectal cancer (mCRC) or non‐colorectal cancer (CRC).3, 4 The Food and Drug Administration granted accelerated approval to nivolumab for use in patients with dMMR mCRC and to pembrolizumab for use in adult and paediatric patients with dMMR solid tumours, including mCRC. Considering that patients with advanced solid tumours are more frequently undergoing PD‐1 blockade treatment according to these new indications, the development of a simple, sensitive and inexpensive method to assess MMR deficiency is desired.
The National Cancer Institute workshop in 2002 concluded that dinucleotide repeat markers (D2S123, D5S346 and D17S250) are less sensitive for the detection of tumours with MMR deficiencies than mononucleotide markers.5 A more sensitive and easy‐to‐analyse panel was developed for MSI testing (Promega, Madison, WI, USA),6 and it consists of five mononucleotide markers (NR‐21, BAT‐25, MONO‐27, NR‐24 and BAT‐26). As these mononucleotide markers are known to have few variant alleles in Caucasians and Asians,7 the lengths of PCR products from normal DNA are almost confined within the quasi‐monomorphic variation range (QMVR), suggesting that the mononucleotide maker panel can determine MSI status without normal DNA. In fact, some researchers have reported that the MSI status of Caucasian CRC patients could be retrospectively determined using the QMVR, with high sensitivity and specificity.8, 9 According to three large Japanese cohorts, the frequencies of variant alleles for the five mononucleotide markers are very low (Table S1).10
A pilot study using blood samples from 149 healthy Japanese individuals suggested that the QMVR of each marker was generated in base pairs (bps) as the mean size of each marker ± 3 bps, which is consistent with the findings in previous studies (Table S2).9, 10 This suggests that the QMVR might be useful for determining the MSI status in not only Caucasian patients but also Asian patients. However, the clinical utility of the QMVR has not been verified prospectively in patients with unresectable mCRC. The present study aimed to assess the clinical utility of the QMVR for determining the MSI status in patients with unresectable mCRC.
2. MATERIAL AND METHODS
2.1. Patients
Since February 2016, the multi‐institutional joint study called GI‐SCREEN CRC‐MSI is being conducted to investigate the MSI status of Japanese patients with mCRC (UMIN000020437). Promega MSI tests using DNA isolated from both tumour tissue and paired normal tissue were performed as a standard method in the GI‐SCREEN CRC‐MSI study.10
This sub‐study of GI SCREEN CRC‐MSI included patients who were enrolled in the GI‐SCREEN CRC‐MSI study and fulfilled the following conditions: (a) agreement for the secondary use of specimens was obtained when the patients were enrolled in the GI‐SCREEN CRC‐MSI study; (b) the MSI status was determined by a standard method using both tumour and normal DNA and (c) sufficient DNA from tumour tissue remained for MSI testing.
The protocol of this sub‐study was approved by the Institutional Review Board at the National Cancer Centre. All procedures related to the study were in accordance with the Helsinki Declaration of 1964 and later versions.
2.2. MSI analysis with paired normal DNA
Standard MSI analysis in the GI‐SCREEN CRC‐MSI study was performed using tumour and normal DNA from formalin‐fixed paraffin embedded (FFPE) patient tissues with the MSI Analysis System Version 1.2 (Promega), which includes fluorescent‐labelled primers for co‐amplification of five mononucleotide markers (NR‐21, BAT‐25, MONO‐27, NR‐24 and BAT‐26), according to the manufacturer's instructions. By comparing the sizes of the PCR products between the tumour and normal samples, the presence of new alleles in the tumour sample that were not present in the corresponding normal sample was confirmed. Using the entire five‐marker panel, tumours exhibiting two or more microsatellite unstable markers were classified as MSI‐H, tumours exhibiting one unstable marker were classified as MSI‐L and tumours without any unstable marker were classified as MSS.
2.3. MSI determination using the QMVR
Testing using the QMVR was performed with a new MSI kit including the primer mixture of the Promega panel and an amplification enzyme mixture. Briefly, 20 ng of tumour DNA from FFPE tissue was amplified with the Veriti Dx Thermal Cycler (Life Technologies, Carlsbad, CA, USA) using the following cycling profile: 1 cycle at 96°C for 1 minute; 30 cycles at 94°C for 30 seconds, 59°C for 2 minutes, and 72°C for 90 seconds; 60°C for 45 minutes with a hold at 4°C. The PCR products were separated by capillary electrophoresis using the 3500xl Dx Genetic Analyzer (Life Technologies) and were analysed using GeneMapper software (Life Technologies) to determine the MSI status. The QMVR of each marker generated from 149 healthy Japanese individuals was used as the normal reference range for determining the MSI status in the testing method of the sub‐study (Table S2). Using the entire five‐marker panel and the QMVR of each marker, tumours exhibiting two or more markers outside the corresponding QMVR were classified as MSI‐H, tumours exhibiting one marker outside the QMVR were classified as MSI‐L and tumours without any marker outside the QMVR were classified as MSS. New MSI kits using the QMVR as the reference of the five mononucleotide markers of the Promega panel were manufactured under the Quality Management System for in vitro diagnostics.
As all registration numbers for the GI‐SCREEN CRC‐MSI study were double encoded before this sub‐study, the testing method using only tumour samples was conducted blindly without any information with regard to the MSI status by the standard method.
2.4. Statistical analysis
Endpoints were analysed by creating a 2 × 2 cross‐tabulation table for the number of specimens in which the MSI status was determined to be MSI‐H (positive) and MSS/MSI‐L (negative) using the testing method and the standard method. The primary endpoints of this sub‐study were the sensitivity (positive conformity rate) and specificity (negative conformity rate) of the testing method when compared with the standard method. The testing method would be considered effective if the decision rule previously established with regard to sensitivity is fulfilled and if specificity is ≥90% (Table S3). As secondary endpoints, the concordance rate, positive‐predictive value and negative‐predictive value were evaluated.
2.5. Repeated PCR examination
As a post hoc analysis, both the standard and testing MSI methods were performed again by using the same tumour and normal DNA samples as in the first evaluation, when the discordant markers were observed.
3. RESULTS
3.1. Patient characteristics
Between February 2016 and November 2016, the results of MSI testing were obtained from 474 patients in the GI‐SCREEN CRC‐MSI study. Of these patients, 38 did not fulfil the eligibility criteria, and they were excluded. Thus, 436 patients were eligible for the GI‐SCREEN‐CRC MSI sub‐study. However, one patient was excluded because MSI could not be analysed using the newly developed MSI kit. Finally, 435 patients were eligible for the analysis set (Figure 1).
Figure 1.
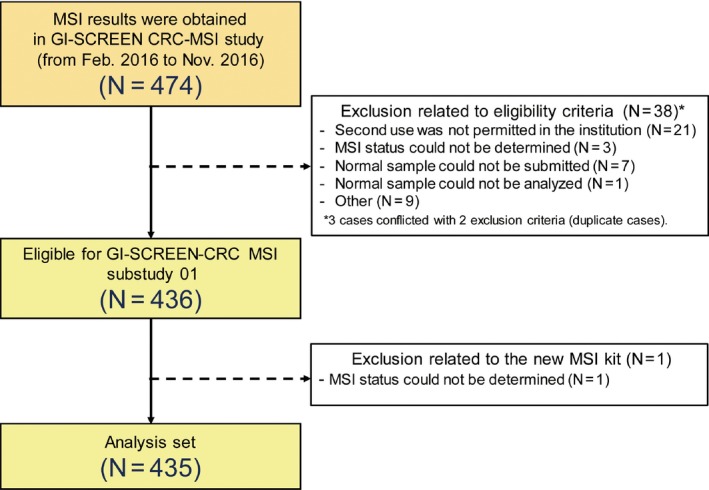
Results of Microsatellite instability (MSI) testing were obtained from 474 patients in the GI‐SCREEN CRC‐MSI study. Of these patients, 38 did not fulfil the eligibility criteria, and they were excluded. Thus, 436 patients were eligible for the GI‐SCREEN‐CRC MSI sub‐study. However, one patient was excluded because MSI could not be analysed using the newly developed MSI kit. Finally, 435 patients were eligible for the analysis set
The median patient age was 66 years, and 248 (57.0%) patients were male. The analysis involved 368 (84.6%) primary and 67 (15.4%) metastatic specimens (Tables 1, S4).
Table 1.
Patient characteristics
| Age | |
| Median (range) | 66 (29‐94) |
| Sex | |
| Male | 248 (57%) |
| Female | 187 (43%) |
| Histology | |
| tub1/tub2 | 386 (88.7%) |
| por1/por2 | 19 (4.4%) |
| pap | 2 (0.5%) |
| muc | 18 (4.1%) |
| Other | 10 (2.3%) |
| Sites of sample collection | |
| Primary | 368 (84.5%) |
| Right‐sided colon | 112 (25.7%) |
| Left‐sided colon | 161 (37.0%) |
| Rectum | 94 (21.6%) |
| Unknown | 1 (0.2%) |
| Metastatic | 67 (15.4%) |
| Liver | 27 (6.2%) |
| Lung | 10 (2.3%) |
| Peritoneum | 3 (0.7%) |
| Lymph node | 7 (1.6%) |
| Other | 20 (29.9%) |
3.2. Comparison of the MSI status between the standard and testing methods
Among the 435 patients, 11 (2.5%) had tumours classified as MSI‐H with the standard method, 10 of which were in right side colon and 1 were in rectum, respectively. These tumours were also classified as MSI‐H with the testing method. The remaining 424 patients had tumours classified as MSS/MSI‐L with the standard method. These tumours were also classified as MSS/MSI‐L with the testing method. Therefore, the sensitivity and specificity of the MSI status with the testing method were both 100%. The concordance rate, positive‐predictive value and negative‐predictive value were all 100% (Table 2).
Table 2.
Cross‐tabulation table (2 × 2)
| Standard method | |||
|---|---|---|---|
| Negative (MSS/MSI‐L) | Positive (MSI‐H) | ||
| Testing method | Negative (MSS/MSI‐L) | 424 | 0 |
| Positive (MSI‐H) | 0 | 11 | |
3.3. Comparison of the five mononucleotide markers between the standard and testing methods
Five patients had only one discordance in their markers. Four showed one microsatellite unstable marker (NR‐21 in four and BAT‐25 in one) in only the testing method. On the other hand, one patient showed 1 microsatellite unstable markers (BAT‐25 in one) in only the standard method. No patient had two or more unstable markers. As they were classified as “MSI‐L”, the decision was not affected (either MSI‐H [positive] or MSS/MSI‐L [negative]). For BAT‐26, NR‐24 and MONO‐27, all patients showed concordance (Table 3).
Table 3.
Concordance with each microsatellite marker
| NR21 | Standard method | |||
|---|---|---|---|---|
| Stable | Unstable | Total | ||
| Testing method | Stable | 421 | 0 | 421 |
| Unstable | 3 | 11 | 14 | |
| Total | 424 | 11 | 435 | |
| BAT25 | Standard method | |||
|---|---|---|---|---|
| Stable | Unstable | Total | ||
| Testing method | Stable | 422 | 1 | 423 |
| Unstable | 1 | 11 | 12 | |
| Total | 423 | 12 | 435 | |
| MONO27 | Standard method | |||
|---|---|---|---|---|
| Stable | Unstable | Total | ||
| Testing method | Stable | 424 | 0 | 421 |
| Unstable | 0 | 11 | 11 | |
| Total | 424 | 11 | 435 | |
| BAT26a | Standard method | |||
|---|---|---|---|---|
| Stable | Unstable | Total | ||
| Testing method | Stable | 423 | 0 | 423 |
| Unstable | 0 | 11 | 11 | |
| Total | 423 | 11 | 434 | |
| NR24 | Standard method | |||
|---|---|---|---|---|
| Stable | Unstable | Total | ||
| Testing method | Stable | 424 | 0 | 421 |
| Unstable | 0 | 11 | 11 | |
| Total | 424 | 11 | 435 | |
In one patient, the BAT‐26 status was not determined.
In the post hoc analysis, we re‐evaluated microsatellite unstable markers in the five discordant cases using the same tumour and normal DNA samples. The same microsatellite alterations were observed in all five cases.
4. DISCUSSION
We verified the clinical utility of the QMVR in patients with mCRC. The sensitivity and specificity of the testing method and standard method were perfectly concordant. As germline variants involving the five assessed mononucleotide markers are very rare in Japanese individuals, the QMVR of each marker was useful as a reference in MSI testing.
Five patients had only one discordant marker (NR‐21 in three and BAT‐25 in two) among the 2 methods. Repeated tests showed the same results, suggesting that the discordant results were not associated with artefacts. Of the five patients, four had different sizes of microsatellites outside the QMVR in the germline. In these patients, the testing method showed false positive. According to the frequencies of variant alleles in NR21 and BAT25 in Japanese cohorts (Table S1), these discordant rates can be considered reasonable. On the other hand, one patient showed microsatellite instability for one marker in the standard method but not in the testing method. The reason for this finding is unclear, but genetic heterogeneity of tumour tissue may have influenced the result.
According to the results in the pilot study, the QMVRs among Japanese individuals were 98.4‐104.4 bps for NR‐21, 111.4‐117.4 bps for BAT‐26, 121.0‐127.0 bps for BAT‐25, 129.5‐135.5 bps for NR‐24 and 149.9‐155.9 bps for MONO‐27, which are almost the same as those in Caucasians (Table S2).9, 10 These results suggest that MSI kits using the same QMVRs could be applied in both Asian and Caucasian patients. However, further studies will be needed to clarify the clinical utility of the new MSI‐kit for Caucasian patients.
The present study has some limitations. Immunohistochemistry (IHC) testing was not performed to compare PCR‐based testing. Comparisons between the results with the new MSI kits and those with IHC testing may support the robustness of the newly developed PCR‐based testing approach, as 13% of MSI‐H cases were reported to have an indefinite/equivocal staining pattern in IHC testing.11 Furthermore, as MSI‐H cases are rare among patients with mCRC, only 11 (2.5%) cases were included in this sub‐study. Before starting this sub‐study, we estimated the approximately 2%‐4% of MSI‐H cases would be included. For this situation, even if the sensitivity of the testing method is low, the concordance rate is likely to be high. As we consider evaluation using the concordance rate is inappropriate to accurately evaluate the clinical performance, both the sensitivity and specificity of the testing method were decided as primary endpoint. We also made ‘the decision rule for sensitivity’ before starting this sub‐study to accurately evaluate the sensitivity using small number of MSI‐H cases (Table S3). Although the small number of positive cases might have affected the evaluation of the clinical performance of the new MSI kits, we can conclude the testing method is effective because the decision rule regarding sensitivity is fulfilled and specificity is ≥90%. Considering the application of MSI testing for various types of inoperable advanced solid tumours, a simple and inexpensive method is desired. The QMVR can be used as a reference of MSI testing and can help reduce the cost of PCR examination.
In conclusion, by using the QMVR, the MSI status of patients with mCRC can be determined without matched normal DNA, and the same QMVR might be applicable to both Asian and Caucasian patients.
CONFLICT OF INTEREST
HB reports research funding from AstraZeneca and Sysmex. WO reports research funding from MSD. TF reports employment by FALCO biosystems during the conduct of the study. TY reports research funding from Taiho Pharmaceutical and Takeda; honoraria from Taiho Pharmaceutical, Takeda, Chugai Pharmaceutical, and Boehringer Ingelheim; and advisory role from AstraZeneca, Daiichi Sankyo, Gilead Sciences, Sysmex, and HUYA Bioscience. KA reports research funding from MSD. TY reports research funding from GlaxoSmithKline and Nippon Boehringer Ingelheim and honoraria from Taiho Pharmaceutical, Eli Lilly Japan, and Chugai Pharmaceutical.
AUTHORS’ CONTRIBUTIONS
Conception and design: HB, WO, TY, KA, and TY. Collection and assembly of data: HB, WO, TY, KA, and TY. MSI testing: TF. Data analysis and interpretation: All authors. Manuscript writing: All authors. Final approval of manuscript: All authors.
Supporting information
ACKNOWLEDGEMENTS
We would like to express special thanks to all participating patients, their families, and all participating investigators.
Bando H, Okamoto W, Fukui T, Yamanaka T, Akagi K, Yoshino T. Utility of the quasi‐monomorphic variation range in unresectable metastatic colorectal cancer patients. Cancer Sci. 2018;109:3411–3415. 10.1111/cas.13774
Funding information
This research was supported by Japan Agency for Medical Research and Development (AMED) (Grant Number JP18kk0205004), MSD, and FALCO biosystems.
Clinical trial registration: UMIN000024144.
REFERENCES
- 1. Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260(5109):816‐819. [DOI] [PubMed] [Google Scholar]
- 2. Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363(6429):558‐561. [DOI] [PubMed] [Google Scholar]
- 3. Le DT, Durham JN, Smith KN, et al. PD‐1 blockade in tumors with mismatch‐repair deficiency. N Engl J Med. 2015;372(26):2509‐2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Overman MJ, McDermott R, Leach JL, et al. Nivolumab in patients with metastatic DNA mismatch repair‐deficient or microsatellite instability‐high colorectal cancer (CheckMate 142): an open‐label, multicentre, phase 2 study. Lancet Oncol. 2017;18(9):1182‐1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96(4):261‐268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bacher JW, Flanagan LA, Smalley RL, et al. Development of a fluorescent multiplex assay for detection of MSI‐High tumors. Dis Markers. 2004;20(4–5):237‐250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Buhard O, Cattaneo F, Wong YF, et al. Multipopulation analysis of polymorphisms in five mononucleotide repeats used to determine the microsatellite instability status of human tumors. J Clin Oncol. 2006;24(2):241‐251. [DOI] [PubMed] [Google Scholar]
- 8. Goel A, Nagasaka T, Hamelin R, Boland CR. An optimized pentaplex PCR for detecting DNA mismatch repair‐deficient colorectal cancers. PLoS ONE. 2010;5(2):e9393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Patil DT, Bronner MP, Portier BP, Fraser CR, Plesec TP, Liu X. A five‐marker panel in a multiplex PCR accurately detects microsatellite instability‐high colorectal tumors without control DNA. Diagn Mol Pathol. 2012;21(3):127‐133. [DOI] [PubMed] [Google Scholar]
- 10. Bando H, Okamoto W, Fukui T, Yamanaka T, Akagi K, Yoshino T. Clinical utility of quasimonomorphic variation range (QMVR) on the determination of microsatellite instability (MSI) status in Japanese patients (pts) with colorectal cancer (CRC): GI‐SCREEN‐CRC‐MSI sub‐study 01. J Clin Oncol, 2017;35(suppl 4S):TPS808. [Google Scholar]
- 11. Overbeek LI, Ligtenberg MJ, Willems RW, et al. Interpretation of immunohistochemistry for mismatch repair proteins is only reliable in a specialized setting. Am J Surg Pathol. 2008;32(8):1246‐1251. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


