Abstract
Purpose
Increasing evidence indicates that members of forkhead transcription factor family (FOXO) play key roles in cell proliferation and apoptosis in multiple cancers, including prostate cancer. However, the underlying mechanism of FOXO6 was not yet known. The aim of our work is to investigate the function of FOXO6 in breast cancer.
Methods
In the present study, quantitative real-time polymerase chain reaction and Western blotting analyses were used to detect the expression of FOXO6 in breast cancer tissues and cell lines.
Results
The results revealed that FOXO6 was downregulated in breast cancer tissues and cell lines, compared with adjacent normal tissues and MCF-10A cells, respectively. Moreover, the expression of FOXO6 was associated with the expression of epithelial–mesenchymal transition (EMT) indicator proteins, such as E-cadherin and N-cadherin. Additionally, our findings suggested that FOXO6 expression was negatively associated with tumor size (p=0.002), pathological grade (p=0.018) and lymph node metastasis (p=0.003). Sirt6 has been found to promote cell proliferation and metastasis in several cancers, and quantitative chromatin immunoprecipitation and luciferase reporter assays indicated FOXO6 transcriptionally regulated Sirt6 expression. Furthermore, various functional experiments, including wound healing assay, transwell invasion assay, colony formation assay and Cell Counting Kit-8 assay, revealed that FOXO6 suppressed cell migration, invasion, and proliferation of breast cancer cells.
Conclusion
In conclusion, FOXO6 serves as a tumor suppressor in breast cancer, and suppresses EMT through regulation of Sirt6.
Keywords: FOXO6, EMT, Sirt6, migration, breast cancer
Introduction
Forkhead transcription factor family (FOXO) consists of four members, namely FOXO1, 3, 4 and 6.1 Recent evidence reveal that FOXO family is an attractive target for cancer therapy.2 The members of FOXO family are considered as tumor suppressors that inhibit cell proliferation and promote apoptosis in multiple cancers, including prostate cancer,3 rhabdomyosarcoma,4 lymphoma,5 gastric cancer,6 hepatocellular carcinoma,7 breast cancer,8 lung cancer,9 and leukemia.10 FOXO6, a member of FOXO family, has been found to play a key role in oxidative stress inhibiting cell proliferation.11,12 Additionally, previous data suggest that FOXO6 regulates hepatic glucose homeostasis and synaptic function in mice,13,14 and facilitates oncogenicity of gastric cancer through upregulation of C-myc signal pathway.15 However, the function of FOXO6 in breast cancer has not yet known.
Cancer metastasis results in more than 90% of cancer-related deaths.16 A complex series of events result in the metastasis of cancer cells, including unrestrained cell proliferation, loss of cell–cell adhesion, migration and invasion.17,18
Epithelial–mesenchymal transition (EMT) is the initial step of migration.19 The main characteristic of EMT is the loss of epithelial cell–cell adhesion and the gain of mesenchymal-like morphology.20,21 EMT can improve the invasive ability of tumor cells, thereby enabling them to leave their primary sites.22 Previous studies have shown that different signaling molecules can trigger EMT, such as hepatocyte growth factor, epidermal growth factor (EGF), Notch and Wnt proteins, transforming growth factor-β and bone morphogenetic proteins.23,24
Moreover, matrix metalloproteinases (MMPs) have been found to involve in the breakdown of the extracellular matrix components, thereby promoting tumor progression and metastasis.18,25,26 Upregulation of MMP9 is observed in malignant tumors as well as in cancers with poor outcomes, such as breast cancer.27,28 Furthermore, expression of MMP9 predicts a distant metastasis in breast cancer patients.29
The purpose of our study is to investigate the mechanism of breast cancer development, and explore the function of FOXO6 in breast cancer. Our findings reveal that FOXO6 serves as a tumor suppressor in breast cancer and downregulation of FOXO6 promotes EMT, thereby facilitating cancer cell migration and invasion. Moreover, the results confirm that FOXO6 transcriptionally regulates Sirt6, suppressing MMP9 secretion. We highlight that FOXO6 can serve as a novel molecular therapeutic target for breast cancer.
Materials and methods
Patient samples
A total of 110 pairs of breast cancer tissues and adjacent normal tissues were obtained from the patients admitted to Linyi Central Hospital of Shandong during August 2013 to April 2016. All the patients provided signed informed consent. The University Ethical Committee of Linyi Central Hospital of Shandong approved the experiments of tumor tissues. The tissues were prepared for the study and stored immediately at −80°C.
Cell culture
Human normal breast cell line MCF-10A and human breast cancer cell lines MCF-7 and MDA-MB-231 were obtained from ATCC (Manassas, VA, USA). MCF-7 and MDA-MB-231 cells were maintained in DMEM (HyClone, Logan, UT, USA) supplemented with 1% penicillin/strepto mycin (Sigma-Aldrich, St. Louis, MO, USA) and 10% FBS (HyClone) at 37°C in a humidified 5% CO2 atmosphere. MCF-10A cells were maintained in DMEM/F12 medium supplemented with heat-inactivated horse serum (5%), cholera toxin (1 μg/mL), EGF (10 ng/mL), insulin (10 μg/mL) and hydrocortisone (1 μg/mL) at 37°C in a humidified 5% CO2 atmosphere.
Transfection
The MCF-7 and MDA-MB-231 cells were grown in a six-well plate to almost 70%–80% confluence, and transfected with 2.5 μg empty vector (pCMV-Tag 2B) or FLAG-FOXO6 plasmid, or 50 nM scramble siRNA (SCR) or FOXO6 siRNA using lipo 2000 reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. After transfection for 48 h, cells were harvested and used for subsequent experiments. The sequences of the siRNA used were as follows: FOXO6 siRNA: 5′-GCGACGAAAUGGACUUCAACU-3′ and scramble siRNA: 5′-UUCUCCGAACGUGUCACGU-3′. All the siRNAs were synthesized by GeneChem Inc. (Shanghai, People’s Republic of China).
Western blotting analysis
Western blotting was performed as previously described.30,31 In brief, tissues and cells were lysed in RIPA buffer (Beyotime, Haimen, People’s Republic of China), and after centrifugation at 13,000 g for 15 min, the concentration of protein in the supernatant was measured using BCA kit (Pierce, Waltham, MA, USA). Approximately 50 μg protein was resolved by 10% SDS-PAGE and transferred onto PVDF membranes (EMD Millipore, Billerica, MA, USA). Followed by incubating with 5% skim milk at room temperature for 1 h, the membranes were incubated with antibodies at 4°C overnight. The membranes were washed with PBST three times and incubated with indicated HRP-conjugated secondary antibodies at room temperature for 1 h. Finally, the membranes were washed with PBST three times, and the blots were visualized by enhanced chemiluminescence-based method. The anti-FOXO6 polyclonal antibody (Ab487306) and anti-Sirt6 antibody (Ab62739) were purchased from Abcam (Cambridge, UK), and the anti-β-actin antibody (A3854) was purchased from Sigma-Aldrich.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA of the tissues and cultured cells was extracted using TRIzol reagent according to the manufacturer’s instruction (Thermo Fisher Scientific). Two micrograms of RNA sample was used for reverse transcription using ABScript II cDNA First-Strand Synthesis Kit (ABclonal, Woburn, MA, USA) according to the manufacturer’s instructions. Subsequently, qRT-PCR was performed with SYBR Green I (Solarbio, Beijing, People’s Republic of China). The primer sequences utilized were the following: FOXO6 sense: 5′-AGCGACAGGTTAAGGTTGAG-3′ and antisense: 5′-GTGCTGTTGATGGCGAATTG-3′; E-cadherin forward: 5′-TGCCCAGAAAATGAAAAAGG-3′ and reverse: 5′-GTGTATGTGGCAATGCGTTC-3′; α-catenin forward: 5′-CTCTGTCCTCAGGTTATTAATGC-3′ and reverse: 5′-TGAGGTGACTACGTGAATGAC-3′; N-cadherin forward: 5′-GTCATCACAGTGACAGATGTC-3′ and reverse: 5′-TTCAAAGTCGATTGGTTTGACC-3′; vimentin forward: 5′-GAGAACTTTGCCGTTGAAGC-3′ and reverse: 5′-CTCAATGTCAAGGGCCATCT-3′; and GAPDH forward: 5′-ACGGATTTGGTCGTATTGGGCG-3′ and reverse: 5′-CTCCTGGAAGATGGTGATGG-3′. GAPDH was used as an internal control. Experiments were performed at least three times.
Wound healing assay
Approximately 5 × 106 cells were placed into six-well plates and transfected with 2.5 μg vector or FOXO6 plasmid, or 50 nM siControl or siFOXO6, respectively. When cells were grown to 90%–100% confluence, the cell monolayer was scratched with a 20 μL pipette tip, and washed three times with cold PBS to remove the detached cells. Subsequently, cells were maintained in serum-free DMEM. The wounded areas were imaged under a light microscope at 0 and 24 h after scratches, respectively. The relative distance of cell migration was measured by comparing the difference in wound healing areas using ImageJ (National Institute of Mental Health, Bethesda, MA, USA). Experiments were performed at least three times.
Transwell invasion assay
Transwell invasion assay was performed as described previously.30 In brief, after transfection for 48 h, ~2×104 transfected MDA-MB-231 cells were plated onto transwell chamber (8 μm microporous filters; Corning Costar, Corning, NY, USA) coated with 80 μL Matrigel (1:16 dilution; BD Biosciences, San Jose, CA, USA) and incubated for 16–18 h at 37°C in a humidified 5% CO2 atmosphere. Cells in the upper filters were removed by swab cotton, and invaded cells were fixed in 4% paraformaldehyde at room temperature for 15 min, followed by staining with 0.5% (w/v) crystal violet at room temperature for 10 min. The number of invaded cells was counted in six random optical fields (100× magnification) under a light microscope. Experiments were performed at least three times.
Quantitative chromatin immunoprecipitation (qChIP) assay
Chromatin Immunoprecipitation Assay Kit (Beyotime) was used to perform qChIP assay according to the manufacturer’s protocol. Briefly, MCF-7 or MDA-MB-231 cells were grown to 80%–100% confluence, and washed with cold PBS at least three times and chemically cross-linked with 1% formaldehyde at 37°C for 25 min. Subsequently, cells were lysed with 3 mL lysis buffer at 4°C for 2 h and sonicated for four cycles at 4°C, with each cycle involving lysis and sonication for 20 times. Four micrograms of anti-rabbit IgG or FOXO6 antibody was added to the lysis solution and incubated at 4°C overnight. Following incubation, 70 μL Protein A beads (PBS 1:1 dilution) was used to isolate IgG- or FOXO6-interacted DNA fragments. PCR Purification Kit (Qiagen NV, Venlo, the Netherlands) was used to solute the binding chromatin, and subsequently, qRT-PCR was performed. The primers of sIRT6 used were as follows: forward: 5′-AATAAGAAGGGCCTGATGGC-3′ and reverse: 5′-TGTATGTGGGAGAAAGAAGC-3′. Experiments were performed at least three times.
Luciferase assay
The promoter region of Sirt6 (−2,000 to +100) was cloned into pGL3-basic plasmid. MCF-7 and MDA-MB-231 cells were co-transfected with pGL3-Sirt6, Renilla, vector or FOXO6 plasmids. After transfection for 24 h, cells were collected and lysed, and the luciferase activity was detected by dual-luciferase reporter assay kit (Promega, Madison, WI, USA) according to the manufacturer’s protocol. Experiments were performed at least three times.
Colony formation assay
After transfection for 48 h, ~1 × 104 MCF-7 or MDA-MB-231 cells were placed into six-well plates, and cultured with serum-free DMEM. After incubation for 14 days, cells were fixed with 4% paraformaldehyde solution at room temperature for 10 min and stained with 0.1% (w/v) crystal violet (Sigma) at room temperature for 10 min. The number of colonies was counted under a light microscope. Experiments were performed at least three times.
Cell Counting Kit-8 (CCK-8) assay
CCK-8 assay was used to determine the effect of FOXO6 on cell proliferation. In brief, after transfection for 48 h, ~4,000 MCF-7 or MDA-MB-231 cells were resuspended in 250 μL DMEM and placed into 96-well plates. Then, 25 μL CCK-8 reagent (Beyotime) was added into each well and cultured at 37°C for 1.5 h. The absorbance at OD 450 nm was measured in a microplate reader. Experiments were performed at least three times.
Statistical analyses
All data were analyzed using SPSS 18.0 software (SPSS, Inc., Chicago, IL, USA). The comparison of FOXO6 expression and clinicopathological characteristics of the patients was performed using the Chi-square test. Survival data were analyzed using Kaplan–Meier analysis followed by log-rank test. The correlations between the expression of EMT indicator proteins and FOXO6 were analyzed using Spearman rank correlation coefficients. The difference between two groups was analyzed using Student’s t-test (two-tailed). In the analyses, p < 0.05 was considered as statistically significant.
Results
FOXO6 is downregulated in breast cancer tissues and associated with EMT-associated proteins E-cadherin and N-cadherin
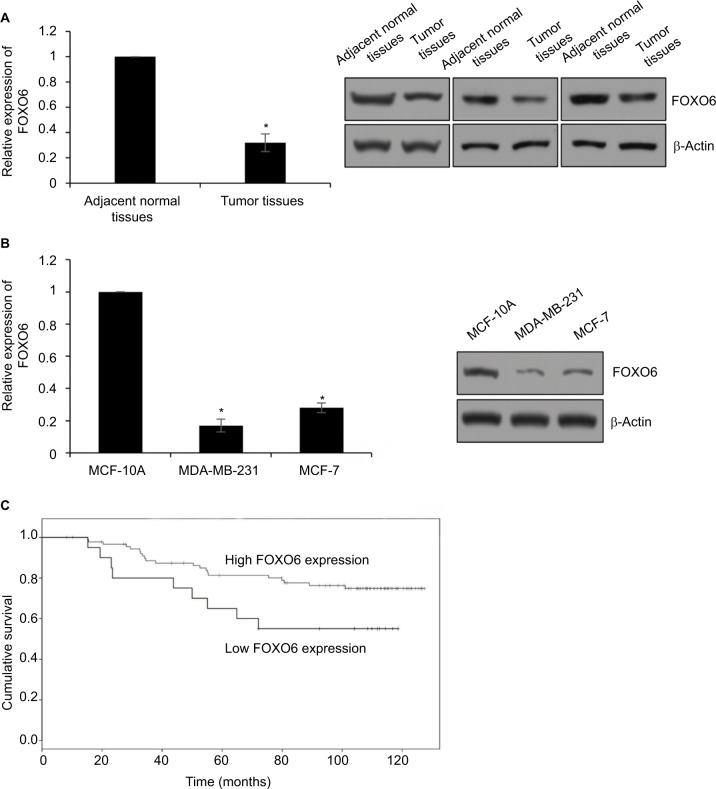
To decipher the function of FOXO6 in breast cancer, we first detected the expression of FOXO6 in breast cancer tissues, using the adjacent normal tissues as control. As shown in Figure 1A, we found the expression of FOXO6 in tumor tissues was less than that in adjacent normal tissues (Figure 1A). We also found FOXO6 was downregulated in breast cancer cell lines MCF-7 and MDA-MB-231, using the human normal breast MCF-10A cells as control (Figure 1B). Subsequently, we analyzed the correlation of FOXO6 expression and clinicopathological parameters. As shown in Table 1, we found that FOXO6 expression was significantly negatively associated with tumor size (p=0.002), pathological grade (p=0.018) and lymph node metastasis (p=0.003), suggesting FOXO6 plays an important role in tumor proliferation and invasion in breast cancer (Table 1). Furthermore, Kaplan–Meier analysis of survival data showed the patients with low expression of FOXO6 had a poor prognosis (p < 0.05, log-rank test; Figure 1C). EMT is a complex process which contributes to cancer cell metastasis and results in poor prognosis. To explore the correlation between FOXO6 expression and EMT indicator proteins in breast cancer tissues, Spearman correlation analysis was performed. As shown in Table 2, low expression of FOXO6 was found to associate with low expression of E-cadherin (r=−2.80, p=0.03) and α-catenin (r=−0.210, p=0.027) and high expression of N-cadherin (r=0.280, p=0.003) and vimentin (r=0.232, p=0.015) in the breast cancer tissues. These results suggest that FOXO6 is downregulated in breast cancer and the expression of FOXO6 is closely associated with EMT in the breast cancer tissues.
Figure 1.
FOXO6 is downregulated in breast cancer tissues and associated with EMT-associated proteins E-cadherin and N-cadherin.
Notes: (A) The expression of FOXO6 in tumor tissues and adjacent normal tissues was detected by qRT-PCR and Western blotting analysis, respectively. Tumor tissues vs adjacent normal tissues, *p < 0.05. (B) The expression of FOXO6 in breast cancer cell lines was detected by qRT-PCR and Western blotting analysis, respectively. Human normal breast MCF-10A cells were used as control. MDA-MB-231 or MCF-7 vs MCF-10A, *p < 0.05. (C) Kaplan–Meier analysis was used to analyze overall survival according to the expression status of FOXO6 (log-rank).
Abbreviations: EMT, epithelial–mesenchymal transition; qRT-PCR, quantitative real-time polymerase chain reaction; FOXO, forkhead transcription factor family.
Table 1.
Clinicopathological data of the 110 breast cancer patients
| Variables | n | FOXO6 protein expression
|
P value | |
|---|---|---|---|---|
| Low | High | |||
| Age | ||||
| <40 | 58 | 33 | 25 | 0.621 |
| ≥40 | 52 | 32 | 20 | |
| Tumor size (diameter) | ||||
| Small (<3 cm) | 64 | 30 | 34 | 0.002 |
| Large (≥3 cm) | 46 | 35 | 11 | |
| Pathological grade | ||||
| I–II | 56 | 27 | 29 | 0.018 |
| III–IV | 54 | 38 | 16 | |
| Metastasis | ||||
| Yes | 53 | 39 | 14 | 0.003 |
| No | 57 | 26 | 31 | |
Abbreviation: FOXO, forkhead transcription factor family.
Table 2.
Spearman correlation analysis between expression of FOXO6 and EMT indicator proteins in breast cancer
| Variables | n | FOXO6 expression
|
P value | r value | |
|---|---|---|---|---|---|
| Low | High | ||||
| E-cadherin expression | 65 | 45 | |||
| High | 58 | 22 | 28 | 0.03 | −2.80 |
| Low | 52 | 43 | 17 | ||
| α-Catenin expression | |||||
| High | 28 | 29 | 0.027 | −0.210 | |
| Low | 37 | 16 | |||
| N-cadherin expression | |||||
| High | 43 | 17 | 0.003 | 0.280 | |
| Low | 22 | 28 | |||
| Vimentin expression | |||||
| High | 53 | 37 | 15 | 0.015 | 0.232 |
| Low | 57 | 28 | 30 | ||
Abbreviations: FOXO, forkhead transcription factor family; EMT, epithelial–mesenchymal transition.
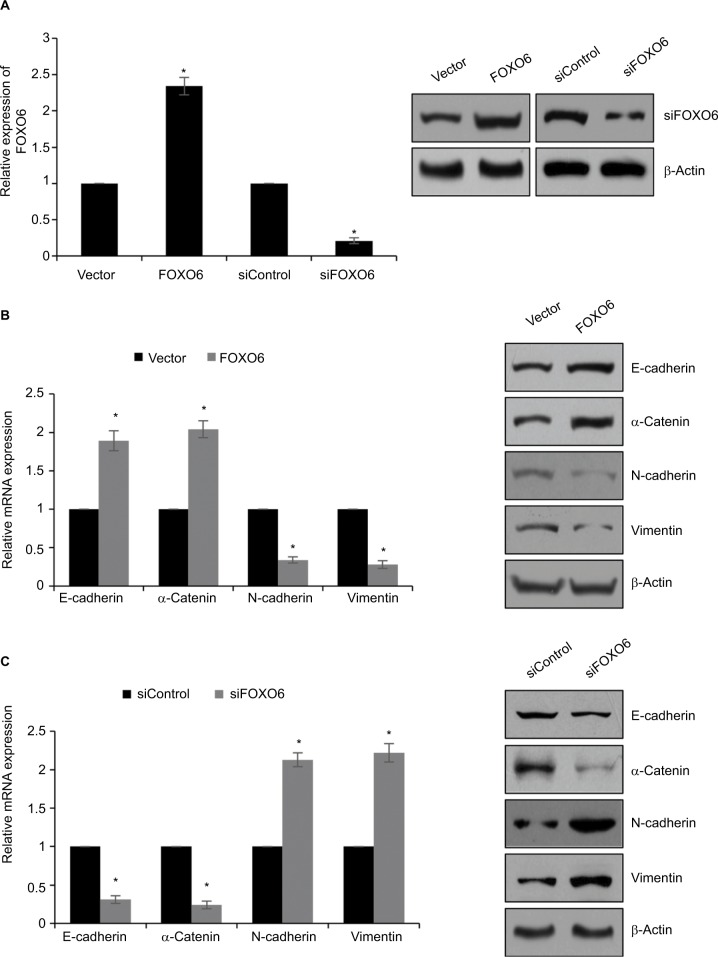
FOXO6 suppresses EMT in breast cancer cells
Our findings revealed that low expression of FOXO6 is negatively associated with EMT indicator proteins in breast cancer. To further detect the effect of FOXO6 on EMT, human breast cancer MCF-7 cells were transfected with vector or FOXO6 plasmid, or scramble siRNA or FOXO6 siRNA, respectively. The expression of FOXO6 was determined by qRT-PCR and Western blotting analyses. The expression of FOXO6 was significantly increased 2.3-fold or decreased by FOXO6 plasmid or FOXO6 siRNA, respectively (Figure 2A). Subsequently, the expression of EMT-associated proteins was detected. As shown in Figure 2B, ectopic expression of FOXO6 led to increase in both mRNA and protein levels of E-cadherin and α-catenin, but the expression of N-cadherin and vimentin was decreased (Figure 2B). On the contrary, inhibition of FOXO6 obviously decreased the expression of E-cadherin and α-catenin, and increased the expression of N-cadherin and vimentin (Figure 2C). Together, FOXO6 suppresses EMT in breast cancer cells.
Figure 2.
FOXO6 suppresses EMT in breast cancer cells.
Notes: (A) MCF-7 cells were transfected with vector or FOXO6, or siControl or siFOXO6, respectively. After transfection for 48 h, the expression of FOXO6 was determined by qRT-PCR and Western blotting analyses. FOXO6 vs vector, siFOXO6 vs siControl, *p < 0.05. (B) FOXO6 was overexpressed in MCF-7 cells. The mRNA and protein levels of EMT-associated proteins were detected by qRT-PCR and Western blotting analyses. FOXO6 vs vector, *p < 0.05. (C) FOXO6 was knocked down in MCF-7 cells. The mRNA and protein levels of EMT-associated proteins were detected by qRT-PCR and Western blotting analyses. siFOXO6 vs siControl, *p < 0.05.
Abbreviations: FOXO, forkhead transcription factor family; EMT, epithelial–mesenchymal transition; qRT-PCR, quantitative real-time polymerase chain reaction.
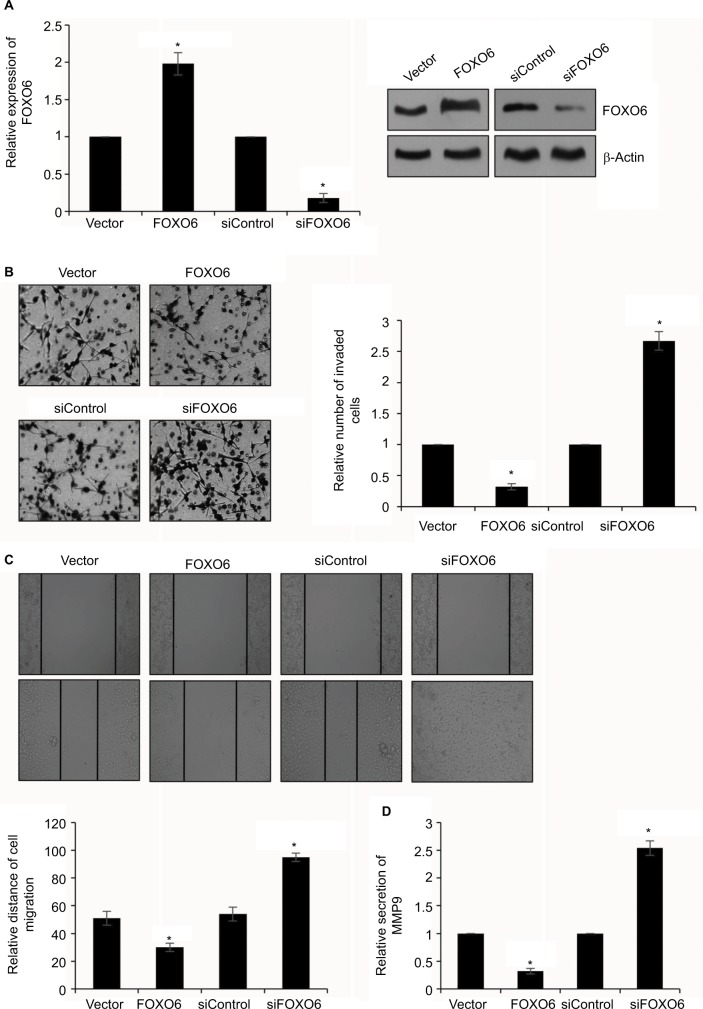
FOXO6 inhibits breast cancer cells migration and invasion
To further characterize the effect of FOXO6 on breast cancer cell migration and invasion, highly invasive human breast cancer MDA-MB-231 cells were transfected with vector or FOXO6 plasmid, or control siRNA or FOXO6 siRNA, respectively. The expression of FOXO6 was determined by qRT-PCR and Western blotting analyses. The expression of FOXO6 was significantly increased 2-fold or decreased by FOXO6 plasmid or FOXO6 siRNA, respectively (Figure 3A). Following transfection, transwell invasion assay and wound healing assay were performed. The result of transwell invasion assay suggested that elevated expression of FOXO6 decreased the number of invasive cells compared with control group; however, knockdown of FOXO6 obviously increased almost 2.5-fold the number of invasive cells (Figure 3B). Similar results were observed in wound healing assay, revealing that elevated expression of FOXO6 reduced the relative distance of migration; however, knockdown of FOXO6 increased the relative distance of migration (Figure 3C). A previous work revealed MMP9 was associated with distant metastasis in breast cancer patients.29 So, we next detected the effect of FOXO6 on MMP9 secretion using ELISA, and found that the elevated expression of FOXO6 suppressed MMP9 secretion; however, inhibition of FOXO6 facilitated MMP9 secretion (Figure 3D). These data suggest that FOXO6 facilitates breast cancer cells migration and invasion.
Figure 3.
FOXO6 inhibits breast cancer cells migration and invasion.
Notes: (A) MDA-MB-231 cells were transfected with vector or FOXO6, or siControl or siFOXO6, respectively. After transfection for 48 h, the expression of FOXO6 was determined by qRT-PCR and Western blotting analyses. FOXO6 vs vector, siFOXO6 vs siControl, *p < 0.05. (B) FOXO6 was overexpressed or knocked down in MDA-MB-231 cells. The effect of FOXO6 on cell invasion was determined by transwell invasion assay. FOXO6 vs vector, siFOXO6 vs siControl, *p < 0.05. (C) FOXO6 was overexpressed or knocked down in MDA-MB-231 cells. The effect of FOXO6 on cell migration was determined by wound healing assay. FOXO6 vs vector, siFOXO6 vs siControl, *p < 0.05. (D) FOXO6 was overexpressed or knocked down in MDA-MB-231 cells. The secretion of MMP9 was detected by ELISA. FOXO6 vs vector, siFOXO6 vs siControl, *p < 0.05.
Abbreviations: FOXO, forkhead transcription factor family; qRT-PCR, quantitative real-time polymerase chain reaction.
Sirt6 is a direct transcriptional target of FOXO6
A recent report has showed that Sirt6 promotes metastasis of non-small-cell lung cancer via the ERK1/2/MMP9 pathway.32 Our works indicated that FOXO6 suppressed MMP9 secretion, and we assumed that FOXO6 might regulate MMP9 secretion through regulation of Sirt6. To verify our hypothesis, FOXO6 was overexpressed or knocked down in MCF-7 and MDA-MB-231 cells, and the expression of Sirt6 was determined by qRT-PCR and Western blotting analyses. The results revealed that ectopic expression of FOXO6 significantly inhibited the expression of Sirt6, and inhibition of FOXO6 improved the expression of Sirt6 (Figure 4A and B). Both mRNA and protein levels of Sirt6 were regulated by FOXO6, suggesting FOXO6 might transcriptionally regulate Sirt6. Subsequently, qChIP assay was performed in MCF-7 and MDA-MB-231 cells, which revealed the Sirt6 promoter region exhibited obvious enrichment after immunoprecipitation with an anti-FOXO6 antibody (Figure 4C). Subsequently, we cloned the promoter region of Sirt6 into pGL3-basic plasmid and performed luciferase reporter assay. As shown in Figure 4D, FOXO6 could stimulate Sirt6 activity, and the relative luciferase activity was increased by FOXO6 in a dose-dependent manner (Figure 4D). Taken together, these data demonstrate that FOXO6 transcriptionally suppresses Sirt6.
Figure 4.
Sirt6 is a direct transcriptional target of FOXO6.
Notes: (A) FOXO6 was overexpressed or knocked down in MCF-7 and MDA-MB-231 cells. The mRNA level of Sirt6 was assessed by qRT-PCR assay. FOXO6 vs vector, siFOXO6 vs siControl, *p < 0.05. (B) FOXO6 was overexpressed or knocked down in MCF-7 and MDA-MB-231 cells. The protein level of Sirt6 was assessed by Western blotting assay. (C) qChIP sassy was performed using anti-FOXO6 antibody. FOXO6 vs IgG, *p < 0.05. (D) FOXO6 was overexpressed or knocked down in MCF-7 and MDA-MB-231 cells. After transfection for 24 h, relative luciferase activity was detected by dual-luciferase reporter assay kit. *p < 0.05.
Abbreviations: FOXO, forkhead transcription factor family; qRT-PCR, quantitative real-time polymerase chain reaction; qChIP, quantitative chromatin immunoprecipitation.
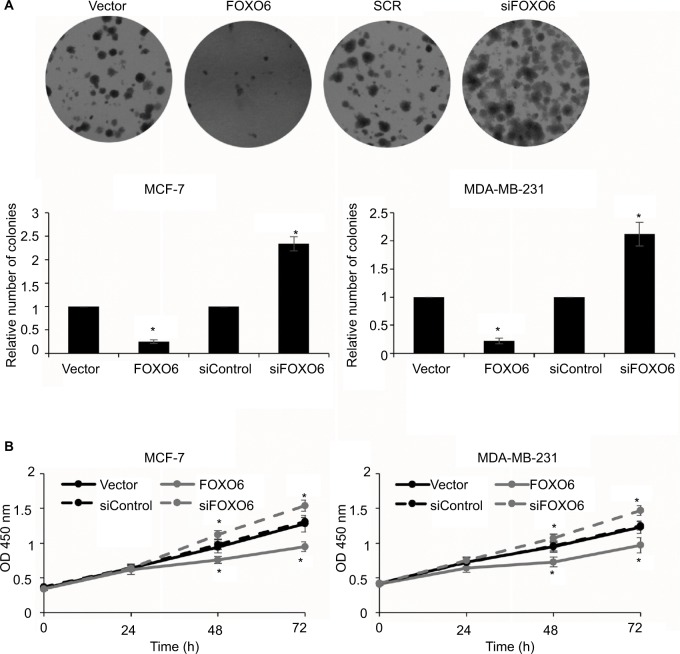
FOXO6 inhibits the proliferation of breast cancer cells
As shown in Table 1, the expression of FOXO6 was associated with tumor size, so we assumed that FOXO6 might inhibit cell proliferation in breast cancer. To verify our hypothesis, colony formation assay and CCK-8 assay were performed. The result of colony formation sassy demonstrated that the elevated expression of FOXO6 markedly decreased the number of colonies, but knockdown of FOXO6 significantly increased the number of colonies (Figure 5A). Meanwhile, the result of CCK-8 assay suggested that ectopic expression of FOXO6 suppressed the proliferation rate of MCF-7 and MDA-MB-231 cells; however, inhibition of FOXO6 enhanced the proliferation rate of MCF-7 and MDA-MB-231 cells (Figure 5B). These results suggest FOXO6 inhibits the proliferation of breast cancer cells.
Figure 5.
FOXO6 inhibits the proliferation of breast cancer cells.
Notes: (A) FOXO6 was overexpressed or knocked down in MCF-7 and MDA-MB-231 cells. Colony formation assay was used to detect the effect of FOXO6 on cell proliferation. FOXO6 vs vector, siFOXO6 vs siControl, *p < 0.05. (B) FOXO6 was overexpressed or knocked down in MCF-7 and MDA-MB-231 cells. CCK-8 assay was used to detect the effect of FOXO6 on cell proliferation. FOXO6 vs vector, siFOXO6 vs siControl, *p < 0.05.
Abbreviation: CCK-8, Cell Counting Kit-8; FOXO, forkhead transcription factor family.
Discussion
Breast cancer has become one of the most common cancers in women, and the incidence of breast carcinoma is steadily increasing. Therefore, it is necessary to further investigate the mechanism of breast cancer development and identify a new therapeutic target.
Our present work reported that FOXO6 was downregulated in breast cancer tissues, and its expression was negatively associated with tumor size (p=0.002), pathological grade (p=0.018) and metastasis (p=0.003). Low FOXO6 expression was also closely correlated with EMT in breast cancer tissues. Interestingly, we found that low expression of FOXO6 predicted a poor prognosis. These results indicated that FOXO6 might serve as a tumor suppressor in breast cancer. Our findings were also consistent with a previous report.33
Multiple reports have shown that the members of FOXO family regulate EMT.33,34,35 So, we investigated the function of FOXO6 in EMT using human breast cancer MCF-7 cells. To our surprise, we found that FOXO6 upregulated E-cadherin and α-catenin, and downregulated N-cadherin and vimentin. Notably, Liu et al have reported that FOXO3a suppresses EMT in prostate cancer cells.3 These results suggest that FOXO6 expression is associated with a poor prognosis by regulation of EMT in breast cancer. Further, we found FOXO6 suppressed cell migration and invasion. MMP9 has been found to regulate cancer cell migration and invasion. Our findings showed that FOXO6 inhibited MMP9 secretion in MDA-MB-231 cells. A previous report has shown that Sirt6 promotes metastasis of non-small-cell lung cancer via the ERK1/2/MMP9 pathway.32 Subsequently, qChIP and luciferase reporter assays demonstrated that FOXO6 transcriptionally regulates Sirt6 in breast cancer cells, thereby suppressing MMP9 secretion.
In summary, our results revealed that FOXO6 expression is correlated with a poor prognosis and EMT in breast cancer. The cell culture experiments, including wound healing assay and transwell invasion assay, indicated ectopic expression of FOXO6 promotes cell migration and invasion by inducing the EMT of breast cancer cells. Moreover, FOXO6 also suppressed breast cell proliferation. Thus, the present study indicates the possibility that FOXO6 may serve as a novel molecular therapeutic target for breast cancer.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Kim CG, Lee H, Gupta N, et al. Role of Forkhead Box Class O proteins in cancer progression and metastasis. Semin Cancer Biol. 2018;50:142–151. doi: 10.1016/j.semcancer.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coomans de Brachène A, Demoulin JB. FOXO transcription factors in cancer development and therapy. Cell Mol Life Sci. 2016;73(6):1159–1172. doi: 10.1007/s00018-015-2112-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu H, Yin J, Wang H, et al. FOXO3a modulates WNT/β-catenin signaling and suppresses epithelial-to-mesenchymal transition in prostate cancer cells. Cell Signal. 2015;27(3):510–518. doi: 10.1016/j.cellsig.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Schmitt-Ney M, Camussi G. The PAX3-FOXO1 fusion protein present in rhabdomyosarcoma interferes with normal FOXO activity and the TGF-β pathway. PLoS One. 2015;10(3):e0121474. doi: 10.1371/journal.pone.0121474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vandenberg CJ, Motoyama N, Cory S. FoxO3 suppresses Myc-driven lymphomagenesis. Cell Death Dis. 2016;6:e2046. doi: 10.1038/cddis.2015.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su L, Liu X, Chai N, et al. The transcription factor FOXO4 is down-regulated and inhibits tumor proliferation and metastasis in gastric cancer. BMC Cancer. 2014;14:378. doi: 10.1186/1471-2407-14-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamaguchi F, Hirata Y, Akram H, et al. FOXO/TXNIP pathway is involved in the suppression of hepatocellular carcinoma growth by glutamate antagonist MK-801. BMC Cancer. 2013;13:468. doi: 10.1186/1471-2407-13-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JJ, Lee HJ, Son BH, et al. Expression of FOXM1 and related proteins in breast cancer molecular subtypes. Int J Exp Pathol. 2016;97(2):170–177. doi: 10.1111/iep.12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu H, Yin J, Wang C, Gu Y, Deng M, He Z. FOXO3a mediates the cytotoxic effects of cisplatin in lung cancer cells. Anticancer Drugs. 2014;25(8):898–907. doi: 10.1097/CAD.0000000000000117. [DOI] [PubMed] [Google Scholar]
- 10.Zhu H. Targeting forkhead box transcription factors FOXM1 and FOXO in leukemia (Review) Oncol Rep. 2014;32(4):1327–1334. doi: 10.3892/or.2014.3357. [DOI] [PubMed] [Google Scholar]
- 11.Estall JL. The Foxo family: partners in crime or silent heroes. Endocrinology. 2012;153(2):549–551. doi: 10.1210/en.2011-2080. [DOI] [PubMed] [Google Scholar]
- 12.Kim DH, Park MH, Chung KW, et al. The essential role of FoxO6 phosphorylation in aging and calorie restriction. Age (Dordr) 2014;36(4):9679. doi: 10.1007/s11357-014-9679-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim DH, Perdomo G, Zhang T, et al. FoxO6 integrates insulin signaling with gluconeogenesis in the liver. Diabetes. 2011;60(11):2763–2774. doi: 10.2337/db11-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salih DA, Rashid AJ, Colas D, et al. FoxO6 regulates memory consolidation and synaptic function. Genes Dev. 2012;26(24):2780–2801. doi: 10.1101/gad.208926.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qinyu L, Long C, Zhen-dong D, et al. FOXO6 promotes gastric cancer cell tumorigenicity via upregulation of C-myc. FEBS Lett. 2013;587(14):2105–2111. doi: 10.1016/j.febslet.2013.05.027. [DOI] [PubMed] [Google Scholar]
- 16.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331(6024):1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 17.Yilmaz M, Christofori G, Lehembre F. Distinct mechanisms of tumor invasion and metastasis. Trends Mol Med. 2007;13(12):535–541. doi: 10.1016/j.molmed.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 18.Chabottaux V, Noel A. Breast cancer progression: insights into multifaceted matrix metalloproteinases. Clin Exp Metastasis. 2007;24(8):647–656. doi: 10.1007/s10585-007-9113-7. [DOI] [PubMed] [Google Scholar]
- 19.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thiery JP. Epithelial-mesenchymal transitions in development and pathologies. Curr Opin Cell Biol. 2003;15(6):740–746. doi: 10.1016/j.ceb.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 22.Yao D, Dai C, Peng S. Mechanism of the mesenchymal-epithelial transition and its relationship with metastatic tumor formation. Mol Cancer Res. 2011;9(12):1608–1620. doi: 10.1158/1541-7786.MCR-10-0568. [DOI] [PubMed] [Google Scholar]
- 23.Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009;19(2):156–172. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132(14):3151–3161. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- 25.Sato H, Takino T, Okada Y, et al. A matrix metalloproteinase expressed on the surface of invasive tumour cells. Nature. 1994;370(6484):61–65. doi: 10.1038/370061a0. [DOI] [PubMed] [Google Scholar]
- 26.Hidalgo M, Eckhardt SG. Development of matrix metalloproteinase inhibitors in cancer therapy. J Natl Cancer Inst. 2001;93(3):178–193. doi: 10.1093/jnci/93.3.178. [DOI] [PubMed] [Google Scholar]
- 27.Radisky ES, Radisky DC. Matrix metalloproteinase-induced epithelial-mesenchymal transition in breast cancer. J Mammary Gland Biol Neoplasia. 2010;15(2):201–212. doi: 10.1007/s10911-010-9177-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Köhrmann A, Kammerer U, Kapp M, Dietl J, Anacker J. Expression of matrix metalloproteinases (MMPs) in primary human breast cancer and breast cancer cell lines: new findings and review of the literature. BMC Cancer. 2009;9:188. doi: 10.1186/1471-2407-9-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van’t Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 30.Lin G, Sun L, Wang R, Guo Y, Xie C. Overexpression of muscarinic receptor 3 promotes metastasis and predicts poor prognosis in non-small-cell lung cancer. J Thorac Oncol. 2014;9(2):170–178. doi: 10.1097/JTO.0000000000000066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun L, Bai L, Lin G, et al. CUEDC2 down-regulation is associated with tumor growth and poor prognosis in lung adenocarcinoma. Oncotarget. 2015;6(24):20685–20696. doi: 10.18632/oncotarget.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bai L, Lin G, Sun L, et al. Upregulation of SIRT6 predicts poor prognosis and promotes metastasis of non-small cell lung cancer via the ERK1/2/MMP9 pathway. Oncotarget. 2016;7(26):40377–40386. doi: 10.18632/oncotarget.9750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu HJ, Zhang LG, Wang ZH, Guo XX. FoxO6 inhibits cell proliferation in lung carcinoma through up-regulation of USP7. Mol Med Rep. 2015;12(1):575–580. doi: 10.3892/mmr.2015.3362. [DOI] [PubMed] [Google Scholar]
- 34.Lo PK, Lee JS, Liang X, Sukumar S. The dual role of FOXF2 in regulation of DNA replication and the epithelial-mesenchymal transition in breast cancer progression. Cell Signal. 2016;28(10):1502–1519. doi: 10.1016/j.cellsig.2016.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cai J, Tian AX, Wang QS, et al. FOXF2 suppresses the FOXC2-mediated epithelial-mesenchymal transition and multidrug resistance of basal-like breast cancer. Cancer Lett. 2015;367(2):129–137. doi: 10.1016/j.canlet.2015.07.001. [DOI] [PubMed] [Google Scholar]