Abstract
Aim: BAP1 is frequently mutated in clear cell renal cell carcinoma (ccRCC) with a definitive role still unclear.
Methods: In silico analysis of BAP1-mutant and wild-type gene enrichment and functional annotation in TCGA-KIRC dataset was performed. Target gene was studied based on functional clustering and was knowledge-based. Validation using in-house pathological sections were performed immunohistochemically. In vitro and in vivo studies on target gene were performed.
Results: The TCGA ccRCC dataset included 534 ccRCC samples. BAP1 was frequently mutated and more frequently downregulated in ccRCC compared to normal kidney tissue or benign renal tumors. In the analysis between samples with BAP1 mutation (N = 33) and pan-negative (N = 33), we found that cancers with BAP1 mutation was significantly enriched for 14 pathways, of which 3 were DNA repair pathways, in which EZH2 played a role. CcRCC patients with lower BAP1 expression had poor prognosis and showed higher EZH2 expression, which also conferred worsened survival. Genetic and pharmaceutical inhibition of EZH2 not only inhibited BAP1-mutatn ccRCC cell viability and invasion but also abrogated genetic replenishing of BAP1 expression. Validation cohort encompassing 62 ccRCC samples confirmed the worsened phenotype for cases with higher EZH2 expression and significant positive correlation between expressions of EZH2 and BAP1. EZH2 inhibitor also inhibited tumor growth in xenograft mouse model with BAP1-mutated ccRCC cells with unremarkable toxicity.
Conclusion: CcRCC with decreased BAP1 level has poor prognosis and is associated with higher EZH2 expression. Inhibition of EZH2 in BAP1-mutated entity holds promise for further investigation.
Keywords: EZH2 Expression, BAP1-mutant
Introduction
Renal cell carcinoma (RCC), or kidney cancer, takes up 2%-3% of all malignant tumours. The age standardized incidence for RCC is 5.8/100,000 in developed regions and the mortality is 1.4/100,000 1. RCC is the most common solid mass in kidney and comprises of 90% of all kidney cancers and clear cell renal cell carcinoma (ccRCC) is the predominant subtype of RCC. Recent next-generation sequencing has revealed predominant genetic disposition for distinctive histological subtypes including frequent mutations in VHL, PBRM1, SETD2, BAP1, in some series KDM5C 2, 3.
Unlike the definitive signalling axis involving VHL/HIF axis in ccRCC, the latter 4 genes were suggested to maintain genomic stability and among which, BAP1, functions as both a tumour suppressor and metastasis suppressor. BAP1 has the deubiquitinase activity and is the member of polycomb-group proteins that maintains long-term silencing of genes in charge of cell-fate determination and stem cell pluripotency 4, 5. Mutation in BAP1 has been reported in a variety of cancers. Somatic alteration in BAP1 has been noted in breast cancer and lung cancer, where the gene was first identified as a tumour suppressor 4. Later in uveal melanomas (UM), BAP1 is also reported to be mutated in ~50% of cases with functional impairment in case of missense mutation within UCH and ULD deubiquitinase domain 6. Mutated BAP1 in UM is associated with a more aggressive phenotype. Similar mutational spectrum is also observed in mesothelioma 7. In ccRCC, BAP1 loss is characterized with both mutation and loss of heterozygosity, the latter owing to its proximity to 3p25 region that is often deleted in kidney cancer. It is now suggested that loss of VHL function by mutation with and without LOH or methylation occurs in almost if not all ccRCC and VHL loss is accepted as a truncal genetic alteration in ccRCC 8. PBRM1 and BAP1 mutations are shown to be mutually exclusive in ccRCC and are suggested to be later-on events due to selectivity 9. CcRCC patients with BAP1 mutations have worse prognosis compared with the rest 7. A comprehensive understanding of the genetic features of BAP1-mutated ccRCC remains unclear and is urgently needed.
Here we performed in silico analysis of BAP1-mutant and wild-type gene enrichment and functional annotation in TCGA-KIRC dataset was performed. Target gene was studied based on functional clustering and was knowledge-based. In vitro and in vivo studies on target gene were performed.
Materials and methods
We have here performed an in silico analysis by reproducing TCGA database using the online cBioPortal platform, as previously reports 10-12. We confirm that our reproduction and report of the TCGA kidney cancer dataset and relevant data met no restriction 3. By analysing the TCGA Kidney Cancer Provisional dataset on the cBioPortal dataset, we first captured 538 samples with genomic and clinicopathological data. We then captured samples with BAP1 mutation and excluded samples with mutations of any of the genes including VHL, PBRM1, SETD2 and KDM5C, generating a 'pan-negative' cohort (Control 1). Also, cases solely without BAP1 mutation were set as Control 2. For both groups, survivals were plotted using Kaplan-Meier method. For Control 1, the enrichment was studied at mutation, copy number variance (CNV), expression and protein levels. In the expressional enrichment analysis, we extracted differentially expressed genes that passed both P and Q (false discover rate, FDR) thresholds. Genes were code-converted (http://biit.cs.ut.ee/gprofiler/gconvert.cgi) to EntrezGene ID and input to the DAVID platform (https://david.ncifcrf.gov/) for pathway analysis and functional clustering. The KEGG dataset was designated.
Tissue microarrays (TMAs) and immunohistochemistry (IHC)
For TMAs construction, slides were deparaffinized and heated in citrate buffer pH6 for antigenic retrieval. The primary antibodies were EZH2 (Abcam, ab186006, Shanghai, China), BAP1 (sc-28383, Santa Cruz Biotechnology, OR, USA), VHL (sc-135657, Santa Cruz Biotechnology, OR, USA), PBRM1 (SAB1409486, Sigma-Aldrich), SETD2 (AV47617, Sigma-Aldrich), and KDM5C (SAB4301634, Sigma-Aldrich). A validation study was conducted to confirm findings in silico. Six ccRCC samples and 3 normal kidney samples were stained for BAP1 using IHC. Another 62 formalin-fixed, paraffin embedded tissue blocks of ccRCC were used. All patients underwent partial or radical nephrectomy at Nanjing Medical University Affiliated Suzhou Hospital and informed consent was acquired from each patient. Immunohistochemistry was performed using the streptavidin-biotin-peroxidase method with diaminobenzidine as the chromogen (KitLSAB, Dakocytomotion, Glostrup, Denmark). Negative controls were obtained after omission of the primary antibody or incubation with an irrelevant antibody. IHC staining readouts and assessment were graded according to the intensity and extensity of the positively stained cells semi-quantatively 13-16.
Publicly available gene expression data sets and clinical data sets
In this article, we used IHC and prognosis data of BAP1 in human renal tissues and cancer tissues from The Human Protein Atlas portal (http://www.proteinatlas.org). All data were available directly online. We made use of some separate RCC clinical data sets that were publicly usable. These contained the data from ONCOMINE and data reported in GDS2880.
Cell line and viral transduction
The 769-P and 786-O renal cell carcinoma cell lines were originally obtained from ATCC cell bank. Cells were cultured in complete DMEM media supplemented with 10% of fetal bovine serum. Lentiviral overexpression of BAP1 was constructed using the cDNA clone of BAP1 purchased from OriGene (Rockville, MD). A standard recombination protocol was followed for the generation of a BAP1-bearing lentivirus, which was subsequently produced in abundance using the 293A cells. A 100 MOI of efficiency was used in all assays. For shRNA construction, the target sequence was referenced from TRC (http://portals.broadinstitute.org/gpp/public/), as follows: TRCN0000007371 as BAP1 shRNA#1, and TRCN0000007370 as BAP1 shRNA#2. ShRNA against EZH2 was generated similarly. Vectors with puromycin resistance were used and were transected in to cells using the Fugene system. Stably transfected cells were selected using puromycin at 1:5000 of dilution.
Quantitative PCR and western blotting
For quantitative PCR, a standard protocol was followed. Total RNA was extracted with Trizol and was converted to cDNA. Primers for BAP1 were 5'-GCT CGT GGA AGA TTT CGG TGT-3' as forward, and 5'- TCA TCA ATC ACG GAC GTA TCA TC-3' as reverse. GAPDH was used as internal reference. cDNA was subject to the ABI 7500 for quantitative PCR procedure and the program was per manufacturer's instruction of SYBR Green system. The expression of BAP1 was calculated according to internal references and expressed as folds over the control group.
For western blotting, total protein of cell lysates was extracted and equal protein amount of 25 μg was loaded onto 10% sodium dodecyl sulphate polyacrylamide gel. Gels were subsequently transferred to nitrocellulose membrane. The membranes were blockedd with 5% non-fat milk. Primary anti-human BAP1 (Cell Signaling), EZH2 (Abcam), H3K27me3 (Abcam), and Actin (Abcam) antibodies were then added at the dose recommended by the manufacturers and membranes were kept at 4°C overnight. Procedure was finalized by enhanced chemiluminescence.
Proliferation assay
Proliferation was studies using the crystal violet assay. Cells were cultured in the 96-well plates and depleted of media at different time points (24 h, 48 h, and 72 h). Cells were then dyed using crystal violet and subsequently dissolved using methanol. Plates were read on a plate reader with absorbance at 540 nm of wavelength.
Transwell and colony formation assays
Transwell assays were performed to profile cell invasion and migration. Briefly, the inserts were coated with or without Matrigel for invasion and migration assays, respectively. Cells were cultured in media without serum and were seeded in the inserts, which were then placed in the lower chamber filled with complete media. After 72 h, inserts were cleared for inner contents and the outer membranes were dyed with crystal violet and counted for cell number microscopically.
The colony formation was performed to profile anchorage-independent growth of cells. Briefly, 6-well plates were paved with 3 layers of media. The lowest were mixture of 0.6% agar and complete media. On top of that were mixture of 0.4% agar and complete media, in which ~1000 cells were seeded. The uppermost layer was 1 mL of compete media with 100 μm of EPZ011989 (Selleck Chemicals) replaced every 3 days. Two weeks later, the plate was dyed using crystal violet and tubers were counted microscopically.
Animal model
Twenty nude mice at 6 weeks of age were injected with 107 of 769-P cells at dorsal axillary region subcutaneously. Tumors were monitored for calibration every 3 days. On day 5 after implantation, mice were treated with either vehicle or 500 mg/kg BID EPZ011989. Mice were euthanized on week 5.
Statistical analyses
The Stata version 12.0 software was used to analyze the data and graphs were generated directly from cBioPortal and Human Protein Atlas platforms. Correlation of gene expressions was analyzed using the Spearman test. Survival analyses were studied between Group 1 and Group 2, which were plotted using the Kaplan-Meier curve and compared using the Log-rank test. The two-tailed t test or Mann-Whitney test was used for comparison of means between 2 groups for parametric or non-parametric variables, respectively. Correlation was studied using the Spearman test. The P value of < 0.05 was accepted as statistically significant.
Results
Kidney cancer has decreased BAP1 level which relates to worsened prognosis
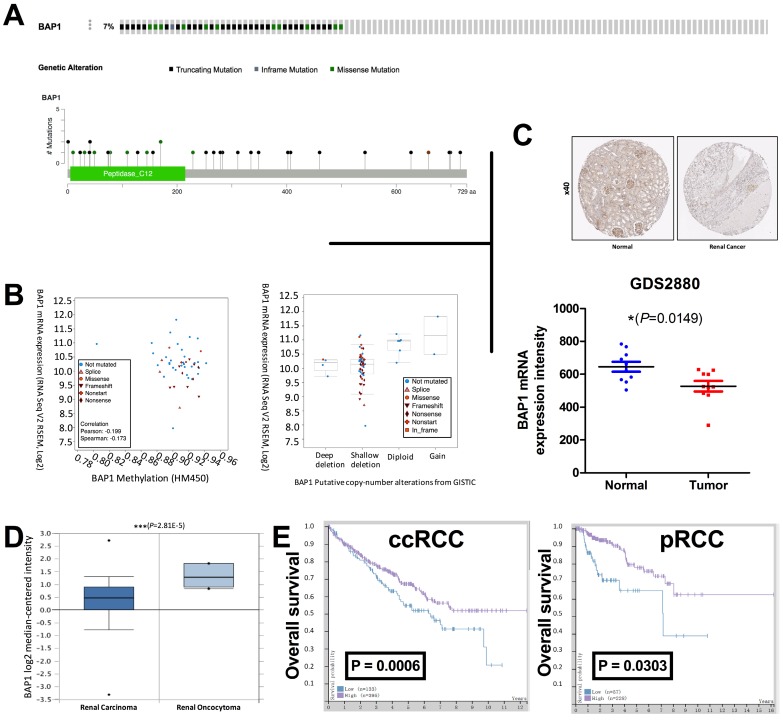
The TCGA renal cell carcinoma dataset included 534 ccRCC samples with complete mRNA expression data detected using RNA seq. We captured 33 cases with BAP1 mutation while excluding cases only with deep copy number losses. Control 1 (pan-negative) included 33 cases with no mutation or copy number loss in VHL, PBRM1, SETD2 or KDM5C. Control 2 included, literally 501 cases in the dataset. BAP1 mutation occurred in approximately 7% of all cases (Fig 1A). Mutation types included truncating, inframe, and missense mutations spanning over entire gene. In general, cases with and without BAP1 mutation shared similar levels of DNA methylation while mutated cases showed lower mRNA expression (Fig 1B). Analysis of CNV revealed that most mutated cases had simultaneous shallow focal deletion at 3p with no mutated case copy-neutral or -amplified (Fig 1B). To demonstrate the downregulation of BAP1's protein level in ccRCC, we tested BAP1 expression by IHC in ccRCC tissues and matched non-tumor tissues, and found the expression level of BAP1 in ccRCC tissues was significantly lower than non-tumor tissues (Fig 1C). To further determine whether decreased BAP1 product was RCC-associated, we analyzed the data from GDS2880, ONCOMINE and The Human Protein Atlas portal (HPA). Immunohistochemistry (IHC) and mRNA results showed that the expression of BAP1 was lower in ccRCC tissues (Fig 1C-D). Survival data from HPA also indicate that patients with a lower BAP1 level in Renal Clear Cell Carcinoma and Renal Papillary Cell Carcinoma tumor tissues showed much poorer prognosis (Fig 1E). All these results indicate that the low level of BAP1 positively correlates with ccRCC progression and poor prognosis in ccRCC patients. All 62 cases in the validation cohort were eligible for IHC assessment. EZH2 expression was significantly higher in patients with progressed stage, grade, nodal invasion, and metastasis (Table 1). EZH2 expression was not associated with age (Table 1). Expression of EZH2 was significantly and positively correlated with expression of BAP1 but not with that of VHL, PBRM1, SETD2, or KDM5C (Table 2).
Fig 1.
Loss of BAP1 is associated with renal cell carcinoma and prognosis. (A) Mutation frequency and types of BAP1 in clear cell renal cell carcinoma (ccRCC) reproduced from the cancer Genome Atlas (TCGA) database; (B) correlation between BAP methylation and mRNA expression; and B) relation between BAP1 copy number change and mRNA expression; Data from (C) HPA (left) and GDS2880 (right) and (D) ONCOMINE showing that mRNA level of BAP1 is significantly downregulated in human ccRCC. Representative immunohistochemistry (IHC) of BAP1 on benign renal and ccRCC tissues; (E) Overall survival between cases with higher or lower BAP1 expression in ccRCC and pRCC reproduced from TCGA database.
Table 1.
EZH2 expression in ccRCC and association with clinicopathological parameters in validation cohort.
| Parameter | Validation | ||||
|---|---|---|---|---|---|
| Breakdown | N | LRP1 Expression | P | ||
| Mean | SD | ||||
| T | T1 | 29 | 1.97 | 0.186 | <0.0001 |
| T2 | 22 | 1.09 | 0.426 | ||
| T3 | 6 | 2.33 | 0.816 | ||
| T4 | 5 | 2.8 | 0.447 | ||
| N | N0 | 56 | 1.66 | 0.611 | 0.0006 |
| N1 | 6 | 2.67 | 0.516 | ||
| M | M0 | 57 | 1.67 | 0.607 | 0.0002 |
| M1 | 5 | 2.8 | 0.447 | ||
| Age | 62 | 58.9 | 6.87 | 0.0003 | |
| Gender | Male | 57 | 1.67 | 0.607 | 0.4639 |
| Female | 22 | 1.59 | 0.503 | ||
| Grade | I | 13 | 2 | 0 | <0.0001 |
| II | 35 | 1.4 | 0.604 | ||
| III | 8 | 2 | 0 | ||
| IV | 6 | 3 | 0 | ||
Table 2.
Correlations between expressions of EZH2 and other significantly mutated genes in ccRCC in validation cohort.
| EZH2vs.BAP1 | EZH2vs.VHL | EZH2vs.PBRM1 | EZH2vs.SETD2 | EZH2vs.KDM5C | |
|---|---|---|---|---|---|
| Pearson r | |||||
| r | 0.6852 | 0.03872 | 0.08798 | -0.1038 | 0.0505 |
| 95% confidence interval | 0.5253 to 0.7983 | -0.2131 to 0.2857 | -0.1654 to 0.3305 | -0.3446 to 0.1499 | -0.2018 to 0.2965 |
| R squared | 0.4694 | 0.001499 | 0.007741 | 0.01077 | 0.00255 |
| P (two-tailed) | <0.0001 | 0.7651 | 0.4965 | 0.4221 | 0.6967 |
Functional clustering of BAP1-mutated ccRCC
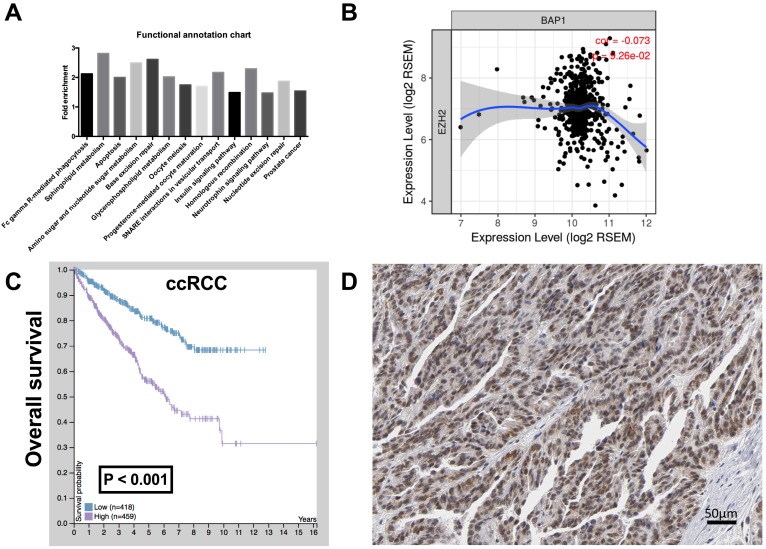
To investigate the function of BAP1 in ccRCC, by analyzing the enrichment of mRNA expression between BAP1 mutated cases and pan-negative cases, we acquired 2,132 genes that were significantly enriched according to both P and Q value thresholds. Further functional clustering revealed 5 significant clusters, including Cluster 1 (Nucleotide excision repair, DNA replication, Mismatch repair), Cluster 2 (Prostate cancer, Glioma, Pancreatic cancer, B cell receptor signaling pathway, Colorectal cancer, Non-small cell lung cancer, Endometrial cancer, Melanoma, T cell receptor signaling pathway, Small cell lung cancer, Chronic myeloid leukemia, Acute myeloid leukemia, ErbB signaling pathway, Renal cell carcinoma), Cluster 3 (GnRH signaling pathway, Long-term potentiation, Vascular smooth muscle contraction), Cluster 4 (VEGF signaling pathway, Fc epsilon RI signaling pathway, ErbB signaling pathway), and Cluster 5 (Dilated cardiomyopathy, Arrhythmogenic right ventricular cardiomyopathy, Hypertrophic cardiomyopathy). We then analyzed functional annotation of the enriched genes and found that the significant enriched pathway were Fc gamma R-mediated phagocytosis, Sphingolipid metabolism, Apoptosis, Amino sugar and nucleotide sugar metabolism, Base excision repair, Glycerophospholipid metabolism, Oocyte meiosis, Progesterone-mediated oocyte maturation, SNARE interactions in vesicular transport, Insulin signaling pathway, Homologous recombination, Neurotrophin signaling pathway, Nucleotide excision repair, and Prostate cancer. According to change in fold enrichment, the top 3 rankings of the enriched pathways were Sphingolipid metabolism, Base excision repair, and Amino sugar and nucleotide sugar metabolism (Fig 2A).
Fig 2.
EZH 2 was upregulated in ccRCC with low BAP1 level. (A) Functional annotation and pathway analysis of enriched genes in BAP1 mutated ccRCC compared with pan-negative cases, reproduced from the cancer Genome Atlas (TCGA) database, showing 14 significantly changed pathways; (B) Correlation between expressions of BAP1 and EZH2 in TCGA-KIRC database; (C) Overall survival between cases with higher or lower EZH2 expression in ccRCC reproduced from TCGA database; (D) Reproduced from HPA, showing typical EZH1 positive cells in IHC.
The Cluster 1 functional annotation was in strong association with genomic instability, a hallmark of genomic alteration ccRCC. We therefore queried whether Enhancer of zeste homolog 2 (EZH2), a histone-lysine N-methyltransferase enzyme that plays an important role in chromatin modification, was enriched in BAP1-mutatn setting. By reproducing TCGA RNA sequencing data, we found that expressions of BAP1 and EZH2 showed a negative correlation in ccRCC with a marginal significance (Fig 2B). CcRCC patients with higher EZH2 expression or positive IHC staining also showed significantly worsened prognosis (Fig 2C-D).
BAP1-mutated ccRCC are characterized with high EZH2 level
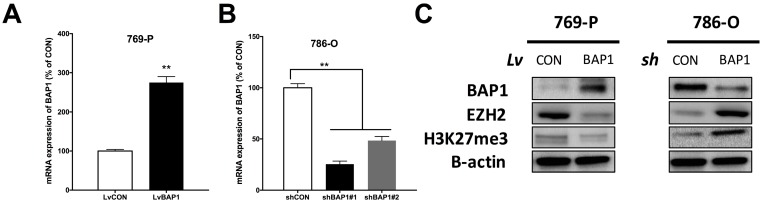
Thus far, it has not been reported whether EZH2 expression was associated with BAP1 status. To address our hypothesis, we used 769-P ccRCC cell line that harbored BAP1 mutation and 786-O ccRCC cell line that was BAP-wildtype (WT) 17. We conducted genetic replenishment of BAP1 using lentivirus (lv) in 769-P cells and applied genetic knockdown (KD) of BAP1 using shRNA (sh) in 786-O cells (Fig 3A-B). In both cell lines, replenishment of BAP1 level induced decreased EZH2 level and vice versa (Fig 3C). The alteration of EZH2 protein level also had impact on H3K37me level though not as contrasting (Fig 3C).
Fig 3.
Expression of EZH2 was higher in BAP1-mutant ccRCC cell line. (A) Efficacy of lentiviral delivery of BAP1 overexpression in 769-P ccRCC cells; (B) Efficacy of shRNA against BAP1 in 786-O ccRCC cells; (C) Protein levels of BAP1, EZH2, H3K27me3 in 769-P and 786-O cells with genetic manipulation of BAP1 status (**P < 0.01).
EZH2-KD inhibited oncogenesis of BAP1-loss in ccRCC
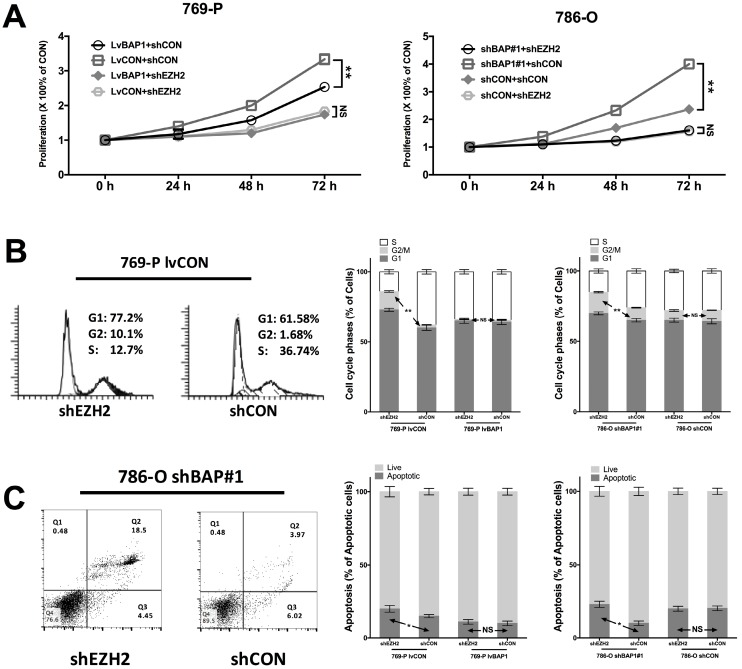
To determine the regulatory relation between BAP1 and EZH2 in ccRCC, we used different combinations of genetic manipulations of BAP1 in both 769-P and 786-O cells. EZH2-KD significantly inhibited proliferation in both 769-P and 786-O cells selectively in BAPI-KD status (Fig 4A). EZH2-KD also induced significant cell population in both G1 and G2 phases of cell cycle (Fig 4B). Such effect was solely when BAP1 mutation was absent (Fig 4B). Similarly, EZH2-KD induced significantly increased cell apoptosis in both cell lines when BAP1-loss was present (Fig 4C). These findings supported the notion that BAP1-loss ccRCC were at least in part addictive to EZH2 overexpression which warrant further pharmaceutical investigation.
Fig 4.
EZH2-KD inhibited oncogenesis of BAP1-loss in ccRCC. (A) EZH2-KD selectively effective in decreasing cell proliferation in 769-P cells harboring BAP1 mutation or 786-O cells with BAP1-KD, with similar selective effects in (B) cell cycle arrest and (C) cell apoptosis (*P<0.05, **P < 0.01).
Pharmaceutic inhibition of EZH2 inhibited tumorigenesis of BAP1-mutant ccRCC
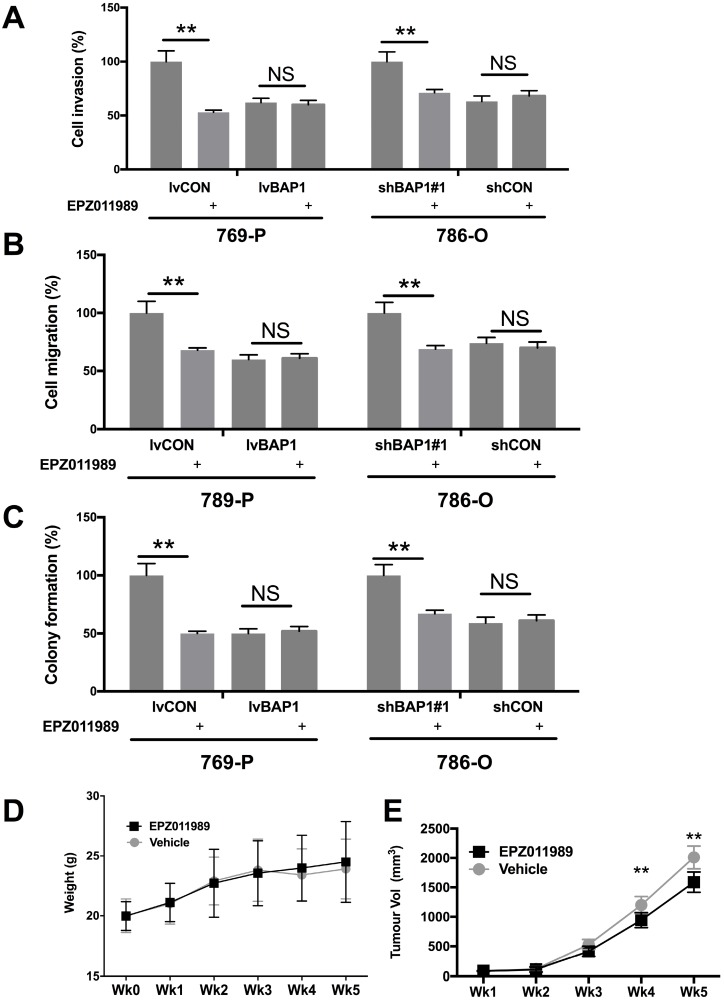
EZH2 inhibition has now entered several early phase clinical trials. We therefore explored the EZH2 inhibitor EPZ011989 at concentration of 0.625 μM. EPZ011989 significantly and selectively inhibited cell invasion, migration, and anchorage-independent growth in ccRCC cells with BAP1 mutation (Fig 5A-C). EPZ011989 at 500 mg/kg twice a day showed tolerable toxicity in all mice and significant inhibition of tumor growth of BAP1-mutant tumors (Fig 5D).
Fig 5.
Pharmaceutical inhibition of EZH2 selectively inhibited ccRCC with BAP1 mutation. EZH2 inhibitor EPZ011989 significantly and selectively inhibited (A) cell invasion, (B) migration, (C) colony formation, and (D) animal weight, and (E) tumor growth of ccRCC with BAP1 mutation or KD (*P<0.05, **P < 0.01).
Discussion
BAP1 is a tumor suppressor gene that encodes a deubiquitinating enzyme (DUB), regulating key cellular pathways, including cell cycle, cellular differentiation, transcription and DNA damage response 18. As germline BAP1 mutation is associated with increased risk and earlier onset of a variety of cancers and somatic BAP1 mutation is noted in a series of cancers, BAP1-defficient tumors are now recognized as a specific entity that characterize a unique tumor profile. In kidney cancer, germline BAP1 mutation is not mainstay. Contemporary understanding of the kidney tumorigenesis points out that VHL mutations are truncal and its inactivation and subsequent hyperactivity of HIF2 axis is almost ubiquitous in ccRCC, other frequently mutated genes including BAP1 are considered branch events in response to selection19.
Many studies have shown that BAP1 mutation confers worsened prognosis in ccRCC. Nonetheless, those studies did not distinguish the effect of other 3p genes that may bias substantially. For example, PBRM1 is reported to be mutually exclusive to BAP1 and is associated with better prognosis 9. In the current study, we found that BAP1 mutated cases did not have worsened prognosis compared with pan-negative cases. Our results strongly indicate that the prognostic contribution of BAP1 is biased by PBRM1 in most studies. Also, as there is a small proportion of ccRCC that is VHL/HIF2 independent, which are expected to fit our pan-negative category, the similar prognosis of such to BAP1 mutated cases also suggest that BAP1 deficient tumor is a standalone entity.
Functional annotation of BAP1 is the essence of our study. Previously, there has been a dearth of studies addressing the role of BAP1 in ccRCC. It is suggested that stabilization of MCRS1 by BAP1 prevents chromosome instability in ccRCC 20. Our annotation showed notably that BAP1 mutation is associated with Fc gamma R-mediated phagocytosis, Apoptosis, Amino sugar and nucleotide sugar metabolism, Base excision repair, Glycerophospholipid metabolism, Insulin signaling pathway, Homologous recombination, and Nucleotide excision repair. Those results are quite indicative of potential BAP1 functions and therapeutic targets. For example, the immunomodulation annotation may indicate that BAP1 mutated ccRCC is specially associated with tumor infiltrated immunoresponse and may predict the effect of current novel PD1/PD-L1 therapies. Association between BAP1 and metabolism has not been reported either. Metabolism plays a critical role in ccRCC 21. Therefore, association between BAP1 and metabolism warrants further studies as this may provide a variety of therapeutic targets. Finally, BAP1 as a histone modifier is closely related with DNA replication and repair. Though direct evidence that BAP1 mediates DNA repair is lacking, we can still imply from the other 3p gene SETD2 that also belongs to histone modifier. SETD2 is now associated with homologous recombination and mismatch repair, both of which are critical DNA repair mechanisms 22, 23. Our functional annotation supports a similar role of BAP1 in ccRCC. DNA repair is a highly conservative mechanism and impairment of such function may trigger tumorigenesis. Establishment of transgenic animal model of ccRCC is currently elusive. Unlike in human, sole inactivation in mouse is insufficient to trigger malignant nephropathy. The most promising report of developing a transgenic kidney cancer model is by simultaneous inactivation of VHL an BAP1 24. Despite so, how VHL and BAP1 cooperate with each other remains unknown.
Relation between EZH2 and BAP1 has been reported in some cancers yet not in ccRCC. Shinozaki-Ushiku investigated Diagnostic utility of BAP1 and EZH2 expression in malignant mesothelioma. They found that BAP1 loss and high EZH2 expression were highly specific to malignant mesothelioma in differentiating it from benign mesothelial proliferations, and the combination of these two markers improved the diagnostic accuracy 25. Morel et al investigated Targeting chromatin defects in selected solid tumors based on oncogene addiction, synthetic lethality and epigenetic antagonism. They found that Epigenetic-targeting drugs have historically failed proving efficacy in solid malignancies when used broadly, but novel mechanism-based approaches in molecularly selected patient populations have facilitated recent successes in proof-of-concept studies in solid tumors 26. LaFave et al reported Loss of BAP1 function leads to EZH2-dependent transformation, showing mesothelioma cells that lack BAP1 are sensitive to EZH2 pharmacologic inhibition, suggesting a novel therapeutic approach for BAP1-mutant malignancies 27. Schoumacher et al however reported that Uveal melanoma cells are resistant to EZH2 inhibition regardless of BAP1 status 28. Those seminal studies lead to a conclusion that targeted therapy against chromatin-related pathways are strictly cell context -dependent and is hard to be extrapolated form one study to another.
Conclusion
BAP1 is frequently mutated in ccRCC and its function remains unknown. We performed in silico analysis of TCGA ccRCC database for expressional enrichments between BAP1 mutated and pan-negative cases. CcRCC with decreased BAP1 level has poor prognosis and is associated with higher EZH2 expression. Inhibition of EZH2 in BAP1-mutated entity holds promise for further investigation.
Acknowledgments
This study was supported by Youth Elites Scientific Funding of Shanghai Municipal Health Bureau (grant no. 20144Y0111), Shanghai Municipal Natural Science Foundation (14ZR1432300) and National Natural Science Foundation (NSFC81000311).
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA: a cancer journal for clinicians. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Kovacs G, Akhtar M, Beckwith BJ, Bugert P, Cooper CS, Delahunt B, Eble JN, Fleming S, Ljungberg B, Medeiros LJ, Moch H, Reuter VE, Ritz E, Roos G, Schmidt D, Srigley JR. et al. The Heidelberg classification of renal cell tumours. The Journal of pathology. 1997;183(2):131–133. doi: 10.1002/(SICI)1096-9896(199710)183:2<131::AID-PATH931>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jensen DE, Proctor M, Marquis ST, Gardner HP, Ha SI, Chodosh LA, Ishov AM, Tommerup N, Vissing H, Sekido Y, Minna J, Borodovsky A, Schultz DC, Wilkinson KD, Maul GG, Barlev N. et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16(9):1097–1112. doi: 10.1038/sj.onc.1201861. [DOI] [PubMed] [Google Scholar]
- 5.Gaytan de Ayala Alonso A, Gutierrez L, Fritsch C, Papp B, Beuchle D, Muller J. A genetic screen identifies novel polycomb group genes in Drosophila. Genetics. 2007;176(4):2099–2108. doi: 10.1534/genetics.107.075739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bott M, Brevet M, Taylor BS, Shimizu S, Ito T, Wang L, Creaney J, Lake RA, Zakowski MF, Reva B, Sander C, Delsite R, Powell S, Zhou Q, Shen R, Olshen A. et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat Genet. 2011;43(7):668–672. doi: 10.1038/ng.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pena-Llopis S, Vega-Rubin-de-Celis S, Liao A, Leng N, Pavia-Jimenez A, Wang S, Yamasaki T, Zhrebker L, Sivanand S, Spence P, Kinch L, Hambuch T, Jain S, Lotan Y, Margulis V, Sagalowsky AI. et al. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet. 2012;44(7):751–759. doi: 10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gossage L, Eisen T, Maher ER. VHL, the story of a tumour suppressor gene. Nat Rev Cancer. 2015;15(1):55–64. doi: 10.1038/nrc3844. [DOI] [PubMed] [Google Scholar]
- 9.Joseph RW, Kapur P, Serie DJ, Parasramka M, Ho TH, Cheville JC, Frenkel E, Parker AS, Brugarolas J. Clear Cell Renal Cell Carcinoma Subtypes Identified by BAP1 and PBRM1 Expression. J Urol. 2016;195(1):180–187. doi: 10.1016/j.juro.2015.07.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng C, Ding G, Jiang H, Ding Q, Wen H. Loss of MLH1 confers resistance to PI3Kbeta inhibitors in renal clear cell carcinoma with SETD2 mutation. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36(5):3457–3464. doi: 10.1007/s13277-014-2981-y. [DOI] [PubMed] [Google Scholar]
- 11.Feng C, Sun Y, Ding G, Wu Z, Jiang H, Wang L, Ding Q, Wen H. PI3Kbeta inhibitor TGX221 selectively inhibits renal cell carcinoma cells with both VHL and SETD2 mutations and links multiple pathways. Sci Rep. 2015;5:9465. doi: 10.1038/srep09465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng C, Xiong Z, Jiang H, Ding Q, Fang Z, Hui W. Genetic alteration in notch pathway is associated with better prognosis in renal cell carcinoma. Biofactors. 2016;42(1):41–48. doi: 10.1002/biof.1250. [DOI] [PubMed] [Google Scholar]
- 13.Chen K, Xiao H, Zeng J, Yu G, Zhou H, Huang C, Yao W, Xiao W, Hu J, Guan W, Wu L, Huang J, Huang Q, Xu H, Ye Z. Alternative Splicing of EZH2 pre-mRNA by SF3B3 Contributes to the Tumorigenic Potential of Renal Cancer. Clinical Cancer Research. 2017;23(13):3428–3441. doi: 10.1158/1078-0432.CCR-16-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Samant R, Jiang W, Dulaimi E, Devarajan K, Parsons T, Wang Q, Liao L, Cho E-A, O'Neill R, Solomides C, Peiper SC, Testa JR, Uzzo R, Yang H. Immunohistochemistry Successfully Uncovers Intratumoral Heterogeneity and Widespread Co-Losses of Chromatin Regulators in Clear Cell Renal Cell Carcinoma. Plos One. 2016;11(10):e0164554. doi: 10.1371/journal.pone.0164554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oka S, Inoshita N, Miura Y, Oki R, Miyama Y, Nagamoto S, Ogawa K, Sakaguchi K, Kondoh C, Kurosawa K, Urakami S, Takano T, Okaneya T. The loss of BAP1 protein expression predicts poor prognosis in patients with nonmetastatic clear cell renal cell carcinoma with inferior vena cava tumor thrombosis. Urologic Oncology: Seminars and Original Investigations; 2018. [DOI] [PubMed] [Google Scholar]
- 16.Liu L, Guo R, Zhang X, Liang Y, Kong F, Wang J, Xu Z. Loss of SETD2, but not H3K36me3, correlates with aggressive clinicopathological features of clear cell renal cell carcinoma patients. BioScience Trends. 2017;11(2):214–220. doi: 10.5582/bst.2016.01228. [DOI] [PubMed] [Google Scholar]
- 17.Sinha R, Winer AG, Chevinsky M, Jakubowski C, Chen YB, Dong Y, Tickoo SK, Reuter VE, Russo P, Coleman JA, Sander C, Hsieh JJ, Hakimi AA. Analysis of renal cancer cell lines from two major resources enables genomics-guided cell line selection. Nat Commun. 2017;8:15165. doi: 10.1038/ncomms15165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang A, Papneja A, Hyrcza M, Al-Habeeb A, Ghazarian D. Gene of the month: BAP1. J Clin Pathol. 2016;69(9):750–753. doi: 10.1136/jclinpath-2016-203866. [DOI] [PubMed] [Google Scholar]
- 19.Chen W, Hill H, Christie A, Kim MS, Holloman E, Pavia-Jimenez A, Homayoun F, Ma Y, Patel N, Yell P, Hao G, Yousuf Q, Joyce A, Pedrosa I, Geiger H, Zhang H. et al. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature. 2016;539(7627):112–117. doi: 10.1038/nature19796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng J, Ma J, Li W, Mo R, Zhang P, Gao K, Jin X, Xiao J, Wang C, Fan J. Stabilization of MCRS1 by BAP1 prevents chromosome instability in renal cell carcinoma. Cancer Lett. 2015;369(1):167–174. doi: 10.1016/j.canlet.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 21.Li B, Qiu B, Lee DS, Walton ZE, Ochocki JD, Mathew LK, Mancuso A, Gade TP, Keith B, Nissim I, Simon MC. Fructose-1,6-bisphosphatase opposes renal carcinoma progression. Nature. 2014;513(7517):251–255. doi: 10.1038/nature13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanu N, Gronroos E, Martinez P, Burrell RA, Yi Goh X, Bartkova J, Maya-Mendoza A, Mistrik M, Rowan AJ, Patel H, Rabinowitz A, East P, Wilson G, Santos CR, McGranahan N, Gulati S. et al. SETD2 loss-of-function promotes renal cancer branched evolution through replication stress and impaired DNA repair. Oncogene. 2015;34(46):5699–5708. doi: 10.1038/onc.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfister SX, Ahrabi S, Zalmas LP, Sarkar S, Aymard F, Bachrati CZ, Helleday T, Legube G, La Thangue NB, Porter AC, Humphrey TC. SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep. 2014;7(6):2006–2018. doi: 10.1016/j.celrep.2014.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang SS, Gu YF, Wolff N, Stefanius K, Christie A, Dey A, Hammer RE, Xie XJ, Rakheja D, Pedrosa I, Carroll T, McKay RM, Kapur P, Brugarolas J. Bap1 is essential for kidney function and cooperates with Vhl in renal tumorigenesis. Proc Natl Acad Sci U S A. 2014;111(46):16538–16543. doi: 10.1073/pnas.1414789111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shinozaki-Ushiku A, Ushiku T, Morita S, Anraku M, Nakajima J, Fukayama M. Diagnostic utility of BAP1 and EZH2 expression in malignant mesothelioma. Histopathology. 2017;70(5):722–733. doi: 10.1111/his.13123. [DOI] [PubMed] [Google Scholar]
- 26.Morel D, Almouzni G, Soria JC, Postel-Vinay S. Targeting chromatin defects in selected solid tumors based on oncogene addiction, synthetic lethality and epigenetic antagonism. Ann Oncol. 2017;28(2):254–269. doi: 10.1093/annonc/mdw552. [DOI] [PubMed] [Google Scholar]
- 27.LaFave LM, Beguelin W, Koche R, Teater M, Spitzer B, Chramiec A, Papalexi E, Keller MD, Hricik T, Konstantinoff K, Micol JB, Durham B, Knutson SK, Campbell JE, Blum G, Shi X. et al. Loss of BAP1 function leads to EZH2-dependent transformation. Nat Med. 2015;21(11):1344–1349. doi: 10.1038/nm.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schoumacher M, Le Corre S, Houy A, Mulugeta E, Stern MH, Roman-Roman S, Margueron R. Uveal melanoma cells are resistant to EZH2 inhibition regardless of BAP1 status. Nat Med. 2016;22(6):577–578. doi: 10.1038/nm.4098. [DOI] [PubMed] [Google Scholar]