Abstract
Background: Esophageal squamous cell carcinoma (ESCC) is one of the most lethal cancers in China. Recently, a study identified that cullin 3 (CUL3) was significantly mutated and deleted in ESCC. We then hypothesis that germline variants in CUL3 may also associated with the susceptibility of ESCC. Variants in the gene 3'-untranslated region (3'-UTR) may associate with gene expression by altering miRNAs binding.
Material and Methods: We systematically searched for variants in the 3'-UTR of CUL3 using the Ensembl database. Taqman SNP Genotyping Assay was performed in 638 ESCC cases and 546 controls to examine the association between the rs2396092 and the risk of ESCC. The eQTL analysis for CUL3 were conducted by using the GTEx database.
Results: We identified that the rs2396092 was significantly associated with the susceptibility of ESCC. Compared with the TT genotype carriers, the CT genotype and CC genotype carriers were correlated with risk of ESCC with odds ratio being 1.33 (95% CI: 1.04-1.70, P=0.0222) and 1.63 (95% CI: 1.07-2.50, P=0.0241), respectively. Different genotypes of rs2396092 was also shown to be correlated with altered CUL3 expression.
Conclusion: The results emphasize the importance of CUL3 in the development of ESCC and may contribute to the personalized prevention of this cancer in the future.
Keywords: Esophageal squamous cell carcinoma, SNP, Susceptibility, CUL3, 3'UTR
Background
Esophageal squamous cell carcinoma (ESCC) is the fourth leading cause of cancer death in China and approximately half of the world's new ESCC cases each year occur in China 1. Epidemiology studies suggest that alcohol and tobacco consumption, nutrient deficiency and carcinogen exposure are the risk factors of this type of cancer 2. Previous genome-wide association study (GWAS) found several genetic loci that was associated with ESCC susceptibility 3-5. However, GWAS-identified variants could only explain a small fraction of the heritability 6. More functional susceptibility loci still need to be further explored.
Recently, a genomic analysis identified that CUL3 was significantly mutated and deleted in ESCC 7. The cullin 3 (CUL3) was an ubiquitin ligase and was significantly correlated with cancer development, progression, and therapeutic response 8. However, the relationship between germline variation in CUL3 and ESCC susceptibility has not been investigated. MicroRNAs (miRNA) are a class of small, noncoding RNA that regulate gene expression by binding the 3'-untranslated region (3′-UTR) of mRNAs of their target genes, resulting in mRNA cleavage or translation repression 9,10. Germline variants in the 3′-UTR of miRNA target genes might alter miRNA binding and thus associated with risk of cancer by affecting gene expression 11.
In the present study, we searched for variants in the 3'-UTR of CUL3 and performed a case-control study to test whether these variants are associated with ESCC susceptibility.
Materials and Methods
Study subjects
In this study, we enrolled 638 ESCC patients and 546 healthy controls. ESCC patients were enrolled from the People's Hospital of Zhengzhou University and Henan Provincial People's Hospital, Zhengzhou, China between January 1st, 2014 to November 30th, 2016. Health controls were selected from a community cancer screening program for early detection conducted in the same region during the same period as cases were collected. All participants were unrelated Han Chinese descent. The informed consent was obtained from every participant at recruitment. The demographic characteristics such as sex, age and ethnicity were obtained from the medical records. The study was approved by the Institutional Review Board of Zhengzhou University.
SNP selection and genotyping
We investigated all variants in the 3'UTR of CUL3 by using Ensembl (http://asia.ensembl.org/). There are 13 variants with global MAF > 0.05 and 6 variants with significant eQTL for CUL3 in GTex database. These 6 variants were in perfect LD with each other and the rs2396092 was selected for genotyping. The variants were genotyped by Taqman SNP Genotyping Assay (Applied Biosystems, USA).
Statistical analysis
Odds ratios (ORs) and 95% confidence intervals (CIs) adjusted for sex and age were used to assess the strength of associations between selected polymorphisms and ESCC risk by using unconditional multivariate logistic regression analysis. All statistical analyses were performed using SAS software (version 9.1; SAS Institute, Cary, NC), with a significance level of 0.05. All tests were two-sided.
Results
Characteristic of Study Subjects
The details of selected demographic characteristics including sex and age of the ESCC cases and controls were summarized in Table 1. The present study included 638 ESCC patients and 546 healthy controls. There are 79.9% males and 20.0% females in the cases, while there are 73.4% males and 26.6% females in controls. The average ages were 59.4 (±9.8) for cases and 63.2 (±8.2) for controls, respectively.
Table 1.
Summary of characteristics of study subjects
| Cases (n=638) | Controls(n=546) | |
|---|---|---|
| Age (years), mean±S.D. | 59.4±9.8 | 63.2±8.2 |
| Gender, n (%) | ||
| Male | 510 (79.9) | 401 (73.4) |
| Female | 128 (20.0) | 145 (26.6) |
The rs2396092 in CUL3 was associated with the susceptibility of ESCC
The CUL3 3'-UTR variants with global minor allele frequency (MAF)>0.05 were listed (Table 2). Among these variants, 6 variants were associated with CUL3 expression in GTex database and they were in perfect LD. Therefore, we selected rs2396092 as a TagSNP for genotyping. The association between rs2396092 and ESCC risk were shown in the Table 3. Compared with TT genotype carriers, CT genotype and CC genotype carriers were associated with risk of ESCC with OR being 1.33 (95% CI: 1.04-1.70, P=0.0222) and 1.63 (95% CI: 1.07-2.50, P=0.0241), respectively (Table 3). When combing the CT genotype and CC genotype carriers, the association was more significant with P values being 0.0070 (Table 3).
Table 2.
Genetic variants in the 3'-UTR of CUL3 with global minor allele frequency >0.05
| Chr | SNP | Position | Gene | Location | Alleles | Global MAF | GTex eQTL |
|---|---|---|---|---|---|---|---|
| 2 | rs12470077 | 225334971 | CUL3 | 3'-UTR | T/C | 0.308 | - |
| 2 | rs5839066 | 225335226 | CUL3 | 3'-UTR | A/- | 0.233 | - |
| 2 | rs2396092 | 225335290 | CUL3 | 3'-UTR | A/G | 0.233 | CUL3 |
| 2 | rs4674908 | 225335595 | CUL3 | 3'-UTR | A/G | 0.453 | - |
| 2 | rs17479770 | 225335814 | CUL3 | 3'-UTR | T/C | 0.250 | - |
| 2 | rs3768899 | 225336309 | CUL3 | 3'-UTR | C/T | 0.233 | CUL3 |
| 2 | rs10498160 | 225336531 | CUL3 | 3'-UTR | C/A | 0.233 | CUL3 |
| 2 | rs73993899 | 225336769 | CUL3 | 3'-UTR | C/T | 0.080 | - |
| 2 | rs3768898 | 225336770 | CUL3 | 3'-UTR | A/G | 0.233 | CUL3 |
| 2 | rs10498161 | 225336889 | CUL3 | 3'-UTR | T/C | 0.233 | CUL3 |
| 2 | rs13016316 | 225337188 | CUL3 | 3'-UTR | C/A | 0.140 | - |
| 2 | rs3768897 | 225338022 | CUL3 | 3'-UTR | G/A/C | 0.234 | CUL3 |
Note: SNP, single nucleotide polymorphism; MAF, minor allele frequency.
Table 3.
Association between rs2396092 and risk of ESCC in a Chinese population
| SNP | Chr | Position | Gene | Location | Genotype | Cases No. (%) | Controls No. (%) | OR (95% CI)a | Pa |
|---|---|---|---|---|---|---|---|---|---|
| rs2396092 | 2 | 225335290 | CUL3 | 3'-UTR | TT | 266 (41.7) | 272 (49.8) | 1.00 (Reference) | |
| CT | 300 (47.0) | 232 (42.5) | 1.33 (1.04-1.70) | 0.0222 | |||||
| CC | 72 (11.3) | 42 (7.7) | 1.63 (1.07-2.50) | 0.0241 | |||||
| CT+CC | 372 (58.3) | 274 (50.2) | 1.38 (1.09-1.75) | 0.0070 |
aCalculated by logistic regression model adjusted for sex and age.
The rs2396092 was significantly associated with CUL3 expression
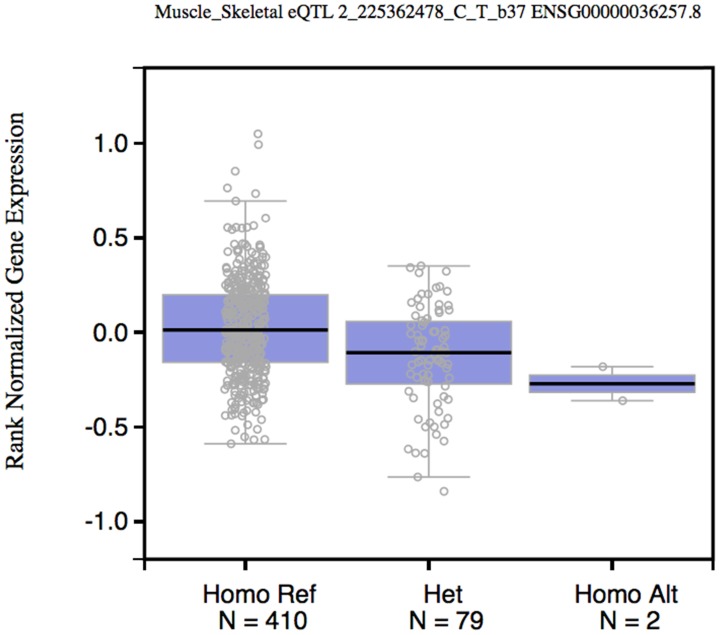
To test whether rs2396092 (tagSNP of 6 variants in the 3'-UTR of CUL3) was an expression Quantitative Trait Loci (eQTL) of CUL3, we obtained the rs3738952 data (In perfect LD with rs2396092) from the GTex database (https://gtexportal.org) 12. The CUL3 expression CC or CT genotype was significantly lower than the TT genotype with P value being in 2×10-7 in the “Muscle - Skeletal” tissue (Figure 1). The rs2396092 was also significantly associated with CUL3 expression in whole blood in the Westra et al study 13.
Figure 1.
Box plot shows the correlation between rs2396092 with CUL3 expression. Data were from 492 muscle samples in the GTEx database.
Discussion
In this case-control study, we interrogated the association between variants in the 3'-UTR of CUL3 and ESCC susceptibility in 638 ESCC patients and 546 controls. We identified the frequency of the TC/CC genotypes of rs2396092 were significantly more than that of TT genotypes. We also found that the CUL3 expression was significantly associated with rs2396092 genotypes in two studies. The results showed that the rs2396092 T>C variant in CUL3 was correlated with the risk of ESCC and indicated that the CUL3 played an important role in the ESCC carcinogenesis.
The CUL3 gene locates in chromosome 2q36.2 and encodes a member of the cullin protein family. CUL3 plays a critical role in the polyubiquitination and subsequent degradation of specific protein substrates as the core component and scaffold protein of an E3 ubiquitin ligase complex 14,15. CUL3 also plays a role in the oxidative stress response pathway and was identified to function as a tumor suppressor 16. Mutations in CUL3 have been identified to associate with multiple complex diseases, including cancer 17-20. In this study, we identified six variants in 3'-UTR of CUL3 and correlated with CUL3 expression. The genotype correlated with lower CUL3 expression was also associated with increased risk of ESCC, which is concordant with previous findings that CUL3 is an important tumor suppressor.
There are some limitations for this study. First, the association still needs to be replicated in other samples. Second, the identified rs2396092 variant was only a tagSNP and further functional analysis should be conducted to identify the real functional SNP.
Conclusion
In summary, through a case-control study in a Chinese Han population, we find a significant association between CUL3 3'-UTR variants and ESCC susceptibility. These results expand our insights of ESCC carcinogenesis and provide more evidence for the precision medicine of this disease.
Funding
This work was supported by grants from Henan Provincial Science and Technique Program (162102310014) and Henan Provincial Medical Science and Technique Program (201602218).
Author contribution
Conception or design: JL Hu, SD Cang; Acquisition, analysis, or interpretation of data: JL Hu; XL Hu; CX Lu; XJ Chen; L F; Q H; Draft the manuscript: JL Hu; Review and final approval of the manuscript: All authors.
References
- 1.Chen W, Zheng R, Baade PD. et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Morita M, Kumashiro R, Kubo N. et al. Alcohol drinking, cigarette smoking, and the development of squamous cell carcinoma of the esophagus: epidemiology, clinical findings, and prevention. Int J Clin Oncol. 2010;15:126–34. doi: 10.1007/s10147-010-0056-7. [DOI] [PubMed] [Google Scholar]
- 3.Wu C, Hu Z, He Z. et al. Genome-wide association study identifies three new susceptibility loci for esophageal squamous-cell carcinoma in Chinese populations. Nat Genet. 2011;43:679–84. doi: 10.1038/ng.849. [DOI] [PubMed] [Google Scholar]
- 4.Wu C, Kraft P, Zhai K. et al. Genome-wide association analyses of esophageal squamous cell carcinoma in Chinese identify multiple susceptibility loci and gene-environment interactions. Nat Genet. 2012;44:1090–7. doi: 10.1038/ng.2411. [DOI] [PubMed] [Google Scholar]
- 5.Chang J, Zhong R, Tian J. et al. Exome-wide analyses identify low-frequency variant in CYP26B1 and additional coding variants associated with esophageal squamous cell carcinoma. Nat Genet. 2018;50:338–343. doi: 10.1038/s41588-018-0045-8. [DOI] [PubMed] [Google Scholar]
- 6.Dai J, Shen W, Wen W. et al. Estimation of heritability for nine common cancers using data from genome-wide association studies in Chinese population. Int J Cancer. 2017;140:329–336. doi: 10.1002/ijc.30447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang J, Tan W, Ling Z. et al. Genomic analysis of oesophageal squamous-cell carcinoma identifies alcohol drinking-related mutation signature and genomic alterations. Nat Commun. 2017;8:15290. doi: 10.1038/ncomms15290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen HY, Chen RH. Cullin 3 Ubiquitin Ligases in Cancer Biology: Functions and Therapeutic Implications. Front Oncol. 2016;6:113. doi: 10.3389/fonc.2016.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Carthew RW. Gene regulation by microRNAs. Curr Opin Genet Dev. 2006;16:203–8. doi: 10.1016/j.gde.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 11.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–60. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Westra HJ, Peters MJ, Esko T. et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–43. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin L, Pahuja KB, Wickliffe KE. et al. Ubiquitin-dependent regulation of COPII coat size and function. Nature. 2012;482:495–500. doi: 10.1038/nature10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu L, Wei Y, Reboul J. et al. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature. 2003;425:316–21. doi: 10.1038/nature01985. [DOI] [PubMed] [Google Scholar]
- 16.Dorr C, Janik C, Weg M. et al. Transposon Mutagenesis Screen Identifies Potential Lung Cancer Drivers and CUL3 as a Tumor Suppressor. Mol Cancer Res. 2015;13:1238–47. doi: 10.1158/1541-7786.MCR-14-0674-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyden LM, Choi M, Choate KA. et al. Mutations in kelch-like 3 and cullin 3 cause hypertension and electrolyte abnormalities. Nature. 2012;482:98–102. doi: 10.1038/nature10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agbor LN, Ibeawuchi SC, Hu C. et al. Cullin-3 mutation causes arterial stiffness and hypertension through a vascular smooth muscle mechanism. JCI Insight. 2016;1:e91015. doi: 10.1172/jci.insight.91015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Fan H, Fang S. et al. Mutations and expression of the NFE2L2/KEAP1/CUL3 pathway in Chinese patients with lung squamous cell carcinoma. J Thorac Dis. 2016;8:1639–44. doi: 10.21037/jtd.2016.06.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kovac M, Navas C, Horswell S. et al. Recurrent chromosomal gains and heterogeneous driver mutations characterise papillary renal cancer evolution. Nat Commun. 2015;6:6336. doi: 10.1038/ncomms7336. [DOI] [PMC free article] [PubMed] [Google Scholar]