Abstract
Whole-exome or whole-genome sequencing is becoming routine in clinical situations for identifying mutations underlying presumed genetic causes of disease including infertility. While this is a powerful approach for implicating polymorphisms or de novo mutations in genes plausibly related to the phenotype, a greater challenge is to definitively prove causality. This is a crucial requisite for treatment, especially for infertility, in which validation options are limited. In this study, we created a mouse model of a putative infertility allele, DMC1M200V. DMC1 encodes a RecA homolog essential for meiotic recombination and fertility in mice. This allele was originally implicated as being responsible for the sterility of a homozygous African woman, a conclusion supported by subsequent biochemical analyses of the mutant protein and by studies of yeast with the orthologous amino acid change. Here, we found that Dmc1M200V/M200V male and female mice are fully fertile and do not exhibit any gonadal abnormalities. Detailed immunocytological analysis of meiosis revealed no defects suggestive of compromised fertility. This study serves as a cautionary tale for making conclusions about consequences of genetic variants, especially with respect to infertility, and emphasizes the importance of conducting relevant biological assays for making accurate diagnoses in the era of genomic medicine.
Introduction
Surveys estimate that 10–15% of the United States population experiences some form of infertility (1), and it is believed that as many as half of infertility cases have an underlying genetic basis. Over the past decade, candidate gene sequencing and whole-exome/genome re-sequencing have become increasingly used clinically for identifying potential causes of presumed genetic disorders including infertility. Several studies have employed the general approach of identifying a candidate variant/mutation in a sterile patient(s), sequencing the suspect gene in a number of normal patients, and then suggesting causation of the candidate mutation if it is unique (or homozygous in) only the infertile people (2–5). However, such correlation is not proof, as illustrated in the case of a purported globozoospermia allele of Spata16 (6) that was shown by mouse modeling to not impact fertility (7).
A major strategy for identifying and implicating putative disease mutations is the use of algorithms designed to predict the impact of mutations on protein function. Popular algorithms such as Polyphen-2, SIFT and FATHMM utilize direct or machine-learning strategies to assess protein sequence, structure and amino acid properties to generate predictions (8–10). Nonetheless, recent work has found that the predictions of damaging alleles are frequently inaccurate, and many predicted null mutations cause subtle deficiencies at best (11,12). In addition, studies specifically aimed at fertility effects in vivo found that as low as 25% of predicted high-damaging variants cause any detectable phenotype (13). Although computational approaches can be useful, these studies emphasize that current algorithms may not be sufficiently reliable for high-confidence assessment of the physiological impact of variants, especially in a clinical context.
In this present study, we took a closer look at a variant of human DMC1, a methionine-to-valine change at amino acid position 200 (Dmc1M200V). Dmc1 is a meiosis-specific homolog of Escherichia coli RecA and is essential for the repair of SPO11/TOP6BL-induced double-strand breaks (DSBs) by homologous recombination (14–17), a process that is essential for pairing of chromosome homologs. Dmc1 knockout mice are sterile due to arrest of meiocytes stemming from failed homologous chromosome synapsis (14,18) and activation of the meiotic DNA damage checkpoint (19). DMC1M200V was implicated as being responsible for premature ovarian failure in an African woman who was homozygous for this allele (20,21). Follow-up studies of this allele including crystallographic characterization, enzymatic assays and phenotypic analysis of yeast bearing the orthologous amino acid change demonstrated impaired function of the protein (21). We sought to expand upon these findings by introducing the M200V allele in the mouse ortholog of Dmc1. Here, we report that Dmc1M200V/M200V male and female mice are fertile, have normal fecundity and gamete numbers and have no meiotic defects. Our findings underscore the potential difficulties in reliably assessing allele pathogenicity and argue for the use of relevant in vivo functional assays to guide therapeutic actions.
Results
Dmc1M200V/M200V mice are fertile and phenotypically similar to wild type
To generate mice modeling the human DMC1M200V allele, CRISPR/Cas9-mediated genome editing in single-celled zygotes was performed. As diagrammed in Figure 1A and B, a single-stranded oligodeoxynucleotide (ssODN) was used as a template to introduce two nucleotide changes into codon 200 via homologous recombination, causing this codon to encode valine instead of methionine. Founder mice with the desired mutation were identified (Fig. 1C), backcrossed into FVB/NJ for at least two generations, then intercrossed to generate heterozygous and homozygous mice for phenotypic analysis.
Figure 1.
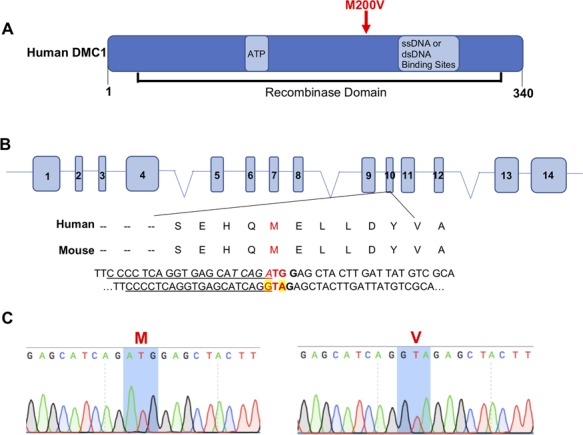
CRISPR/Cas9-mediated generation of Dmc1 M200V mice. (A) Diagram of human DMC1 protein labeled with known functional domains and binding sites. The M200V amino acid change is encoded by SNP rs2227914. (B) CRISPR-Cas9 genome editing strategy to introduce the M200V amino acid change. Underlined is the sgRNA sequence, in bold is the PAM site, highlighted in yellow are nucleotide changes for Val and italicized is the restriction enzyme site for Hpy188I (TCN/GA). (C) Sanger sequencing chromatograms from WT (left) and Dmc1M200V/M200V mouse (right).
Male and female Dmc1M200V/M200V mice did not show gross phenotypic abnormalities. To assess the fertility status of these mice, 8-week-old Dmc1M200V/M200V male and female mice were housed with wild-type (WT) mates over the course of 4–8 months. Litter sizes of these mutants versus controls were not significantly different (Fig. 2A). Thus, the Dmc1M200V allele does not impair fertility or fecundity in mice.
Figure 2.
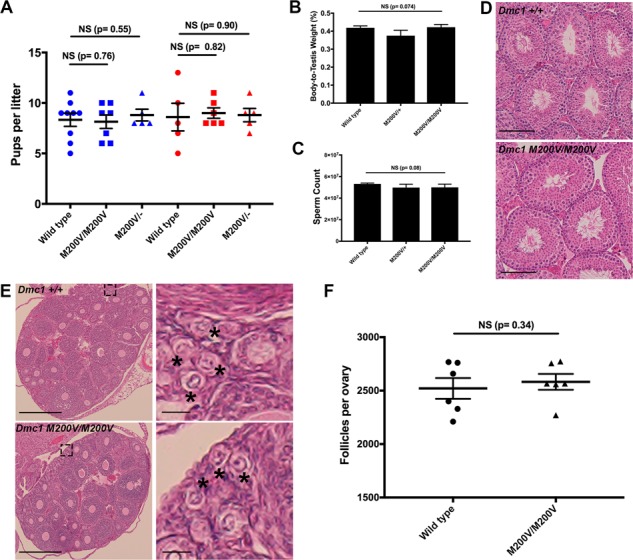
Dmc1 M200V/+ and Dmc1 M200V/M200V mice are fertile and show no gross abnormalities. (A) Fertility testing. Mice of each genotype listed were bred to a WT mate starting at 8 weeks of age. Litter sizes are shown. The three genotypes on the left (blue) refer to the father’s genotype and the three genotypes on the right (red) refes to the mother’s genotype. WT males (n = 3) sired an average of 8.3 ± 0.65 (SEM) pups and WT females (n = 3) had average litters of 8.6 ± 1.4. The average litters of Dmc1M200V/M200V males (n = 3) and females (n = 3) were 8.2 ± 0.67 and 9 ± 1.52, respectively. The average litters of Dmc1M200V/- males (n = 2) and females (n = 2) mice were 8.8 ± 0.58 and 8.8 ± 0.66, respectively. P-values were from the Student’s t-tests. NS = not significant. (B) The mass of 8-week-old testes were measured and averaged, then the ratio was calculated relative to the respective animal’s body mass. Average values ± SEM were 0.41 ± 0.0097 for WT (n = 6), 0.38 ± 0.036 for Dmc1M200V/+ (n = 5) and 0.42 ± 0.015 for Dmc1M200V/M200V (n = 4). (C) Sperm counts from 8-week-old males. Average values ± SEM were 53 150 000 ± 802 808 for WT (n = 6); 49 806 667 ± 3 097 300 for Dmc1M200V/+ (n = 5) and 50 025 ,000 ± 2 947 987 for Dmc1M200V/M200V (n = 4). (D) Testicular histology of WT and mutant. H&E stained testis cross-sections were from 8-week-old animals, and imaged at 40x magnification. Size bar, 75 μm. (E) Ovarian histology of WT and mutant. Paraffin-embedded and H&E stained cross-sections were from 3-week-old WT and Dmc1M200V/M200V ovaries. The left panels were imaged at 10x (size bar, 500 μm). The right panels are higher magnifications from the insets in the left panels (size bar, 50 μm). Black asterisks mark primordial follicles. (F) Quantification of primordial follicles in 3-week-old ovaries. Averages for each ovary are 2516 ± 144 follicles in WT (n = 3; SEM) and 2581 ± 74.2 follicles in Dmc1M200V/M200V (n = 3) and Dmc1M200V/M200V (n = 3).
To determine if the mutation caused subclinical gonadal defects, gross and histological analyses of testes and ovaries were performed. Eight-week old Dmc1M200V/+ and Dmc1M200V/M200V testis weights and sperm counts were indistinguishable from WT (Fig. 2B, C). Similarly, mutant testis histology revealed no abnormalities in spermatogenesis (Fig. 2D).
Although females had normal fecundity, it is possible that the mutation caused reduction in the ovarian reserve. Because DMC1 is required for DSB repair, any deficiency in its function would result in activation of the DNA damage checkpoint, eliminating oocytes perinatally before folliculogenesis (19,22). WT and Dmc1M200V/M200V ovaries were histologically indistinguishable (Fig. 2E). We serially sectioned mutant and WT ovaries and quantified the total number of primordial follicles in 3-week-old ovaries, revealing no significant difference (Fig. 2F). These results are consistent with the breeding studies and provide no evidence of any compromise to gametogenesis in either sex.
DMC1M200V does not disrupt meiotic chromosome behavior in mice
Despite having normal fertility, fecundity and histology, it is possible that the M200V amino acid change has a mild effect only detectable at the cellular level. Dmc1 is a meiosis-specific recombinase that acts at sites of meiotically programmed DSBs to catalyze D-loop formation and strand exchange between chromosomes, ultimately promoting chromosome pairing, synapsis and recombination (14,15,18,23). As mentioned in the Introduction, in vitro assays showed that DMC1M200V had altered biochemical activities under certain conditions, namely reduced ATPase activity at higher temperatures and reduced D-loop formation at low magnesium concentrations. Furthermore, the orthologous amino acid change in the fission yeast Schizosaccharomyces pombe caused a 50% reduction in recombination at two loci (21). To detect any anomalies that may reflect defective function of DMC1M200V during mouse meiosis, we conducted immunocytological analyses of surface-spread chromosomes from WT and Dmc1M200V/M200V spermatocytes, using markers diagnostic for certain key events of meiosis. First, we tested if DMC1M200V localizes normally to meiotically programmed DSBs. There was no significant difference in DMC1 focus numbers between WT and Dmc1M200V/M200V during the peak stages of DSB formation: leptonema and zygonema (Fig. 3A, B). As meiotic prophase I progresses and DSBs are repaired by recombination, DSBs (marked by DMC1 foci) along the lateral/axial elements of the synaptonemal complex (SC; marked by SYCP3) (24) decrease in zygonema and pachynema, ultimately disappearing in late pachynema. This was the case with Dmc1M200V/M200V spermatocytes. These results indicate that DMC1M200V is properly recruited to sites of meiotic DSBs, catalyzes homologous recombination repair of these DSBs which is essential for pairing of homologous chromosomes and is properly released from processed DSBs following repair.
Figure 3.
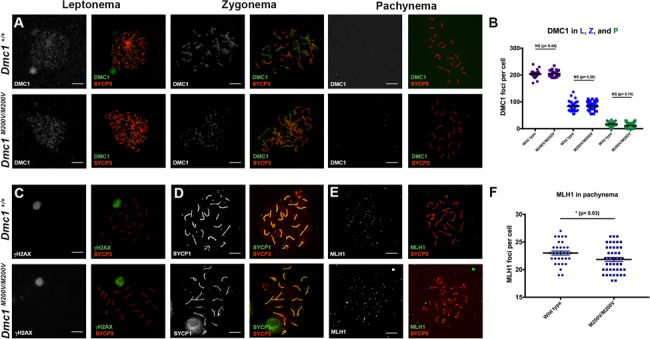
Immunocytological analysis of meiotic chromosomes in Dmc1 M200V/M200V spermatocytes. (A) Immunolabeling of WT and Dmc1M200V/M200V surface-spread leptotene, zygotene and pachytene spermatocyte nuclei with indicated antibodies. Size bar, 20 μm. (B) Quantification of DMC1 foci from A. L, leptonema; Z, zygonema; P, pachynema. The average numbers of DMC1 foci/cell ± SEM were 206 ± 3.2 (25 cells), 79 ± 3.7 (30 cells) and 16 ± 1.4 (30 cells) in leptonema, zygonema and pachynema, respectively. The average ± SEM DMC1 foci in Dmc1M200V/M200V were 208 ± 2.9 (45 cells), 83 ± 3.2 (40 cells) and 11 ± 1.9 (38 cells) in leptonema, zygonema and pachynema, respectively. The cells scored were from three WT males and three Dmc1M200V/M200V males. (C) Dmc1M200V/M200V pachytene spermatocytes exhibit normal-appearing XY bodies (marked by intense γH2AX staining). They also lack residual meiotic γH2AX—marked DSB-containing regions over autosomes. Testes from three males of each genotype were surveyed. Size bar, 20 μm. (D) Dmc1M200V/M200V spermatocytes exhibit complete synapsis. SYCP1, an SC central element protein, labels regions of synapsed chromosomes. The only asynapsed region in both genotypes corresponds to the non-homologous parts of the X and Y, which are stained red (SYCP3), rather than yellow (the composite of SYCP1/3). WT (n = 3) and Dmc1M200V/M200V (n = 3). Size bar, 20 μm. (E) Quantification of COs using the surrogate marker, MLH1. Representative pachytene cells are stained for MLH1 (green) and SYCP3 (red). Size bar, 20 μm. (F) Quantification of MLH1 foci in pachytene spermatocytes. Average ± SEM MLH1 foci in WT was 23 ± 2.0 (n = 3; 96 cells) and 22 ± 2.4 (n = 4; 120 cells) in Dmc1M200V/M200V. P-value is from the two-tailed Student’s t-test.
Next, we immunolabeled spreads with γH2AX, a histone variant phosphorylated at sites of DNA damage, which also serves as a marker of the transcriptionally silenced and heterochromatinized XY body that forms in pachynema (25). Similar to WT, leptotene and zygotene Dmc1M200V/M200V spermatocytes had high levels of γH2AX along chromosomes (data not shown) and by pachynema, γH2AX was exclusive to the X and Y chromosomes, indicative of complete DSB repair on autosomes and XY body formation (Fig. 3C). We then immunolabeled chromosome spreads for SYCP3 and the transverse element protein SYCP1, which marks regions of complete synapsis between homologs (24). In the absence of DMC1, repair of meiotic DSBs does not occur, and homologous chromosomes do not synapse (14,18). SC formation occurred normally in Dmc1M200V/M200V spermatocytes as indicated by SYCP3/SYCP1 staining of pachytene chromosomes (Fig. 3D). This indicates that DMC1M200V does not affect homologous chromosome synapsis.
Meiotic DSBs are repaired by one of two mechanisms: the majority (∼90%) as non-crossovers (NCOs) and the rest as crossovers (COs). While both types of events are important for driving homolog pairing and synapsis, chiasmata formed by CO events are essential for proper chromosome segregation at the first meiotic division. They ensure that homologs correctly align at the metaphase plate and segregate to opposite poles when the COs are resolved. To test whether CO formation is affected in Dmc1M200V/M200V mutants, we quantified MLH1 foci—a proxy for the predominant class of COs in mice—in pachytene spermatocyte nuclei (Fig. 3E). Interestingly, whereas WT spermatocytes had 23 ± 2.0 MLH1 foci/cell (average ± SEM, n = 3, 96 cells), Dmc1M200V/M200V had 22 ± 2.4 MLH1 foci/cell (average ± SEM, n = 4, 120 cells), which is a small but statistically significant difference (P-value = 0.03; Fig. 3F).
One copy of Dmc1M200V is sufficient for normal meiosis
Although fertility, fecundity and meiotic prophase I chromosome behavior was normal in Dmc1M200V/M200V mice, we considered the possibility that the mutant protein might be compromised in a subtle way that would only manifest under more challenging conditions. Accordingly, we bred mice hemizygous for Dmc1M200V (Dmc1M200V/-) by crossing our Dmc1M200V/M200V to mice heterozygous for a functional Dmc1 allele (Dmc1tm1Jcs). The testis weights and sperm counts of 8-week-old Dmc1M200V/- males were no different than WT (Fig. 4A, B), and testis histology was unremarkable (Fig. 4C). Likewise, females at 3 weeks of age had primordial follicle counts similar to WT (Fig. 4D). Lastly, breeding Dmc1M200V/- animals to WT mates yielded normal sized litters; males sired an average of 8.8 pups/litter and females birthed 8.8 pups/litter (P-value = 0.55 and P-value = 0.90, respectively; Fig. 2A). These results provide further evidence that under in vivo conditions, DMC1M200V has no substantial impact on fertility, fecundity or gametogenesis.
Figure 4.
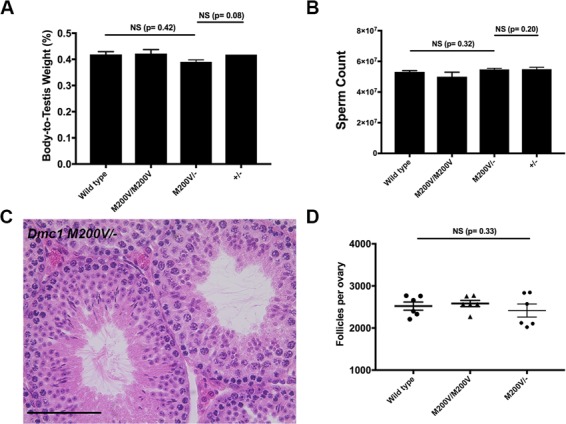
One copy of the Dmc1 M200V allele is sufficient for normal meiosis and fertility in mice. (A) Testis weight is unaffected by the Dmc1M200V allele in a hemizygous state. The chart includes some data from Figure 2B. Mice were 8 weeks old. Average body-to-testis weight values (± SEM) not presented in Figure 2 were Dmc1M200V/- (n = 4; 0.39 ± 0.0075) and Dmc1+/- (n = 2, average ± SEM = 0.42 ± 0.00003). P-values were from the Student’s t-test. (B) Sperm count is unaffected by the Dmc1M200V allele in a hemizygous state. The chart includes some data from Figure 2C. Mice were 8 weeks old. Average values (± SEM) not presented in Figure 2 were Dmc1M200V/- (n = 4; 54 300 000 ± 703 562) and Dmc1+/- (n = 2; average ± SEM = 56 100 000 ± 600 000). P-values were from the Student’s t-test. (C) Representative image of paraffin-embedded and H&E stained testis cross-section from an 8-week-old Dmc1M200V/- male. Size bar, 75 μm. (D) Primordial follicle counts from three-week old Dmc1M200V/- ovaries. The chart includes some data from Figure 2F. The average ± SEM for Dmc1M200V/- was 2 416 ± 155 follicles/ovary (n = 3 animals and 6 ovaries).
Discussion
Remarkable advances in genomics capabilities in the past few years are revolutionizing medicine. As DNA sequencing costs have dropped precipitously, it is becoming more routine to use whole-exome or whole-genome sequencing as clinical tests for certain common (namely, cancer) or rare diseases (such as undiagnosed developmental disorders). This will certainly become more commonplace for identifying potential genetic causes of idiopathic infertility in individual patients. Coupled with the advent of genome editing technology, it is increasingly plausible (scientifically) to envision genetic correction of infertility-causing alleles, particularly in males where spermatogonial stem cells are present and can be cultured, manipulated and returned into the patient. Of course such interventions, should they indeed be deemed as safe and acceptable, are absolutely dependent upon unequivocal identification of the infertility-causing mutation or variant.
The work presented here is part of an NIH-funded project (see acknowledgements) we have undertaken to specifically address this issue (13), and our results serve to highlight the serious nature of the problem. The Dmc1M200V allele was identified as a potentially causative infertility variant 10 years ago (20,21) and associated work provided compelling evidence that the allele encoded a protein with altered biochemical activities that might be consistent with disrupted meiosis (21). However, our in vivo modeling of this allele showed that this allele, in mice, did not impact fertility or fecundity at all.
While we are of the opinion that in vivo modeling in mice is a more physiologically relevant biological test than in vitro biochemical assays, there are still potential caveats. One is that it is possible that the M200V alteration is tolerated in mice but not humans. We consider this unlikely because the mouse and human DMC1 proteins are 97.5% identical, and there are no differences from position M200 to the C terminus of both proteins (position 340). The nearest difference is conservative (ASP versus GLU), 18 AAs N terminal. Nevertheless, we checked the octameric ring crystal structure (21) to see if M200 interacts with a polymorphic distant residue in the folded state. The Met200 residue on helix alpha-11 of one DMC1 monomer appears to interact with MET249 on alpha helix 13 of the adjacent DMC1 monomer. This amino acid, as well as the entire AA sequence of both alpha helices are identical between mouse and human. Finally, there are no Dmc1 paralogs unique to mice, and the mouse gene is absolutely required for meiosis and fertility in mice.
It is also conceivable that the gonadal environment in humans is such that the M200V change is catalytically more unstable in human meiocytes than in mouse meiocytes. For example, we found that in Dmc1M200V/M200V spermatocytes, there was a slight decrease (<5%) in the number of MLH1 foci. However, the number of MLH1 foci varies among inbred strains (26), and since the founder mice were of mixed genetic background, it is possible that the variation is due to these differences, possibly linked to the Dmc1 locus. Nevertheless, the impact, if any, was negligible in terms of all other reproductive parameters (e.g. germ cell numbers, litter sizes etc.).
A final caveat is the possibility that the mixed genetic background used in this study somehow suppressed the biochemical consequences of the Dmc1M200V mutation, and that if placed in other backgrounds (say, a completely inbred background), that a hypomorphic phenotype might be revealed. However, there have been no reports of anything but catastrophic germ cell loss in Dmc1 null mouse studies (14,18) in numerous laboratories (including our own, where we produced the first null allele, and have maintained it in the C57BL/6J background).
The case of Dmc1M200V is emblematic of the challenges facing genetic diagnosis of infertility in a clinical situation. Computational algorithms are useful for estimating the potential damage of a variant on protein function, but should not serve as an endpoint during the validation process due to their insufficient reliability (11,12), particularly if the information was to be used for treatment actions. The DMC1M200V allele was originally cited as being ‘probably damaging’ by the widely used Polyphen algorithm (20). However, the updated Polyphen-2 now predicts the variant as benign, as does FATHMM, SNPs & GO and SIFT. In contrast, PANTHER and Mutation Assessor score the allele as being ‘possibly damaging’ and ‘medium functional impact’, respectively. Current algorithms measure different criteria with different emphases, which can result in contradictory conclusions. However, technology is dynamic, and with increasing numbers of in vivo studies and training data sets, these algorithms may become more accurate with time. Another indicator of whether an allele might cause infertility is allele frequency. As noted by He et al. (27), the Dmc1M200V has a high allelic frequency in African populations (12% according to the gnomAD database), which seems incompatible with it being an infertility allele.
In conclusion, this study highlights the importance of in vivo modeling to make accurate conclusions about putative infertility-associated polymorphisms or de novo mutations before taking clinical actions. While mice are currently the most accurate models for this (7,13,28), we anticipate that improvements to in vitro gametogenesis, especially with human cells, will be important for development of higher-throughput and lower-cost analysis for diagnostics of genetic causes of human infertility.
Materials and Methods
Generation of Dmc1M200V mice by CRISPR-Cas9 genome editing
An optimal guide sequence was selected using online software at mit.crispr.edu. The crRNA and CRISPR-Cas9 tracrRNA was synthesized by IDT (ALT-R service) and the ssODN was also synthesized by IDT’s (Ultramer service). Prior to pronuclear injection, the crRNA (25 ng/μl) and tracrRNA (25 ng/μl) were co-incubated to form a ribonucleoprotein complex according to manufacturer’s instructions. The ssODN (50 ng/μl) and additional CAS9 protein (1000 ng/μl; PNA Bio) were added, and all materials were co-injected into zygotes (F1 hybrids between strains FVB/NJ and B6(Cg)-Tyrc-2J/J), then transferred into the oviduct of pseudopregnant females. Founders bearing at least one copy of the desired alteration were identified by PCR with primers flanking the SNP (Supplementary Material, Table I), then backcrossed into FVB/NJ. Initial phenotyping was performed after one backcross generation followed by intercrossing, then additional phenotyping was done with animals backcrossed additional two or more generations. No phenotypic differences were found among different generations.
All sequences used for CRISPR-Cas9 editing and mouse genotyping are in Supplementary Material, Table I.
Mice
The Dmc1tm1Jcs allele (referred to in the text as Dmc1-) was previously described (18).
All animal use was conducted under protocol (2004-0038) to J.C.S. and approved by Cornell University’s Institutional Animal Use and Care Committee.
Genotyping of Dmc1M200V and Dmc1+/- mice
Toes or ear clips were collected from pups at 8–14 days of age. A crude DNA lysate was made as described (29). PCR was performed using EconoTaq and associated reagents (Lucigen), following the manufacturer’s protocol with 3 μl of crude DNA lysate per reaction. Primers were obtained from IDT and sequences are listed in Supplementary Material, Table I. The PCR cycle used for both Dmc1M200V and Dmc1+/- mice PCR was initial denature at 95°C for 5 min, 30 cycles of 95°C for 30 seconds, 58°C for 30 seconds, 72°C for 30 seconds, and final elongation at 72°C for 5 min. For identification of Dmc1+/- mice, we used agarose gel electrophoresis to analyze the presence of WT (167 bp) and/or mutant PCR (250 bp) products amplified by respective primers. For identification of Dmc1M200V mice, PCR samples were digested with restriction enzyme Hpy188I (NEB) at 37°C for 2 h then analyzed using agarose gel electrophoresis. The WT allele is detected as two bands (177 bp and 31 bp) while the Dmc1M200V allele remains intact (208 bp).
Histology and primordial follicle quantification
Testes were harvested from 8-week-old males and fixed in Bouin’s for 24 h, washed in 70% ethanol for 24 h and embedded in paraffin. Sections of 6 μm were made and stained with hematoxylin and eosin (H&E). Ovaries were harvested from 3-week-old females and prepared in the same way, then serial sectioned at 6 μm. Primordial follicles were counted in every fifth section and final follicle counts were calculated as previously described (30). Statistical analysis was done with a two-tailed Student’s t-test on Prism 7 (GraphPad).
Sperm counts
Epidydymides were isolated from 8-week-old males. The tissue was minced in 5 ml of MEM media and incubated at 37°C for 15 min, allowing spermatozoa to swim out. The spermatozoa were diluted 1:2 in MEM and counted using a hemacytometer
Immunocytochemistry of meiotic chromosomes
We used a published protocol (31). In brief, testes were isolated from 8- to 12-week-old males, detunicated and minced in MEM media. Spermatocytes were hypotonically swollen in 4.5% sucrose solution, lysed in 0.1% Triton X-100, 0.02% SDS and 2% formalin. Slides were washed and stained immediately or stored at -80°C. Blocking buffer used was 5% goat serum in PBS, 0.1% Tween20 and slides were blocked for 1 h at room temperature. Primary antibodies and dilutions used were anti-SYCP3 (1:600, Abcam, #ab15093), anti-SYCP3 (1:600, Abcam, #ab97672), anti-SYCP1 (1:400, Abcam, #ab15090), anti-DMC1 (1:100, Abcam, #ab11054), anti-MLH1 (1:100, BD Pharmingen, #554073) and anti-phospho-H2A.X (1:1000, Millipore, #16-193). Primary staining of chromosome surface spreads was done at 37°C and incubated overnight. Secondary antibodies used were goat anti-mouse IgG AlexaFluor 488 (1:1000, ThermoFisher Scientific, #R37120) and goat anti-rabbit IgG AlexaFluor 594 (1:800, ThermoFisher Scientific, #R37117). Secondary antibodies were incubated for 1 h at room temperature. Images were acquired with an Olympus microscope with 40x lens using cellSens software (Olympus). Foci were quantified using ImageJ with plugins Cell Counter (Kurt De Vos) and Nucleus Counter. All data were analyzed in Prism 7 (GraphPad).
Supplementary Material
Acknowledgements
We thank Robert Munroe and Christian Abratte at Cornell University’s Stem Cell and Transgenic Core Facility (supported by contract C029155 from the New York State Stem Cell program) for performing the CRISPR-Cas9 microinjections.
Conflict of Interest statement. None declared.
Funding
National Institute of Child Health and Human Development (R01HD082568, T32HD052471); SUNY Graduate Diversity Fellowship.
References
- 1. Chandra A., Copen C.E. and Stephen E.H. (2013) Infertility and impaired fecundity in the United States, 1982-2010: data from the National Survey of Family Growth. Natl. Health Stat. Reports, 67, 1–19. [PubMed] [Google Scholar]
- 2. Sato H., Miyamoto T., Yogev L., Namiki M., Koh E., Hayashi H., Sasaki Y., Ishikawa M., Lamb D.J., Matsumoto N. et al. (2006) Polymorphic alleles of the human MEI1 gene are associated with human azoospermia by meiotic arrest. J. Hum. Genet., 51, 533–540. [DOI] [PubMed] [Google Scholar]
- 3. Miyamoto T., Koh E., Sakugawa N., Sato H., Hayashi H., Namiki M. and Sengoku K. (2008) Two single nucleotide polymorphisms in PRDM9 (MEISETZ) gene may be a genetic risk factor for Japanese patients with azoospermia by meiotic arrest. J. Assist. Reprod. Genet., 25, 553–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Miyamoto T., Tsujimura A., Miyagawa Y., Koh E., Sakugawa N., Miyakawa H., Sato H., Namiki M., Okuyama A. and Sengoku K. (2009) A single nucleotide polymorphism in SPATA17 may be a genetic risk factor for Japanese patients with meiotic arrest. Asian J. Androl., 11, 623–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. França M.M., Lerario A.M., Funari M.F.A., Nishi M.Y., Narcizo A.M., Mello M.P., Guerra-Junior G., Maciel-Guerra A.T. and Mendonça B.B. (2017) A novel homozygous missense FSHR variant associated with hypergonadotropic hypogonadism in two siblings from a Brazilian family. Sex Dev., 11, 137–142. [DOI] [PubMed] [Google Scholar]
- 6. Dam A.H.D.M., Koscinski I., Kremer J.A.M., Moutou C., Jaeger A.-S., Oudakker A.R., Tournaye H., Charlet N., Lagier-Tourenne C., Bokhoven H. et al. (2007) Homozygous mutation in SPATA16 is associated with male infertility in human globozoospermia. Am. J. Hum. Genet., 81, 813–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fujihara Y., Oji A., Larasati T., Kojima-Kita K. and Ikawa M. (2017) Human globozoospermia-related gene Spata16 is required for sperm formation revealed by CRISPR/Cas9-mediated mouse models. Int. J. Mol. Sci., 18(10) pii: E2208. doi: 10.3390/ijms18102208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kumar P., Henikoff S. and Ng P.C. (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc., 4, 1073–1081. [DOI] [PubMed] [Google Scholar]
- 9. Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S. and Sunyaev S.R. (2010) A method and server for predicting damaging missense mutations. Nat. Methods, 7, 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shihab H.A., Gough J., Cooper D.N., Stenson P.D., Barker G.L.A., Edwards K.J., Day I.N.M. and Gaunt T.R. (2013) Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum. Mutat., 34, 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang T., Bu C.H., Hildebrand S., Jia G., Siggs O.M., Lyon S., Pratt D., Scott L., Russell J., Ludwig S. et al. (2018) Probability of phenotypically detectable protein damage by ENU-induced mutations in the Mutagenetix database. Nat Commun, 9, 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Miosge L.A., Field M.A., Sontani Y., Cho V., Johnson S., Palkova A., Balakishnan B., Liang R., Zhang Y., Lyon S. et al. (2015) Comparison of predicted and actual consequences of missense mutations. Proc. Natl. Acad. Sci. USA, 112, E5189–E5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Singh P. and Schimenti J.C. (2015) The genetics of human infertility by functional interrogation of SNPs in mice. Proc. Natl. Acad. Sci. USA, 112, 10431–10436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yoshida K., Kondoh G., Matsuda Y., Habu T., Nishimune Y. and Morita T. (1998) The mouse RecA-like gene Dmc1 is required for homologous chromosome synapsis during meiosis. Mol. Cell, 1, 707–718. [DOI] [PubMed] [Google Scholar]
- 15. Bishop D.K., Park D., Xu L. and Kleckner N. (1992) DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell, 69, 439–456. [DOI] [PubMed] [Google Scholar]
- 16. Robert T., Nore A., Brun C., Maffre C., Crimi B., Bourbon H.M. and Massy B. (2016) The TopoVIB-Like protein family is required for meiotic DNA double-strand break formation. Science, 351, 943–949. [DOI] [PubMed] [Google Scholar]
- 17. Keeney S., Giroux C.N. and Kleckner N. (1997) Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell, 88, 375–384. [DOI] [PubMed] [Google Scholar]
- 18. Pittman D., Cobb J., Schimenti K., Wilson L., Cooper D., Brignull E., Handel M.A. and Schimenti J. (1998) Meiotic prophase arrest with failure of chromosome pairing and synapsis in mice deficient for. Mol. Cell, 1, 697–705. [DOI] [PubMed] [Google Scholar]
- 19. Bolcun-Filas E., Rinaldi V.D., White M.E. and Schimenti J.C. (2014) Reversal of female infertility by Chk2 ablation reveals the oocyte DNA damage checkpoint pathway. Science, 343, 533–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mandon-Pépin B., Touraine P., Kuttenn F., Derbois C., Rouxel A., Matsuda F., Nicolas A., Cotinot C. and Fellous M. (2008) Genetic investigation of four meiotic genes in women with premature ovarian failure. Eur. J. Endocrinol., 158,107–115. [DOI] [PubMed] [Google Scholar]
- 21. Hikiba J., Hirota K., Kagawa W., Ikawa S., Kinebuchi T., Sakane I., Takizawa Y., Yokoyama S., Mandon-Pépin B., Nicolas A. et al. (2008) Structural and functional analyses of the DMC1-M200V polymorphism found in the human population. Nucleic Acids Res., 36, 4181–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Di Giacomo M., Barchi M., Baudat F., Edelmann W., Keeney S. and Jasin M. (2005) Distinct DNA-damage-dependent and -independent responses drive the loss of oocytes in recombination-defective mouse mutants. Proc. Natl. Acad. Sci. USA, 102, 737–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schwacha A. and Kleckner N. (1997) Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell, 90, 1123–1135. [DOI] [PubMed] [Google Scholar]
- 24. Dobson M.J., Pearlman R.E., Karaiskakis A., Spyropoulos B. and Moens P.B. (1994) Synaptonemal complex proteins: occurrence, epitope mapping and chromosome disjunction. J. Cell Sci., 107 (Pt 10), 2749–2760. [DOI] [PubMed] [Google Scholar]
- 25. Burma S., Chen B.P., Murphy M., Kurimasa A. and Chen D.J. (2001) ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem., 276, 42462–42467. [DOI] [PubMed] [Google Scholar]
- 26. Anderson L.K., Reeves A., Webb L.M. and Ashley T. (1999) Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics, 151, 1569–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. He W.-B., Tu C.-F., Liu Q., Meng L.-L., Yuan S.-M., Luo A.-X., He F.-S., Shen J., Li W., Du J. et al. (2018) DMC1 mutation that causes human non-obstructive azoospermia and premature ovarian insufficiency identified by whole-exome sequencing. J. Med. Genet., 55, 198–204. [DOI] [PubMed] [Google Scholar]
- 28. Kherraf Z.-E., Conne B., Amiri-Yekta A., Kent M.C., Coutton C., Escoffier J., Nef S., Arnoult C. and Ray P.F. (2018) Creation of knock out and knock in mice by CRISPR/Cas9 to validate candidate genes for human male infertility, interest, difficulties and feasibility. Mol. Cell. Endocrinol. 268, 70–80, 10.1016/j.mce.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 29. Truett G.E., Heeger P., Mynatt R.L., Truett A.A., Walker J.A. and Warman M.L. (2000) Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). BioTechniques, 29, 52–54. [DOI] [PubMed] [Google Scholar]
- 30. Myers M., Britt K.L., Wreford N.G.M., Ebling F.J.P. and Kerr J.B. (2004) Methods for quantifying follicular numbers within the mouse ovary. Reproduction, 127, 569–580. [DOI] [PubMed] [Google Scholar]
- 31. McNairn A.J., Rinaldi V.D. and Schimenti J.C. (2017) Repair of meiotic DNA breaks and homolog pairing in mouse meiosis requires a minichromosome maintenance (MCM) paralog. Genetics, 205, 529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


