Abstract
Despite growing evidence linking Drosophila melanogaster tweety‐homologue 1 (Ttyh1) to normal mammalian brain development and cell proliferation, its exact role has not yet been determined. Here, we show that Ttyh1 is required for the maintenance of neural stem cell (NSC) properties as assessed by neurosphere formation and in vivo analyses of cell localization after in utero electroporation. We find that enhanced Ttyh1‐dependent stemness of NSCs is caused by enhanced γ‐secretase activity resulting in increased levels of Notch intracellular domain (NICD) production and activation of Notch targets. This is a unique function of Ttyh1 among all other Ttyh family members. Molecular analyses revealed that Ttyh1 binds to the regulator of γ‐secretase activity Rer1 in the endoplasmic reticulum and thereby destabilizes Rer1 protein levels. This is the key step for Ttyh1‐dependent enhancement of γ‐secretase activity, as Rer1 overexpression completely abolishes the effects of Ttyh1 on NSC maintenance. Taken together, these findings indicate that Ttyh1 plays an important role during mammalian brain development by positively regulating the Notch signaling pathway through the downregulation of Rer1.
Keywords: central nervous system development, embryonic neural stem cells, Notch, Ttyh1, γ‐secretase
Subject Categories: Development & Differentiation, Signal Transduction, Stem Cells
Introduction
Mammalian brain development requires the precise regulation of adequate production and organization of diverse neural cells such as neurons, astrocytes, and oligodendrocytes 1. Embryonic neural cell fate specification, which determines neuronal or glial cell types, is controlled by the cell microenvironment where multiple signaling cues act. For example, spatiotemporally regulated cell signaling pathways such as the Notch, Sonic hedgehog (Shh), and Wnt pathways maintain the balance between the number of neural stem cells and newly generated differentiated progeny cells 2. During the early stages of brain development, neuroepithelial cells mostly divide symmetrically to amplify the neural stem cell pool. As development progresses, the neurogenic period begins and neural stem cells undergo asymmetric divisions to produce intermediate progenitors or postmitotic neurons as well as neural stem cells 3.
Notch receptors are cell surface transmembrane proteins which regulate important cellular events such as cell fate decision, proliferation, and apoptosis in numerous tissues and organs 4. In vertebrates, there are four mammalian Notch receptors (Notch1 to Notch4), and although they share similar structures and ligands, they perform distinct functional roles because of spatiotemporally different expression patterns in the adult as well as embryonic tissues 5. During transport to the membrane, Notch is cleaved by furin convertase activity at site 1 (S1), producing the heterodimeric mature receptor 6. Initiation of signaling cascades is triggered by direct cell‐to‐cell contact which enables interaction between Notch receptors and its ligands such as Delta‐like 1 (Dll1), Dll3, Dll4, Jagged1 (Jag1), or Jag2. Ligand binding induces a conformational change in Notch receptors followed by a second cut at S2 by ADAM metalloproteases. The Notch intracellular region is then further cleaved by γ‐secretase activity at S3 in proximity to the transmembrane domain to liberate the Notch intracellular domain (NICD). NICD then moves to the nucleus and associates with Notch coactivational members including CBF1 and Mastermind‐like 1 (MAML1) to activate Notch target gene expression.
The Drosophila melanogaster tweety (tty) gene was originally isolated as a transcription unit adjacent to the flightless 1 gene 7. Subsequently, three mammalian tty homologues, Ttyh1, Ttyh2, and Ttyh3, were identified on the basis of sequence homology to the Drosophila Tty protein 8, 9, 10. Protein sequence analysis predicted that the Ttyh family are integral membrane proteins containing five transmembrane domains, and that they may act as chloride channels 8, 10. Our interest in the Ttyh family members stemmed from previous works in which Ttyh2 was identified as a gene upregulated in renal and colon cancers 9, 11, implicating Ttyh as a potent regulator of cell proliferation. Recently, this hypothesis was further supported by a study reporting that Ttyh1 was important for glioma growth and Ttyh1 downregulation resulted in reduced tumor progression 12. More relevant to our study, Ttyh1 was found to be strongly expressed in the ventricular zone (VZ) 13, 14, 15, where neural stem cells reside and active cell proliferation occurs in the mammalian developing brain. Later, taking advantage of this finding, another group used Ttyh1 as a marker for embryonic apical neural progenitor cells 16. However, the exact role and the underlying mechanisms of Ttyh1 action in the brain were yet to be elucidated.
In this study, we show that Ttyh1 enhances embryonic neural stem cell properties by facilitating the Notch signaling pathway. Furthermore, we find that retention in endoplasmic reticulum 1 (Rer1) is the direct target of Ttyh1 action, providing a potential mechanism by which Ttyh1 influences the Notch signaling pathway and thereby brain development. Rer1 has been known to exist in the ER and cis‐Golgi and have roles in the regulation of γ‐secretase activity 17, 18, 19, intracellular rhodopsin trafficking 20, alpha‐synuclein degradation 21, ER retention of peripheral myelin protein 22 (PMP22) 22, and the quality control of some voltage‐gated sodium channels 23. Since Notch has been known to be implicated in tumor progression in the adult, our study should also help better understanding of the molecular basis involved in Ttyh1‐implicated phenotypic outcomes in other cellular events such as oncogenesis.
Results and Discussion
Ttyh1 enhances embryonic neural stem cell properties
The most prominent evidence for the functional role of Ttyh1 came from a study reporting the early embryonic lethality of Ttyh1‐deficient mice 15. This study also suggested the involvement of Ttyh1 in brain development by showing that Ttyh1 is highly expressed in the VZ, the germinal layer of the embryonic brain, during the mid‐neurogenic period. However, so far, the exact functional contribution of Ttyh1 to brain development remains to be established. In an effort to understand the importance of embryonic expression of Ttyh1 in the VZ, we first tested the effects of Ttyh1 on embryonic neural progenitor cells by the neurosphere assay. The neurosphere assay is the most widely used method to assess the stem cell capacity of isolated neural cells 24, 25. Mouse neural progenitor cells isolated from the brain tissues of embryonic day 14.5 (E14.5) embryos were transduced with a Ttyh1‐expressing retroviral vector MSIG 26 and cultured in defined neural stem cell media. Under this experimental condition, only cells with neural stem cell properties can grow into cell clusters, called neurospheres. MSIG contains the murine stem cell virus long terminal repeat (LTR) that drives transgene expression and an internal ribosome entry sequence (IRES) which enables GFP reporter gene expression. MSIG‐Ttyh1 retroviral transduction increased Ttyh1 mRNA levels in the transduced cells up to 15‐fold compared to control MSIG‐transduced cells (Fig EV1A). At 7 days posttransduction, Ttyh1 expression gave rise to an increase both in the number and size of neurospheres compared to the control (Fig 1A), suggesting that Ttyh1 enhances the self‐renewal and proliferative activities of neural progenitor cells, respectively. A cell proliferation assay using the tetrazolium salt XTT also showed increased neural progenitor cell growth upon Ttyh1 expression (Fig 1B). Consistently, Ttyh1 knockdown by short hairpin RNA against Ttyh1 (shTtyh1) significantly reduced the number and size of neurospheres (Fig 1C), and neural progenitor cell growth assessed by the XTT assay (Fig 1D). In in vivo settings, we found that a significantly higher fraction of cells overexpressing Ttyh1 was detected in the VZ, the embryonic neural stem cell region, and subventricular zone (SVZ) at 2 days post‐in utero electroporation (Fig 1E and F), and expressed the neural stem cell marker Sox2 (Fig 1G and H). Ttyh1 knockdown produced opposite results; shTtyh1 expression increased cell exit from the VZ and SVZ toward SVZ and IZ, respectively, at only a day post‐electroporation, and these cell position changes by shTtyh1 were restored toward the VZ by coexpression of a shRNA‐resistant Ttyh1 mutant gene (Ttyh1res; Fig 1I and J), excluding the possibility of nonspecific off‐target effects of the shTtyh1 used in this study. Taken together, these results suggest that Ttyh1 enhances embryonic neural stem cell properties in vivo as well as in vitro.
Figure EV1. mRNA levels of Ttyh1 after gene manipulation in primary mouse neural progenitor cells in vitro .
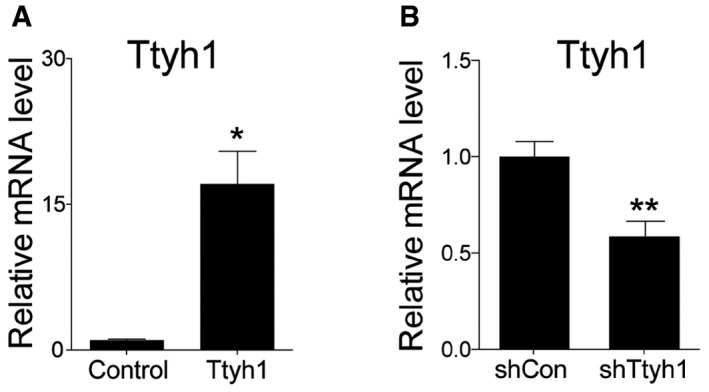
-
A, BTwo days after E14.5 primary neural progenitor cells were transduced with retroviral vectors expressing (A) Ttyh1 or (B) shTtyh1, mRNA expression levels of Ttyh1 were measured by qPCR. Error bars represent SD. Student's t‐test was used to determine statistical significance. *P < 0.05, **P < 0.01, n = 3 for (A), n = 4 for (B).
Figure 1. Ttyh1 enhances embryonic neural stem cell properties.
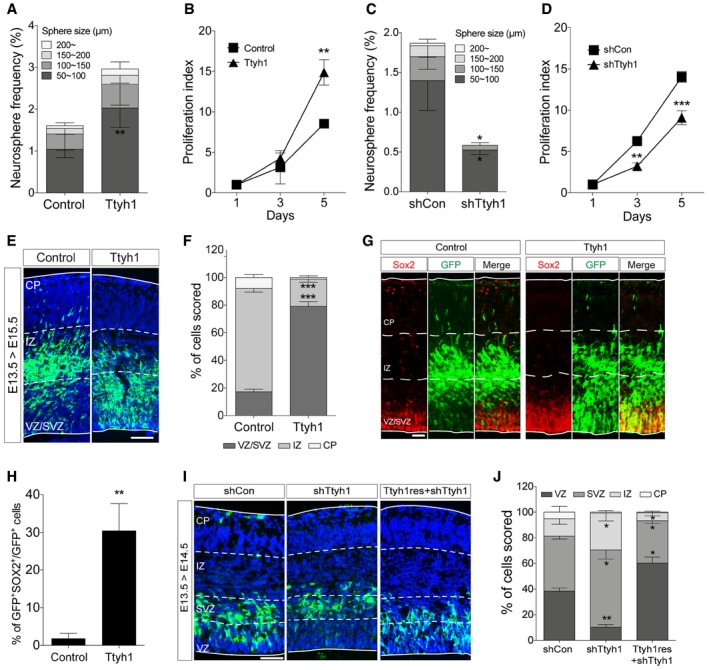
-
A–DNeurosphere (A, C) and XTT (B, D) assays were performed using mouse E14.5 primary neural progenitors transduced with a retroviral vector expressing Ttyh1 or short hairpin RNA specific to Ttyh1 (shTtyh1) (n = 4 for A and C, n = 3 for B and D).
-
ETtyh1‐expressing cells in E15.5 embryonic brains that were intraventricularly electroporated at E13.5 were immunostained using an anti‐GFP primary (reporter gene) and Alexa Fluor 488‐conjugated secondary antibodies. GFP immunofluorescence (green) was merged with DAPI‐counterstained images (blue). VZ, ventricular zone; SVZ, subventricular zone; IZ, intermediate zone; CP, cortical plate. Scale bar: 100 μm.
-
FQuantification of (E) (n = 4).
-
GDouble‐immunolabeling of E15.5 brain sections electroporated in utero with Ttyh1‐expressing plasmid at E13.5 using anti‐GFP (green) and Sox2 (red) antibodies, and Alexa Fluor 488‐ and 555‐conjugated secondary antibodies. Scale bar: 50 μm.
-
HQuantification of (G) (n = 3).
-
IImmunolabeling of E14.5 brain sections electroporated in utero with shTtyh1 alone or in combination with a shTtyh1‐resistant Ttyh1 (Ttyh1res) expression vector at E13.5 using anti‐GFP and Alexa Fluor 488‐conjugated secondary antibodies. The DAPI nuclear counterstain is shown in blue. Scale bar: 100 μm.
-
JQuantification of (I) (n = 3).
Ttyh1 increases Notch target gene expression
Next, we searched for an intracellular signaling pathway associated with the Ttyh1‐enhanced neural stem cell characteristics. Using quantitative real‐time PCR (qPCR) analysis of downstream target genes of various stem cell‐related signaling pathways, we found that Ttyh1 upregulated the expression of Notch target genes. Ttyh1 expression increased mRNA expression levels of the Hes1, Hes5, and Hey1 genes in primary neural progenitor cells (Fig 2A). Furthermore, shTtyh1 reduced the mRNA levels of Notch target genes, and these effects were reversed by coexpression of Ttyh1res (Fig 2B). MAML1 is one of the core Notch coactivational complex members, and a dominant negative form of MAML1 (dnMAML1) has been used to block Notch signaling pathway 27, 28, 29. Coexpression of a dnMAML1 was found to completely abolish the ability of Ttyh1 to enhance Notch target gene expression in the primary neural progenitors (Fig 2C). In vivo, coexpression of dnMAML1 also efficiently disrupted Ttyh1‐induced cell localization in the VZ/SVZ (Fig 2D and E). Furthermore, the cell position changes upon Ttyh1 knockdown were reverted toward the VZ by coexpression of NICD (Fig 2F and G). These data indicate that Ttyh1 actions in neural stem cell regulation were accomplished through enhancing the Notch signaling pathway. In light of previous findings showing that Ttyh1 is a direct transcriptional target of Notch 30, our findings complete the identification of a positive feedback loop mechanism between Ttyh1 expression and Notch signaling pathway, where mutual increasement would occur during the neurogenic period.
Figure 2. Ttyh1 increases Notch target gene expression.
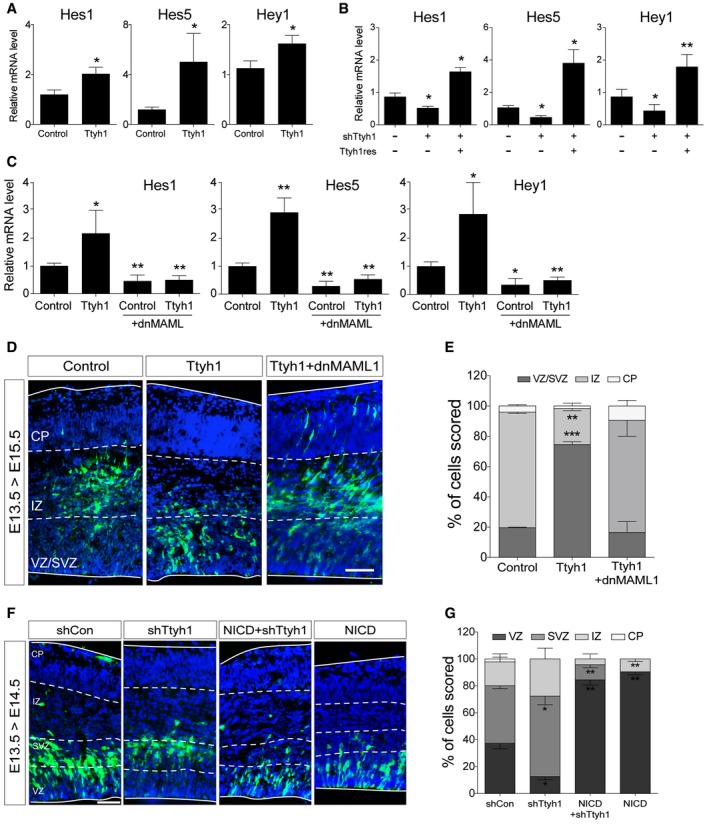
-
A, BTwo days after E14.5 primary neural progenitor cells were transduced with retroviral vectors expressing (A) Ttyh1 or (B) shTtyh1, mRNA expression levels of each indicated Notch target gene were measured by qPCR (n = 4 for A, n = 3 for B). To confirm specificity of the Ttyh1 knockdown, rescue experiments were performed by cotransducing retroviral vectors expressing shTtyh1 and Ttyh1res.
-
CEffects of dominant negative MAML1 (dnMAML1) on Ttyh1‐enhanced Notch target gene expression were tested by qPCR using E14.5 primary neural progenitors at 2 days posttransduction (n = 4).
-
DThe dnMAML1 plasmid was introduced with the Ttyh1 vector into embryonic brains at E13.5, and cell localization was analyzed at E15.5 by anti‐GFP immunostaining. GFP and DAPI signals are shown in green and blue, respectively. VZ, ventricular zone; SVZ, subventricular zone; IZ, intermediate zone; CP, cortical plate. Scale bar: 100 μm.
-
EQuantification of GFP+ cell positions in (D) (n = 3).
-
FThe NICD plasmid was electroporated with the Ttyh1 knockdown vector (shTtyh1) into embryonic brains at E13.5, and cell localization was analyzed at E14.5 by anti‐GFP immunostaining. Scale bar: 100 μm.
-
GQuantification of GFP+ cell positions in (F) (n = 3).
Ttyh1 upregulates γ‐secretase activity
We then investigated which step of the Notch pathway was affected by Ttyh1. Ligand‐receptor binding induces a series of proteolytic Notch receptor cleavages that release the NICD. NICD then translocates into the nucleus where it forms a Notch coactivational complex. qPCR (Fig EV2A), Western blot (Fig EV2B), and fluorescence‐activated cell sorting (FACS) analysis (Fig EV2C) showed that Notch mRNA, protein, and cell surface levels of the Notch receptor, respectively, were not changed by Ttyh1 expression in primary neural progenitor cells, suggesting that the enhanced transactivation of Notch target genes caused by Ttyh1 expression was not due to increased expression of Notch receptor or enhanced Notch trafficking to the membrane. Rather, Ttyh1 expression was found to increase the amount of NICD in neural progenitor cells (Fig 3A and B), and Ttyh1 knockdown produced the opposite effects (Fig 3C and D). Promotion of NICD generation without changes in the expression of the Notch receptor implicated the γ‐secretase, which is responsible for the intramembranous cleavage of Notch. To test this possibility, we examined the level of γ‐secretase activity by performing a γ‐secretase luciferase reporter assay using primary neural progenitor cells. One γ‐secretase reporter construct contains a membrane‐tethered form of truncated Notch into which a Gal4 DNA‐binding/VP16 transactivation (GVP) domain was inserted (Fig 3E) 31. Upon γ‐secretase cleavage, the liberated GVP domain translocates into the nucleus and transactivates luciferase expression from the UAS promoter. This assay has been used as a specific and sensitive method for measurement of γ‐secretase activity. As shown in Fig 3F, Ttyh1 expression significantly increased luciferase expression, and this effect was blocked by a γ‐secretase inhibitor, N‐[N‐(3,5‐difluorophenacetyl)‐L‐alanyl]‐S‐phenylglycine t‐butyl ester (DAPT). shRNA‐mediated Ttyh1 knockdown also significantly reduced luciferase expression (Fig 3G). Consistently, Ttyh1‐enhanced NICD production and Notch target gene expression were reduced by DAPT down to DAPT‐treated control levels (Fig 3H and I), suggesting a role for γ‐secretase in Ttyh1‐increased Notch activity. Generation of NICD requires the S2 cleavage by ADAM10 metalloprotease prior to the S3 cleavage by γ‐secretase 6, 32. As shown in Fig EV3A, an ADAM10 inhibitor, GI254023, also efficiently inhibited Ttyh1‐increased NICD generation. Presenilin 1 (PS1) is one of the four core proteins in the γ‐secretase complex, and a dominant negative form of PS1 (dnPS1) effectively abolishes γ‐secretase activity 33. The importance of Ttyh1‐augmented γ‐secretase activity in Ttyh1‐enhanced neural stem cell properties was further supported by in vivo experiments employing cointroduction of Ttyh1 and dnPS1 into the developing brain. As expected, coexpression of dnPS1 restores the position of Ttyh1‐expressing cells from the VZ/SVS to intermediate zone (IZ) to control levels in vivo (Fig 3J and K). Taken together, these data indicate that Ttyh1 enhances neural stem cell properties by strengthening the Notch signaling pathway through increasing γ‐secretase activity.
Figure EV2. Notch1 receptor expression was not changed by Ttyh1 overexpression.
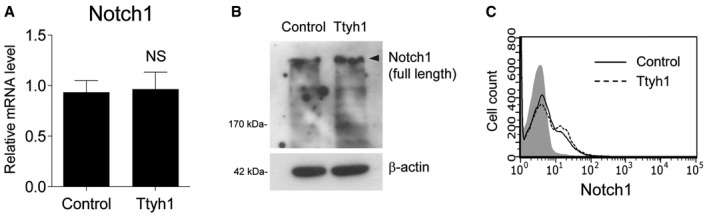
-
A, BTwo days after E14.5 primary neural progenitor cells were transduced with retroviral vectors expressing Ttyh1, Notch1 mRNA (A) and protein (B) levels were assessed by qPCR. Error bars represent SD. Student's t‐test was used to determine statistical significance. NS, not significant. n = 3 for (A).
-
CE14.5 neural progenitor cells were transduced with Ttyh1 retroviruses. At 2 days posttransduction, a representative FACS histogram showing cell surface levels of Notch1 was obtained using an anti‐Notch1 antibody (Invitrogen, MA5‐11961). The lightly shaded curve represents the secondary antibody‐only control.
Source data are available online for this figure.
Figure 3. Ttyh1 increases Notch intracellular domain (NICD) production by enhancing γ‐secretase activity.
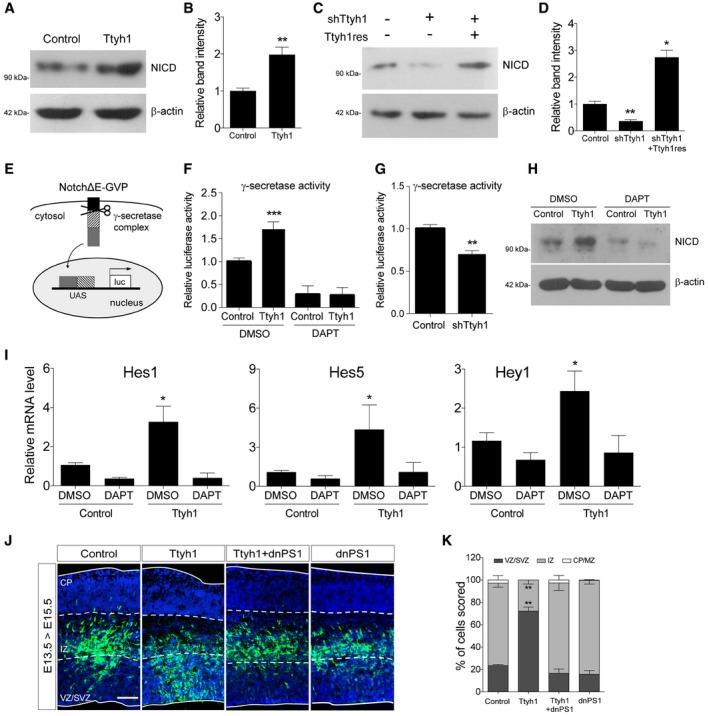
-
A–DEffects of Ttyh1 expression (A) or Ttyh1 knockdown (C) on the generation of NICD at 2 days posttransduction of E14.5 neural progenitor cells were analyzed by Western blotting using an anti‐NICD antibody. (B, D) Quantification of band intensities in (A, C) (n = 4 for B, n = 3 for D).
-
ESchematic representation of the γ‐secretase reporter assay. Following cleavage at the γ‐secretase site in NotchΔE‐Gal4‐VP16 (NotchΔE‐GVP), the intracellular domain containing the GVP domain translocates into the nucleus and triggers luciferase expression driven by the UAS promoter (Figure redrawn from Karlström et al 31).
-
F, G(F) Ttyh1‐transduced E14.5 neural progenitors were incubated with DAPT for 2 days, then cells were harvested and lysates were used for luciferase assay (n = 3). (G) Effects of Ttyh1 knockdown on γ‐secretase activity were also assessed using E14.5 neural progenitor cells (n = 4 for F, n = 3 for G).
-
H, ITtyh1‐transduced E14.5 neural progenitors were incubated for 2 days in the presence or absence of DAPT, and samples were prepared for Western blotting using anti‐NICD antibody (H) or qPCR for Notch target genes (I) (n = 3).
-
J, K(J) The indicated plasmids were electroporated into E13.5 brains, and brain samples were harvested at E15.5 for anti‐GFP immunolabeling. GFP and DAPI signals are shown in green and blue, respectively. (K) Quantification of GFP+ cell localization in (J) (n = 3). dnPS1, dominant negative presenilin 1; VZ, ventricular zone; SVZ, subventricular zone; IZ, intermediate zone; CP, cortical plate. Scale bar: 100 μm.
Figure EV3. Effects of ADAM10 inhibition on Ttyh1‐enhanced NICD production and effects of synoviolin (Syvn) inhibition on Ttyh1‐induced Rer1 reduction.
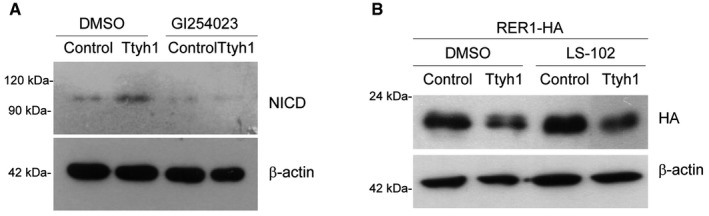
- Ttyh1‐transduced E14.5 neural progenitors were incubated with or without an ADAM10 inhibitor, GI254023, for 2 days, then cells were harvested for Western blot analysis using anti‐NICD antibody.
- HEK293T cells were transfected with Ttyh1‐Myc and Rer1‐HA expression plasmids and treated with a Syvn inhibitor LS102. At 2 days posttransfection, cells were harvested and analyzed by Western blotting using anti‐HA antibody.
Source data are available online for this figure.
Ttyh1 enhances Notch signaling in a chloride channel activity‐independent manner
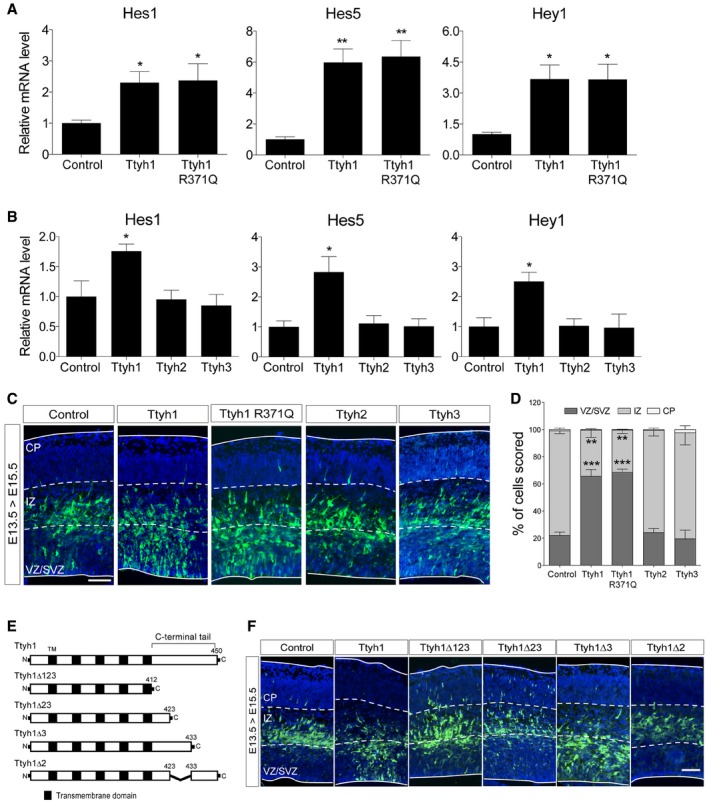
Since the only currently known mechanistic function of Ttyh1 is a maxi‐Cl− ion channel 10, we tested if the Cl− ion channel activity contributed to Notch signaling pathway. Reyes et al 34 showed that the R371 residue of Ttyh1 provides the electrostatic basis for the selectivity filter preferring anions over cations, and the R371Q mutation resulted in a pore more permeable to cations. We found that a Ttyh1 R371Q mutant increased Notch target gene expression in vitro and induced cell localization in the VZ/SVZ in vivo comparable to wild type, suggesting that the Notch‐enhancing activity of Ttyh1 is not linked to Cl− channel activity (Fig 4A and C). We also tested other Ttyh family members, Ttyh2 and Ttyh3, which are also known as maxi‐Cl− channels. As shown in Fig 4B and C, similar effects to Ttyh1 were not observed with these Ttyh family proteins in vitro and in vivo.
Figure 4. Cl− channel property of Ttyh1 is not associated with Notch pathway‐enhancing activity.
-
A, BEffects of the ion selectivity‐perturbing R371Q mutation in (A) Ttyh1 and (B) other Ttyh family members on Notch‐dependent transcription. E14.5 neural progenitor cells were transduced with retroviral vectors expressing each indicated gene, and 2 days posttransduction, cells were harvested for qPCR analyses of Notch target mRNAs (n = 3).
-
C, D(C) Immunolabeling of E15.5 brain sections electroporated in utero with indicated plasmids at E13.5 using anti‐GFP and Alexa Fluor 488‐conjugated secondary antibodies. GFP immunofluorescence (green) was merged with DAPI‐counterstained images (blue). (D) Quantification of GFP+ cell localization in (C) (n = 3). VZ, ventricular zone; SVZ, subventricular zone; IZ, intermediate zone; CP, cortical plate. Scale bar: 100 μm.
-
E, F(E) Schematic representation of a series of Ttyh1 mutants with deletions in the C‐terminal cytoplasmic tail region (Ttyh1 gene structure was not drawn in proportion to the actual domain size), and (F) their effects on cell distribution assessed by intraventricular in utero electroporation at E13.5 followed by immunolabeling at E15.5 using anti‐GFP antibody. Scale bar: 100 μm.
Ttyh1 is composed of a short N‐terminal extracellular domain, five membrane‐spanning segments, and C‐terminal cytoplasmic tail with less than 40 amino acid residues 8, 9. Since Notch receptor cleavage by γ‐secretase occurs inside cells, we hypothesized that the C‐terminal cytoplasmic region of Ttyh1 might be involved in the regulation of γ‐secretase activity. To test this hypothesis, we divided the Ttyh1 C‐terminal cytoplasmic region into three parts, generated four Ttyh1 deletion mutants (Fig 4E), and analyzed the outcomes of each deletion mutation. In vivo analysis showed that a deletion of a short 10 amino acid‐long midpart of the cytoplasmic tail (Ttyh1Δ2) was sufficient to completely disrupt the ability of Ttyh1 to induce cell localization in the VZ/SVZ (Fig 4F). Taken all together, our results indicate that Ttyh1 facilitates Notch signaling pathway in a Cl− channel activity‐independent manner, and that the C‐terminal cytoplasmic tail of Ttyh1 is important for Ttyh1 action in upregulating the Notch signaling pathway.
Despite sequence differences, Ttyh family member proteins were presumed to share some similar properties including Cl− channel activity 10. However, Ttyh1‐ablation alone in mice induced early embryonic lethality 15 suggesting two possibilities: First, distinct spatiotemporal expression patterns of Ttyh family member genes may define tissue‐specific and age‐specific functions of individual Ttyh members. Second, Ttyh2 and Ttyh3 may have been unable to compensate for the absence of Ttyh1 due to the unique roles of Ttyh1. The former possibility is supported by RNA in situ hybridization results showing that Ttyh2 and Ttyh3 expression was not detected in the embryonic tissues (http://www.informatics.jax.org). Our study strongly supports the latter possibility by providing an example of functional nonredundancy between Ttyh proteins with respect to Notch activation. In addition, although mammalian Ttyh1 C‐terminal sequences are highly conserved across species (Fig EV4A), the highest level of sequence variation among mouse Ttyh family proteins is observed at the C‐terminal tail region (Fig EV4B), which was expected to specify the differential functions of these proteins 9. Indeed, we showed that a small deletion of the cytoplasmic C‐terminal region could effectively abrogate the Ttyh1‐dependent phenotypes observed in this study.
Figure EV4. Protein sequence comparison of mammalian Ttyh C‐terminal tail regions.

- Alignment of Ttyh1 C‐terminal tail regions from cattle (GenBank accession number NM_001077015), chimpanzee (XM_512893), dog (XM_857514), horse (XM_023650408), human (NM_001005367), mouse (NM_001109765), and rat (NM_001106225).
- Protein sequence analysis of C‐terminal tail regions of mouse Ttyh1 (NM_001109765), Ttyh2 (NM_053273), and Ttyh3 (NM_175274).
Rer1 reduction is a key step for Ttyh1‐enhanced Notch pathway activation
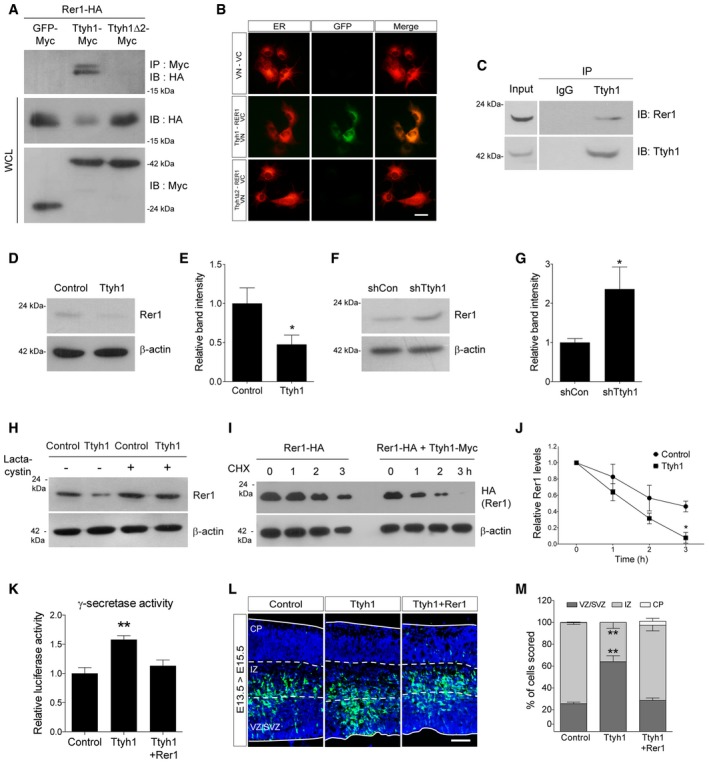
Finally, we attempted to further identify the molecular mechanism by which Ttyh1 increases γ‐secretase activity. We focused on a report in which Rer1 was screened as a Ttyh1 binding protein by biotin‐streptavidin pull‐down assay 35. In independent studies, Rer1 was also identified to negatively regulate γ‐secretase by altering γ‐secretase trafficking 17, 18, 19. These results led us to ask whether Rer1 is implicated in Ttyh1‐regulated Notch pathway activation. By coimmunoprecipitation (coIP) analysis of HEK293T cells expressing Myc‐tagged Ttyh1 and HA‐tagged Rer1, we confirmed Ttyh1 binding to Rer1 (Fig 5A). Interestingly, Rer1 was not coimmunoprecipitated with Ttyh1Δ2. We also analyzed the interaction between Ttyh1 and Rer1 using the Venus fluorescent protein‐based bimolecular fluorescence complementation (BiFC) assay 36. We fused Ttyh1 and Rer1 to amino‐ and carboxy‐terminal fragments of the Venus protein (VN and VC), respectively, and coexpressed them in the COS‐7 cells. As shown in Fig 5B, Venus complementation was observed only in the cells cotransfected with Ttyh1‐VN and Rer1‐VC, confirming that the two proteins indeed interact in living cells. An important advantage of the BiFC assays is that it provides information on the subcellular localization of protein interaction 37. By using a DsRed2 gene fused with the endoplasmic reticulum (ER) targeting sequence of calreticulin (DsRed2‐ER) 38, we identified that Venus complementation occurred in the ER. Since both Ttyh1 and Rer1 were previously identified as ER‐ and Golgi‐resident proteins 15, 18, 19, 39, 40, the first step of Ttyh1 action, namely binding to Rer1, would be expected to occur in these subcellular organelles. Consistent with this prediction, BiFC analysis proved that Ttyh1 interacts with Rer1 in the ER. More importantly, the endogenous interaction between Ttyh1 and Rer1 proteins was further confirmed in a human glioblastoma cell line U373MG by coIP assays using anti‐Ttyh1 and anti‐Rer1 antibodies (Fig 5C). In addition to association of Ttyh1 with Rer1, the coIP assay produced the unexpected important finding that Rer1 protein levels were dramatically decreased by Ttyh1 coexpression (Fig 5A lane 2 on 2nd blot). Ttyh1Δ2 did not reduce Rer1 protein levels implying that the ability of Ttyh1 to bind to Rer1 is tightly linked with the reduction of Rer1 proteins. Using primary neural progenitor cells, we confirmed that endogenous levels of Rer1 proteins were significantly decreased by Ttyh1 expression (Fig 5D and E) and increased by shTtyh1 transduction up to levels twofold of the control (Fig 5F and G). Considering that mRNA knockdown efficiency of the shTtyh1 used in this study is less than 50% (Fig EV1B), this degree of increase strongly indicates that endogenous Ttyh1 efficiently reduces the amounts of Rer1 proteins under physiological conditions. In addition, the Ttyh1‐dependent reduction of Rer1 levels was restored to the control level by treatment with the proteasome inhibitor lactacystin (Fig 5H) or MG132 (Fig EV5A), suggesting that proteasome function is required for Rer1 reduction. To further investigate if the Rer1 reduction reflects a decrease in the stability of Rer1 protein, we performed Western blot analysis following cycloheximide block of de novo protein synthesis. As shown in Fig 5I and J, Ttyh1 coexpression resulted in a marked faster decrease of Rer1 signals compared with the control. The half‐life of the Rer1 protein decreased from approximately 3 h in control cells to 1.5 h in cells with Ttyh1 expression, whereas Rer1 mRNA levels were not affected by Ttyh1 expression (Fig EV5B). These findings indicate that Ttyh1 decreases Rer1 protein levels by reducing its stability. Since synoviolin (Syvn) was also previously reported to be able to interact with and degrade Rer1 via the proteasomal or lysosomal pathways 41, we tested if Syvn was implicated in Ttyh1‐induced Rer1 degradation. As shown in Fig EV3B, a Syvn inhibitor, LS‐102, did not affect the Ttyh1‐mediated Rer1 reduction, suggesting that Rer1 protein levels are independently controlled by Ttyh1 and Syvn.
Figure 5. Ttyh1 downregulates Rer1, and Rer1 expression counteracts Ttyh1‐induced enhancement of neural stem cell properties.
-
ARepresentative Western blots showing coimmunoprecipitation (coIP) of HA‐tagged Rer1 with Myc‐tagged Ttyh1 using the HEK293T cell line. WCL, whole‐cell lysates.
-
BVenus fluorescent protein‐based bimolecular fluorescence complementation analysis of Ttyh1 and Rer1 interaction (green) in living COS‐7 cells by fluorescence microscopy. Ttyh1 and Rer1 were tagged with N‐ and C‐terminal fragments of the Venus fluorescent protein (VN and VC), respectively. DsRed2‐ER was cotransfected to label the endoplasmic reticulum (ER, red). Scale bar: 25 μm.
-
CCoIP assays were performed with anti‐Ttyh1 and anti‐Rer1 antibodies to show endogenous interaction between Ttyh1 and Rer1 in U373MG cells.
-
D–GWestern blot analyses of endogenous Rer1 proteins were performed at 2 days posttransduction using E14.5 primary neural progenitor cells transduced with retroviral vectors expressing (D) Ttyh1 or (F) shTtyh1. (E, G) Quantification of relative band intensities in (D, F), respectively (n = 3).
-
HCell extracts from primary neural progenitor cells transduced with Ttyh1 retroviruses in the presence of DMSO vehicle or a proteasome inhibitor lactacystin (10 μM) were immunoblotted with an antibody to Rer1.
-
I, J(I) HEK293T cells were transfected with Ttyh1‐Myc and Rer1‐HA expression plasmids, and after 24 h, cells were treated with cycloheximide (CHX) (100 μg/ml) and then harvested at the indicated time intervals. Cell lysates were analyzed by Western blotting using anti‐HA antibody. (J) Quantification of band intensities in (I) (n = 3).
-
Kγ‐secretase‐dependent luciferase activity was measured using primary neural progenitor cells transduced with Ttyh1 with or without Rer1 retroviral vectors at 2 days posttransduction (n = 3).
-
L, M(L) Effects of Rer1 expression on Ttyh1 activity in vivo were examined by in utero electroporation into E13.5 brains and subsequent GFP immunofluorescence of E15.5 brain sections. GFP and DAPI signals are shown in green and blue, respectively. (M) Distribution of GFP+ cells in each brain layer in (L) was quantified (n = 3). VZ, ventricular zone; SVZ, subventricular zone; IZ, intermediate zone; CP, cortical plate. Scale bar: 100 μm.
Figure Figure EV5. Ttyh1 downregulates Rer1, and Rer1 expression abolishes Ttyh1‐induced enhancement of the Notch signaling pathway.
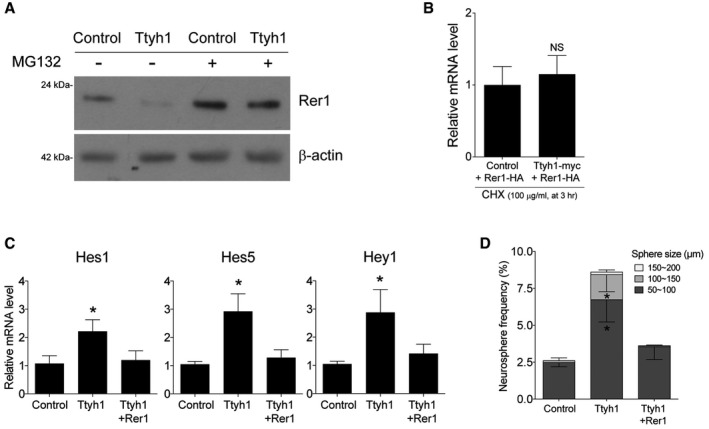
-
ACell extracts from primary neural progenitor cells transduced with Ttyh1 retroviruses in the presence of the DMSO vehicle or the proteasome inhibitor MG132 (5 μM) were immunoblotted with an antibody to Rer1.
-
BqPCR analysis for Rer1‐HA mRNA was performed using samples corresponding to lane 4 and lane 8 in Fig 5I (n = 4).
-
C, DE14.5 neural progenitor cells transduced with the indicated retroviruses were used for (C) qPCR analysis of Notch target mRNAs at 2 days posttransduction or (D) cultured for 7 days for the neurosphere assay. n = 4 for (C) and n = 3 for (D)
To test if Rer1 was the causal target of Ttyh1 for augmenting Notch pathway activation, we coexpressed Ttyh1 and Rer1 in primary embryonic neural progenitor cells and examined the effects on γ‐secretase activity and neural stem cell characteristics. As shown in Figs 5K and EV5C, Rer1 expression completely abolished Ttyh1‐enhanced γ‐secretase activity and Notch target gene expression. Consistently, Ttyh1‐increased neurosphere formation frequency and neurosphere size were decreased by Rer1 expression down to control levels (Fig EV5D). In vivo experiments also revealed that Rer1 coexpression disrupted the Ttyh1‐induced VZ/SVZ‐localizing cell distribution pattern (Fig 5L and M). These results strongly indicate that Rer1 downregulation is a key cellular process associated with the Ttyh1‐induced Notch pathway activation and enhancement of neural stem cell properties.
In this study, we focused on the role of Ttyh1 during embryonic development. However, Ttyh1 is also highly likely to play a regulatory role in γ‐secretase‐related cellular events in the postnatal stages. Particularly, Ttyh1 may be involved in adult neurogenesis by regulation of Notch activity because it is also detected in the adult hippocampus 42, an important adult neural stem cell niche. In addition to Notch, amyloid precursor protein (APP) is also a well‐known substrate of γ‐secretase, by which APP is cleaved to produce the hydrophobic amyloid beta peptide which is considered a major underlying causal factor of Alzheimer's disease 43. It is of note that Ttyh1 is also found to be expressed widely at significant levels in the adult brain (Allen Brain Atlas, http://www.brain-map.org) including the cerebrum 15. Therefore, future studies addressing whether Ttyh1 levels change as the brain ages and whether Ttyh1 is associated with amyloid beta‐caused neurodegenerative diseases in the aged brain should be of interest.
Materials and Methods
Plasmid construction
To construct expression vectors of the Ttyh family genes and Rer1, total RNA was isolated from primary mouse neural progenitor cells using TRIzol reagent (Invitrogen). The isolated RNA (2 μg) and oligo (dT) primers (Promega) were utilized to synthesize single‐stranded cDNA using a reverse transcription kit (Promega) according to the manufacturer's instructions. 1 μl of the cDNA product was amplified by PCR using 125 nM Ttyh‐ or Rer1‐specific primers: Ttyh1‐F (5′‐AACGCGTGCCACCATGGGGGCACCCCCGGGCT‐3′), Ttyh1‐R (5′‐TGGATCCTCACAAGTCCTCTTCAGAAATGAGCTTGATGGAAGACTGCCACTG‐3′), Ttyh2‐F (5′‐AACGCGTGCCACCATGCCGGCGGCGCGAGTG‐3′), Ttyh2‐R (5′‐TGGATCCCTACAAGTCCTCTGAAATGAGCTTGGACGGGAACTCGTAGTG‐3′), Ttyh3‐F (5′‐AACGCGTGCCACCATGGCCGGGGTCAGCTAC‐3′), Ttyh3‐R (5′‐TACGCGTCTACAAGTCCTCTTCAGAAATGAGCTTGTGCCCGCTGCCGCTGGA‐3′), Rer1‐F (5′‐AACGCGTGCCACCATGTCTGAAGGGGACAG‐3′), and Rer1‐R (5′‐TAGATCTCTAAGCGTAATCTGGAACATCGTATGGGTAACTAGCAAATGTCTTGCC‐3′). Amplified sequences were initially cloned into the pGem‐T Easy vector (Promega) and then introduced into the BamHI site of the retroviral vector MSIG 26. For Ttyh1 knockdown studies, the Ttyh1 target sequences 5′‐AGACACTGAATGTGACAGAA‐3′ were cloned into the pSIREN‐RetroQ vector (Clontech). Ttyh1 mutant constructs were generated using the QuikChange II Site‐Directed Mutagenesis Kit (Stratagene) and synthetic oligonucleotides: Ttyh1 R371Q Up (5′‐GGTGGCGCTGCTGCACTGCCAGAGTCTGCACAAGGACTATGG‐3′), Ttyh1 R371Q Down (5′‐CCATAGTCCTTGTGCAGACTCTGGCAGTGCAGCAGCGCCACC‐3′), Ttyh1Δ2 Up (5′‐GCCCTCTTCCCACCCAGTACTCCTTTCAACCCTCAGGAA‐3′), Ttyh1Δ2 Down (5′‐TTCCTGAGGGTTGAAAGGAGTACTGGGTGGGAAGAGGGC‐3′), Ttyh1Δ3 Up (5′‐ACACAGATGACGATGACAAGCTCATTTCTGAAGAGGACTTGTGAGGATCCA‐3′), Ttyh1Δ3 Down (5′‐TGGATCCTCACAAGTCCTCTTCAGAAATGAGCTTGTCATCGTCATCTGTGT‐3′), Ttyh1Δ23 Up (5′‐CCCTCTTCCCACCCAGTAAGCTCATTTCTGAAGAGGACTTGTGAGGATCCA‐3′), Ttyh1Δ23 Down (5′‐TGGATCCTCACAAGTCCTCTTCAGAAATGAGCTTACTGGGTGGGAAGAGGG‐3′), Ttyh1Δ123 Up (5′‐CCACCACTCTGTGCAGCAAGCTCATTTCTGAAGAGGACTTGTGAGGATCCA‐3′), and Ttyh1Δ123 Down (5′‐TGGATCCTCACAAGTCCTCTTCAGAAATGAGCTTGCTGCACAGAGTGGTGG‐3′). For bimolecular fluorescence complementation assays (BiFC), Ttyh1 and Ttyh1Δ2 fused to the N‐terminal half of the Venus fluorescent protein (VN) were obtained by cloning the corresponding PCR fragments into the BglII and SalI sites of pBiFC‐VN173 (Addgene, #22010). To generate Rer1 fused to the C‐terminal half of the Venus fluorescent protein (VC), the PCR‐amplified Rer1 coding sequence was inserted between the SalI and BglII sites of pBiFC‐VC155 (Addgene, #22011).
Coimmunoprecipitation (coIP) assay
For immunoprecipitations of tagged proteins, transfected HEK293T cells were lysed in immunoprecipitation lysis buffer [50 mM pH 7.5 Tris–HCl, 150 mM NaCl, 0.5% Triton X‐100, 1 mM EDTA] with protease inhibitor (Sigma). Total proteins were incubated with primary antibody against the Myc tag (Santa Cruz Biotechnology, sc‐40; 1:1,000) and protein G‐conjugated beads (Invitrogen) at 4°C overnight. Beads were washed three times with lysis buffer and 2× sample buffer was added. Samples were then boiled for 10 min, and beads were removed by centrifuging at 17,000 × g for 30 min. Proteins were resolved by SDS–PAGE and subjected to immunoblotting using an anti‐HA tag antibody (Abcam, ab16918; 1:1,000). Immunoprecipitations of endogenous Ttyh1 with anti‐Ttyh1 antibody (Sigma, WH0057348M4; 1:50) using a human glioblastoma cell line U373MG and subsequent immunoblotting with anti‐Rer1 antibody (Sigma, R4407; 1:200) were performed in a similar manner. The NIH ImageJ software (http://rsb.info.nih.gov/ij/) was used to quantify band intensity of the immunoblot.
Animals, neurosphere assay, and in utero electroporation
All animal procedures were approved by the Institutional Animal Care and Use Committee of Sungkyunkwan University under the animal protocol No. 2016‐04‐0005‐2. All experiments were conducted in accordance with the institutional guidelines and regulations for animal experiments. Primary neural progenitor cells were prepared from E14.5 timed‐pregnant CD1 mice (Orient Bio) embryos. Dissected brain tissue was minced, washed three times with PBS, and incubated in 0.25% trypsin (Invitrogen) at 37°C for 5 min. DNase I and ovomucoid trypsin inhibitor (both from Worthington) were added, and samples were triturated using a fire‐polished Pasteur pipette. Cells were washed twice with DMEM/F12 media, resuspended in PBS, and run through a 40 μm cell strainer (Falcon). For neurosphere assays, neural progenitors were transduced with concentrated retroviral vectors. After 48 h, aggregated cells were mechanically dissociated, and 5,000 cells were seeded and incubated in DMEM/F12 media supplemented with B27/N2 (Invitrogen) and FGF2 (Peprotech). After 7 days of incubation, total neurosphere numbers were counted under a light microscope (Eclipse TS100, Nikon). Coexpression by in vitro transduction was achieved by mixing two concentrated retroviral vectors at a ratio of 1:1. For in utero electroporation, timed‐pregnant CD1 mice were anesthetized with pentobarbital sodium (Hanlim Pharm). 1–2 μl of DNA solution (2–4.0 μg/μl, depending on the construct) in PBS with 0.01% fast green dye (Sigma) was injected into the lateral ventricle of E13.5 embryos and five square electric pulses (33 V) were delivered at one pulse per second (50 ms pulse followed by 950 ms gap) to embryos through the uterus with forceps‐type electrodes (CUY650P5, 5‐mm diameter platinum round plates; Nepagene). Coexpression by in utero electroporation was achieved by mixing two effector plasmids at a ratio of 1:1.
Immunofluorescence
Standard immunofluorescence procedures were used for visualization of target gene expression in electroporated animals. Briefly, gene‐transferred embryonic brains were fixed in 4% paraformaldehyde, dehydrated in 30% sucrose, and cryosectioned. Sections were washed in PBS, then blocked for 1 h with PBS containing 10% fetal bovine serum and 0.2% Triton X‐100. Samples were then incubated overnight at 4°C with anti‐GFP (Invitrogen, A‐11122; 1:1,000) with or without anti‐Sox2 (Chemicon, AB5603, 1:1,000) primary antibodies, washed three times in PBS, and incubated for 3 h at room temperature with Alexa Fluor 488‐ or 555‐conjugated secondary antibodies (Invitrogen) diluted in blocking solution. Samples were further counterstained with 4′,6‐diamidino‐2‐phenylindole (DAPI; Sigma). Fluorescent images were acquired using an upright microscope (Eclipse 80i, Nikon).
Western blot
Primary neural progenitor cells were lysed using the RIPA buffer (Sigma) with a protease and phosphatase inhibitor cocktail (Pierce Biotechnology, Rockford, IL). Equal amounts of protein (20~40 μg, depending on target protein) were resolved in 8~10% (w/v) sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and transferred to polyvinylidene fluoride (PVDF) membranes (PALL, Port Washington, NY). The membranes were blocked with TBST [150 mM NaCl, 10 mM Tris–HCl, 0.1% (v/v) Tween 20, pH 8.0] containing 5% (w/v) skim milk and analyzed with the following primary antibodies: anti‐Notch1 (Santa Cruz Biotechnology, sc‐9170; 1:200), anti‐cleaved Notch1 (Val1744; Cell Signaling Technology, 2421s; 1:300), anti‐Rer1 (Sigma, R4407; 1:200), anti‐Myc tag (Santa Cruz Biotechnology, sc‐40X; 1:1,000), anti‐HA tag (Abcam, ab16918; 1:1,000), and anti‐β‐actin (Sigma, A3854; 1:15,000). The protein bands were visualized with an enhanced chemiluminescence system (Atto, Tokyo, Japan) and X‐Omat film (Kodak, Rochester, NY). All blots were incubated overnight with a primary antibody at 4°C and with horseradish peroxidase‐conjugated anti‐mouse or anti‐rabbit secondary antibodies (1:10,000; Pierce Biotechnology) for 2 h at room temperature.
Quantitative real‐time PCR
Total RNA was prepared from mouse neural progenitor cells or HEK293T cells using TRIzol reagent (Invitrogen), and cDNAs were synthesized from 500 ng of each RNA sample by using an oligo (dT) primer and M‐MLV reverse transcription enzyme (Promega). Quantitative real‐time PCR (qPCR) was performed according to the Smart Cycler System (Takara) protocol using the following primers: Hes1 forward (5′‐TACCCCAGCCAGTGTCAACA‐3′), Hes1 reverse (5′‐TCTTGCCCTTCGCCTCTTC‐3′), Hes5 forward (5′‐CAAGGAGAAAAACCGACTGC‐3′), Hes5 reverse (5′‐GCTGGAAGTGGTAAAGCAGC‐3′), Hey1 forward (5′‐TGAGCTGAGAAGGCTGGTAC‐3′), Hey1 reverse (5′‐ACCCCAAACTCCGATAGTCC‐3′), Rer1 forward (5′‐GGCTGGACAAGTCTACCCC‐3′), Rer1 reverse (5′‐GCGTAGGTCACAATGTACCAAC‐3′) β‐actin forward (5′‐CAAAAGCCACCCCCACTCCTAAGA‐3′), and β‐actin reverse (5′‐GCCCTGGCTGCCTCAACACCTC‐3′).
BiFC assay
Different combinations of BiFC plasmids together with pDsRed2‐ER vector (Clontech) were transfected into COS‐7 cells using Lipofectamine 2000 (Invitrogen). Two days posttransfection, fluorescence signals from living COS‐7 cells were examined with an inverted microscope (Eclipse Ti, Nikon) equipped with GFP and Cy3 filter sets. Image analysis was conducted with the Nikon Elements software.
Rer1 protein stability assay
Six‐well plates of HEK293T cells were transfected with plasmids expressing HA‐tagged Rer1 alone or in combination with the Ttyh1 vector. After 24 h, 100 μg/ml cycloheximide (Sigma) was added to the medium, and cells were harvested at the indicated time points. Total cell extracts were prepared, and equivalent amounts of proteins were run on SDS–PAGE and analyzed by Western blotting using an anti‐HA antibody (Abcam, ab16918; 1:1,000). ImageJ software (http://rsb.info.nih.gov/ij/) was used to quantify band intensity of the immunoblot.
Statistical analysis
Statistical tests were performed using the Prism 6 software (GraphPad). Statistical analyses were performed using unpaired two‐tailed Student's t‐test. All data represent three or more independent experiments.
Author contributions
JK, DH, S‐HB, and MK carried out experiments. JK, DH, and JYC analyzed data. KY designed the study and wrote the manuscript with contribution from SJP.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Source Data for Expanded View
Review Process File
Source Data for Figure 3
Source Data for Figure 5
Acknowledgements
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (2018R1A2B2001076).
EMBO Reports (2018) 19: e45472
References
- 1. Jiang X, Nardelli J (2016) Cellular and molecular introduction to brain development. Neurobiol Dis 92: 3–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sansom SN, Griffiths DS, Faedo A, Kleinjan DJ, Ruan Y, Smith J, van Heyningen V, Rubenstein JL, Livesey FJ (2009) The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self‐renewal and neurogenesis. PLoS Genet 5: e1000511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Götz M, Huttner WB (2005) The cell biology of neurogenesis. Nat Rev Mol Cell Biol 6: 777–788 [DOI] [PubMed] [Google Scholar]
- 4. Yoon K, Gaiano N (2005) Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat Neurosci 8: 709–715 [DOI] [PubMed] [Google Scholar]
- 5. Wu J, Bresnick EH (2007) Bare rudiments of notch signaling: how receptor levels are regulated. Trends Biochem Sci 32: 477–485 [DOI] [PubMed] [Google Scholar]
- 6. Kopan R, Ilagan MX (2009) The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137: 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Campbell HD, Schimansky T, Claudianos C, Ozsarac N, Kasprzak AB, Cotsell JN, Young IG, de Couet HG, Miklos GL (1993) The Drosophila melanogaster flightless‐I gene involved in gastrulation and muscle degeneration encodes gelsolin‐like and leucine‐rich repeat domains and is conserved in Caenorhabditis elegans and humans. Proc Natl Acad Sci USA 90: 11386–11390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Campbell HD, Kamei M, Claudianos C, Woollatt E, Sutherland GR, Suzuki Y, Hida M, Sugano S, Young IG (2000) Human and mouse homologues of the Drosophila melanogaster tweety (tty) gene: a novel gene family encoding predicted transmembrane proteins. Genomics 68: 89–92 [DOI] [PubMed] [Google Scholar]
- 9. Rae FK, Hooper JD, Eyre HJ, Sutherland GR, Nicol DL, Clements JA (2001) TTYH2, a human homologue of the Drosophila melanogaster gene tweety, is located on 17q24 and upregulated in renal cell carcinoma. Genomics 77: 200–207 [DOI] [PubMed] [Google Scholar]
- 10. Suzuki M, Mizuno A (2004) A novel human Cl(‐) channel family related to Drosophila flightless locus. J Biol Chem 279: 22461–22468 [DOI] [PubMed] [Google Scholar]
- 11. Toiyama Y, Mizoguchi A, Kimura K, Hiro J, Inoue Y, Tutumi T, Miki C, Kusunoki M (2007) TTYH2, a human homologue of the Drosophila melanogaster gene tweety, is up‐regulated in colon carcinoma and involved in cell proliferation and cell aggregation. World J Gastroenterol 13: 2717–2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jung E, Osswald M, Blaes J, Wiestler B, Sahm F, Schmenger T, Solecki G, Deumelandt K, Kurz FT, Xie R et al (2017) Tweety‐homolog 1 drives brain colonization of gliomas. J Neurosci 37: 6837–6850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Abramova N, Charniga C, Goderie SK, Temple S (2005) Stage‐specific changes in gene expression in acutely isolated mouse CNS progenitor cells. Dev Biol 283: 269–281 [DOI] [PubMed] [Google Scholar]
- 14. Kawaguchi A, Ikawa T, Kasukawa T, Ueda HR, Kurimoto K, Saitou M, Matsuzaki F (2008) Single‐cell gene profiling defines differential progenitor subclasses in mammalian neurogenesis. Development 135: 3113–3124 [DOI] [PubMed] [Google Scholar]
- 15. Kumada T, Yamanaka Y, Kitano A, Shibata M, Awaya T, Kato T, Okawa K, Abe T, Oshima N, Nakahata T et al (2010) Ttyh1, a Ca(2+)‐binding protein localized to the endoplasmic reticulum, is required for early embryonic development. Dev Dyn 239: 2233–2245 [DOI] [PubMed] [Google Scholar]
- 16. Okamoto M, Miyata T, Konno D, Ueda HR, Kasukawa T, Hashimoto M, Matsuzaki F, Kawaguchi A (2016) Cell‐cycle‐independent transitions in temporal identity of mammalian neural progenitor cells. Nat Commun 7: 11349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Spasic D, Raemaekers T, Dillen K, Declerck I, Baert V, Serneels L, Füllekrug J, Annaert W (2007) Rer1p competes with APH‐1 for binding to nicastrin and regulates γ‐secretase complex assembly in the early secretory pathway. J Cell Biol 176: 629–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Park HJ, Shabashvili D, Nekorchuk MD, Shyqyriu E, Jung JI, Ladd TB, Moore BD, Felsenstein KM, Golde TE, Kim SH (2012) Retention in endoplasmic reticulum 1 (RER1) modulates amyloid‐β (Aβ) production by altering trafficking of γ‐secretase and amyloid precursor protein (APP). J Biol Chem 287: 40629–40640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jurisch‐Yaksi N, Rose AJ, Lu H, Raemaekers T, Munck S, Baatsen P, Baert V, Vermeire W, Scales SJ, Verleyen D et al (2013) Rer1p maintains ciliary length and signaling by regulating γ‐secretase activity and Foxj1a levels. J Cell Biol 200: 709–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yamasaki A, Hara T, Maejima I, Sato M, Sato K, Sato K (2014) Rer1p regulates the ER retention of immature rhodopsin and modulates its intracellular trafficking. Sci Rep 4: 5973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Park H‐J, Ryu D, Parmar M, Giasson BI, McFarland NR (2017) The ER retention protein RER1 promotes alpha‐synuclein degradation via the proteasome. PLoS One 12: e0184262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hara T, Hashimoto Y, Akuzawa T, Hirai R, Kobayashi H, Sato K (2014) Rer1 and calnexin regulate endoplasmic reticulum retention of a peripheral myelin protein 22 mutant that causes type 1A Charcot‐Marie‐Tooth disease. Sci Rep 4: 6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Valkova C, Liebmann L, Krämer A, Hübner CA, Kaether C (2017) The sorting receptor Rer1 controls Purkinje cell function via voltage gated sodium channels. Sci Rep 7: 41248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Reynolds B, Weiss S (1992) Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science 255: 1707–1710 [DOI] [PubMed] [Google Scholar]
- 25. Singec I, Knoth R, Meyer RP, Maciaczyk J, Volk B, Nikkhah G, Frotscher M, Snyder EY (2006) Defining the actual sensitivity and specificity of the neurosphere assay in stem cell biology. Nat Methods 3: 801–806 [DOI] [PubMed] [Google Scholar]
- 26. Byun SH, Kim J, Han D, Kwon M, Cho JY, Ng HX, Pleasure SJ, Yoon K (2017) TRBP maintains mammalian embryonic neural stem cell properties by acting as a novel transcriptional coactivator of the Notch signaling pathway. Development 144: 778–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu L, Aster JC, Blacklow SC, Lake R, Artavanis‐Tsakonas S, Griffin JD (2000) MAML1, a human homologue of Drosophila mastermind, is a transcriptional co‐activator for NOTCH receptors. Nat Genet 26: 484–489 [DOI] [PubMed] [Google Scholar]
- 28. Weng AP, Nam Y, Wolfe MS, Pear WS, Griffin JD, Blacklow SC, Aster JC (2003) Growth suppression of pre‐T acute lymphoblastic leukemia cells by inhibition of notch signaling. Mol Cell Biol 23: 655–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moellering RE, Cornejo M, Davis TN, Del Bianco C, Aster JC, Blacklow SC, Kung AL, Gilliland DG, Verdine GL, Bradner JE (2009) Direct inhibition of the NOTCH transcription factor complex. Nature 462: 182–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li Y, Hibbs MA, Gard AL, Shylo NA, Yun K (2012) Genome‐wide analysis of N1ICD/RBPJ targets in vivo reveals direct transcriptional regulation of Wnt, SHH, and hippo pathway effectors by Notch1. Stem Cells 30: 741–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Karlström H, Bergman A, Lendahl U, Näslund J, Lundkvist J (2002) A sensitive and quantitative assay for measuring cleavage of presenilin substrates. J Biol Chem 277: 6763–6766 [DOI] [PubMed] [Google Scholar]
- 32. Groot AJ, Habets R, Yahyanejad S, Hodin CM, Reiss K, Saftig P, Theys J, Vooijs M (2014) Regulated proteolysis of NOTCH2 and NOTCH3 receptors by ADAM10 and presenilins. Mol Cell Biol 34: 2822–2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Park CS, Kim OS, Yun SM, Jo SA, Jo I, Koh YH (2008) Presenilin 1/γ‐secretase is associated with cadmium‐induced E‐cadherin cleavage and COX‐2 gene expression in T47D breast cancer cells. Toxicol Sci 106: 413–422 [DOI] [PubMed] [Google Scholar]
- 34. Reyes J, Castillo‐Hernández J, Maldonado‐Cervantes M, Maldonado‐Cervantes E, López AR, Hernández AV, García‐Rangel M (2016) Delimitation of the pore in tweety homolog 1 channels: a model‐guided approach. J Adv Biol Biotechnol 10: 1–12 [Google Scholar]
- 35. Stefaniuk M, Swiech L, Dzwonek J, Lukasiuk K (2010) Expression of Ttyh1, a member of the Tweety family in neurons in vitro and in vivo and its potential role in brain pathology. J Neurochem 115: 1183–1194 [DOI] [PubMed] [Google Scholar]
- 36. Hu CD, Chinenov Y, Kerppola TK (2002) Visualization of interactions among bZip and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 9: 789–798 [DOI] [PubMed] [Google Scholar]
- 37. Wang L, Carnegie GK (2013) Flow cytometric analysis of bimolecular fluorescence complementation: a high throughput quantitative method to study protein‐protein interaction. J Vis Exp 78: 50529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Milner RE, Baksh S, Shemanko C, Carpenter MR, Smillie L, Vance JE, Opas M, Michalak M (1991) Calreticulin, and not calsequestrin, is the major calcium binding protein of smooth muscle sarcoplasmic reticulum and liver endoplasmic reticulum. J Biol Chem 266: 7155–7165 [PubMed] [Google Scholar]
- 39. Hua Z, Graham TR (2003) Requirement for Neo1p in retrograde transport from the Golgi Complex to the endoplasmic reticulum. Mol Biol Cell 14: 4971–4983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wiernasz E, Kaliszewska A, Brutkowski W, Bednarczyk J, Gorniak M, Kaza B, Lukasiuk K (2014) Ttyh1 protein is expressed in glia in vitro and shows elevated expression in activated astrocytes following status epilepticus. Neurochem Res 39: 2516–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tanabe C, Maeda T, Zou K, Liu J, Liu S, Nakajima T, Komano H (2012) The ubiquitin ligase synoviolin up‐regulates amyloid β production by targeting a negative regulator of γ‐secretase, Rer1, for degradation. J Biol Chem 287: 44203–44211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Matthews CA, Shaw JE, Hooper JA, Young IG, Crouch MF, Campbell HD (2007) Expression and evolution of the mammalian brain gene Ttyh1. J Neurochem 100: 693–707 [DOI] [PubMed] [Google Scholar]
- 43. Murphy MP, LeVine H III (2010) Alzheimer's disease and the β‐amyloid peptide. J Alzheimers Dis 19: 311–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Source Data for Expanded View
Review Process File
Source Data for Figure 3
Source Data for Figure 5


